Abstract
The function of the REP27 protein (GenBank accession no. EF127650) in the photosystem II (PSII) repair process was elucidated. REP27 is a nucleus-encoded and chloroplast-targeted protein containing two tetratricopeptide repeat (TPR) motifs, two putative transmembrane domains, and an extended carboxyl (C)-terminal region. Cell fractionation and western-blot analysis localized the REP27 protein in the Chlamydomonas reinhardtii chloroplast thylakoids. A folding model for REP27 suggested chloroplast stroma localization for amino- and C-terminal regions as well as the two TPRs. A REP27 gene knockout strain of Chlamydomonas, termed the rep27 mutant, was employed for complementation studies. The rep27 mutant was aberrant in the PSII-repair process and had substantially lower than wild-type levels of D1 protein. Truncated REP27 cDNA constructs were made for complementation of rep27, whereby TPR1, TPR2, TPR1+TPR2, or the C-terminal domains were deleted. rep27-complemented strains minus the TPR motifs showed elevated levels of D1 in thylakoids, comparable to those in the wild type, but the PSII photochemical efficiency of these strains was not restored, suggesting that the functionality of the PSII reaction center could not be recovered in the absence of the TPR motifs. It is suggested that TPR motifs play a role in the functional activation of the newly integrated D1 protein in the PSII reaction center. rep27-complemented strains missing the C-terminal domain showed low levels of D1 protein in thylakoids as well as low PSII photochemical efficiency, comparable to those in the rep27 mutant. Therefore, the C-terminal domain is needed for a de novo biosynthesis and/or assembly of D1 in the photodamaged PSII template. We conclude that REP27 plays a dual role in the regulation of D1 protein turnover by facilitating cotranslational biosynthesis insertion (C-terminal domain) and activation (TPR motifs) of the nascent D1 during the PSII repair process.
The unicellular green alga Chlamydomonas reinhardtii is a good model system to study the regulation of photosynthesis at the molecular level, since the chloroplast development and differentiation can take place under autotrophic, photo heterotrophic, or dark heterotrophic conditions. The chloroplast biogenesis and most of the photosynthetic apparatus assembly can occur in the dark, when cells are supplied with organic carbon such as acetate (Guenther et al., 1990). Vegetative cells are haploid, permitting ready phenotypic manifestation of mutations, or genetic lesions. Photosynthesis-deficient mutants can thus be isolated and investigated, conferring to Chlamydomonas a significant advantage over other model plant systems. Measurement of the chlorophyll fluorescence with intact cells offers a noninvasive approach to assess the functionality of PSII and of the electron transport process in the thylakoid membrane of photosynthesis. Thus, a number of photosynthesis mutants with defects in the biogenesis and assembly of thylakoid membrane complexes were generated and isolated (Zhang et al., 1997; Wollman et al., 1999; Minai et al., 2006), providing valuable information about the corresponding processes and leading to the isolation and characterization of genes and proteins.
The PSII repair cycle (Guenther and Melis, 1990) is a process essential to photosynthesis and plant growth, occurring in all organisms of oxygenic photosynthesis, and serving to restore the functional status of PSII from a frequently occurring photodamage. Repair entails the unique selective degradation and replacement of the D1/32-kD PSII reaction center protein from the massive (>1,000 kD) PSII holocomplex (Mattoo and Edelman, 1987). The PSII damage and repair mechanism is highly conserved in all organisms of oxygenic photosynthesis, as it maintains the activity of photosynthesis by selectively degrading and replacing the PSII D1/32-kD reaction center protein (Melis, 1991; Aro et al., 1993). The rate constant of photodamage is a linear function of light intensity (Baroli and Melis, 1996; Tyystjärvi and Aro, 1996), ranging between 0 in the dark to about 1.2 h−1 under bright sunlight and physiological growth conditions. In contrast, the enzymatic repair process occurs with a light intensity-independent rate constant, equal to about 0.7 h−1 (Neidhardt et al., 1998; Ohnishi et al., 2005; Yokthongwattana and Melis, 2006). Under bright sunlight conditions, the rate of photodamage can be faster than the rate of repair, resulting in the accumulation of inactive D1 proteins, loss of photosynthetic yield, and loss of chloroplast productivity (Adir et al., 1990; Bailey et al., 2002). The repair entails D1 activation (Guenther et al., 1990; Neale and Melis, 1991) and posttranslational modifications to restore the PSII water-splitting activity (Diner et al., 1988; Bowyer et al., 1992).
Biogenesis of the photosynthetic apparatus is a process involving the coordinated expression of genes leading to the biosynthesis and assembly of both chloroplast- and nucleus-encoded proteins. The chloroplast genome of the unicellular green alga Chlamydomonas encodes approximately 100 genes, required for protein synthesis of the photosynthetic apparatus and carbon-fixing machinery (Maul et al., 2002). Genetic and biochemical studies of Chlamydomonas revealed the involvement of numerous nucleus-encoded factors acting in the transcription/translation or in several posttranscriptional events of chloroplast gene expression, such as mRNA processing, stability, and translation into proteins (Barkan and Goldschmidt-Clermont, 2000; Somanchi and Mayfield, 2001). Compared with the information available at present on the rapid light-dependent turnover of the D1 protein in PSII (Aro et al., 1993; Yokthongwattana and Melis, 2006), our understanding of the regulation of the PSII repair mechanism is very limited, either at the level of protein translation or posttranslational steps leading to a functional PSII. The de novo synthesis, membrane insertion, and assembly of D1 processes are most likely to require the participation of nucleus-encoded auxiliary proteins.
In earlier studies from this laboratory (Zhang et al., 1997; Park et al., 2007), DNA insertional mutagenesis in the model organism Chlamydomonas was applied for the isolation of mutants defective in photoautotrophic growth. Isolated from this screening protocol, the rep27 strain was found to grow normally in the presence of acetate but displayed low photosynthetic activity even under low-light conditions. Under weak growth irradiance, rep27 showed a limited water oxidation and oxygen evolution capacity, whereas under moderate to high irradiance, PSII activity ceased to exist, indicating an inability of the chloroplast to perform the D1 protein turnover. Gene cloning and biochemical analysis of the rep27 strain resulted in the identification of the REP27 nuclear gene, which was deleted from the rep27 mutant. The REP27 cDNA sequence was isolated and used for complementation of the rep27 mutant, permitting the recovery of the wild-type phenotype (Park et al., 2007). From these preliminary results, it was suggested that REP27 is a nuclear gene involved in the rapid light-dependent turnover of the D1 protein during the PSII repair process.
This work investigated and elucidated the mechanism of REP27 protein action in D1 protein turnover and PSII repair from photodamage. It was concluded that REP27 plays a dual role in the regulation of D1 protein turnover by facilitating cotranslational biosynthesis insertion (C-terminal domain) and activation (tetratricopeptide repeat [TPR] motifs) of the nascent D1 during the PSII repair process.
RESULTS
Blue-Native- and SDS-PAGE Analysis of the Wild Type and the rep27 Mutant
DNA insertional mutagenesis with the model organism C. reinhardtii was applied for the isolation and characterization of putative PSII repair mutants (Zhang et al., 1997; Park et al., 2007). The rep27 strain, defective in photoautotrophic growth, was isolated from this screening library. It was found that rep27 grew in the presence of acetate, but it displayed a lower level of photosynthetic activity under low-light conditions. Under moderate to high light intensity, PSII photochemical activity was absent in this mutant. Further physiological and biochemical analyses showed lower steady-state levels of the D1/32-kD reaction center protein in rep27 than in the wild type, while other subunits of the PSII holocomplex, such as D2 and CP47, were only somewhat reduced. Peripheral subunits of PSII, such as the PsbO proteins, were in equivalent amounts in the wild type and rep27. Subunits of non-PSII complexes (cytochrome b6f, PSI, and ATP synthase) also occurred in equivalent amounts in the wild type and rep27 (Park et al., 2007). Results from the earlier work supported a working hypothesis whereby de novo biogenesis/assembly of the PSII holocomplex occurred more or less normally in the rep27 mutant; however, replacement (turnover) of the photodamaged D1/32-kD protein was severely impaired.
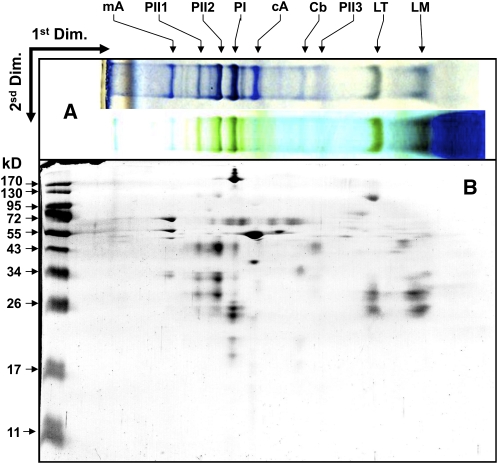
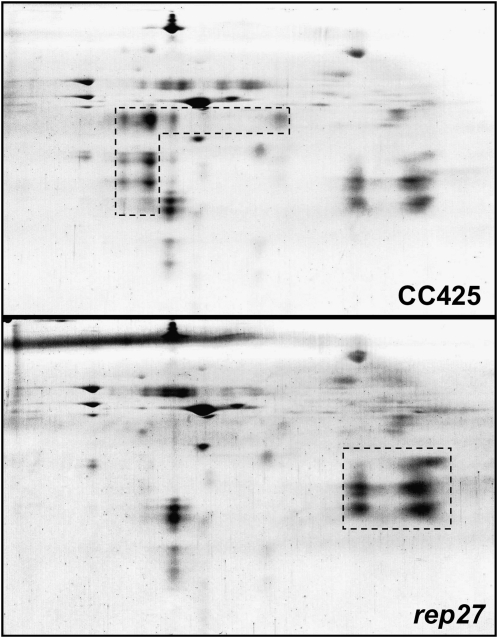
A comparative thylakoid membrane protein profile analysis of the wild type and the rep27 mutant was undertaken, aiming to identify biochemical and functional differences between the two strains. For better resolution of the integral and hydrophobic components of the thylakoid membrane, holocomplexes were separated in a first dimension via nondenaturing PAGE. Figure 1A (top lane) shows a Blue-Native (BN) gel of wild-type thylakoid membrane proteins compared with the green native gel of the same sample (Fig. 1A, bottom lane). The native form of the resolved holocomplexes was then separated into their constituent subunits upon running in a second dimension by denaturing SDS-PAGE and visualized by silver staining (Fig. 1B). The combination of BN-PAGE and SDS-PAGE was previously used to identify specific thylakoid membrane proteins of Chlamydomonas by peptide mass fingerprinting and matrix-assisted laser-desorption ionization time of flight mass spectrometry (Rexroth et al., 2003). This analysis permitted separation and identification of the main PSII core subunits (D1, D2, CP43, and CP47 proteins), PSI proteins (PsaA, PsaB, PsaF, and light-harvesting complex I [LHCI] protein), the LHCs of PSII in trimeric and monomeric forms, cytochrome b6f complex proteins, and chloroplast ATP synthase complex (CF0F1) proteins (Rexroth et al., 2003). A selective comparison of the two-dimensional gel electrophoresis proteome of photomixotrophically grown Chlamydomonas wild type and rep27 mutant is given in Figure 2. Demarcated by the dashed box are subunits of the PSII complex (Fig. 2, CC425). In the rep27 mutant, these proteins appeared to occur at substantially lower levels (Fig. 2, rep27) relative to the wild type. The two-dimensional electrophoresis gel also showed a higher relative amount of LHCII proteins in the rep27 mutant relative to the wild type, demarcated by the dashed box in Figure 2 (rep27). Since the overall chlorophyll-cell ratio in the rep27 mutant was similar to that in the wild type (Zhang et al., 1997; Park et al., 2007) and the two-dimensional gels were loaded on the basis of equal chlorophyll, the results suggest a higher LHCII/reaction center II ratio in the rep27 mutant compared with the wild type. This is a consequence of the rep27 mutation adversely affecting the reaction center II and PSII core complexes but not the LHCII.
Figure 1.
Solubilized thylakoid membranes of Chlamydomonas wild type (CC425) were subjected to PAGE analyses. A (top lane), BN-PAGE of thylakoid membrane proteins stained with Coomassie Brilliant Blue. A (bottom lane), green native gel of the same sample. B, Two-dimensional gel electrophoresis of the wild type, where BN-PAGE was used in the first dimension (Dim.) and 12% acrylamide SDS-PAGE was used in the second dimension. The two-dimensional gel was visualized by silver staining. mA, Mitochondrial H+-ATP synthase; PII1 to PII3, PSII reaction center subunits (D1, D2, CP43, and CP47 proteins); PI, PSI complex (PsaA, PsaB, and PsaF) and LHCI protein; cA, chloroplast ATP synthase (CF0F1); Cb, cytochrome b6f; LT, LHCII trimer; LM, LHCII monomer.
Figure 2.
Comparison of silver-stained two-dimensional gels between the wild type (CC425) and the rep27 mutant. The dashed lines demarcate the expected locations of the PSII core complex (CC425) and the LHCII (rep27). For more details, see legend to Figure 1.
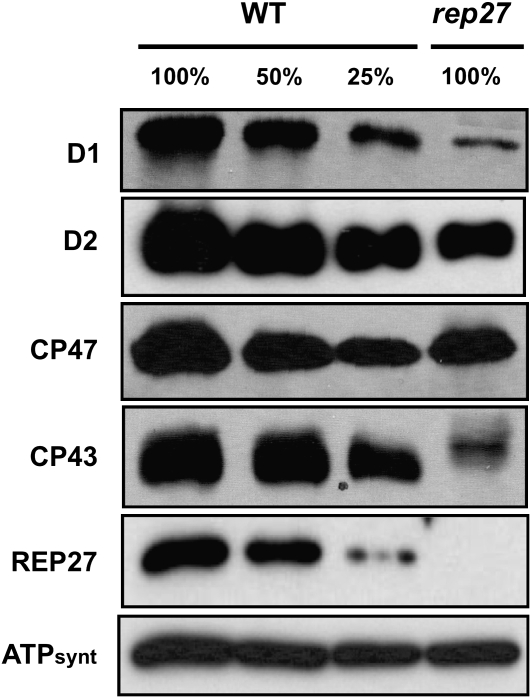
Quantitative western-blot analysis was employed to more directly compare levels of the PSII core and reaction center proteins in the wild type and the rep27 mutant. Figure 3 shows that D1 and CP43 were depleted from the thylakoid membrane of the rep27 mutant, whereas D2 and CP47 occurred in comparable quantities in the wild type and rep27. These results suggest that PSII core and reaction center complexes are not stable in the rep27 mutant but become dissociated easily, even under the mild detergent conditions employed in the BN gel experiments of Figures 1 and 2. This observation would explain the greatly reduced amounts of PSII core and reaction center proteins in Figure 2 (rep27).
Figure 3.
Western-blot analysis of Chlamydomonas wild-type (WT) and rep27 thylakoid membrane proteins. Cells were grown under continuous irradiance of 100 μmol m−2 s−1 incident intensity. Steady-state levels of PSII subunits were determined from the intensity of the antibody cross-reaction on western blots with specific polyclonal antibodies generated against D1, D2, CP47, CP43, REP27, and the ATP synthase (ATPsynt). Note the lower D1 and CP43 levels in rep27 and the absence of REP27 in the mutant.
The above proteome-based results are consistent with previous spectrophotometric and western-blot analyses probing the steady-state levels of PSII and D1 reaction center protein in Chlamydomonas wild type and rep27 mutant (Zhang et al., 1997; Park et al., 2007).
Occurrence of REP27 in the Wild Type and Photochemical Apparatus Mutants
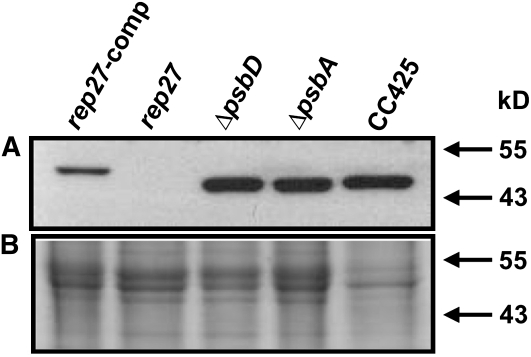
The cDNA sequence of REP27 (GenBank accession no. EF127650) was used for complementation of the rep27 mutant, which permitted recovery of the wild-type phenotype. Based on these findings, specific polyclonal antibodies were generated in rabbit against a portion of the REP27 protein, defined by amino acids Phe-273 to Leu-367. SDS-PAGE and western-blot analyses were then applied to probe the occurrence and steady-state levels of REP27 proteins in Chlamydomonas wild type and selected photochemical apparatus mutants. Results from such comparative analyses are shown in Figure 4, where a specific cross-reaction was detected between the REP27 antibodies and a protein band migrating to 47 kD (Fig. 4, CC425). The REP27 molecular mass obtained by this western-blot analysis was similar to that deduced from the amino acid composition of the protein (45.6 kD; Park et al., 2007). The REP27-specific polyclonal antibodies failed to recognize any protein band in extracts from the rep27 mutant (Fig. 4, rep27), consistent with the notion that the latter is a REP27 knockout. Conversely, rep27-complemented strains showed a distinct protein band in the approximately 48-kD region (Fig. 4, rep27-comp; the recombinant REP27 is larger by about 2 kD due to the presence of a 2×MYC tag). It is of interest that both the D1-less (ΔpsbA) and D2-less (ΔpsbD) mutant strains of Chlamydomonas showed an equal to wild type abundance for the REP27 protein (Fig. 4), in spite of the fact that these strains have no PSII reaction center complex and display no variable chlorophyll fluorescence or oxygen evolution (Bennoun et al., 1986; Erickson et al., 1986; Minai et al., 2006) and, therefore, do not undergo a photodamage-and-repair cycle. It is suggested that the REP27 gene is expressed constitutively in Chlamydomonas under all growth conditions. Moreover, it appears that REP27 gene transcription and translation are not down-regulated by the absence of the D1/D2 reaction center proteins of PSII. It may be concluded that a lack of de novo biogenesis/assembly of the PSII reaction center does not adversely affect the synthesis of the REP27 protein.
Figure 4.
A, Western-blot analysis of total proteins from Chlamydomonas wild type (CC425), rep27, rep27-comp, D1-less (ΔpsbA), and D2-less (ΔpsbD) mutant strains probed with specific polyclonal antibodies raised against the REP27 protein. B, SDS-PAGE of proteins (2 μg of chlorophyll loaded) separated onto 12% acrylamide and visualized with Coomassie Brilliant Blue. The slightly slower electrophoretic mobility of the REP27 protein in the complemented strain (rep27-comp) is due to the presence of the 2×MYC tag, introduced into the plasmid construct used for complementation.
Thylakoid Membrane Fractionation and Localization of REP27
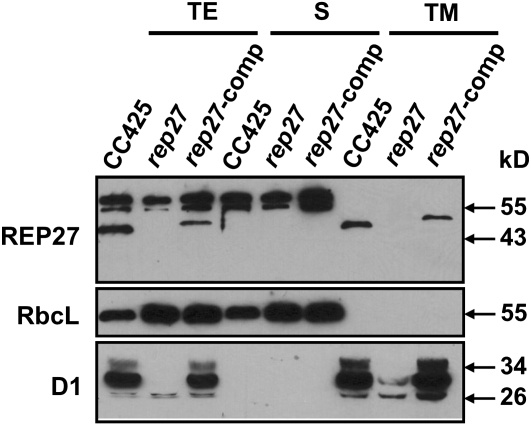
It was previously suggested that REP27 is a putative chloroplast-targeted protein (Park et al., 2007), as the REP27 amino acid sequence analysis (Emanuelsson et al., 1999) predicted a chloroplast transit peptide in the precursor protein. Moreover, two putative transmembrane helices (amino acid regions TMH1 and TMH2, including amino acids 180–202 and 217–239, respectively) were also predicted by HMMTOP version 2.0 software analysis (Tusnady and Simon, 2001). It was hypothesized that REP27 is integral to the thylakoid membrane. To experimentally test this hypothesis, a cellular fractionation and analysis of soluble and membrane-bound proteins was undertaken. Figure 5 shows a western-blot analysis of wild-type (CC425), rep27 mutant, and rep27-complemented strains, probed with specific polyclonal antibodies generated against the REP27, RbcL, and D1 proteins. Total cell extract and soluble and total membrane fractions were assayed. RbcL- and D1-specific polyclonal antibodies served in testing for the enrichment of the soluble and total membrane fractions. Results showed absence of the REP27 protein from the total protein extract of the rep27 mutant (Fig. 5, TE) but presence in the CC425 and rep27-complemented strains (Fig. 5, REP27). Importantly, the REP27 protein was found in exclusive association with the total membrane extract of the CC425 and rep27-complemented strains (Fig. 5, TM) and not with the soluble protein fraction (Fig. 5, S). Resolved thylakoid membranes and isolation of a light membrane fraction, including the chloroplast envelopes, was achieved by differential ultracentrifugation, as recently done in this laboratory (Lindberg and Melis, 2008). Western-blot analysis of these membrane fractions with REP27-specific antibodies showed presence of the REP27 protein in the thylakoid membrane fraction and absence from the lighter envelope-containing fraction (data not shown). These results clearly suggest integral thylakoid membrane localization for the REP27 protein, probably spanning the thylakoid lipid bilayer through the TMH1 and TMH2 regions.
Figure 5.
Western-blot analysis of total proteins from Chlamydomonas wild-type (CC425), rep27, and rep27-comp strains. Cells were fractionated into soluble (S) and total membrane (TM) portions. Total extract (TE) was obtained upon breaking the cells with acid-washed glass beads. The total membrane fraction was obtained by centrifugation at 20,000g for 60 min. Lanes were loaded with 2 μg of chlorophyll. The supernatant was used as the soluble fraction. Relative protein content was estimated from the intensity of the antibody cross-reaction on the western blots. Specific polyclonal antibodies were used, generated against REP27, RbcL, or D1.
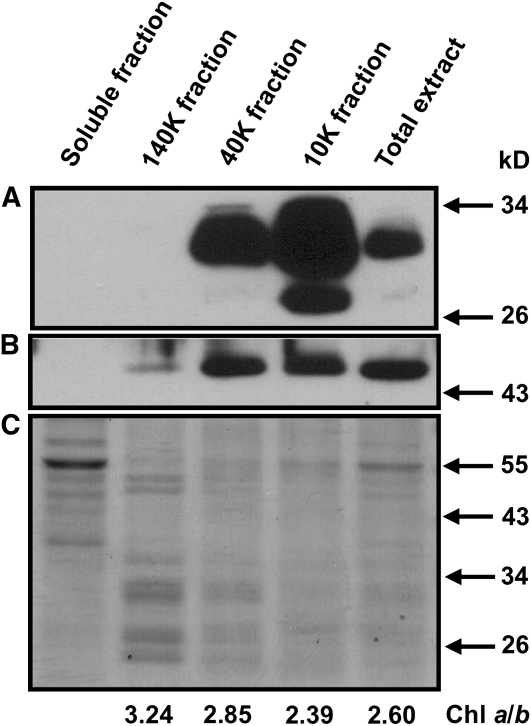
In an effort to more precisely localize the REP27 in the thylakoid membrane of photosynthesis, appressed and nonappressed thylakoid membrane domains were isolated upon mechanical fractionation of Chlamydomonas wild type (Neale and Melis, 1991). It was shown previously that the 10k fraction is enriched in thylakoid grana membranes and the 140k fraction is enriched in stroma-exposed membranes, as evidenced from direct measurements of PSI and PSII contents using P700, QA, and pheophytin quantitations (Neale and Melis, 1991). The differential distribution of the photosystems among such thylakoid membrane domains was also reflected in the chlorophyll a/b ratio of the fractions, thus providing an easy to measure assay and convenient marker. Figure 6 shows that the chlorophyll a/b ratio was 2.60 in the total thylakoid extract, was lower (2.39) in the heavy grana-enriched 10k fraction, and was higher (3.24) in the light stroma-exposed thylakoid containing PSI-enriched 140 k fraction. Figure 6 also shows western-blot analysis of the various thylakoid membrane fractions with D1-specific (Fig. 6A) and REP27-specific (Fig. 6B) polyclonal antibodies. Measurement of the REP27-D1 ratio showed that the REP27 protein is relatively more prevalent in the 40k than in the 10k or 140 k fraction, suggesting that the membrane domain of the REP27 protein is intermediate to the fully appressed and stroma-exposed thylakoids. These could be the “fret” domains of the thylakoid membrane (Morrissey et al., 1986), where the PSII repair takes place, localized adjacent to the appressed grana membranes, yet exposed to the stroma medium so as to permit the D1 protein turnover.
Figure 6.
Western-blot analysis of proteins from thylakoid membrane fractions isolated upon sonication and differential centrifugation at 10,000g (10k), 40,000g (40k), and 140,000g (140k). Lanes were loaded with 2 μg of chlorophyll (Chl). Proteins were probed with specific polyclonal antibodies generated against D1 (A) and REP27 (B). C shows a Coomassie Brilliant Blue-stained SDS-PAGE protein profile of the different thylakoid membrane fractions.
Functional Role of the REP27 TPR Motifs and C-Terminal Domain
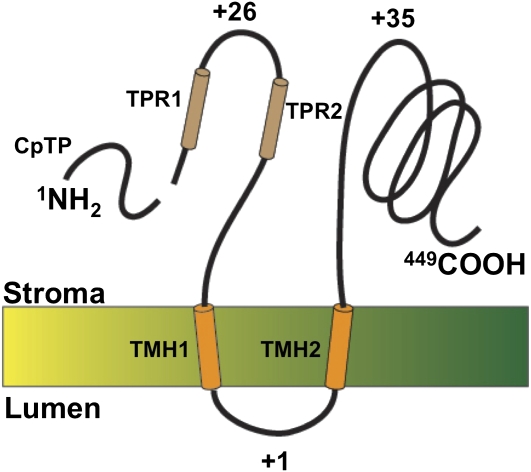
Using InterProScan 13.1 software analysis (Quevillon et al., 2005), two distinct TPR motifs were also identified in REP27 (Park et al., 2007). These were termed TPR1A/TPR1B and TPR2A/TPR2B and were shown to occur near the N-terminal end of the protein. TPR motifs form a compact unit of two helices interacting with each other in the antiparallel direction (Blatch and Lässle, 1999). Accordingly, the structure of the REP27 protein entails the N terminus, followed by two TPR motifs (TPR1 and TPR2), the two transmembrane helices, and the long C-terminal hydrophilic portion of the protein. On the basis of topology analysis, using the TMAP transmembrane program and the positive inside rule (Persson and Argos, 1994), we propose that the N and C termini, including the two TPR motifs, are exposed to the chloroplast stroma phase. A folding model, depicting the structural association of REP27 with the thylakoid membrane, was thus constructed (Fig. 7).
Figure 7.
Folding model of the REP27 protein in the chloroplast thylakoid membrane. This protein contains 449 amino acids from the N to the C terminus. The number of positively charged amino acids is indicated for each domain of the protein. Two transmembrane helices (TMH1 and TMH2) are shown spanning the thylakoid membrane such that N and C termini are exposed in the chloroplast stroma phase. The model further shows two TPR motifs (TPR1 and TPR2) occurring near the N terminus and the transit peptide that is cleaved upon chloroplast import. REP27 displays a rather extensive C-terminal portion, which is exposed in the chloroplast stroma. [See online article for color version of this figure.]
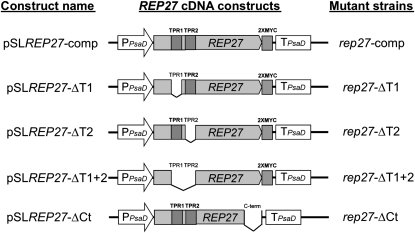
To gain a better understanding of the function of REP27 during the D1/32-kD PSII reaction center protein turnover, the roles of the TPR motifs and of the C-terminal region were investigated. It has been suggested that TPR motifs are involved in a variety of critical protein-protein interactions in the living cell (Blatch and Lässle, 1999), and this is consistent with a putative functional role of REP27 in the unique D1 replacement process. Therefore, REP27 cDNA constructs missing the TPR1 (pSLREP27-ΔT1), TPR2 (pSLREP27-ΔT2), both TPR1 and TPR2 (pSLREP27-ΔT1+2), or the C-terminal region (pSLREP27-ΔCt) were made (Fig. 8; see “Materials and Methods”). These were used for transformation of the rep27 mutant strain. Complementation of the rep27 mutant (rep27-comp) with a full-length REP27 cDNA construct (pSLREP27-comp) was used as a control. The paramomycin resistance cassette was used as the first selectable marker for the isolation of transformants. Putative rep27-ΔT1, rep27-ΔT2, rep27-ΔT1+2, and rep27-ΔCt transformant strains were screened further via western-blot analysis with specific polyclonal antibodies generated against the REP27 protein. Only strains with a positive expression of the modified REP27 protein were selected for further analysis.
Figure 8.
REP27 cDNA constructs used for complementation of the rep27 mutant. On the left, the name of each construct is given. In the middle, a map of the structural organization of the respective construct is shown. On the right, transformant strains obtained from the transformation of rep27 mutant by these cDNA constructs are shown.
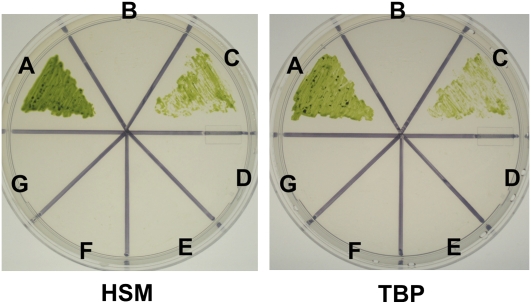
Positive transformant strains rep27-ΔT1, rep27-ΔT2, rep27-ΔT1+2, and rep27-ΔCt were tested for photoautotrophic growth on both high-salt medium (HSM) and Tris-bicarbonate phosphate (TBP) minimal medium (Fig. 9). It is known that a functional REP27 protein is needed for autotrophic growth of Chlamydomonas in minimal medium (Park et al., 2007). Accordingly, the wild type (Fig. 9A) and rep27-comp (Fig. 9C) grew viable colonies on both HSM and TBP plates. Conversely, the rep27 mutant (Fig. 9B) and transformant strains rep27-ΔT1 (Fig. 9D), rep27-ΔT2 (Fig. 9E), rep27-ΔT1+2 (Fig. 9F), and rep27-ΔCt (Fig. 9G) all failed to rescue the acetate-requiring phenotype of the rep27 mutant. It is concluded that both TPR motifs and the C-terminal portion of the REP27 protein are needed for the completion of the PSII repair cycle and the survival of Chlamydomonas under photoautotrophic growth conditions.
Figure 9.
Photoautotrophic growth of Chlamydomonas wild type, rep27 mutant, and rep27 transformants grown on HSM and TBP media. A, CC125 wild type; B, rep27 mutant; C, rep27-comp; D, rep27-ΔT1; E, rep27-ΔT2; F, rep27-ΔT1+2; G, rep27-ΔCt. [See online article for color version of this figure.]
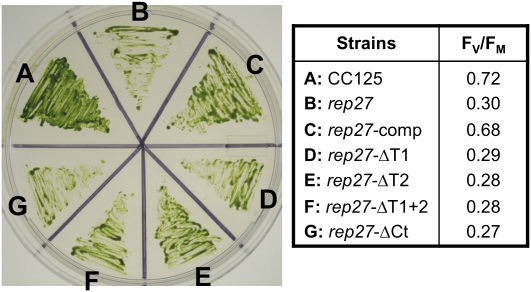
To probe the status of the truncated REP27 protein in transformant strains rep27-ΔT1, rep27-ΔT2, rep27-ΔT1+2, and rep27-ΔCt, these were grown on Tris-acetate phosphate (TAP) medium, followed by biophysical and biochemical analysis of the function of PSII. Growth in the presence of acetate (TAP) under weak illumination enables biosynthesis and assembly of functional PSII (Zhang et al., 1997; Park et al., 2007), that is, under conditions when the rate of photodamage is rather slow (Melis, 1999), thereby permitting accumulation of a smaller-than-wild-type fraction of functional PSII reaction centers. Figure 10 (left) shows that wild-type, rep27 mutant, and transformant strains employed in this work all grew viable colonies and greened normally on TAP plates. To probe the functional properties of PSII in these strains, the fluorescence yield Fv/Fm ratio (for maximum photochemical efficiency of PSII in the dark-adapted state) was measured. The Fv/Fm ratio provides a measure of the photochemical charge separation efficiency of the PSII reaction centers (Kitajima and Butler, 1975); therefore, it offers an indication of the proportion of functional PSII reaction centers under in vivo conditions. The Fv/Fm ratio of the wild type was equal to 0.72, whereas Fv/Fm for the rep27 mutant (0.30) was only about 40% of that in the wild type, suggesting that only a fraction of the PSII reaction centers were functional in the rep27 mutant (growth on TAP). As a positive control for the recovery of the REP27 function, the rep27-comp strain showed an elevated Fv/Fm ratio (0.68) similar to that of the Chlamydomonas wild type. The rep27-ΔT1, rep27-ΔT2, rep27-ΔT1+2, and rep27-ΔCt transformants all showed low Fv/Fm ratios similar to that of the rep27 mutant (Fig. 10, right). It is concluded that the TPR motifs and C-terminal region of the REP27 protein are all necessary to restore the proportion of functional PSII reaction centers and electron transport capacity in the chloroplast of this green microalga.
Figure 10.
Growth (left) and efficiency of PSII primary photochemistry (Fv/Fm; right) of CC125 wild type, rep27 mutant, and rep27 transformant strains grown on TAP medium. Fv/Fm values shown are averages from three independent experiments. [See online article for color version of this figure.]
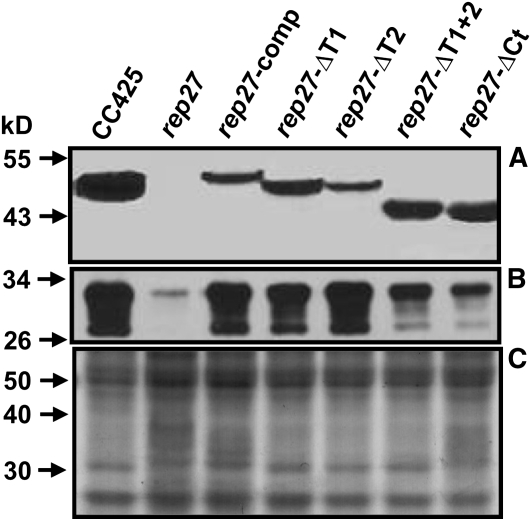
Western-blot analysis was employed with the wild-type, rep27, rep27-comp, rep27-ΔT1, rep27-ΔT2, rep27-ΔT1+2, and rep27-ΔCt transformant strains to test for the presence and relative steady-state amounts of the REP27 and D1 proteins (Fig. 11). Consistent with earlier results (Park et al., 2007), the rep27 mutant lacked the REP27 protein and showed low levels of the D1 protein in its thylakoids (Fig. 11, rep27). The rep27-comp strain offered positive evidence of the REP27 protein and substantially enhanced levels of the D1 protein, which were similar to those in the wild type (Fig. 11, rep27-comp). This is consistent with the elevated Fv/Fm ratio (0.68) in the latter. Interestingly, all truncated REP27 transformant strains (Fig. 11, rep27-ΔT1, rep27-ΔT2, rep27-ΔT1+2, and rep27-ΔCt) clearly showed the presence of the modified REP27 protein in the cell. Moreover, they also showed elevated amounts of the D1 protein in the chloroplast thylakoid membranes. Specifically, strains rep27-ΔT1 and rep27-ΔT2 displayed levels of the D1 protein that were nearly equivalent to those in the wild type and rep27-comp. Strains rep27-ΔT1+2 and rep27-ΔCt contained substantial amounts of the truncated REP27 protein; however, they did not accumulate D1 protein to levels equivalent to that in the other transformants.
Figure 11.
Western-blot analysis of total proteins from Chlamydomonas wild type (CC425), rep27 mutant, and rep27-comp, rep27-ΔCt, rep27-ΔT1, rep27-ΔT2, and rep27-ΔT1+2 transformant strains probed with specific polyclonal antibodies against REP27 (A) or D1 (B) proteins. Also shown is the Coomassie Brilliant Blue-stained SDS-PAGE profile of the proteins from the various samples (C). Four micrograms of chlorophyll was loaded onto 12% acrylamide for the SDS-PAGE and western-blot analyses.
These results clearly showed that removal of TPR1, TPR2, TPR1+TPR2, or 61 amino acids from the C-terminal domain did not interfere with the synthesis and assembly of the REP27 protein in the thylakoid membrane. Moreover, these transformants had elevated steady-state amounts of the D1 protein in the respective thylakoid membranes. However, from the low Fv/Fm ratio and inability to grow on minimal media, it may be concluded that synthesis and assembly of the truncated REP27 to the thylakoid membrane is not sufficient by itself to restore function in these transformants. The presence of both TPR motifs and of the C-terminal portion of the protein is required for the completion of the D1 protein turnover during the PSII repair process. It is hypothesized (1) that the C-terminal portion of the REP27 protein is needed for the insertion of nascent D1 proteins in the D1-less PSII template, and (2) that TPR motifs are required for the “activation” of newly inserted and bound D1 proteins, which, in the case of the truncated REP27 transformants, remained inactive. Such an activation step was inferred earlier from biophysical studies (Guenther et al., 1990; Neale and Melis, 1990) and could involve a posttranslational modification and/or proper membrane deployment/folding of the nascent D1 protein during its assembly within the PSII template and subsequent maturation. Since the TPR-less strains had a low PSII photochemical charge separation efficiency and lack of photoautotrophic growth capacity, it appears that TPR domains are needed to ensure a full integration of nascent D1 proteins into the PSII reaction center template, leading to a functional PSII reaction center.
The rep27-ΔCt mutant showed the lowest steady-state level of D1 present, compared with the other rep27 transformant strains, but the level was still higher than that of the original rep27 mutant. It is most likely that the REP27 C-terminal domain plays an essential role in the de novo D1 biosynthesis at the level of ribosomal psbA mRNA translation and may also be required for the insertion of nascent D1 proteins in the D1-less PSII template. Taken together, these results illuminate, to our knowledge for the first time and in great detail, the role that is played by the REP27 protein in D1 protein turnover and PSII repair from photodamage.
DISCUSSION
The D1 protein is subject to frequent turnover, which far surpasses that of all other thylakoid membrane and PSII subunits. Turnover can take place at all light intensities but is accelerated under increasing irradiance (Melis, 1999). The D1 protein turnover was demonstrated in pulse-chase experiments in which the D1 was preferentially labeled over other thylakoid membrane proteins (Mattoo and Edelman, 1987; Schuster et al., 1988; Adir et al., 1990). In a previous study presenting the isolation and characterization of the rep27 mutant strain (Park et al., 2007), comparative [35S]sulfate-labeling experiments showed that the rep27 mutant accumulated radiolabeled D1 in tandem with the accumulation of the other PSII subunits. However, and opposite to that in the wild type, it did not show the prolific and preferential accumulation of D1 over the other PSII subunits. Since the latter is evidence for active D1 turnover (Park et al., 2007), it was suggested that rep27 has a specific defect in the active turnover of this PSII reaction center protein. Furthermore, steady-state levels of psbA and psbD mRNA, measured by northern-blot analysis, were similar in the wild type and the mutant. From these results, it was proposed that the rep27 mutant is defective in D1 protein turnover and, therefore, unable to complete the PSII repair process. The comparative thylakoid membrane functional and protein analyses of the wild type and the rep27 mutant in this work confirm and strengthen this hypothesis.
Since the REP27 knockout mutant contains functional PSII units when grown in the presence of acetate (Fig. 10) and under weak irradiance, it is likely that the REP27 protein does not play a direct role in the de novo biogenesis and assembly of the PSII holocomplex. This conclusion is strengthened by the finding that REP27 is localized on fully assembled thylakoid membrane domains, in which there is a fully assembled PSII and electron-transport apparatus (Figs. 4–6) and where turnover of the D1 protein is the only function requiring assembly. On the contrary, de novo biosynthesis and assembly of PSII, and of thylakoid membranes, is expected to take place at the chloroplast “polar regions,” where thylakoid membranes begin to emanate, and probably far away from the fully assembled grana complexes. The de novo biogenesis/assembly of PSII was in fact proposed to occur in areas of the chloroplast where there is a low density of membranes in a process of assembly biochemically related to the chloroplast inner envelope (Zerges and Rochaix, 1998). The spatially separated de novo biogenesis/assembly of PSII from the D1 repair machinery explains why substantial residual oxygen-evolving photosynthetic activity is encountered even after the knockout inactivation of REP27. Consistent with this reasoning is also the finding that regulation of psbA translation by autofeedback repression for PSII biogenesis/assembly is clearly distinct from the D1 biosynthesis for PSII repair (Minai et al., 2006). Therefore, we conclude that the rep27 mutant performs biogenesis/assembly of a functional PSII holocomplex but fails to undertake D1 protein turnover, as would be required by the PSII repair process.
Regulation of D1 biosynthesis takes place primarily during the psbA mRNA translation, with translation initiation, elongation, and cotranslational assembly of the D1 protein into PSII all being regulated (Kettunen et al., 1997; Zhang et al., 1999, 2000; Zhang and Aro, 2002). Association of cofactors to the D1 protein during its de novo synthesis has been suggested to stabilize and properly fold the nascent D1 chain (Kim et al., 1994). Adding to this earlier information, results from this work showed how the REP27 protein plays a role in facilitating, and probably regulating, different stages of the de novo D1 biosynthesis, assembly, and activation during the PSII repair process. For example, the REP27 C terminus is essential for the psbA mRNA translation initiation and assembly of the nascent D1. The TPR domains of REP27 are required for the activation of the bound D1 in the PSII reaction center during the PSII repair process.
REP27 is not the only TPR motif-containing chloroplast protein in Chlamydomonas that is implicated in posttranscriptional steps of the chloroplast gene expression. The nucleus-encoded TPR protein Mbb1 is involved in psbB mRNA processing, stability, and translation (Vaistij et al., 2000). Moreover, the Nac2 factor is required for the stabilization, processing, and translation of the psbD mRNA, permitting the proper folding of the D2 protein during the de novo biosynthesis assembly of PSII (Boudreau et al., 2000). It is interesting to observe that REP27 has a similar functionality with the Mbb1 and Nac2 proteins (i.e. mediating protein-protein interactions) during chloroplast mRNA metabolism, even though the three proteins probably act independently in different signaling pathways.
Other possibly significant TPR motif-containing proteins that play a role in photosynthesis include the periplasmic PratA in Synechocystis species PCC 6803 (Klinkert et al., 2004; Schottkowski et al., 2009) and the thylakoid membrane-bound LPA1 in Arabidopsis (Arabidopsis thaliana; Peng et al., 2006). Klinkert et al. (2004) showed that targeted inactivation of PratA resulted in drastically reduced PSII content in the cyanobacterium cell. Additional experimentation further supported the notion of a specific role for the PratA protein in D1 biosynthesis, as this occurred during the de novo assembly of newly synthesized PSII holocomplexes. Consistent with this view was the finding that PratA is a cell periplasmic protein (i.e. occurs in a domain where de novo biosynthesis of photosynthetic complexes is taking place and is not associated with the mature thylakoid membrane, where the repair of PSII occurs). Comparison of the deduced amino acid sequences (Clustal 2.0.8 analysis; Supplemental Fig. S1A) of REP27 with PratA suggested a low similarity, further strengthening the notion of two different proteins.
There is greater similarity between the deduced amino acid sequences of REP27 and LPA1 (Clustal 2.0.8 analysis; Supplemental Fig. S1B). LPA1 has been localized to the thylakoid membrane of Arabidopsis chloroplasts (Peng et al., 2006). However, and somewhat at variance with the conclusions drawn from this work, Peng et al. (2006) assigned a primary function to LPA1 in the de novo biosynthesis and assembly of the PSII holocomplex, rather than to a specific function in the turnover of the D1 protein, occurring during the PSII damage and repair cycle. More work is needed to delineate between these alternative possibilities of the three proteins in the different photosynthetic systems.
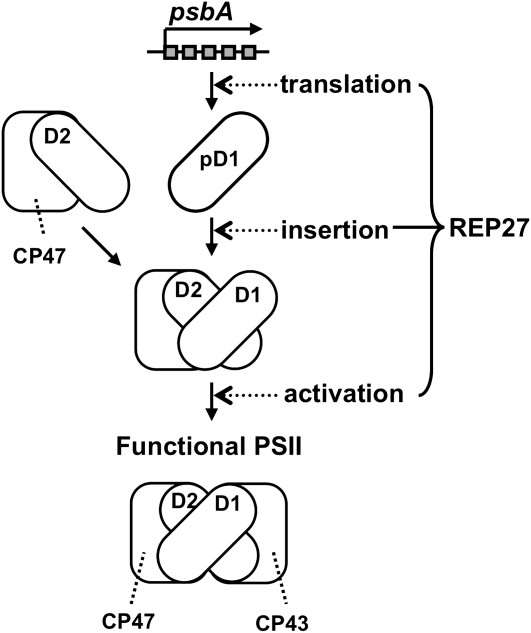
In conclusion, REP27, a nucleus-encoded protein, is essential for D1 reaction center protein turnover, permitting a completion of the translation process, maturation, and activation of D1 into a functional PSII reaction center holocomplex. Figure 12 is a schematic illustration of the role of the REP27 protein during PSII repair from photodamage. It portrays the critical role played by the REP27 protein in the translation of the psbA mRNA, insertion of the nascent D1 protein in the D1-less PSII template, and activation of the newly assembled reaction center complex. According to this proposed mechanism, the REP27 C terminus permits the initiation of ribosomal psbA mRNA translation and protein insertion, whereas the TPR motifs enable functional activation of the newly assembled D1 within the existing PSII template.
Figure 12.
Schematic of the REP27 protein function in D1 protein turnover. The REP27 C terminus is essential for the de novo D1 biosynthesis at the level of ribosomal psbA mRNA translation and initial assembly in the PSII template. The TPR motifs participate in posttranslational modification (D1 activation). Both TPR motifs are needed to activate the nascent D1 and to confer functional status to the PSII holocomplex.
MATERIALS AND METHODS
Growth Media and Culture Conditions
The wild type, rep27 mutant, and related transformants of the green alga Chlamydomonas reinhardtii were grown mixotrophically in acetate-containing TAP medium (Gorman and Levine, 1965) at 25°C under illumination of 50 μmol photons m−2 s−1 provided by cool-white fluorescent lamps. Algal cultures in early exponential growth phase were used for experiments, with cells either in liquid culture or on 1.5% agar plates. To test photoautotrophic growth of strains, cells were grown on TBP minimal medium in which sodium bicarbonate (25 mm, pH 7.4) replaced the acetate as the growth carbon source (Polle et al., 2003). Cells were collected by centrifugation at 5,000g for 5 min at 20°C. Cell pellets were stored at −80°C or used immediately for extraction of total proteins. Chlorophyll concentration was determined in 80% acetone extracts according to Arnon (1949), with equations corrected as by Melis et al. (1987). Each experiment was repeated three times with independently grown cell cultures.
Mutant Strain Generation
Generation of truncated REP27 proteins and the corresponding cDNA constructs was implemented using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. Oligonucleotides (Bioneer) carrying the desired mutations are listed in Table I. Plasmids carrying the targeted mutations were identified by sequencing, isolated, and reintroduced into pSL18 for complementation of the Chlamydomonas rep27 mutant.
Table I.
Primers used in this work
| Use of Primer | Name of Primer | Primer Sequence (5′–3′) | Primer Sequence (5′–3′) |
|---|---|---|---|
| Antibody generation | F273F and L367R | CCCATATGTTCCGCGGCCAGGTCCGGCCG | GTCGGTGCTGGGCGGCCTTGGATCCCC |
| Construction of truncated REP27 | TPR1 | CAGCAATATCAACCGACCAGCATGCAGCCCAACGACGATGAG | CTCATCGTCGTTGGGCTGCATGCTGGTCGGTTGATATTGCTG |
| TPR2 | CCCAACGACGATGAGGCTCGTGGCCTGAAGCTGATTGTGGCT | AGCCACAATCAGCTTCAGGCCACGAGCCTCATCGTCGTTGGG | |
| TPR1+2 | CAGCAATATCAACCGACCAGCGGCCTGAAGCTGATTGTGGCT | AGCCACAATCAGCTTCAGGCCGCTGGTCGGTTGATATTGCTG | |
| Chlamydomonas codon-optimized 2×MYC tag | Myc-F&R | GGGGTCTAGACATATGGAGCAGAAGCTGATCAGCGAGGAGGACCTGGGGGAGCAGAAGCTGATCAGCGAGGAGGACCTGTAA | TTACAGGTCCTCCTCGCTGATCAGCTTCTGCTCCCCCAGGTCCTCCTCGCTGATCAGCTTCTGCTCCATATGTCTAGACCCC |
Deletion of specific amino acids from the mature REP27 protein in the various constructs is as follows: ΔTPR1, Ala-58 to Glu-87; ΔTPR2, Ala-97 to Tyr-126; ΔTPR1+2, Ala-58 to Tyr-126; and ΔCt, Leu-389 to Glu-449.
Complementation Studies
Chlamydomonas rep27 mutant was generated via DNA insertional mutagenesis as described by Park et al. (2007). Chlamydomonas rep27-complemented transformants were generated by complementation of the rep27 mutant with the pSL18 plasmid vector containing wild-type truncated REP27 cDNA via the conventional glass bead transformation protocol (Kindle, 1990). The pSL18 plasmid contains the paramomycin resistance gene (selectable marker) operated under the control of the Chlamydomonas Hsp70A and RbcS2 promoters (Sizova et al., 2001) and linked to the PsaD promoter and terminator that can be used to express open reading frames in Chlamydomonas (Depege et al., 2003). Chlamydomonas transformants were selected on TAP plates containing 5 mg mL−1 paramomycin (Sigma Chemical).
BN-PAGE
Thylakoid membranes were diluted to 0.5 mg mL−1 chlorophyll in BN-PAGE solubilization buffer (50 mm Bis-Tris-HCl, pH 7.0, 750 mm ɛ-amino-n-caproic acid, and 20% glycerol) according to Schägger et al. (1994). Dodecyl-β-d-maltoside was added to a final concentration of 1% (w/v), solubilization was carried out on ice for 40 min, followed by centrifugation at 1,000g for 10 min. The supernatant was supplemented with Serva Blue G-250 from a 5% (w/v) stock in 500 mm ɛ-amino-n-caproic acid to a detergent/Serva Blue G-250 ratio of 4:1 (w/w) and directly loaded onto the gel. BN-PAGE was carried out in a 5% to 12.5% gradient using the Hoefer SE 600 and Hoefer SE 250 electrophoresis apparatuses according to Schägger et al. (1994) with modifications according to Thidholm et al. (2002).
Denaturing SDS-PAGE
For the isolation of total protein, cell biomass equivalent to 100 μg of chlorophyll were resuspended in 400 μL of 0.1 m dithiothreitol and 0.1 m Na2CO3. Following incubation for 5 min, 400 μL of 2× sample solubilization buffer containing 250 mm Tris-HCl (pH adjusted to 6.8), 7% SDS, 20% glycerol, 2 m urea, and 10% β-mercaptoethanol was added and incubated for 1 h at room temperature. Unsolubilized material was removed by centrifugation at 15,000g for 5 min prior to loading samples onto the SDS-PAGE apparatus. Aliquots corresponding to an equal amount of chlorophyll were loaded in the wells of the stacking gel and electrophoresed through the 12.5% SDS-polyacrylamide running gels containing 1 m urea, as described by Tetali et al. (2007).
Generation of REP27-Specific Polyclonal Antibodies and Western-Blot Analysis
Specific polyclonal antibodies were generated in rabbit against the REP27 protein using a C-terminal portion of the REP27 protein as recombinant antigen. Nucleotide fragments corresponding to Phe-273 to Leu-367 of Chlamydomonas REP27 were amplified using the primers listed in Table I and subcloned into the pET15b vector (Novagen), and the resulting constructs were expressed in Escherichia coli BL21 (DE3) (Novagen). Expressed protein fragments with 6× His tag were purified through a nickel-nitrilotriacetic acid agarose column and injected into rabbits according to the standard protocol of ProSci Inc. After three injections, cleared sera were used as antibodies against each antigen. For immunoblot analyses, SDS-PAGE-resolved proteins were transferred onto polyvinylidene difluoride membranes (Millipore). The latter were blocked and subsequently incubated with rabbit immune serum containing specific polyclonal antibodies against the REP27 protein. Cross-reactions were visualized with horseradish peroxidase-conjugated secondary antibodies (Bio-Rad). Specific polyclonal antibodies against the D1/32-kD PSII reaction center protein were also employed, as described by Park and Rodermel (2004). Cross-reactions between protein bands and antibodies were visualized by the Supersignal ECL Detection Kit (Pierce) following the manufacturer's specifications.
Measurement of Photosynthetic Parameters
The maximum quantum yield of primary PSII photochemistry was determined from measurements of the fluorescence yield ratio Fv/Fm [= (Fm − F0)/Fm], performed with a Plant Efficiency Analyzer fluorometer (Hansatech). The Plant Efficiency Analyzer saturating flash was provided by an array of six light-emitting diodes giving a maximum emission at 650 nm with an intensity of 3,000 μmol photons m−2 s−1. The fluorescence yield at 50 μs after the flash was considered as the nonvariable F0 value, and the maximum fluorescence yield attained at later times after the flash was considered as the Fm (Strasser et al., 1995).
Cell and Thylakoid Membrane Fractionations
Algal cells were harvested upon centrifugation (Beckman JA-10 rotor) at 5,000g for 5 min. Pelleted cells were resuspended in sonication buffer containing protease inhibitors (50 mm Tricine/NaOH, pH 7.8, 10 mm NaCl, 5 mm MgC12, 1 mm aminocaproic acid, 1 mm aminobenzamidine, 0.1 mm phenylmethylsufonyl fluoride, and 2 mm Na-ascorbate) and sonicated on ice three times for 60 s in a 50% duty cycle pulse mode, with 60-s cooling intervals between (Branson sonifier). The crude homogenate was then centrifuged at 3,000g for 5 min in order to remove unbroken cells and large cell fragments. To separate membrane-bound from soluble proteins, the crude homogenate was subjected to centrifugation at 20,000g for 60 min. The pellet was used as the total thylakoid membrane fraction. The supernatant was used as the soluble fraction. Appressed and nonappressed thylakoid membrane regions were isolated upon mechanical fractionation and differential centrifugation, as described previously (Neale and Melis, 1991). Thylakoid membrane vesicles were precipitated by differential centrifugation at 10,000g (10k fraction), 40,000g (40k fraction), and 140,000g (140k fraction). The chlorophyll a/b ratio was determined in all fractions to provide a measure of the differential enrichment of grana- and stroma-exposed thylakoid membranes and an indication of the PSII-PSI ratio. The supernatant of the 140k fraction was used as the soluble fraction. Proteins of all subfractions were separated by SDS-PAGE and probed with specific polyclonal antibodies.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EF127650.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignments of REP27, PratA, and LPA1 proteins.
Supplementary Material
Acknowledgments
D.D. acknowledges postdoctoral fellowship support from the Natural Science and Engineering Research Council of Canada. J.G.G.-C. acknowledges a fellowship award by the Lawski foundation and exchange student grants from the Teknisk-Naturvetenskapliga Fakulteten of Umeå University for the visit to the University of California, Berkeley.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Anastasios Melis (melis@nature.berkeley.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adir N, Shochat S, Ohad I (1990) Light-dependent D1 protein synthesis and translocation is regulated by reaction center II: reaction center II serves as an acceptor for the D1 precursor. J Biol Chem 265 12563–12568 [PubMed] [Google Scholar]
- Arnon D (1949) Copper enzymes in isolated chloroplasts: polyphenol oxidase in Beta vulgaris. Plant Physiol 24 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143 113–134 [DOI] [PubMed] [Google Scholar]
- Bailey S, Thompson E, Nixon PJ, Horton P, Mullineaux CW, Robinson C, Mann NH (2002) A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J Biol Chem 277 2006–2011 [DOI] [PubMed] [Google Scholar]
- Barkan A, Goldschmidt-Clermont M (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82 559–572 [DOI] [PubMed] [Google Scholar]
- Baroli I, Melis A (1996) Photoinhibition and repair in Dunaliella salina acclimated to different growth irradiances. Planta 198 640–646 [DOI] [PubMed] [Google Scholar]
- Bennoun P, Spierer-Herz M, Erickson J, Girard-Bascou J, Pierre Y, Delosme M, Rochaix J-D (1986) Characterization of the photosystem II mutants of Chlamydomonas reinhardtii lacking the psbA gene. Plant Mol Biol 6 151–160 [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lässle M (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21 932–939 [DOI] [PubMed] [Google Scholar]
- Boudreau E, Nickelsen J, Lemaire SD, Ossenbuhl F, Rochaix JD (2000) The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J 19 3366–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer JR, Packer JCL, McCormack BA, Whitelegge JP, Robinson C, Taylor M (1992) Carboxyl-terminal processing of the D1 protein and photoactivation of water splitting in photosystem II. J Biol Chem 267 5424–5433 [PubMed] [Google Scholar]
- Depege N, Bellafiore S, Rochaix JD (2003) Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299 1572–1575 [DOI] [PubMed] [Google Scholar]
- Diner BA, Ries DF, Cohen BN, Metz JG (1988) COOH-terminal processing of polypeptide D1 of the photosystem II reaction center of Scenedesmus obliquus is necessary for the assembly of the oxygen-evolving complex. J Biol Chem 263 8972–8980 [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JM, Rahire M, Malnoë P, Girard-Bascou J, Pierre Y, Bennoun P, Rochaix J-D (1986) Lack of the D2 protein in a Chlamydomonas reinhardtii psbD mutant affects photosystem II stability and D1 expression. EMBO J 5 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman DS, Levine RP (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 54 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther JE, Melis A (1990) The physiological significance of photosystem II heterogeneity in chloroplasts. Photosynth Res 23 105–109 [DOI] [PubMed] [Google Scholar]
- Guenther JE, Nemson JA, Melis A (1990) Development of PSII in dark grown Chlamydomonas reinhardtii: a light-dependent conversion of PSII-beta, QB-nonreducing centers to the PSII-alpha, QB-reducing form. Photosynth Res 24 35–46 [DOI] [PubMed] [Google Scholar]
- Kettunen R, Pursiheimo S, Rintamaki E, van Wijk KJ, Aro EM (1997) Transcriptional and translational adjustments of psbA gene expression in mature chloroplasts during photoinhibition and subsequent repair of photosystem II. Eur J Biochem 247 441–448 [DOI] [PubMed] [Google Scholar]
- Kim J, Klein PG, Mullet JE (1994) Synthesis and turnover of photosystem II reaction center protein D1: ribosome pausing increases during chloroplast development. J Biol Chem 269 17918–17923 [PubMed] [Google Scholar]
- Kindle KL (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M, Butler WL (1975) Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. Biochim Biophys Acta 376 105–115 [DOI] [PubMed] [Google Scholar]
- Klinkert B, Ossenbühl F, Sikorski M, Berry S, Eichacker L, Nickelsen J (2004) PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp PCC 6803. J Biol Chem 279 44639–44644 [DOI] [PubMed] [Google Scholar]
- Lindberg P, Melis A (2008) The chloroplast sulfate transport system in the green alga Chlamydomonas reinhardtii. Planta 228 951–961 [DOI] [PubMed] [Google Scholar]
- Mattoo AK, Edelman M (1987) Intramembrane translocation and posttranslational palmitoylation of the chloroplast 32-kD herbicide-binding protein. Proc Natl Acad Sci USA 84 1497–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul JE, Lilly JW, Cui L, DePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A (1991) Dynamics of photosynthetic membrane composition and function. Biochim Biophys Acta 1058 87–106 [Google Scholar]
- Melis A (1999) Photosystem II damage and repair cycle in chloroplasts: what modulates the rate of photodamage? Trends Plant Sci 4 130–135 [DOI] [PubMed] [Google Scholar]
- Melis A, Spangfort M, Andersson B (1987) Light absorption and electron transport balance between photosystem II and photosystem I in spinach chloroplasts. Photochem Photobiol 45 129–136 [Google Scholar]
- Minai L, Wostrikoff K, Wollman FA, Choquet Y (2006) Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey PJ, McCauley SW, Melis A (1986) Differential detergent-solubilization of integral thylakoid membrane complexes in spinach chloroplasts: localization of photosystem II, cytochrome b6-f complex and photosystem I. Eur J Biochem 160 389–393 [DOI] [PubMed] [Google Scholar]
- Neale PJ, Melis A (1990) Activation of a reserve pool of photosystem II in Chlamydomonas reinhardtii counteracts photoinhibition. Plant Physiol 92 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale PJ, Melis A (1991) Dynamics of photosystem II heterogeneity during photoinhibition: depletion of PSIIβ from non-appressed thylakoids during strong-irradiance exposure of Chlamydomonas reinhardtii. Biochim Biophys Acta 1056 195–203 [Google Scholar]
- Neidhardt J, Benemann JR, Zhang L, Melis A (1998) Photosystem II repair and chloroplast recovery from irradiance stress: relationship between chronic photoinhibition, light-harvesting chlorophyll antenna size and photosynthetic productivity in Dunaliella salina (green algae). Photosynth Res 56 175–184 [Google Scholar]
- Ohnishi N, Allakhverdiev SI, Takahashi FS, Higashi S, Watanabe M, Nishiyama Y, Murata N (2005) Two-step mechanism of photodamage to photosystem II: step 1 occurs at the oxygen-evolving complex and step 2 occurs at the photochemical reaction center. Biochemistry 44 8494–8499 [DOI] [PubMed] [Google Scholar]
- Park S, Khamai P, Garcia-Cerdan JG, Melis A (2007) REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol 143 1547–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Rodermel SR (2004) Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc Natl Acad Sci USA 101 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L (2006) LOW PSII ACCUMULATION 1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B, Argos P (1994) Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol 237 182–192 [DOI] [PubMed] [Google Scholar]
- Polle JE, Kanakagiri SD, Melis A (2003) tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 217 49–59 [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroth S, Meyer JMW, Tittingdorf Z, Krause F, Dencher NA, Seelert H (2003) Thylakoid membrane at altered metabolic state: challenging the forgotten realms of the proteome. Electrophoresis 24 2814–2823 [DOI] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217 220–230 [DOI] [PubMed] [Google Scholar]
- Schottkowski M, Gkalympoudis S, Tzekova N, Stelljes C, Schünemann D, Ankele E, Nickelsen J (2009) Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem 284 1813–1819 [DOI] [PubMed] [Google Scholar]
- Schuster G, Timberg R, Ohad I (1988) Turnover of thylakoid photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur J Biochem 177 403–410 [DOI] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277 221–229 [DOI] [PubMed] [Google Scholar]
- Somanchi A, Mayfield SP (2001) Regulation of chloroplast translation. In E-M Aro, B Andersson, eds, Advances in Photosynthesis and Respiration, Vol 11, Regulation of Photosynthesis. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 137–151
- Strasser RJ, Srivastava A, Govindjee A (1995) Polyphasic chlorophyll a fluorescence transient in plants and cyanobacteria. Photochem Photobiol 61 32–42 [Google Scholar]
- Tetali SD, Mitra M, Melis A (2007) Development of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii is regulated by the novel Tla1 gene. Planta 225 813–829 [DOI] [PubMed] [Google Scholar]
- Thidholm E, Lindstrom V, Tissier C, Robinson C, Schroder WP, Funk C (2002) Novel approach reveals localization and assembly pathway of the PsbS and PsbW proteins into the photosystem II dimer. FEBS Lett 513 217–222 [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17 849–850 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi E, Aro EM (1996) The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc Natl Acad Sci USA 93 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Goldschmidt-Clermont M, Wostrikoff K, Rochaix JD (2000) Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J 21 469–482 [DOI] [PubMed] [Google Scholar]
- Wollman F-A, Minai L, Nechustai R (1999) The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim Biophys Acta 1141 21–85 [DOI] [PubMed] [Google Scholar]
- Yokthongwattana K, Melis A (2006) Photoinhibition and recovery in oxygenic photosynthesis: mechanism of a photosystem II damage and repair cycle. In B Demmig-Adams, WW Adams III, AK Mattoo, eds, Photoprotection, Photoinhibition, Gene Regulation and Environment. Springer, Dordrecht, The Netherlands, pp 175–191
- Zerges W, Rochaix JD (1998) Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol 140 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Aro EM (2002) Synthesis, membrane insertion and assembly of the chloroplast-encoded D1 protein into photosystem II. FEBS Lett 512 13–18 [DOI] [PubMed] [Google Scholar]
- Zhang L, Niyogi KK, Baroli I, Nemson JA, Grossman A, Melis A (1997) DNA insertional mutagenesis for the elucidation of a PSII repair process in the green alga Chlamydomonas reinhardtii. Photosynth Res 53 173–184 [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (1999) Co-translational assembly of the D1 protein into photosystem II. J Biol Chem 274 16062–16067 [DOI] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (2000) Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, assembly into photosystem II. Plant Cell 12 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.