Abstract
Low phosphorus (P) availability is a major constraint to crop growth and production, including soybean (Glycine max), on a global scale. However, 50% to 80% of the total P in agricultural soils exists as organic phosphate, which is unavailable to plants unless hydrolyzed to release inorganic phosphate. One strategy for improving crop P nutrition is the enhanced activity of acid phosphatases (APases) to obtain or remobilize inorganic phosphate from organic P sources. In this study, we overexpressed an Arabidopsis (Arabidopsis thaliana) purple APase gene (AtPAP15) containing a carrot (Daucus carota) extracellular targeting peptide in soybean hairy roots and found that the APase activity was increased by 1.5-fold in transgenic hairy roots. We subsequently transformed soybean plants with AtPAP15 and studied three homozygous overexpression lines of AtPAP15. The three transgenic lines exhibited significantly improved P efficiency with 117.8%, 56.5%, and 57.8% increases in plant dry weight, and 90.1%, 18.2%, and 62.6% increases in plant P content, respectively, as compared with wild-type plants grown on sand culture containing phytate as the sole P source. The transgenic soybean lines also exhibited a significant level of APase and phytase activity in leaves and root exudates, respectively. Furthermore, the transgenic lines exhibited improved yields when grown on acid soils, with 35.9%, 41.0%, and 59.0% increases in pod number per plant, and 46.0%, 48.3%, and 66.7% increases in seed number per plant. Taken together, to our knowledge, our study is the first report on the improvement of P efficiency in soybean through constitutive expression of a plant APase gene. These findings could have significant implications for improving crop yield on soils low in available P, which is a serious agricultural limitation worldwide.
Phosphorus (P) is a critical macronutrient for plant growth and development. Terrestrial plants generally take up soil P in its inorganic form (Pi; Marschner, 1995). However, 50% to 80% of the total P in agricultural soils exists as organic phosphate, in which, up to 60% to 80% is myoinositol hexakisphosphate (phytate; Iyamuremye et al., 1996; Turner et al., 2002; George and Richardson, 2008). Since phytate-P is not directly available to plants, low P availability becomes one of the limiting factors to plant growth.
Plants have developed a number of adaptive mechanisms for better growth on low-P soils, including changes in root morphology and architecture, activation of high-affinity Pi transporters, improvement of internal phosphatase activity, and secretion of organic acids and phosphatases (Raghothama, 1999; Vance et al., 2003). Acid phosphatases (APases) are hydrolytic enzymes with acidic pH optima that catalyze the breakdown of P monoesters to release Pi from organic P compounds, and therefore may play an important role in P nutrition (Vincent et al., 1992; Li et al., 2002). APase activity, including extracellular and intracellular APase activity, is generally increased by Pi starvation in higher plants (Duff et al., 1994). Intracellular APases might play a role in internal Pi homeostasis through remobilization of Pi from older leaves and vacuole stores, whereas extracellular APases are believed to be involved in external P acquisition by mobilizing Pi from organic P compounds (Duff et al., 1994). In the last few years, secreted APases have been purified and characterized in some model plants, such as Arabidopsis (Arabidopsis thaliana; Coello, 2002) and tobacco (Nicotiana tabacum; Lung et al., 2008). Furthermore, an Arabidopsis pup3 mutation that underproduced secreted APases in root tissues accumulated 17% less P in shoots when organic P was supplied as the major P source (Tomscha et al., 2004), indicating the possible role of APases during plant growth in response to Pi starvation.
Phytase is a special type of APases with the capability to hydrolyze phytate and its derivatives, which are the predominant inositol phosphates present in seeds and soils. It is generally believed that phytase activation in seeds or resynthesis in plants plays important roles in Pi remobilization through hydrolyzing the phytate into Pi during seed germination (Loewus and Murthy, 2000). Furthermore, phytase in roots and/or root exudation has been demonstrated to be important for utilizing Pi from phytate in the growth medium (Asmar, 1997; Li et al., 1997; Hayes et al., 1999; Richardson et al., 2000).
AtPAP15, a purple APase with confirmed phytase activity from Arabidopsis, can hydrolyze myoinositol hexakisphosphate, yielding myoinositol and Pi (Zhang et al., 2008). Overexpression of AtPAP15 in Arabidopsis significantly decreased phytate content in leaves (Zhang et al., 2008). Sequence analysis indicates that AtPAP15 exhibits 74% similarity to the soybean (Glycine max) phytase gene, GmPhy (Hegeman and Grabau, 2001). It seems likely that the possible involvement of phytase in plant P nutrition might be conserved among different plant species. But it is still unclear whether AtPAP15 or other phytases can be used to directly help crops, including the major agronomic crop, soybean, to acquire P under low-P conditions.
Soybean is one of the most important food crops, accounting for a large segment of the world market in oil crops and also serving as an important protein source for both human consumption and animal feed (Kereszt et al., 2007). Soybean is mainly cultivated in tropic, subtropic, and temperate areas, where the soils are low in P due to intensive erosion, weathering, and strong P fixation by free iron and aluminum oxides (Sample et al., 1980; Stevenson, 1986). Low P availability is especially problematic for soybean, since root nodules responsible for nitrogen fixation have a high P requirement (Robson, 1983; Vance, 2001).
In this study, the Arabidopsis PAP15 gene directed by an extracellular targeting sequence from a carrot (Daucus carota) extensin gene was successfully transformed into both soybean hairy roots and whole soybean plants. Overexpression of AtPAP15 not only increased the secretion of APase from transgenic soybean hairy roots and roots of whole transgenic soybean plants, but also significantly improved APase activity in leaves, as well as P efficiency and yield in the transgenic soybean lines. To the best of our knowledge, this is the first report on the improvement of P efficiency in crop plants through constitutive expression of a plant APase gene. This study could have significant implications for improving crop production on low-P soils, which is a serious agronomic limitation worldwide.
RESULTS
APase Activity in Soybean Hairy Roots Overexpressing AtPAP15
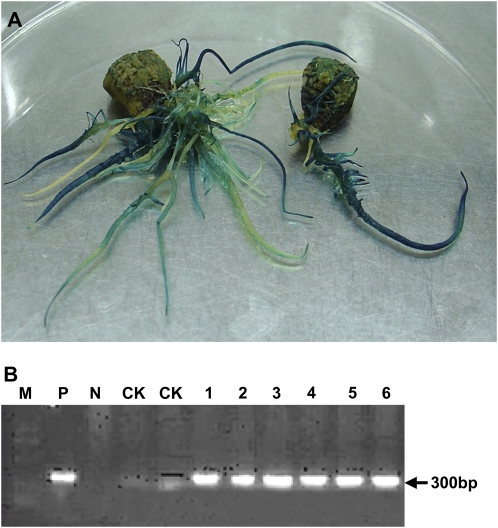
In this study, the binary vector pCAMBIA3301-sp-AtPAP15-GUS was introduced into soybean hairy roots using Agrobacterium rhizogenes-mediated transformation. The transgenic hairy roots were verified by GUS activity staining and genomic PCR amplification (Fig. 1, A and B).
Figure 1.
Expression analyses of the transgenic hairy roots overexpressing AtPAP15. A, GUS histochemical staining of HN66 hairy roots 2 weeks after infection. B, PCR analysis of hairy root DNA using the primers to amplify a 300-bp fragment of the AtPAP15 gene. M, Marker; P, positive control, the AtPAP15 plasmid as template; N, negative control, soybean hairy roots transformed with K599 strain only as templates; CK, soybean hairy roots transformed with the control vector pCAMBIA3301; 1 to 6, individual lines transformed with the binary vector pCAMBIA3301-sp-AtPAP15-GUS.
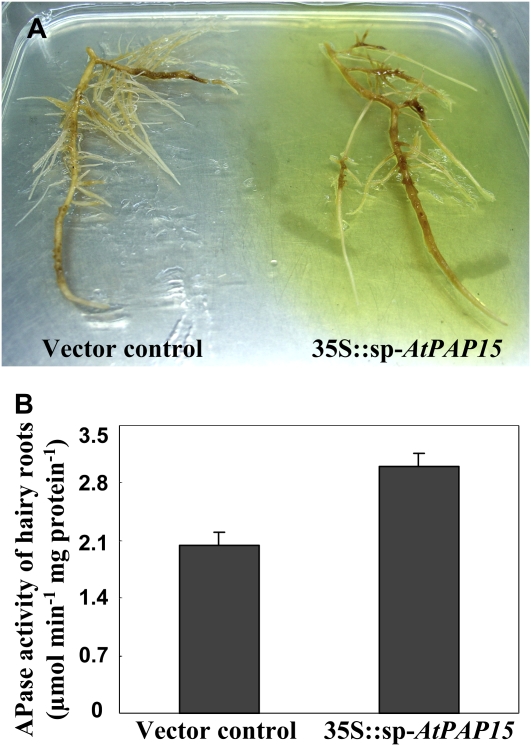
After 20 d of growth on MXB medium (see ?Materials and Methods?), the transgenic hairy roots were transferred to MXB medium containing ρ-nitrophenyl phosphate (ρ-NPP) for another week. The yellow color due to the staining of secreted APase was significantly more intense in the transgenic hairy roots overexpressing AtPAP15 compared with the hairy roots transformed by the vector control (Fig. 2A). APase activity was quantified in hairy roots, and the hairy roots overexpressing AtPAP15 exhibited a 1.5-fold increase in APase activity compared with the hairy roots transformed by the vector control (Fig. 2B). This finding suggested that expression of AtPAP15 in soybean hairy roots indeed increased both APase activity in roots and APase secretion from roots, and thus might improve the P efficiency of whole plants.
Figure 2.
Analyses of APase in the transgenic hairy roots. A, In situ staining for the activity of secreted APases. The yellow color indicates enzyme activity in root exudates. Shown are hairy roots transformed with A. rhizogenes K599 with the binary vector pCAMBIA3301-sp-AtPAP15-GUS (right) or the control vector (left). B, APase activities in the transgenic hairy roots. Data in the figure are the mean of three replicates ±se.
Generation of Transgenic Soybean Lines Overexpressing AtPAP15
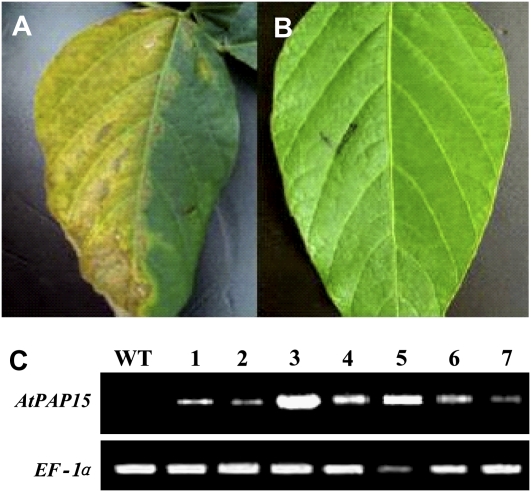
A total of 15 T0 lines were successfully regenerated to yield fertile transgenic plants. Seven stable lines were selected based on the herbicide resistance of the 15 T0 transgenic plants and AtPAP15 expression using semiquanitative reverse transcription-PCR (Fig. 3). The T0 plants were self-pollinated to ultimately obtain T3 transgenic lines for further analysis.
Figure 3.
Expression analyses of AtPAP15 in transgenic soybean plants. A and B, Identification of putative transgenic soybeans by herbicide tolerance. A, Wild-type soybean. B, T0 plants transformed with AtPAP15. C, Expression analysis of AtPAP15 in transgenic soybeans by reverse transcription-PCR. WT, Wild-type soybean; 1 to 7, transformed T0 plants with AtPAP15. Soybean EF-1α gene was used as an internal control.
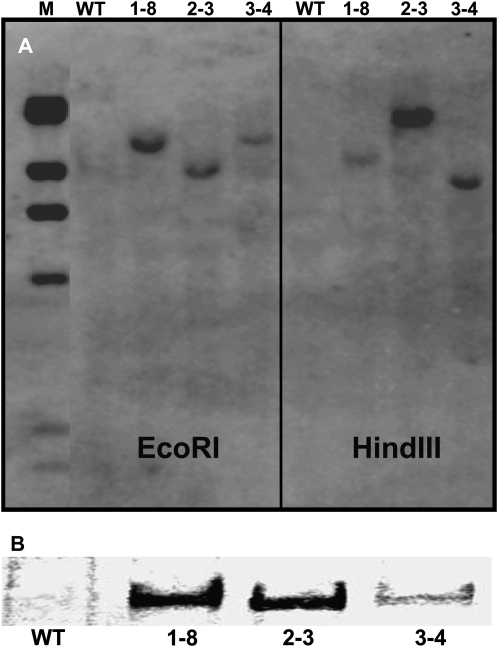
Three soybean transgenic events were subsequently selected for further analyses based on their high expression of AtPAP15. These soybean events were designated 1-8, 2-3, and 3-4. Southern blotting demonstrated that the exogenous AtPAP15 was correctly integrated into the genome of transgenic soybean plants, and transgenic lines 1-8, 2-3, and 3-4 all harbored one transgenic locus (Fig. 4A). The production of AtPAP15 in these three lines was verified by western-blot analysis (Fig. 4B). Immunoreactive bands were observed in all root proteins extracted from the three AtPAP15 transgenic lines, but none were detected in the wild-type root samples.
Figure 4.
Expression analyses of AtPAP15 in the three independent transgenic soybean plants. A, Southern-blot analysis of transgenic lines using the Bar probe. DNA was digested by EcoRI or HindIII. M, Marker; WT, wild-type soybean; 1-8, 2-3, and 3-4, plants transformed with AtPAP15. B, Detection of AtPAP15 protein in the roots of transgenic lines by western-blot analysis. WT, Wild-type soybean; 1-8, 2-3, and 3-4, plants transformed with AtPAP15.
Overexpression and Excretion of AtPAP15 in Soybean Improves APase and Phytase Activity
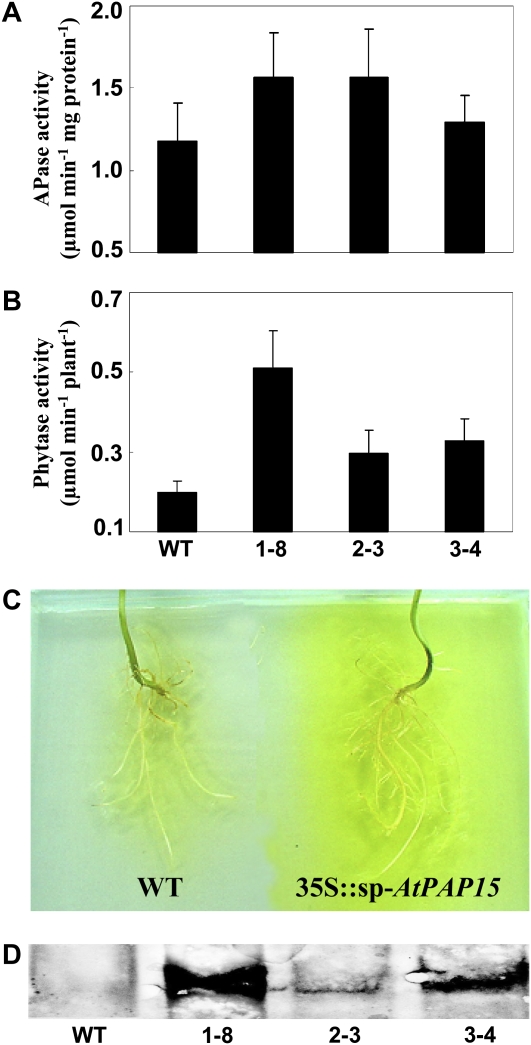
APase activity of leaves and extracellular phytase activity in root exudates was analyzed in the transgenic lines. APase activity in leaves of transgenic plants increased 32.7%, 33.1%, and 9.7% compared to wild-type soybean, respectively (Fig. 5A). Secreted phytase from the transgenic lines increased the extracellular phytase activity by 159.3%, 50.1%, and 66.3% compared to that measured in exudates of wild-type plants, respectively (Fig. 5B). This was further confirmed by staining of transgenic lines with ρ-NPP as substrate and western-blot analysis of root exudates (Fig. 5, C and D).
Figure 5.
APase and phytase activities in transgenic soybean plants. A, APase activity in leaves. B, Phytase activities in root exudates of different transgenic lines. Data in the figure are the mean of three replicates ± se. C, Exudation of APase activity in transgenic (right) and wild-type (left) soybean. The yellow color indicates APase enzyme activity in root exudates. D, Detection of AtPAP15 protein in root exudates by western-blot analysis. WT, Wild-type soybean; 1-8, 2-3, and 3-4, plants transformed with AtPAP15.
Overexpression of AtPAP15 Improves Soybean Biomass and P Accumulation in a Sand Culture Experiment
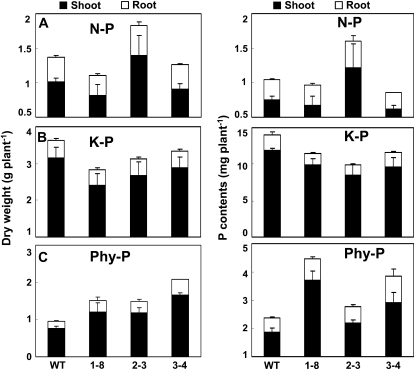
We subsequently performed a sand culture experiment to evaluate the response of transgenic plants to the supply of phytate-P in the growth medium. Phenotypic observations showed that the overall morphology of the three independent transgenic lines was similar to that of the wild-type plants under both N-P and K-P conditions except that transgenic line 2-3 showed a more than 20% increase in dry weight compared to the wild-type plants without P addition (Figs. 6, A and B, and 7, A and B). APase activities in the roots of the three transgenic lines were 81.4%, 42.2%, and 46.1% higher than that of wild-type plants when phy-P was supplied as the sole P source, respectively (Supplemental Fig. S1C). Furthermore, the three transgenic lines grew much better and exhibited significantly improved P efficiency with 117.8%, 56.5%, and 57.8% increases in plant dry weight and 90.1%, 18.2%, and 62.6% increases in plant P content, respectively, as compared with wild-type plants when phytate was the sole P source (Figs. 6C and 7C).
Figure 6.
Effects of different P sources on plant growth in sand culture. Wild-type (WT; far left) and transgenic soybean (next three plants) were grown for 30 d in sand culture with different P sources. A, N-P treatment (no P addition). B, K-P treatment (100 mg P kg−1 sand with KH2PO4). C, Phy-P treatment (100 mg P kg−1 sand with phytate).
Figure 7.
Effects of different P sources on plant biomass and P content of transgenic and wild-type (WT) soybean grown in sand culture for 30 d with different P sources. A, N-P treatment (no P addition). B, K-P treatment (100 mg P kg−1 sand with KH2PO4). C, Phy-P treatment (100 mg P kg−1 sand with phytate). Data in the figure are the mean of three replicates ± se.
Overexpression of AtPAP15 Improves Soybean Yield Potential on Acid Soils
To study whether constitutive overexpression of AtPAP15 could help improve crop yield, a field experiment was carried out on an acidic soil. We found that overexpression of AtPAP15 improved soybean yield potential of the three transgenic soybean lines, 1-8, 2-3, and 3-4, with 35.9%, 41.0%, and 59.0% increases in the pod number per plant, and 46.0%, 48.3%, and 66.7% increases in seed number per plant, respectively, as compared with wild-type plants on acid soils (Table I).
Table I.
Number of pods and seeds for transgenic and wild-type soybean plants grown on acidic red soils
Data are the mean of 30 plants ± se.
| Soybean Lines | Pod No. | Seed No. |
|---|---|---|
| # plant−1 | ||
| Wild type | 39 ± 3.40 | 87 ± 7.11 |
| 1-8 | 53 ± 1.46 | 127 ± 3.71 |
| 2-3 | 55 ± 1.91 | 129 ± 4.58 |
| 3-4 | 62 ± 2.21 | 145 ± 5.71 |
DISCUSSION
In this study, both hairy root and whole-plant transformation systems were used to test the effects on plant P nutrition of overexpressing a heterologous Arabidopsis purple APase gene (AtPAP15) in soybean. The results indicated that both APase activity in hairy roots and APase secretion from hairy roots were increased in AtPAP15 transgenic lines as compared with hairy roots transformed with the control vector (Fig. 2, A and B). In transformed intact soybean plants, significant increases in both APase activity in leaves and phytase activity in root exudates were observed in three independently transformed lines compared with wild-type soybean (Fig. 5, A and B). This enhanced APase and/or phytase activity resulted in improved P efficiency in the three transgenic lines, which exhibited significant increases in plant dry weight as compared with wild-type plants, when phytate was used as the sole P source in sand culture (Fig. 7C). Furthermore, the results from the field experiment indicated that the enhanced APase activity also improved the yield of transgenic soybean lines in the field with both higher pod and seed number per plant grown on acid soils (Table I). These findings indicate this strategy has great potential for improving soybean production on acid soils.
In higher plants, the induction of APase activity has been reported as one of the indicators of plants response to Pi starvation. Some studies showed that APase activities were indeed increased by P stress, indicating that intracellular APase activity may play an important role in P efficiency through internal remobilization of storage P (Helal, 1990; Tian et al., 2003). Our results showed that the APase activity of both old and young leaves under low-P treatment were significantly higher than those under K-P and Phy-P treatments in all the soybean lines, indicating that soybean plants could activate APases and utilize endogenous organic P storage in both young and old leaves when P supply became limited (Supplemental Fig. S1, A and B). Furthermore, the APase activity in leaves was greater in the transgenic lines overexpressing AtPAP15 than that in wild-type plants under both N-P and Phy-P conditions (Supplemental Fig. S1, A and B), indicating that overexpressing AtPAP15 might increase soybean P efficiency by enhanced intracellular APase activity. This was proved by our results showing greater yields for transgenic soybean lines from the field experiment, which was conducted on an acid soil with very low organic P content of only 17.7% of total soil P. These findings suggest that the increased yield of transgenic lines on this field site may be mainly due to enhanced internal P use efficiency through releasing Pi from stored phytate/organic P in the shoots, rather than increased P uptake from the soil. The transgenic line 3-4 had the highest APase activity in both old and young leaves for plants grown on low P in sand culture, and also produced the highest pod and seed number in the field (Supplemental Fig. S1, A and B; Table I). These findings further indicate that enhanced intracellular APase activity could increase the utilization of P in the shoots of the transgenic plants in soils with low organic P content.
Plant roots with higher phosphatase activity may have a greater potential to utilize soil organic P (Helal, 1990). It has been proposed that variations in root phosphatase activity may be useful in the selection of genotypes for greater utilization of soil organic P (Asmar et al., 1995). In this study we found that root APase activities were increased 81.4%, 42.2%, and 46.1% in the three independent transgenic soybean lines when phytate was used as the sole P resource in sand culture (Supplemental Fig. S1C), indicating that the increased P efficiency of transgenic soybean lines could also be due to the enhanced root APase activity.
In native plants, APase and phytase secretion from roots was also increased in response to P deficiency and it was suggested to be a major contributor for plant assimilation of organic P from soils (Li et al., 1997; Hayes et al., 1999, 2000; Lung and Lim, 2006; George and Richardson, 2008). But the studies to date have been tested only in model plant species under sterilized conditions. For example, overexpression of phytase genes from a soil fungus (Aspergillus niger) or a soil bacteria (Bacillus subtilis) in transgenic model plants (such as Arabidopsis and tobacco) significantly increased exudation of phytase from plant roots (Richardson et al., 2001; Mudge et al., 2003; George et al., 2005; Lung et al., 2005), and several studies where the plant purple APase genes such as MtPHY1 and MtPAP1 were expressed in transgenic Arabidopsis and increased phytase exudation was seen (Xiao et al., 2005, 2006). In this study, we transformed a plant APase gene with phytase activity (AtPAP15) into an agriculturally important crop, soybean, and showed significantly increased phytase secretion from soybean roots. Three independent transgenic soybean lines not only exhibited 2.6, 1.5, and 1.7 times more secreted phytase activity than controls in hydroponics, respectively (Fig. 5B), but also significantly increased dry weight and P content in sand culture when phytate was supplied as the sole P source (Fig. 7C). The results from in situ staining using ρ-NPP as substrate for the activity of APase in the transgenic hairy roots (Fig. 2A) and whole-plant roots (Fig. 5C) also provided direct evidence that secreted APases could solublize the organic P in the growth media and release Pi for plant growth. This is advantageous for improving plant P acquisition, because phytate comprises up to 60% to 80% of soil organic P and is poorly utilized by plants (Iyamuremye et al., 1996; Turner et al., 2002; George and Richardson, 2008).
In summary, to our knowledge, this study is the first report of the expression of a plant APase gene in an important crop species, soybean, which led to a significant improvement in P efficiency in sand culture when phytate was supplied as the sole P source, and plant yield in the field on acid soils. The findings reported here should provide important new avenues of research aimed at the development of better crop varieties that are more efficient in P nutrition, representing possibly the best strategy for reducing the use of P fertilizers, expanding agriculture on low-P soils, and achieving more sustainable agriculture.
MATERIALS AND METHODS
Vector Construction
The complete coding sequences of AtPAP15 (At3g07130) was PCR amplified from the cDNA of Arabidopsis (Arabidopsis thaliana) ecotype Columbia, using gene-specific oligonucleotide primers (forward primer: ATATGTCGACATGACGTTTCTACTACTTCTAC, reverse primer: GGACTAGTTCAGTGGTGGTGGTGGTGGTGGCAATGGTTAACAAGGCGGT) that introduced a SalI site and a SpeI site at the 5′ end and the 3′ end, respectively, of the derived PCR clone of AtPAP15 (underlined). The PCR product was subcloned into vector pGEM-T easy vector (Promega).
For soybean (Glycine max) hairy root transformation, the 35S cauliflower mosaic virus (CaMV) AtPAP15 cassette including a carrot (Daucus carota) extensin leader signal peptide (Lung et al., 2005) was subcloned into pCAMBIA3301 (www.cambia.org) with the bar gene as the selective marker and GUS as the reporter gene, to create pCAMBIA3301-sp-AtPAP15-GUS vector.
For soybean whole-plant transformation, pTF101.1-sp-AtPAP15 was created by inserting a HindIII-SacI fragment from the CaMV35S∷sp-AtPAP15 into HindIII-SacI sites of the binary vector pTF101.1. pTF101.1 harbors a bar gene under the control of the 2× 35S CaMV promoter (Paz et al., 2004).
Soybean Transformation
Soybean transformations were made using the HN66 cultivar that was bred in our center and characterized as a P-efficient genotype in field trials. Seeds were surface sterilized for 13.5 h using chlorine gas before germination in B5 medium.
For hairy root transformation mediated by Agrobacterium rhizogenes, plant inoculation was conducted according to Cho et al. (2000) with some modifications. Cotyledons from 5 d seedlings were harvested and wounded with a scalpel previously dipped into an overnight culture of A. rhizogenes strain carrying the binary vectors, and incubated at 25°C in light for 5 d. Then cotyledons were cultured on MXB medium (Murashige and Skoog basal nutrient salts, B5 vitamins, 3% Suc, and 3 g L−1 phytagel [pH 5.7]) at 25°C in dark. Carbenicillin disodium was added to the MXB medium to inhibit overgrowth of A. rhizogenes, and glufosinate was added to select the transgenic hairy roots. After 10 to 14 d, hairy roots formed at the wounded sites were tested for GUS activity. Forty to 50 cotyledons were inoculated with K599 with pCAMBIA3301 as vector control or the pCAMBIA3301-sp-AtPAP15-GUS binary vector.
For whole-plant transformation mediated by A. tumefaciens, the cotyledonary-node method described by Paz et al. (2004) was adopted with some modifications. Transformants were selected on 3.5 mg L−1 glufosinate 4 weeks after shoot initiation, followed by an additional 4 to 6 weeks under 2.5 mg L−1 glufosinate selection for shoot elongation. Primary transformants were established and grown to maturity in the greenhouse.
Identification of Transformants
Transgenic hairy roots were initially analyzed by PCR amplification. Primers 5′-GCTCTAGAATGGGAAGAATTGCTAGAGGCTCAAAAATGAG-3′ and 5′-GAGACCCAGATGGAATCATGATCGGA-3′, were designed to amplify a 300-bp fragment of the AtPAP15 gene. R0 plants were screened with 135 mg L−1 Liberty (AgrEvo) as described by Paz et al. (2004). Glufosinate-resistant R0 plants were grown in the greenhouse to maturity and seeds were harvested.
Histochemical localization of GUS activity was performed using 5-bromo-4-chloro-3-indolyl glucuronide as the chromogenic substrate. A reaction mixture consisting of 1 mm 5-bromo-4-chloro-3-indolyl glucuronide dissolved in 50 mm sodium phosphate buffer (pH 7.2) was used. Tissues were incubated for 12 h at 37°C and pigments were removed by extraction with 100% ethanol prior to observation.
Primary transformants and a subset of individuals from subsequent generations were analyzed by Southern blot. Total genomic DNA was isolated from leaves using the cetyltrimethyl ammonium bromide extraction method. Ten micrograms of genomic DNA digested with EcoRI or HindIII was separated on 0.8% (w/v) agarose gel and transferred to the nylon membranes, which were hybridized with the probes of digoxigenin-labeled PCR product of Bar.
Western blot was carried out to verify the protein expression in root protein extracts and root exudates. Briefly, proteins from plant roots or root exudates were resolved on SDS-PAGE, and electrophoretically transferred to polyvinylidene fluoride membrane (Bio-Rad). The blot was incubated with a rabbit anti-AtPAP15 antibody (1:1,000 dilution) and then with an alkaline-phosphatase-tagged secondary antibody.
Measurement of APase Activities and in Situ Staining
Leaf and root samples were ground with liquid nitrogen and macerated in 1.2 mL extract buffer (45 mm sodium acetate buffer, pH 5.0). APase activities of the supernatant were assayed using ρ-NPP as substrate and phosphatase activity was expressed as μmol substrate hydrolyzed per mg of soluble protein per min.
In situ staining for APase activity was done by culturing transgenic hairy roots or whole-plant roots into the MXB medium with ρ-NPP for 1 week at 25°C.
Measurement of Phytase Activity
Phytase activity was measured as described by Lung et al. (2008) with some modifications. Phytase activity was assayed in 200 μL of 45 mm NaOAc (pH 5.0), using 1 mm C6H6O24P6Na12 (phytate, P3168, Sigma) as substrate. All reactions were carried out at 37°C for 1 h and terminated by an equal volume of 10% (w/v) TCA. The liberated Pi was quantified by molybdenum-blue assay (Murphy and Riley, 1962). Enzyme activity was defined as the activity that released 1 μmol of phosphate per min per plant under the specified assay conditions.
Plant Growth Conditions
For the sand culture experiment, seeds of wild-type and transgenic soybean lines were surface sterilized with 10% (v/v) hydrogen peroxide and planted into the sterilized sand containing 100 mg P kg−1 KH2PO4 or C6H6O24P6Na12 (phytate, P3168, Sigma) as the sole P source. An additional treatment without P was used as a control. The plants were fertilized with the modified one-half-strength Hoagland nutrient solution without P. Thirty days after germination, the plants were harvested for APase activity, P content, and biomass determination. Root samples were extracted for western-blot analysis. P content of shoots and roots was determined by the photocolormetric method (Murphy and Riley, 1962).
In hydroponics, seeds were surface sterilized as described before and germinated in the sterilized sand containing the modified one-half Hoagland nutrient solution with 250 μm KH2PO4. After 5 d, plants were transplanted and cultured into the modified one-half Hoagland nutrient solution with 250 μm KH2PO4 for 10 d. Then the seedlings were transferred to 150 mL modified one-half Hoagland nutrient solution without P for 3 d. The plants were harvested for APase activity determination. The incubated solution was collected and vacuum freeze dried (Labconco) for phytase assays and western-blot analysis.
For the field experiment, wild-type and transgenic soybean lines were sown on acid soils at the Ningxi experimental farm of South China Agricultural University in 2009. Soil P characteristics were as follows: pH, 5.43; available P (Bray I), 50.5 mg P kg−1; total P, 359.1 mg P kg−1; total organic P, 17.7%; organic matter, 1.2%. There were three replicates with 10 soybean plants in each replicate for each plant line. Ninety days after germination, pods were harvested for yield evaluation.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF448726 (AtPAP15) and AF272346 (GmPhy).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. APase activities of transgenic and wild-type soybean grown in sand culture for 30 d under N-P (no P addition), K-P (100 mg P kg−1 sand with KH2PO4), and Phy-P (100 mg P kg−1 sand with phytate).
Supplementary Material
Acknowledgments
We are grateful to Dr. Kan Wang for the generous gift of pTF101.1 vector and Dr. Peter M. Gresshoff for A. rhizogenes strain K599. We thank Dr. Huixia Shou, Jun Fang, Haicui Suo, and Haiqing Wu for technical help.
This work was supported by grants from the National Natural Science Foundation of China (grant no. 30890131) and the National Key Basic Research Special Funds of China (grant no. 2005CB120902).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hong Liao (hliao@scau.edu.cn).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Asmar F (1997) Variation in activity of root extracellular phytase between genotypes of barley. Plant Soil 195 61–64 [Google Scholar]
- Asmar F, Gahoonia TS, Nielsen NE (1995) Barley genotypes differ in activity of soluble extracellular phosphatase and depletion of organic phosphorus in the rhizosphere soil. Plant Soil 172 117–122 [Google Scholar]
- Cho HJ, Farrand SK, Noel GR, Widholm JM (2000) High efficiency induction of soybean hairy roots and progagation of the soybean cyst nematode. Planta 210 195–204 [DOI] [PubMed] [Google Scholar]
- Coello P (2002) Purification and characterization of secreted acid phosphatase in phosphorus-deficient Arabidopsis thaliana. Physiol Plant 116 293–298 [Google Scholar]
- Duff SMG, Gautam S, Plaxtom WC (1994) The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant 90 791–800 [Google Scholar]
- George TS, Richardson AE (2008) Potential and limitations to improving crops for enhanced phosphorus utilization. In PJ White, JP Hammond, eds, Ecophysiology of Plant-Phosphorus Interactions. Springer, Dordrecht, The Netherlands, pp 247–270
- George TS, Simpson RJ, Hadobas PA, Richardson AE (2005) Expression of a fungal phytase gene in Nicotiana tabacum improves phosphorus nutrition of plants grown in amended soils. Plant Biotechnol J 3 129–140 [DOI] [PubMed] [Google Scholar]
- Hayes JE, Richardson AE, Simpson RJ (1999) Phytase and acid phosphatase activities in extracts from roots of temperate pasture grass and legume seedlings. Aust J Plant Physiol 26 801–809 [Google Scholar]
- Hayes JE, Richardson AE, Simpson RJ (2000) Components of organic phosphorus in soil extracts that are hydrolysed by phytase and acid phosphatase. Biol Fertil Soils 32 279–286 [Google Scholar]
- Hegeman CE, Grabau EA (2001) A novel phytase with similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiol 126 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helal HM (1990) Varietal differences in root phosphatase activity as related to the utilization of organic phosphates. Plant Soil 123 161–163 [Google Scholar]
- Iyamuremye F, Dick RP, Baham J (1996) Organic amendment and phosphorus dynamics: II. Distribution of soil phosphorus fractions. Soil Sci 161 436–443 [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CDT, Nontachaiyapoom S, Kinkema M, Gresshoff PM (2007) Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protocols 2 948–952 [DOI] [PubMed] [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D (2002) Purple acid phosphatases of Arabidopsis thaliana: comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277 27772–27781 [DOI] [PubMed] [Google Scholar]
- Li M, Osaki M, Rao IM, Tadano T (1997) Secretion of phytase from the roots of several plant species under phosphorus-deficient conditions. Plant Soil 195 161–169 [Google Scholar]
- Loewus FA, Murthy PPN (2000) myo-Inositol metabolism in plants. Plant Sci 150 1–19 [Google Scholar]
- Lung SC, Chan WL, Yip W, Wang L, Yeung EC, Lim BL (2005) Secretion of beta-propeller phytase from tobacco and Arabidopsis roots enhances phosphorus utilisation. Plant Sci 169 341–349 [Google Scholar]
- Lung SC, Leung A, Kuang R, Wang Y, Leung P, Lim BL (2008) Phytase activity in tobacco (Nicotiana tabacum) root exudates is exhibited by a purple acid phosphatase. Phytochemistry 69 365–373 [DOI] [PubMed] [Google Scholar]
- Lung SC, Lim BL (2006) Assimilation of phytate-phosphorus by the extracellular phytase activity of tobacco (Nicotiana tabacum) is affected by the availability of soluble phytate. Plant Soil 279 187–199 [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants. Academic Press, Harcourt Brace & Company Publishers, London, pp 483–507
- Mudge SR, Smith FW, Richardson AE (2003) Root-specific and phosphate-regulated expression of phytase under the control of a phosphate transporter promoter enables Arabidopsis to grow on phytate as a sole P source. Plant Sci 165 871–878 [Google Scholar]
- Murphy J, Riley J (1962) A modified single solution method for the determination of phosphate in natural water. Anal Chim Acta 27 31–35 [Google Scholar]
- Paz MM, Shou H, Guo Z, Zhang Z, Banerjee AK, Wang K (2004) Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explants. Euphytica 136 167–179 [Google Scholar]
- Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50 665–693 [DOI] [PubMed] [Google Scholar]
- Richardson AE, Hadobas PA, Hayes JE (2000) Acid phosphomonoesterase and phytase activities of wheat (Triticum aestivum L.) roots and utilization of organic phosphorus substrates by seedlings grown in sterile culture. Plant Cell Environ 23 397–405 [Google Scholar]
- Richardson AE, Hadobas PA, Hayes JE (2001) Extracellular secretion of Aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant J 25 641–649 [DOI] [PubMed] [Google Scholar]
- Robson AD (1983) Mineral nutrition. In WJ Broughton, ed, Nitrogen Fixation. Clarendon Press, Oxford, pp 36–55
- Sample EC, Soper RJ, Racz GJ (1980) Reaction of phosphate fertilizers in soils. In FE Khasawneh, EC Sample, EJ Kamprath, eds, The Role of Phosphorus in Agriculture. American Society of Agronomy, Madison, WI, pp 263–310
- Stevenson FJ (1986) Cycles of Soil: Carbon, Nitrogen, Phosphorus, Sulfur, Micronutrients. John Wiley & Sons Inc., New York, pp 350–400
- Tian J, Liao H, Wang X, Yan X (2003) Phosphorus starvation-induced expression of leaf acid phosphatase isoforms in soybean. Acta Bot Sin 45 1037–1042 [Google Scholar]
- Tomscha JL, Trull MC, Deikman J, Lynch JP, Guiltinan MJ (2004) Phosphatase under-producer mutants have altered phosphorus relations. Plant Physiol 135 334–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BL, Paphazy MJ, Haygarth PM, Mckelvie ID (2002) Inositol phosphates in the environment. Philos Trans R Soc Lond B Biol Sci 357 449–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP (2001) Symbiotic nitrogen fixation and phosphorus acquisition: plant nutrition in a world of declining renewable resources. Plant Physiol 127 390–397 [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Uhde-stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157 423–447 [DOI] [PubMed] [Google Scholar]
- Vincent JB, Crowder MW, Averill BA (1992) Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends Biochem Sci 17 105–110 [DOI] [PubMed] [Google Scholar]
- Xiao K, Harrison M, Wang ZY (2005) Transgenic expression of a novel M. truncatula phytase gene results in improved acquisition of organic phosphorus by Arabidopsis. Planta 222 27–36 [DOI] [PubMed] [Google Scholar]
- Xiao K, Katagi H, Harrison M, Wang ZY (2006) Improved phosphorus acquisition and biomass production in Arabidopsis by transgenic expression of a purple acid phosphatase gene from M. truncatula. Plant Sci 170 191–202 [Google Scholar]
- Zhang WY, Gruszewski HA, Chevone BI, Nessler CL (2008) An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol 146 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.