Abstract
Channelrhodopsins are light-gated ion channels involved in the photoresponses of microalgae. Here, we describe the characterization of two channelrhodopsins, Volvox channelrhodopsin-1 (VChR1) and VChR2, from the multicellular green alga Volvox carteri. Both are encoded by nuclear single copy genes and are highly expressed in the small biflagellated somatic cells but not in the asexual reproductive cells (gonidia). Expression of both VChRs increases after cell cleavage and peaks after completion of embryogenesis, when the biosynthesis of the extracellular matrix begins. Likewise, expression of both transcripts increases after addition of the sex-inducer protein, but VChR2 is induced much more than VChR1. The expression of VChR1 is specifically promoted by extended dark periods, and heat stress reduces predominantly VChR1 expression. Expression of both VChRs increased under low light conditions, whereas cold stress and wounding reduced expression. Both VChRs were spectroscopically studied in their purified recombinant forms. VChR2 is similar to the ChR2 counterpart from Chlamydomonas reinhardtii with respect to its absorption maximum (460 nm) and photocycle dynamics. In contrast, VChR1 absorbs maximally at 540 nm at low pH (D540), shifting to 500 nm at high pH (D500). Flash photolysis experiments showed that after light excitation, the D540 dark state bleaches and at least two photoproducts, P600 and P500, are sequentially populated during the photocycle. We hypothesize that VChR2 is a general photoreceptor that is responsible for the avoidance of blue light and might play a key role in sexual development, whereas VChR1 is the main phototaxis photoreceptor under vegetative conditions, as it is more specifically adapted to environmental conditions and the developmental stages of Volvox.
Microbial rhodopsins are photoactive, seven-transmembrane (7TM) helix receptors and use retinal as a chromophore (for review, see Sharma et al., 2006; Spudich, 2006; Jung, 2007; Hegemann, 2008; Klare et al., 2008). The light energy that is absorbed is used either for ion transport or for photosensory purposes. The light-driven proton pump bacteriorhodopsin of the model haloarchaeon Halobacterium salinarum was the first member of this protein family to be identified (Oesterhelt and Stoeckenius, 1973). Shortly thereafter, other members of the family were discovered, such as the chloride pump, halorhodopsin (Matsuno-Yagi and Mukohata, 1977; Schobert and Lanyi, 1982), sensory rhodopsin SRI (Bogomolni and Spudich, 1982), and phoborhodopsin SRII (Takahashi et al., 1985). The increasing number of genomic analyses revealed that microbial rhodopsin-related proteins are also present in eubacteria (Sharma et al., 2006; Jung, 2007), fungi (Brown, 2004), and other lower eukaryotes, including dinoflagellates (Okamoto and Hastings, 2003) and cryptophytes (Sineshchekov et al., 2005). Microbial-type rhodopsins were also found in green algae such as Acetabularia acetabulum (Tsunoda et al., 2006; Jung, 2007) and Chlamydomonas reinhardtii (Hegemann et al., 2001; Nagel et al., 2002, 2003; Sineshchekov et al., 2002). Green algae need sensory photoreceptors for light-dependent regulation of their developmental processes, cell division, photosynthesis, phototactic orientation, and photophobic responses. Algal rhodopsins modulate intracellular acidity, calcium influx, electrical excitability, and, in a more indirect way, transcription, translation, and lipid synthesis (Kirk and Kirk, 1985; Binder and Anderson, 1986; Grossman et al., 2004; Guschina and Harwood, 2006). The microbial-type rhodopsins of Chlamydomonas are integral parts of a primordial visual system, forming, with the pigmented eyespot, a functional eye (Dieckmann, 2003; Schmidt et al., 2006; Hegemann, 2008). In Chlamydomonas, the eye was shown to be a complex sandwich that is composed of the plasma membrane, the outer and inner chloroplast membranes, and several layers of carotenoid-filled lipid granules in the stroma of the chloroplast that are interleaved with thylakoid membranes (Nakamura et al., 1973; Melkonian and Robenek, 1984). The layers of carotenoid-filled granules confer directionality by reflecting light back onto the photoreceptors in the plasma membrane (interference reflection) and by blocking light passing through the cell (Foster and Smyth, 1980; Kreimer and Melkonian, 1990; Harz et al., 1992; Kreimer, 1994; Schaller and Uhl, 1997). The excitation of rhodopsins by light causes transmembrane ion fluxes that initiate a cascade of electrical and other responses (Litvin et al., 1978; Harz and Hegemann, 1991; Braun and Hegemann, 1999) that lead finally to beating changes in the two flagella (Holland et al., 1997).
After it was shown that the animal rhodopsin-related proteins Chlamyopsin-1 (COP1) and COP2 of Chlamydomonas (Beckmann and Hegemann, 1991; Deininger et al., 1995) and Volvoxopsin-1, the Volvox analog of COPs (Ebnet et al., 1999), are not the phototaxis photoreceptors (Fuhrmann et al., 2001, 2003), two microbial-type rhodopsins, now called channelrhodopsin-1 and channelrhodopsin-2 (ChR1 and ChR2), were identified in the Chlamydomonas EST database. Channelrhodopsins are light-gated ion channels that have been shown to function as photoreceptors for phototaxis and photophobic responses. However, it was suggested that ChR1 is the dominant photoreceptor for both responses in the Chlamydomonas strains tested (Sineshchekov et al., 2002; Berthold et al., 2008). ChR1 and ChR2 were studied functionally in host systems including Xenopus oocytes, human embryonic kidney (HEK) cells, and PC12 cells (Nagel et al., 2002, 2003; Hegemann et al., 2005; Ishizuka et al., 2006). They carry the photoreceptor currents and mediate the phototactic and photophobic responses (Sineshchekov et al., 2002; Govorunova et al., 2004; Berthold et al., 2008). The amino acids that form the H+-conducting network in the haloarchaeal bacteriorhodopsin are conserved in channelrhodopsins (Hegemann et al., 2001; Suzuki et al., 2003), although the action spectra of ChR1 and ChR2 are blue shifted and peak at approximately 500 nm (Nagel et al., 2002) and approximately 470 nm (Sineshchekov et al., 2002), respectively. Meanwhile, channelrhodopsins have become widely used tools in neuroscience because the heterologous expression of channelrhodopsins in animal cells allows for a temporally precise photostimulation, and this, in particular, may lead to the development of precise neuromodulation technologies (optogenetics; Miller, 2006). Despite the substantial physiological and biophysical studies, little is known about the expression characteristics of channelrhodopsins. Govorunova et al. (2004) report a 5-fold higher proportion of ChR1 relative to ChR2 in vegetatively grown wild-type Chlamydomonas cells (strain 495), whereas the ChR1-ChR2 ratio was lower in sexually competent gametes. The situation is different in cells of the cell wall-deficient C. reinhardtii mutant CW2, where ChR1 expression increases 2-fold during gametogenesis while ChR2 is reduced from approximately 40% to below 5% of total ChR protein (Berthold et al., 2008).
Channelrhodopsins have also been identified in the multicellular green alga Volvox carteri. However, the photomovement of a multicellular organism is more complicated than that of a unicellular individual. In an adult V. carteri spheroid, approximately 2,000 to 4,000 biflagellate somatic cells are responsible for locomotion, but these cells are no longer connected with each other via cytoplasmic bridges (Fig. 1, A and B). If all of the somatic cells near the surface of the spheroid were identical and acted in the same way, directed swimming would be impossible. Therefore, Volvox shows a distinct anterior-posterior polarity (Hoops, 1984, 1993; Kirk, 1998). This polarity is not only expressed by the direction in which it swims (i.e. with the anterior pole first) but also by morphological characteristics: (1) the spheroid is somewhat more enlarged along the anterior-posterior axis than along the equatorial axis; (2) the daughter spheroids are localized mainly in the posterior hemisphere; (3) the somatic cells near the anterior pole are somewhat larger and more widely spaced than the cells near the posterior pole; (4) the orientation of the flagellar apparatus of each somatic cell is correlated with its position in the spheroid; (5) the eyespots are larger at the anterior pole; (6) in each somatic cell, the eyespot is localized on the side facing toward the posterior pole; and (7) at the anterior pole, the eyespots are located quite a distance from the flagella, whereas the cells in the posterior region have the eyespots closer to the flagella. All of these differences are probably essential for effective organismal motility and the specific photomovement in Volvox (Hoops, 1984, 1993; Kirk, 1998).
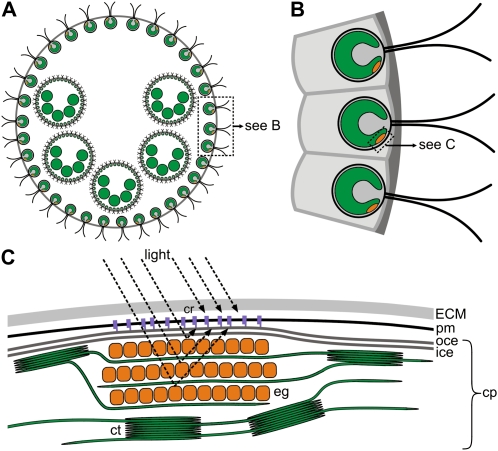
Figure 1.
A schematic representation of the location of the eyespot and the putative localization of channelrhodopsins. A, V. carteri spheroid with daughter spheroids; each daughter spheroid contains large gonidia. For clarity, only some of the small approximately 2,000 to 4,000 biflagellate somatic cells are shown. Likewise, only five of the approximately 16 daughter spheroids and five of the approximately 16 gonidia are shown. More than 95% of the volume of a spheroid consists of a complex, transparent, glycoprotein-rich ECM that holds all of the cells in place. VChR1 and VChR2 are located in the somatic cells. The swimming direction of this Volvox spheroid is from bottom to top (anterior pole first). B, Three somatic cells of the spheroid in A are shown in greater detail. ECM glycoproteins form a honeycomb-like array of cellular compartments that house the somatic cells. Each somatic cell contains a single, large, cup-shaped chloroplast. The eyespot is seen peripherally as a singular orange-red spot. The channelrhodopsins are expected to be localized in the eyespot overlaying part of the plasma membrane. C, A more detailed illustration of the components at and around the eyespot shown in B (according to Schmidt et al., 2006). The eyespot is part of the chloroplast and assembled from the outer and inner chloroplast membranes and several ordered layers of carotenoid-rich, lipid eyespot globules in the stroma of the chloroplast, which are interleaved with thylakoid membranes. In the region of the eyespot, the plasma membrane is closely attached to the chloroplast envelope membranes. Channelrhodopsins are expected to be localized in the plasma membrane patch right above the eyespot globules, and in conjunction with the pigmented eyespot they form a functional eye. cp, Chloroplast; cr, channelrhodopsins; ct, chloroplast thylakoids; eg, eyespot granules; ice, inner chloroplast envelope; oce, outer chloroplast envelope; pm, plasma membrane.
Previously, 7TM fragments of Volvox channelrhodopsins have been expressed in Xenopus oocytes, monkey kidney cells (COS-1), and HEK293 cells (Ernst et al., 2008; Zhang et al., 2008). Preliminary electrical studies and photocycle measurements were carried out first for an artificial hybrid comprising helices 1 and 2 from Volvox channelrhodopsin-1 (VChR1) and helices 3 to 7 from VChR2 (amino acids 1–305, VChR1/2; accession no. DQ094781; Ernst et al., 2008). Later, Zhang et al. (2008) studied cultivated hippocampal neurons expressing a VChR1 7TM fragment (accession no. EU622855). For these studies, the amino acid sequence was retrieved from the ongoing Volvox Joint Genome Institute (JGI) genome project (http://genome.jgi-psf.org/Volca1/Volca1.home.html) and codon humanized for expression. We anticipate that VChRs serve as primary phototaxis photoreceptors in Volvox because the photocurrents of VChR in vitro are similar to those recorded in Volvox cells and because the proteins are closely related to their counterparts in Chlamydomonas.
However, the full-length sequences of the VChRs and the expression pattern of these photoreceptor genes in the different cell types and developmental stages of Volvox have still not been determined. Likewise, nothing is yet known about the expression characteristics in response to external stimuli.
Volvox channelrhodopsins are of particular interest, not only because they may provide additional, useful optogenetic tools for neuroscience but also because they are expected, beyond their role as photoreceptors for behavioral responses, to be key elements in the various light-dependent developmental processes that cannot be studied in unicellular alga. The multicellular alga V. carteri is composed of two cell types, somatic (body) cells and germ (reproductive) cells, and it is capable of both asexual and sexual reproduction. In the asexual mode of reproduction, each adult male and female is a spheroid containing approximately 2,000 to 4,000 biflagellate somatic cells and approximately 16 asexual reproductive cells called gonidia (Starr, 1969, 1970; Kirk, 1998). The gonidia lack visible eyespots (Kirk, 1998). Positive and negative phototaxis of Volvox is controlled by the acceleration or cessation of flagellar movements (Foster and Smyth, 1980; Kirk, 1998). However, blue-green light seems to be essential not only for the regulation of photomovements but also for the different processes of development (e.g. cell differentiation; Kirk and Kirk, 1985). The observed light-induced shifts in protein synthesis are likely to be rhodopsin mediated because the action spectrum is rhodopsin shaped, with a maximum at approximately 520 nm (Kirk and Kirk, 1985).
Here, we report the full-length cDNA and genomic sequences of the microbial-type rhodopsins VChR1 and VChR2 from V. carteri. We show that expression of these channelrhodopsins is extremely cell type specific and that expression of VChR1 and VChR2 is regulated by the light-dark cycle, the temperature, and the stage of development. Addition of the sex inducer of V. carteri leads to increased expression levels, but the stimulation of expression is gradually different for VChR1 and VChR2. Moreover, both channelrhodopsins also respond in a different way to increased temperatures and mechanical wounding. Finally, we reveal spectroscopic properties of VChR1 and VChR2. Those of VChR1, in particular, are different from those of its Chlamydomonas counterpart. The results demonstrate that the spectral properties of VChR2 are quite stable, whereas the spectral properties of VChR1 vary dramatically with changes in pH.
RESULTS
The VChR1 and VChR2 mRNAs and Genes
Searching the V. carteri genome database of the JGI (http://genome.jgi-psf.org/Volca1/Volca1.home.html) revealed sequences that are related to ChR1 and ChR2 from C. reinhardtii (Nagel et al., 2002, 2003). This information was used to obtain the complete coding sequences of the two channelrhodopsins VChR1 and VChR2 from Volvox. The coding sequences were amplified by reverse transcription (RT)-PCR, using total RNA from wild-type females as the templates. The lengths of the mRNAs were 2,694 bp for VChR1 and 2,411 bp for VChR2, with open reading frames of 2,514 bp for VChR1 and 2,244 bp for VChR2 (Fig. 2, A and D). Comparing the mRNA and genomic sequences shows that the VChR1 and VChR2 genes contain 12 and 13 introns, with sizes from 71 to 918 bp (Fig. 2, B and E). Strikingly, there are 79 short sequence repeats (53× ATGGGN, 26× AACGGN) within exons 11 to 13 of VChR1 and 15 repeats (14× ATGGGN, 1× AACGGN) in exons 13 and 14 of VChR2. The two VChR mRNAs had 71% identity in an 817-bp overlap and 74% identity in another 351-bp overlap (Supplemental Fig. S1).
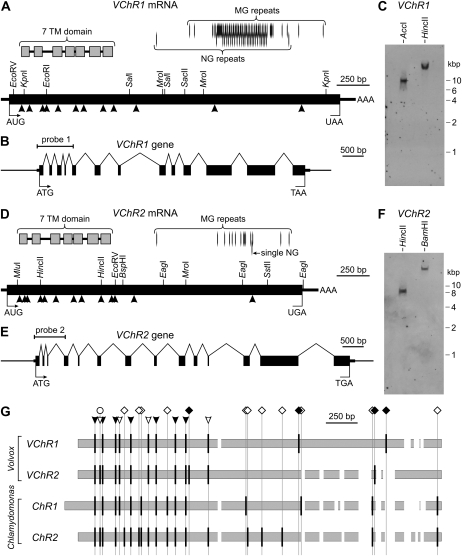
Figure 2.
The VChR1 and VChR2 gene structures and mRNAs. A and D, Physical maps of the VChR1 (A) and VChR2 (D) mRNAs. The gray boxes indicate the conserved regions that code for the 7TM domains. The positions of sequences coding for MG and NG repeats are indicated by vertical lines. Arrowheads show the positions of the introns within the physical maps of the VChR1 and VChR2 mRNAs. AUG, The translation start site; UAA or UGA, the translation stop site; black boxes, the open reading frame; thick horizontal bars, the 5′- and 3′-untranslated regions; AAA, the poly(A) tail. B and E, Physical maps of the VChR1 (B) and VChR2 (E) genes including the exon and intron segments. ATG, The translation start site; TAA and TGA, the translation stop sites; the black boxes represent exons, and the carets represent introns; thick horizontal bars indicate untranslated regions, and the thinner horizontal bars represent upstream and downstream sequences. The positions of the probes used for the DNA gel blot analysis in C and F are indicated. C and F, DNA gel blot analysis of VChR1 (C) and VChR2 (F). Genomic DNA from Volvox females was digested with AccI, HincII, or BamHI. The blots were probed using either VChR1- or VChR2-specific probes. The VChR1 probe (probe 1) is an 867-bp DNA fragment, and the VChR2 probe (probe 2) is a 741-bp DNA fragment. The localization of the gel blot probes is indicated in B and E, respectively. G, A comparison of the intron positions in Volvox and Chlamydomonas channelrhodopsin coding sequences after sequence alignment. Gray boxes represent the aligned coding sequences that are shown in Supplemental Figure S4. Introduced (white) gaps are only given if they are larger than 20 nucleotides. The intron positions are indicated by vertical black bars. The black arrowheads show the seven intron positions that are conserved in all of the channelrhodopsin mRNAs; the four white arrowheads show intron locations that are conserved between both species. Introns that only occur in Volvox are marked with black diamonds; those that only occur in Chlamydomonas are marked with white diamonds; and those that are unique for channelrhodopsin-2 are marked with white circles.
To determine the copy number of channelrhodopsin genes in the Volvox genome, genomic DNA from females was restricted, blotted, and analyzed using an 867-bp VChR1 probe and a 741-bp VChR2 probe located near the 5′ ends (Fig. 2, B and E). Single BamHI and HincII fragments were obtained for both DNAs (Fig. 2, C and F), showing that both genes occur only once in the Volvox genome.
The closest relatives of the VChR genes are, as expected, the channelrhodopsins ChR1 and ChR2 of C. reinhardtii (accession nos. AF508965 and AF508966; Sineshchekov et al., 2002). VChR1 and ChR1 mRNA revealed 70% identity in a 1,432-bp overlap (Supplemental Fig. S2), whereas the similarity between VChR2 and ChR2 mRNA is even higher, with 74% identity in an 804-bp overlap and 70% in another 401-bp overlap (Supplemental Fig. S3). To obtain more insight into the relationship between the genes, the intron positions were compared (Fig. 2G; Supplemental Fig. S4). In total, 26 intron positions were found in at least one of the four channelrhodopsin genes. Remarkably, only seven intron positions are conserved in all four genes; 11 are Chlamydomonas specific, four are Volvox specific, and one is specific for channelrhodopsin-2 of both species. There are also three intron positions where an intron is missing in only one of the four genes. This intron pattern allows the conclusion that the last common ancestor of Volvox and Chlamydomonas already had two channelrhodopsin genes with introns, some of which have been lost and some of which were later acquired.
The Encoded Amino Acid Sequences of VChR1 and VChR2
The open reading frames of VChR1 and VChR2 mRNAs encode polypeptides of 837 and 747 amino acids (Fig. 3, A and B) with expected molecular masses of 86.6 and 77.8 kD, and there is 62% identity over 526 amino acid residues (Supplemental Table S1).
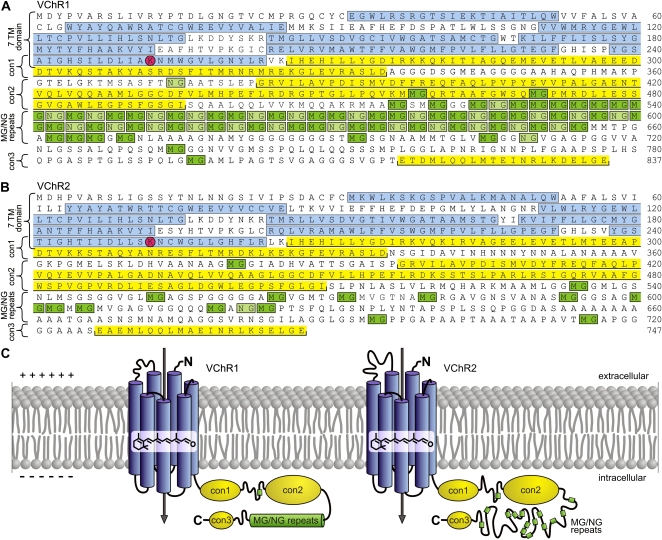
Figure 3.
The amino acid sequences and putative localization of VChR1 and VChR2. A, The amino acid sequence of VChR1. B, The amino acid sequence of VChR2. The seven transmembrane helices are indicated as blue boxes. Three other conserved regions, named con1, con2, and con3, are indicated by yellow boxes. The second halves of both amino acid sequences contain many MG (dark green boxes) and NG (light green boxes) repeats. The retinal-binding Lys (K) residues at position 252 of both VChR1 and VChR2 are circled and shown on a red background. C, A detailed schematic representation of a portion of the eyespot overlaying part of the plasma membrane that is shown in Figure 1C showing the putative transmembrane arrangement of VChR1 and VChR2. The light-gated cation channels VChR1 and VChR2 are composed of seven transmembrane helix domains (blue) with an all-trans-retinal chromophore. Light absorption by channelrhodopsins induces opening of the channel, which results in a cation flux across the plasma membrane.
Amino acid residues 32 to 263 define the putative 7TM-spanning α-helices (Fig. 3), and there is 68% identity between VChR1 and VChR2 in this region. The 7TM sequences are members of the microbial-type rhodopsin family (Sharma et al., 2006), although the similarities between helix 1 and 2 are only weak. Retinal is covalently linked to Lys-252 in both VChR1 and VChR2 (see below), and mutation of this amino acid in the VChR1/2 hybrid resulted in a loss of the chromophore (Ernst et al., 2008).
The long C-terminal extensions of both proteins contain quite a few Met-Gly (MG) and Asn-Gly (NG) repeats. There are 53 MG and 27 NG repeats in VChR1 (Fig. 3A; Table I) and a single NG and 14 MG repeats in VChR2 (Fig. 3B; Table I). These result from the short nucleotide sequence repeats described above (Fig. 2, A and D). Two highly homologous regions are located between the 7TM domain and the MG/NG repeats (Fig. 3, A and B, con1 and con2), and one is located right at the C-terminal end (Fig. 3, A and B, con3), with 72%, 76%, and 80% identity in both proteins (Supplemental Fig. S5). The C-terminal extensions of both channelrhodopsins contain a high percentage (approximately 40%) of the three smallest amino acid residues, Gly, Ala, and Ser (Table I).
Table I.
Characteristics of the long C-terminal extensions of channelrhodopsins
The length of the C-terminal extensions of channelrhodopsins was calculated from the first amino acid residue after helix 7 of the 7TM domain to the last residue of the polypeptide. Percentages and calculations of repeat numbers refer to this C-terminal extension.
| Protein Name | Length of C-Terminal Extension (Amino Acid Residues) | 7TM–con1 Distance (Amino Acid Residues) | con1–con2 Distance (Amino Acid Residues) | con2–con3 Distance (Amino Acid Residues) | No. of MG Repeats | No. of NG Repeats | No. of MG + NG Repeats | Gly | Gly + Ala + Ser |
|---|---|---|---|---|---|---|---|---|---|
| % | % | ||||||||
| VChR1 | 574 | 2 | 47 | 321 | 53 | 27 | 80 | 25 | 39 |
| VChR2 | 484 | 2 | 61 | 217 | 14 | 1 | 15 | 16 | 40 |
| ChR1 | 405 | 2 | 42 | 157 | 21 | 1 | 22 | 16 | 33 |
| ChR2 | 469 | 2 | 57 | 205 | 24 | 19 | 43 | 23 | 42 |
Phylogenetic Analysis of the Channelrhodopsins
Searches in the databases of the National Center for Biotechnology Information revealed many rhodopsins from green algae, fungi, cyanobacteria, cryptomonads, and halobacteria that are related to the VChRs (Supplemental Table S1). Again, the closest relatives of VChRs are ChR1 and ChR2 of Chlamydomonas. VChR1 and ChR1 are 66% identical in the 7TM region at the amino acid level (Supplemental Table S1) and 42% identical in the region covering residues 729 to 837. Likewise, VChR2 and ChR2 are 60% identical in the 7TM region (Supplemental Table S1). Channelrhodopsins are less related to all other rhodopsins from fungi, cyanobacteria, cryptomonads, and halobacteria, as demonstrated by the much lower sequence identity, approximately 20% to 30% (Supplemental Table S1). A multiple sequence alignment of 45 rhodopsins including the Volvox channelrhodopsins (see below) reveals that sequence homology is restricted to the seven-transmembrane helix domain and is most prominent in helices 3 to 7. Figure 4A shows part of the alignment, which includes the seven-transmembrane helix domains of VChR1, VChR2, ChR1, ChR2, and bacteriorhodopsin of H. salinarum. The sequence alignment of the 45 rhodopsins reveals a consensus motif around the retinal-binding Lys in helix 7 (Fig. 4B). There are also consensus motifs around three specific amino acid residues in helices 3 and 7 (Fig. 4B) that are predicted to define the retinal-binding pocket and the counterion complex of the retinal Schiff's base (Spudich, 2006; Zhang et al., 2008). Of the 10 amino acids that are known to be within 5 Å of the retinal chromophore (Henderson et al., 1990; Luecke et al., 1999; Shimono et al., 2002), four are identical or conservatively exchanged in both VChR1 and VChR2 (Fig. 4A). Additionally, several other amino acids are highly conserved within the microbial rhodopsin family and are also present in VChR1 and VChR2 (Fig. 4A).
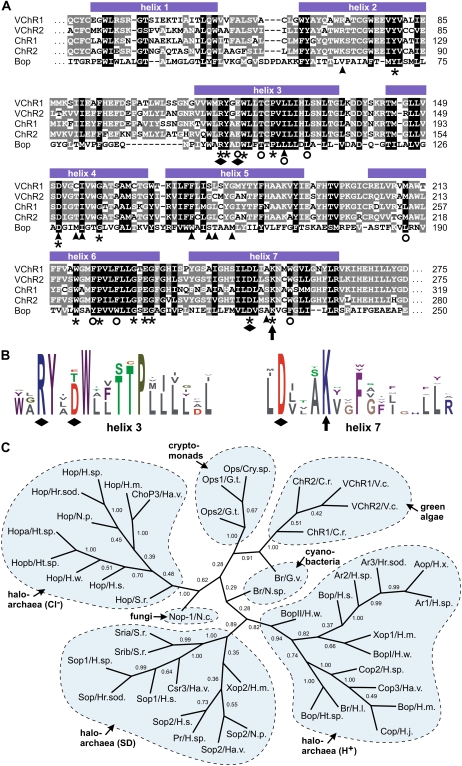
Figure 4.
Protein sequence alignment of the 7TM domain of algal channelrhodopsins with bacteriorhodopsin, and the sequence relationship among 45 rhodopsin-related proteins. A, Protein sequence alignment of the 7TM domain of VChR1 and VChR2 with channelrhodopsins ChR1 and ChR2 of C. reinhardtii and bacteriorhodopsin (Bop) of H. salinarum. Conserved amino acid residues are shaded, with similarity groups enabled. White letters on black backgrounds represent residues that are conserved in 100% of the sequences at the corresponding position; residues that are shown as white letters on gray backgrounds are conserved in more than 60% of the sequences. Amino acid residues that are known from the H. salinarum Bop structure to be within 5 Å of the retinal chromophore (Henderson et al., 1990; Luecke et al., 1999; Shimono et al., 2002) are indicated with arrowheads. Asterisks indicate residues that are conserved in most microbial rhodopsins. Circles highlight conserved positions, including conservative amino acid substitutions. The retinal-binding Lys is indicated with an arrow. Three amino acid residues that are predicted to define the counterion complex of the all-trans-retinal Schiff's base (Spudich, 2006; Zhang et al., 2008) are highlighted with diamonds. B, Consensus motifs of 45 rhodopsins at the hypothetical retinal-binding site and at the positions of residues that define the counterion complex. The relationship of the 45 rhodopsin proteins that were used for the consensus study in B is shown in C. The motifs are displayed as sequences logos (i.e. the higher the one-letter symbol of an amino acid residue at a given position, the better the conservation at that position). The putative retinal-binding Lys in helix 7 is indicated with an arrow. The three amino acid residues within helix 3 and helix 7 that are predicted to define the counterion complex are highlighted with diamonds. The consensus motifs were visualized using LogoBar (Pérez-Bercoff et al., 2006). C, Sequence relationship among 45 rhodopsin-related proteins from green algae, fungi, cyanobacteria, cryptomonads, and haloarchaea. Haloarchaeal rhodopsins are subgrouped into sensory rhodopsins and Cl−- and H+-pumping rhodopsins. The unrooted tree was calculated using the neighbor-joining method (Saitou and Nei, 1987) of PHYLIP (see “Materials and Methods”). Numbers indicate bootstrap analysis values obtained using 10,000 resampled data sets. The analysis is based on the trimmed alignment given in Supplemental Figure S6 and Supplemental Data Set S1. The rhodopsins that were investigated and their source organisms are as follows: VChR1/V.c., channelrhodopsin-1, Volvox carteri; VChR2/V.c., channelrhodopsin-2, Volvox carteri; ChR1/C.r., channelrhodopsin-1, Chlamydomonas reinhardtii; ChR2/C.r., channelrhodopsin-2, Chlamydomonas reinhardtii; Br/G.v., bacterioopsin, Gloeobacter violaceus; Br/N.sp., bacteriorhodopsin, Nostoc species; Ops1/G.t., opsin-1, Guillardia theta; Ops2/G.t., opsin-2, Guillardia theta; Ops/Cry.sp., opsin, Cryptomonas species; Nop-1/N.c., opsin-1, Neurospora crassa; Hop/S.r., halorhodopsin, Salinibacter ruber; ChoP3/Ha.v., cruxhalorhodopsin-3, Haloarcula vallismortis; Hop/H.m., halorhodopsin, Haloarcula marismortui; Hop/N.p., halorhodopsin, Natronomonas pharaonis; Hop/H.sp., halorhodopsin, Halobacterium species; Hop/Hr.sod., halorhodopsin, Halorubrum sodomense; Hopa/Ht.sp., halorhodopsin, Haloterrigena species; Hopb/Ht.sp., halorhodopsin, Haloterrigena species; Hop/H.s., halorhodopsin, Halobacterium salinarum; Hop/H.w., halorhodopsin, Haloquadratum walsbyi; Xop2/H.m., opsin, Haloarcula marismortui; Sop1/H.sp., sensory rhodopsin-1, Halobacterium species; Sop/Hr.sod., rhodopsin, Halorubrum sodomense; Sop1/H.s., sensory rhodopsin-1, Halobacterium salinarum; Csr3/Ha.v., bacterial rhodopsin, Haloarcula vallismortis; Sria/S.r., sensory rhodopsin a, Salinibacter ruber; Srib/S.r., sensory rhodopsin b, Salinibacter ruber; Sop2/H.s., sensory rhodopsin-2, Halobacterium salinarum; Pr/H.sp., phoborhodopsin, Halobacterium species; Sop2/Ha.v., sensory rhodopsin-2, Haloarcula vallismortis; Sop2/N.p., sensory rhodopsin-2, Natronomonas pharaonis; BopII/H.w., bacteriorhodopsin II, Haloquadratum walsbyi; BopI/H.w., bacteriorhodopsin I, Haloquadratum walsbyi; Xop1/H.m., bacteriorhodopsin, Haloarcula marismortui; Bop/H.s., bacteriorhodopsin, Halobacterium salinarum; Ar2/H.sp., archaerhodopsin-2, Halobacterium species; Ar3/Hr.sod., archaerhodopsin-2, Halorubrum sodomense; Ar1/H.sp., archaerhodopsin-1, Halobacterium species; Aop/H.x., archaerhodopsin, Halorubrum xinjiangense; Br/H.l., bacteriorhodopsin, Halobiforma lacisalsi; Bop/Ht.sp., bacteriorhodopsin, Haloterrigena species; Cop2/H.sp., cruxrhodopsin-2, Haloarcula species; Bop/H.m., bacteriorhodopsin, Haloarcula marismortui; Cop/H.j., cruxrhodopsin, Haloarcula japonica; and Cop3/Ha.v., cruxrhodopsin-3, Haloarcula vallismortis. [See online article for color version of this figure.]
VChRs were phylogenetically classified based on trimmed alignments of the 45 rhodopsins (Supplemental Fig. S6; Supplemental Data Set S1). Figure 4C shows an unrooted tree of rhodopsin-related proteins calculated using the neighbor-joining method (Saitou and Nei, 1987). In this tree, the Volvox channelrhodopsins are on the same branch as the Chlamydomonas channelrhodopsins, as expected. Likewise, other rhodopsins branch according to their source group of organisms (i.e. fungi, cyanobacteria, cryptomonads, and haloarchaea). Haloarchaeal rhodopsins build subgroups that are consistent with the type of rhodopsin (i.e. sensory, Cl−-pumping, and H+-pumping rhodopsins).
Expression of VChR1 and VChR2 in the Asexual Life Cycle and after Sex Induction
The expression of VChR1 and VChR2 was investigated at specific developmental stages (Fig. 5). Volvox wild-type females were grown synchronously under vegetative conditions in an 8-h-dark/16-h-light cycle with a life cycle of 48 h (Fig. 5, C and D). Sexual development of V. carteri is triggered by a sex-inducer molecule (also called a sex-inducing pheromone), a 32-kD glycoprotein (Starr, 1970; Starr and Jaenicke, 1974; Tschochner et al., 1987; Mages et al., 1988). This molecule is one of the most potent biological effector molecules known, triggering sexual development at concentrations as low as 10−16 m (Starr, 1970; Gilles et al., 1984). When the sex inducer is added, the females switch to the sexual pathway, resulting in the production of eggs.
Figure 5.
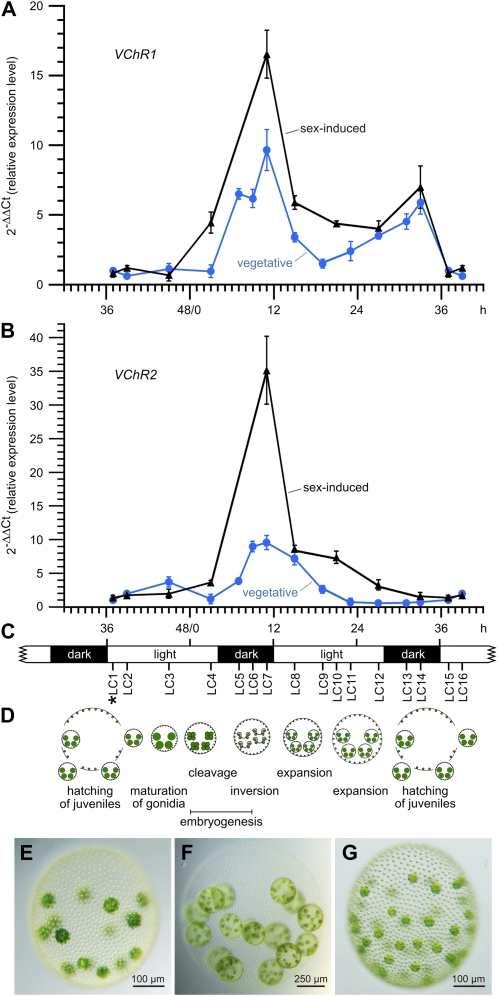
Expression kinetics of VChR1 and VChR2 throughout the life cycle under vegetative conditions and after addition of the sex inducer. A and B, Expression kinetics of VChR1 (A) and VChR2 (B) in Volvox females under vegetative conditions and after the addition of the sex inducer. The expression was analyzed at several time points (LC1–LC16; C) throughout the life cycle of the female wild-type strain, Eve10. The organisms were analyzed under vegetative conditions (vegetative, blue; n = 3) and after addition of the sex inducer (sex-induced, black; n = 3) at LC1 (asterisk). Under the standard 8-h-dark/16-h-light cycle conditions, the generation time of V. carteri is exactly 48 h; the first cleavage division of the gonidium is defined as 0 h. LC1 of vegetatively grown females was used as the reference point (=1) for calculations of the relative expression level. Error bars refer to the sd of the y value. All real-time RT-PCR experiments were performed in triplicate from technical repeats. C, The standard 8-h-dark/16-h-light cycle with the analyzed time points indicated (LC1–LC16). D, A schematic representation of all stages in development in relation to the dark/light cycle. E to G, The phenotypes of vegetative and sexually induced V. carteri females. E, An asexual female after the onset of embryonic cleavage divisions. F, An asexual female immediately before hatching of the juveniles. G, A sexual female approximately 48 h after sex induction. [See online article for color version of this figure.]
Total RNA was isolated at several time points throughout the life cycle of vegetative females: after hatching of the juveniles (LC1 and LC2); shortly before the first cleavage division (LC3), which defines the beginning of embryogenesis; during the embryonic cleavage divisions (LC4); during inversion (LC5), a morphogenetic process at the end of embryogenesis in which embryos turn inside out; after inversion, when the biosynthesis of extracellular matrix (ECM) begins (LC6 and LC7); during expansion of the spheroid by ECM biosynthesis (LC8–LC13); immediately before hatching of the juveniles (LC14); and finally, 3 and 5 h after hatching (LC15 and LC16), corresponding to the first two time points (LC1 and LC2). The photomicrographs in Figure 5, E and F, show asexual females after the onset of embryonic cleavage divisions and immediately before hatching of the juveniles. The expression levels of VChR1 and VChR2 at the different developmental stages were analyzed using quantitative real-time RT-PCR.
The expression of both VChRs in vegetative organisms is quite low after hatching of the juveniles (LC1 and LC2) and remains low before (LC3) and during (LC4) the cleavage divisions (Fig. 5, A and B). The expression increases during embryogenesis (LC4–LC7), with a peak after the completion of embryogenesis when the biosynthesis of ECM begins (LC7), at the end of the first dark phase. Expression of both VChRs reaches a value that is approximately 10-fold above the initial one. In the following light phase, as ECM expansion continues, expression of both VChRs decreases (LC8 and LC9). At the end of the light phase and throughout the second dark phase, VChR1 expression increases again (LC9–LC14), reaching another peak of approximately 6-fold above the initial expression level, shortly before hatching of the juveniles (LC14). In contrast, VChR2 expression drops further and remains low during the same time period (LC9–LC14). In the following light phase, VChR1 expression also decreases and reaches the initial expression level at LC15 and LC16. The fact that VChR1 expression rises during both dark phases, whereas the expression of VChR2 increases only during the first dark period, suggests that both channelrhodopsins are cell cycle dependent; in addition, VChR1 is under dark/light cycle control (see below).
To analyze the expression of the VChRs during sexual development, the sex inducer was added to a vegetatively grown female population shortly before hatching of the juveniles at stage LC1 (Fig. 5, A and B). The sex inducer must be added at this point because, despite its high potency, uncleaved gonidia have to be exposed to it for at least 6 h before they become committed to the sexual cleavage program (Gilles et al., 1984). The photomicrograph in Figure 5G shows a sexual female with approximately 36 eggs approximately 48 h after sex induction for illustration. In principle, the sex inducer does not modify the timing of expression, but the switch to sexual development leads to a moderately increased expression of VChR1 (+70%) and to a strong increase in VChR2 expression (+370%) at LC7 (Fig. 5, A and B).
Cell Type-Specific Expression of VChR1 and VChR2
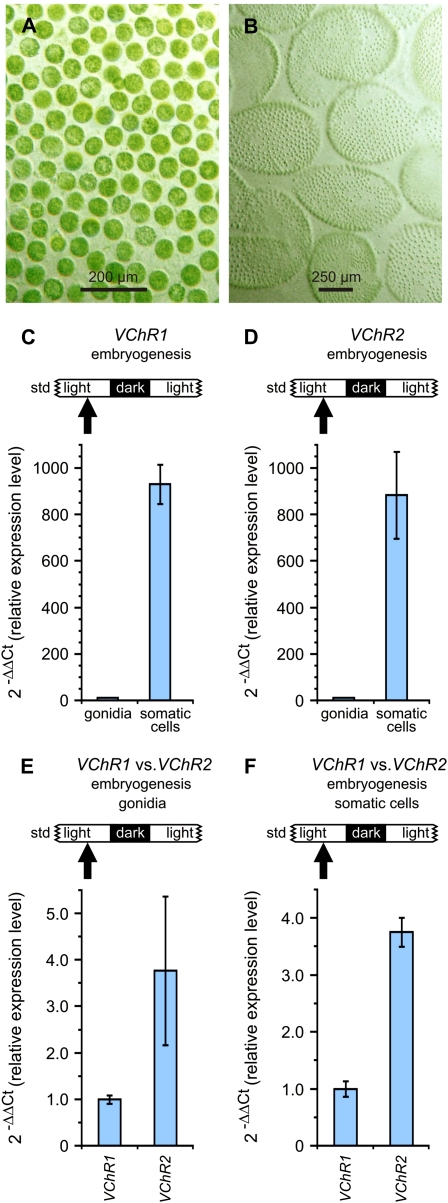
Earlier studies demonstrated that the transcriptome of V. carteri is compartmentalized between the somatic cells and gonidia (Tam and Kirk, 1991; Nematollahi et al., 2006). To determine the expression of VChRs in individual cell types, somatic cells were separated from gonidia prior to the first cleavage division (Fig. 6, A and B), RNA was isolated separately, and expression of the VChRs was analyzed again by quantitative real-time RT-PCR (Figs. 5D and 6C). Both RNAs were extremely cell type specific: the expression of VChR1 and VChR2 in the somatic cells was approximately 930-fold and approximately 880-fold higher compared with the gonidia. This is a higher cell type specificity than has been found for any other gene in previous studies in Volvox (Tam and Kirk, 1991; Nematollahi et al., 2006). Thus, VChRs are de facto not expressed in gonidia.
Figure 6.
Quantitative analysis of the cell type-specific expression of VChR1 and VChR2. A and B, The appearance of the separated cell types of V. carteri females. A, Isolated gonidia. B, Isolated somatic cell sheets. C, The expression analysis of VChR1 in gonidia versus the somatic cells of the parent spheroids. D, The expression analysis of VChR2 in gonidia versus the somatic cells of the parent spheroids. In C and D, expression in gonidia was used as the reference point (=1) for calculations of the relative expression level. The gonidia were in the stage that occurs shortly before the first cleavage division. E, The expression analysis of VChR1 versus VChR2 in gonidia. The gonidia were in the stage that occurs shortly before the first cleavage division. F, The expression analysis of VChR1 versus VChR2 in somatic cells. E and F, The expression of VChR1 was used as the reference point (=1) for calculations of the relative expression level. In C to F, the time point at which the mRNA was isolated is indicated (arrow) in relation to the 8-h-dark/16-h-light cycle, which is shown in more detail in Figure 5C. Because there are two dark phases within the 48-h life cycle (Fig. 5D), at embryogenesis and during hatching of the juveniles, the respective dark phase is indicated by “embryogenesis.” The error bars refer to the sd of the y value. All real-time RT-PCR experiments were performed in triplicate from technical repeats. [See online article for color version of this figure.]
The comparison between the expression of VChR1 and VChR2 revealed that, as shown for the developmental stage shortly before the onset of cell cleavage, VChR2 expression is approximately 4-fold higher than that of VChR1 (Fig. 6, E and F).
The Influence of Light and Stress Conditions on the Expression of VChR1 and VChR2
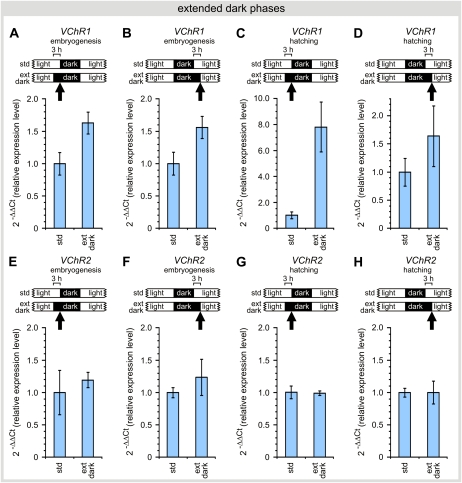
Because VChR1 is expressed in both dark phases whereas VChR2 is only expressed in the first one, we studied different light/dark regimes while minimizing the influence on the overall cell cycle. One subpopulation of Volvox was grown in the standard dark/light cycle and a second population was grown under identical conditions, except that one dark phase had been extended by 3 h. The extension of the dark phase was done in two different ways: either the dark phase began 3 h earlier than in the standard conditions or the dark phase ended 3 h later. We isolated the RNA and tested for VChR expression using quantitative real-time RT-PCR.
Any extension of the dark phase stimulated the expression of VChR1, no matter whether the light was switched off earlier or switched on later, and this was valid in embryogenesis (first dark phase; Fig. 7, A and B) and at the stage of hatching (second dark phase; Fig. 7, C and D). The largest stimulation was observed upon advanced darkness during hatching of the juveniles (Fig. 7C), when VChR1 expression was approximately 8-fold above the standard value. Because the relative effect of the extended dark phases varies in Figure 7, A to D, additional effectors other than light, such as the developmental stage, probably exist. In contrast, the expression of VChR2 varies only to a small extent during the dark period extension (Fig. 7, E–H).
Figure 7.
The influence of extended (ext) dark periods on the expression of VChR1 and VChR2. The dark phases began earlier (A, C, E, and G) or lasted longer than normal (B, D, F, and H) during the early embryogenesis (A, B, E, and F) or during hatching (C, D, G, and H). The expression levels were compared with those of Volvox that were grown under standard dark/light conditions as a reference. The error bars refer to the sd (n = 3). Arrows indicate the time points at which the mRNA was isolated during the 8-h-dark/16-h-light cycle; the respective dark phase is indicated by “embryogenesis” or “hatching.” [See online article for color version of this figure.]
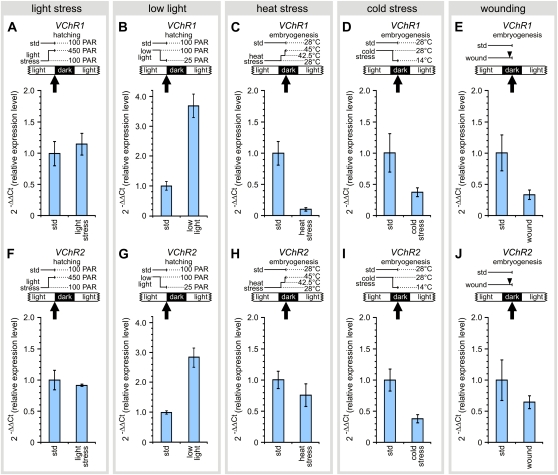
In natural habitats, light intensities vary greatly, but our wild-type strains have been kept in an 8-h-dark/16-h-light cycle with approximately 100 μmol photons m−2 s−1 during the light phase for many years, because these light conditions were considered to be ideal for the cultivation of V. carteri and also because they have been demonstrated to be appropriate for synchronous growth in a 48-h life cycle (Starr and Jaenicke, 1974). To generate a light stress situation, we increased the light intensity from approximately 100 μmol photons m−2 s−1 to approximately 450 μmol photons m−2 s−1 for 3 h. Neither VChR1 nor VChR2 showed any significant change in the expression level, suggesting that the lower expression in the light was not caused by light stress (Fig. 8, A and F). We also investigated the influence of low light intensities by reducing the standard light intensity of approximately 100 μmol photons m−2 s−1 to approximately 25 μmol photons m−2 s−1 for 3 h. Under these conditions, the expression of both VChR1 and VChR2 increased significantly (i.e. approximately 3.8-fold for VChR1 and approximately 2.8-fold for VChR2) relative to those obtained under standard conditions (Fig. 8, B and G), indicating that Volvox compensates for reduced light intensities by increasing the number of photoreceptors.
Figure 8.
The influence of light intensity, heat or cold stress, and wounding on the expression of VChR1 and VChR2. A and F, Expression of VChR1 and VChR2 was analyzed after 3 h at high light intensities of approximately 450 μmol photons m−2 s−1 PAR (light stress, 450 PAR) and compared with standard conditions with approximately 100 μmol photons m−2 s−1 PAR (std, 100 PAR). B and G, Expression of VChR1 and VChR2 was analyzed after 3 h at low light intensities of approximately 25 μmol photons m−2 s−1 PAR (low light, 25 PAR) and compared with standard conditions with approximately 100 μmol photons m−2 s−1 PAR (std, 100 PAR). C and H, Expression of VChR1 and VChR2 was tested after a heat stress and compared with standard conditions (std, 28°C). For heat stress conditions, the culture temperature was elevated from 28°C to 42.5°C for 100 min and then to 45°C for 20 min. D and I, Expression of VChR1 and VChR2 was analyzed after a cold stress and compared with standard conditions (std, 28°C). For cold stress conditions, the culture temperature was reduced from 28°C to 14°C for 120 min. E and J, Expression of VChR1 and VChR2 was tested after mechanical wounding (wound; arrowhead) in comparison with standard conditions (std). The Volvox females were wounded 1 h before RNA isolation. Error bars refer to the sd (n = 3). Arrows indicate the time points at which the mRNA was isolated during the 8-h-dark/16-h-light cycle; the respective dark phase is indicated by “embryogenesis” or “hatching.” [See online article for color version of this figure.]
We then studied the influence of temperature on the expression of VChR, because Volvox photomovement responses are clearly temperature dependent (Sakaguchi and Tawada, 1977; Sakaguchi and Iwasa, 1979). Heat stress may also lead to expression and release of the sex inducer (Kirk and Kirk, 1986), but this should have no influence on the results, as the expression of VChR is tested (after 2 h) long before the sex inducer causes modification of the VChR expression profile (after 8 h; Fig. 5, A and B). First, the culture temperature was elevated from 28°C to 42.5°C for 100 min, and then to 45°C for 20 min, as described previously (Kirk and Kirk, 1986). The heat stress treatment depressed VChR1 expression dramatically to approximately 10% of the normal expression level (Fig. 8C), whereas VChR2 expression declined only slightly to approximately 76% of the reference (Fig. 8H). Second, the culture temperature was reduced from 28°C to 14°C for 120 min, which reduced the expression of VChR1 and VChR2 almost equally to approximately 38% of the normal expression level at 28°C (Fig. 8, D and I).
It is known that the expression of several Volvox genes is promoted not only by the sex inducer but also by mechanical wounding (Amon et al., 1998; Ender et al., 1999; Hallmann et al., 2001; Hallmann, 2006, 2007). To investigate the influence of mechanical wounding on the expression of VChR, Volvox females were wounded and incubated for 1 h under standard conditions. The RNA was immediately isolated and the VChR RNA levels were compared with those in unharmed females. The expression was lowered for both VChRs, to approximately 33% for VChR1 and to approximately 65% for VChR2, relative to standard conditions (Fig. 8, E and J). In contrast to the genes investigated previously, both VChR genes were up-regulated by the sex inducer but down-regulated in response to wounding.
Spectroscopic Characterization of VChR1 and VChR2
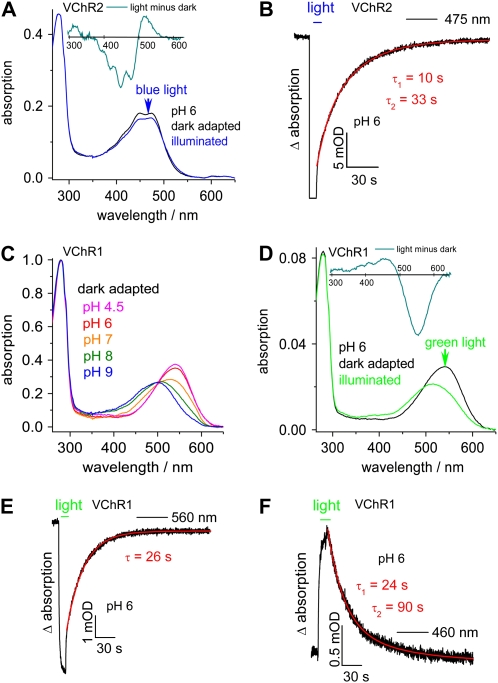
Both VChRs were expressed in green monkey COS cells, and the proteins were affinity purified as described for the VChR1/2 hybrid (Ernst et al., 2008) and for ChR1 and ChR2 (Berthold et al., 2008; Ritter et al., 2008). The UV/visible spectrum of dark-adapted VChR2 is shown in Figure 9A (black line) and is nearly identical under acidic and alkaline conditions (data not shown). The spectrum is fine structured and has maxima at 445 and 475 nm and a shoulder at 413 nm. The mean of the adjoining submaxima is 460 nm, which is regarded as the (averaged) absorption maximum. Accordingly, the dark species was named D460. The maximum and the fine structure indicate that the Schiff base linkage between the chromophore and the protein is protonated and rigidly embedded in the protein. Upon continuous illumination with blue light (5 s, 456 nm), the fine structure is diminished and a red shift of the maximum occurs (Fig. 9A, blue line). This is most obvious from the difference spectrum shown as an inset in Figure 9A. The shift is caused by the accumulation of a late photocycle intermediate that is named P480 in the case of either VChR1/2 or ChR2 (Ernst et al., 2008; Ritter et al., 2008). The dark state D460 recovers biexponentially with τ values of 11 and 33 s as monitored at 475 nm (Fig. 9B). Since absorption, bleaching, and recovery of the dark state are very similar to those found with ChR2 from C. reinhardtii, the photocycle properties were not studied in detail. More interesting were the properties of VChR1. At pH 6, VChR1 absorbs in the visible region maximally at 540 nm; accordingly, the dark species was named D540. The absorption is red shifted compared with the absorption of all other channelrhodopsins known so far. However, the spectrum shifts back to approximately 500 nm at alkaline pH (the D500 isoform), with a pK value of approximately 7 (Fig. 9C). Illumination of VChR1-D540 again shifts the spectrum toward blue (Fig. 9D) due to accumulation of a late photocycle intermediate. This intermediate is named P500 according to its assumed maximal absorption. The dark state recovers very slowly with τ = 26 s (Fig. 9E), which is consistent with the disappearance of the photointermediate P500 as monitored at 460 nm (τ = 24 s; Fig. 9F). A small contribution (approximately 17%) of a second, even slower component with τ = 90 s was only observed at 460 nm. The extinction coefficient of the acidic dark state D540 is at least 25% higher than that of the alkaline form D500 and the late photointermediate P500 (light-adapted state; Fig. 9, C and D).
Figure 9.
Stationary and time-resolved UV/visible spectroscopy of the dark-adapted and photostationary states of VChR1 and VChR2. A, Absorbance spectra of recombinant VChR2 (in dodecyl maltoside solution) in the dark-adapted state (black line) and after 5 s of illumination with blue light of 456 nm at pH 6 (blue line) normalized to the absorption at 280 nm. Inset, The light-minus-dark difference spectrum of VChR2. B, Bleaching and recovery of the dark-adapted VChR2 (at pH 6). The absorbance changes at 475 nm after 5 s of illumination with 456 nm are shown as a function of time. The biphasic recovery process showed time constants of τ1 = 10 s and τ2 = 33 s. C, Absorbance spectra of recombinant VChR1 (in dodecyl maltoside solution) in the dark-adapted state at pH 4.5 (pink line), pH 6 (red line), pH 7 (orange line), pH 8 (green line), and pH 9 (blue line) normalized to the absorption at 280 nm. D, Absorbance spectra of recombinant VChR1 (in dodecyl maltoside solution) in the dark-adapted state (black line) and after 5 s of illumination with green light (540 nm; green line) normalized to the absorption at 280 nm. Inset, The light-minus-dark difference spectrum of VChR1. E, Bleaching and recovery of the dark-adapted VChR1. The absorbance changes at 560 nm after illumination with a 540-nm light-emitting diode are shown as a function of time (τ = 26 s). F, The formation and decay of the late photoproduct, VChR1-P500, was monitored at 460 nm after 5 s of illumination using a 540-nm light-emitting diode. The biphasic disappearance process showed time constants of τ1 = 24 s and τ2 = 90 s. Spectra D to F were measured at pH 6.
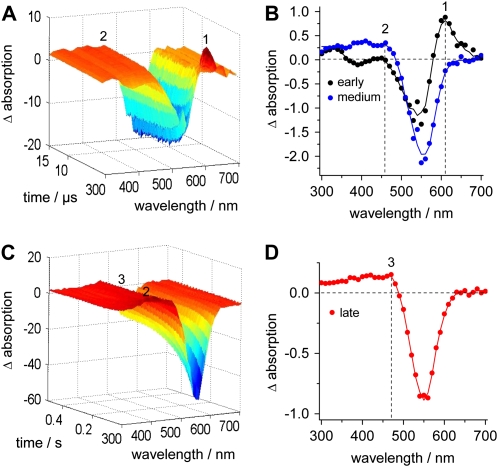
To further characterize the photocycle intermediates, we excited purified VChR1 with 10-ns laser flashes (456 nm) and recorded time-resolved UV/visible spectra. The three-dimensional plots in Figure 10, A and C, show flash-induced absorbance changes of VChR1 at pH 6 in the spectral range between 300 and 700 nm, which visualize the formation and decay of the photocycle intermediates. The spectra were recorded with a resolution of 10 nm over a period of 15 μs after the actinic flash in Figure 10A and over 500 ms in Figure 10C. The plots were reconstructed using the two most significant components that were obtained by singular value decomposition (see “Materials and Methods”). The most significant difference spectra are shown in Figure 10, B and D. An immediate absorbance increase was found at 610 nm on a time scale of nanoseconds (Fig. 10A; early spectrum in Fig. 10B). Generally, the maxima of difference spectra are shifted compared with the absolute spectra when the bands of the initial state and the final state overlap. Therefore, the 610-nm absorption increase that was observed in the early difference spectra corresponds to a photointermediate that absorbs maximally at around 600 nm; accordingly, it was termed P600. Within microseconds, P600 converts into two species that have maxima around 390 nm (P390) and 460 nm (P480; medium spectrum in Fig. 10B). P480 is particularly difficult to see, as the absorption increase is small (due to its small extinction coefficient) and the absorption strongly overlaps with the bleaching of the dark state, D540. Subsequently, both P390 and P480 convert to an intermediate termed P500 (Fig. 10D), which accumulates under steady-state illumination and decays only on a second time scale, as seen in Figure 9, E and F. However, low-temperature experiments are needed to separate these species more clearly.
Figure 10.
Time-resolved UV/visible spectroscopy of VChR1. A and C, Three-dimensional reconstructions of time-resolved absorbance changes of VChR1 at pH 6 after a 10-ns, 540-nm laser flash in the time range of 0.8 to 15 μs (A) and 10 to 500 ms (C) after the flash. B and D, The most significant difference spectra for the time range of 0.8 to 15 μs (B) and 10 to 500 ms (D). B, The early intermediate P600, which is formed immediately after the flash (τ = 1.5 μs; black), is seen as a difference maximum at 610 nm (1), whereas the medium intermediate P480 (τ = 2 μs; blue) is seen as a difference maximum at 460 nm (2). D, The late intermediate P500 (τ = 117 ms; red) shows a broad difference maximum with a difference peak at around 470 nm (3).
DISCUSSION
VChR Contributions to Photomovement Responses
We have shown that the sequences of the analyzed channelrhodopsins, VChR1 and VChR2 from V. carteri, are in several respects closely related to the channelrhodopsins of C. reinhardtii. Based on (1) the high sequence similarity between the channelrhodopsins from Volvox and Chlamydomonas, (2) the photocurrents that are observed in VChR1- and VChR2-expressing host systems, and (3) the photocurrents recorded from eye-containing somatic Volvox cells (Braun and Hegemann, 1999), we conclude that VChR1 and VChR2 are both true photoreceptors that control photomovement responses in Volvox. Despite the fact that the photomovement of a spheroidal, multicellular organism is more complicated than that of a unicellular individual, the structure of the eye in the somatic cells of Volvox is similar to the structure of the eye in Chlamydomonas cells (see introduction; Fig. 1C). The channelrhodopsins VChR1 and VChR2 are only expressed in the somatic cells (Fig. 6), and these somatic cells resemble Chlamydomonas cells. The flagellated somatic cells are responsible for phototaxis and photophobic responses, and they are able to detect the direction and intensity of light using their eyes (Hoops, 1993). As in Chlamydomonas, the channelrhodopsins of Volvox are expected to be localized in a plasma membrane patch right above the eyespot globules (Figs. 1C and 3C). This location hypothesis is supported by the fact that intramembrane particles have been observed in the plasma membrane above the eyespot globules (Melkonian and Robenek, 1984), and consistently, in Volvox the photoreceptor currents are restricted to the eyespot region of somatic cells (Braun and Hegemann, 1999). These photocurrents are detectable not only from intact cells but also from excised eyes.
In Volvox, the photoreceptor currents are slower than in the single-celled Chlamydomonas (Braun and Hegemann, 1999). However, the time scale on which the flagella of Volvox have to respond to a light stimulus is quite different from that of Chlamydomonas: the Volvox spheroid rotates only at 0.45 Hz (Schletz, 1976; Sakaguchi and Iwasa, 1979), whereas Chlamydomonas rotates at 2 Hz. Therefore, there is no need for a faster signaling system in Volvox. However, the time scale of the photocurrents in Volvox allows, in principle, the involvement of a signal transducer mediating between the rhodopsin and the channel, as has been suggested earlier (Braun and Hegemann, 1999).
Changes in light intensity modulate the activity of light-gated ion channels, the cation flux into the eyespot area, and the plasma membrane potential. Small photoreceptor currents modulate the flagellar beating plane and frequency in Chlamydomonas and Volvox. Large photoreceptor currents activate action potential-like flagellar currents in unicellular Chlorophyceae such as Haematococcus pluvialis (Litvin et al., 1978) and C. reinhardtii (Harz and Hegemann, 1991). The massive Ca2+ influx switches the flagellar beating mode from forward swimming (breast stroke style) to slow backward swimming (undulation), resulting in a photophobic response (Schmidt and Eckert, 1976; Holland et al., 1997). In contrast, the flagella of Volvox somatic cells beat synchronously and in parallel, and the light response is graded from normal beating to flagellar arrest. However, both the action potential-like flagellar currents (Braun and Hegemann, 1999) and the switching of the beating mode to an undulation mode do not occur (Sakaguchi and Iwasa, 1979; Hoops, 1993, 1997). The anterior cells were the most responsive cells. They have larger eyespots (Hand and Haupt, 1971; Sakaguchi and Iwasa, 1979), and the resulting sensitivity gradient from the anterior to the posterior pole should consequently also involve a gradient in the number of VChR molecules per cell.
The Putative Role of the Long C-Terminal VChR Extensions
The function of the C-terminal extensions of all channelrhodopsin polypeptide chains, which are at least approximately 400 amino acid residues in length (Table I), is unclear, both in Chlamydomonas and in Volvox. The four channelrhodopsins contain quite a few MG repeats (14–53) in this part of the polypeptide, and VChR1 as well as ChR2 also contain many NG repeats (Table I). In contrast, VChR2 and ChR1 contain just a single NG repeat. The C-terminal extensions of all channelrhodopsins also contain a high percentage (33%–42%) of the three smallest amino acid residues, Gly, Ala, and Ser (Table I). The MG/NG repeats and the high percentage of small residues together create areas with low complexity and (probably) a high degree of structural flexibility.
However, there are also three regions in the C-terminal extensions that are highly conserved among the four channelrhodopsins: con1, con2, and con3 (Supplemental Fig. S7). In addition, the distances between these conserved regions are themselves rather conserved (Table I): con1 comes immediately after the 7TM domain, the distance between con1 and con2 is approximately 40 to 60 amino acid residues, and the distance between con2 and con3 is approximately 160 to 320 residues. The sequences between the conserved regions do not show any significant sequence similarity among the four channelrhodopsins or with any other sequence.
Because channelrhodopsins of both Volvox and Chlamydomonas are expected to be localized in the eyespot overlaying part of the plasma membrane (Figs. 1C and 3C), and because the cytoplasmic membrane is a two-dimensional fluid, there must be a mechanism that holds the channelrhodopsins in place. The role of the three conserved C-terminal domains, which seem to be connected via flexible spacers that allow for some molecular motion, might be to attach channelrhodopsins to proteins of the outer chloroplast envelope. These proteins could in turn be connected to proteins of the inner chloroplast envelope, which might in turn be connected to the carotenoid-filled lipid granules in the stroma of the chloroplast. In this way, the complex sandwich construction of the eye (Fig. 1C) might be held together, thus preventing the channelrhodopsins from diffusing within the plasma membrane, out of the eyespot area (Kreimer, 2009).
The Expression Characteristics Suggest That the VChRs Have Different Functions
The next interesting questions are: (1) why does the expression of both VChRs vary so dramatically during the life cycle? and (2) why it is affected by different external stimuli (Figs. 5, 7, and 8)? In embryogenesis, a cohort of new somatic cells arises, and the produced somatic cells have to build and increase their eyes. Therefore, a strong increase of channelrhodopsin synthesis during embryogenesis was expected. However, why is there a second increase in VChR1 expression before the juveniles hatch but no such occurrence in VChR2 expression? In our 8-h-dark/16-h-light cycle, hatching happens at night. Therefore, we assumed that this second increase of VChR1 is an attempt to compensate for low light intensity. The modification of the 8-h-dark/16-h-light cycle demonstrated that the expression of VChR1 responds significantly to changes in the light conditions. Extended dark phases resulted in an increase of the VChR1 transcription level (Fig. 7, A–D), which could reflect an adaptation to a reduced light intensity by an increased synthesis of VChR1 mRNA. This assumption was confirmed by the fact that, at low light intensities, the expression of VChR1 expression strongly increased compared with normal light conditions (Fig. 8B). However, the expression of VChR2 also increased at low light intensities (Fig. 8G), even though the expression of VChR2 does not vary greatly during dark period extensions (Fig. 7, E–H), and the relative effect of the extended dark phases on VChR1 expression varied between the four investigated time points in the life cycle (Fig. 7, A–D). These facts suggest that not only light but also additional factors, such as the developmental stage, affect the expression levels.
The temperature influences the photomovement responses of Volvox. In moderate light and at temperatures between 20°C and 30°C, Volvox shows positive phototaxis. However, negative phototaxis was observed under the same light conditions when the temperature was reduced to 14°C. Changes in the direction of the photomovements in response to temperature have also been reported in the distantly related crustacean Daphnia (Van Uytvanck and De Meester, 1990) and in the insect Galleria (Kavaliers and Macvean, 1980). In Volvox, channelrhodopsins might be involved in the temperature-dependent photomovement responses. The two absorption maxima at 520 and 460 nm would be quite consistent with a larger contribution of VChR1 and a smaller contribution of VChR2 (Sakaguchi and Tawada, 1977; Sakaguchi and Iwasa, 1979), but since the ratio of the maxima is independent of temperature, the inversion from positive to negative phototaxis is not easily explained by the altered VChR1-VChR2 ratios found in the experiments that are reported in Figure 8.
Possible Involvement of VChRs in Developmental Processes
As the photocurrent action spectrum (Braun and Hegemann, 1999), the absorption spectrum of VChR1 at pH 7 (Fig. 9), and the action spectra of both negative and positive Volvox phototaxis (Sakaguchi and Iwasa, 1979) all peak around 520 nm, we conclude that VChR1 is the main phototaxis photoreceptor under vegetative conditions. However, the role of VChR2 is less clear. As mentioned above, light is not only the stimulus for photomovement responses; it is also essential for other cellular and developmental processes. For example, cell differentiation, one of most important developmental processes in Volvox, is regulated by light (Kirk and Kirk, 1985). Juveniles containing presumptive somatic and reproductive cells are produced during the dark, but their cells do not differentiate until the light comes back on. The pattern of protein synthesis was also shown to change rapidly after the light comes back on (Kirk and Kirk, 1985). These changes are not simply a consequence of photosynthetic activity, because the action spectrum for protein synthesis clearly differs from that for photosynthesis: protein synthesis exhibits a clear maximum in the green range, not purple, blue, and yellow, as is expected for photosynthesis (Kirk and Kirk, 1985). Therefore, it was assumed that the light-dependent change in the protein synthesis of somatic cells and gonidia is rhodopsin mediated (Kirk and Kirk, 1985).
Remarkably, the expression of channelrhodopsins peaks right before the light comes back on (LC7; Fig. 5), which seems to allow for maximum responsivity to light at this developmental stage. Moreover, the spectrum of VChR1, having a maximum at approximately 520 nm at pH 7.5 to 8 (Fig. 9C), matches the action spectrum for protein expression quite nicely, suggesting again that VChR1 is the photoreceptor responsible for this process. VChR1 conducts at least some Ca2+ (Zhang et al., 2008), which might be the trigger for protein synthesis. However, it can do this only in somatic cells where it is expressed; thus, another yet unknown light sensor has to accomplish this function in gonidia.
The striking increase in the expression of VChR2 in the presence of the sex inducer (Fig. 5) suggests that VChR2 plays a key role for the sexual development of the alga, and earlier studies showed that in Volvox, the switch from vegetative to sexual development requires not only the sex inducer but also light (Starr et al., 1980). Light is also a critical parameter for the sexual development of Chlamydomonas, promoting conversion of pregametes to mating-competent gametes with an action spectrum that peaks at 450 nm (Gloeckner and Beck, 1995; Pan et al., 1997). However, phototropin has been established as the primary photoreceptor of this process (Pan et al., 1997). Moreover, as the published action spectrum for protein translation (Kirk and Kirk, 1985) is clearly red shifted relative to the absorption of VChR2, VChR2 is not a likely candidate for this process. Instead, it is conceivable that VChR2 serves as a general, protective photoreceptor that guides the alga phototactically away from blue light, which is particularly necessary during division and development of the alga. In H. salinarum, the phoborhodopsin SRII, which has the most blue-shifted absorption of all rhodopsins (λmax = 495 nm), is constitutively expressed under nearly all conditions, whereas bacteriorhodopsin, halorhodopsin, and sensory rhodopsin SRI, which absorb maximally between 568 and 588 nm, are only expressed at low oxygen concentrations (Spudich, 2006).
The Spectral Properties of VChRs
The absorption spectrum of VChR2 (Fig. 9A) is virtually identical with the spectra of ChR2 from C. reinhardtii (Ritter et al., 2008) and VChR1/2 (Ernst et al., 2008), including the fine structure. In the hybrid VChR1/2, the only spectral effect of the exchange of helices 1 and 2 from VChR2 with those from VChR1 is that it exhibits a 10-nm proton-induced red shift like its ChR1/2 counterpart (Tsunoda and Hegemann, 2009), which is not seen in VChR2 (Fig. 9) and ChR2 (Ritter et al., 2008). As the spectral properties and the photocurrent kinetics of VChR1/2, ChR2, and VChR2 are extremely similar, we anticipated the photocycles of these channelrhodopsin variants also to be closely similar. This was corroborated by the late transitions, with τ1 = 10 s and τ2 = 30 s, resembling the τ1 = 6 s and τ2 = 40 s for ChR2 under comparable conditions (Ritter et al., 2008). In contrast to VChR2, VChR1 is quite distinctive, as it has a large pH-dependent shift of the absorption maximum from 500 nm (D500) at high pH to 540 nm (D540) at low pH. The large absorption coefficient of D540 caused strong bleaching at 540 nm, with a small increase in the absorption at other wavelengths. This made it particularly difficult to identify the photocycle intermediates (Fig. 10). Nevertheless, the early absorption increase, assigned to an early intermediate (P600), is the most red-shifted channelrhodopsin species identified so far. Its extinction coefficient is still quite large, since the positive and negative absorption bands are comparable. The conducting state must appear within the time range of the rise and decay of the photocurrent (Zhang et al., 2008) after a light flash is blue shifted compared with the dark state D540, but the maximum can hardly be estimated from the flat increase of the absorption and the small extinction coefficient of this photocycle intermediate (Fig. 10, C and D). The latest, probably nonconducting photocycle intermediate is synonymous with the light-adapted state, the spectrum of which is seen in Figure 9D. This VChR1 spectrum is also blue shifted, due to accumulation of P500, which has a reduced absorption coefficient relative to D540, whereas the spectra of dark- and light-adapted VChR2 are nearly identical, as is the case for Chlamydomonas ChR2 (Ritter et al., 2008). Clearly, the amino acid residues that dominate the large red absorption at low pH are less influential on the conducting state, the late intermediate state, or the light-adapted state.
The biological function of the acid-induced red shift is unknown and even obscure, as the overlap with the absorption of the pigmented eyespot granules is reduced (Kreimer, 2009). We speculated previously that the pH-induced absorption shift of ChR1 from 470 to 500 nm could be involved in the switch between negative and positive phototaxis (Berthold et al., 2008; Hegemann, 2008). This idea might also apply to VChR1, but more physiological experiments and experiments with deleted or modified VChR1 and VChR2 are needed to verify this hypothesis.
Our results provide a starting point for future studies to elucidate light-induced signal transduction and intracellular trafficking of photoreceptors in a multicellular alga. Clearly, more work is necessary to fully understand the role of both channelrhodopsins in this light-related molecular network.
MATERIALS AND METHODS
Strains and Culture Conditions
The wild-type Volvox carteri f. nagariensis strains Eve10 (female) and 69-1b (male) originate from Japan and have been described previously (Starr, 1969, 1970; Adams et al., 1990). Cultures were grown in Volvox medium (Provasoli and Pintner, 1959) at 28°C in a cycle of 8 h of dark/16 h of cool fluorescent white light (Starr and Jaenicke, 1974) at an average of approximately 100 μmol photons m−2 s−1 photosynthetically active radiation (PAR).
Induction of Sexual Development
A population of vegetatively grown V. carteri males was induced to produce and release the sex inducer using heat shock (Kirk and Kirk, 1986). Seventy milliliters of the fluid of this culture was sterile filtered and added to a 10-L culture of vegetatively grown females at the developmental stage, shortly before hatching of juveniles, to induce sexual development.
Mechanical Wounding
A population of vegetatively grown V. carteri females was wounded using a 50-mL Dounce homogenizer with a tight-fitting pestle (B. Braun); the pestle was moved up and down seven times. In this procedure, V. carteri spheroids were slit into hemispheres and smaller fragments.
High- and Low-Intensity Light Treatment
For the high- and low-intensity light treatments, V. carteri females were grown for 3 h at an average of approximately 450 μmol photons m−2 s−1 PAR or approximately 25 μmol photons m−2 s−1 PAR, respectively (cool fluorescent white light). The reference V. carteri females were grown for 3 h under standard light conditions (i.e. at an average of approximately 100 μmol photons m−2 s−1 PAR). The light intensity was measured using an LI-250A light meter (Li-Cor Biosciences).
Primer Design
For all of the PCRs, we designed the oligonucleotide primers using the primer analysis software Oligo 6 (Molecular Biology Insights), DNASIS (version 7.00; Hitachi Software Engineering), or Primer Express (Applied Biosystems).
Isolation of Genomic DNA
Genomic DNA was prepared from Volvox spheroids using the DNeasy Plant Mini Kit (Qiagen). We checked the purity and quantity of the DNA using agarose gel electrophoresis and UV spectrophotometry (Ultrospec 2100 Pro UV/Visible Spectrophotometer; GE Healthcare).
DNA Gel-Blot Analysis
After restriction enzyme digestion (AccI, BamHI, and HincII), genomic DNA fragments were separated on 1% agarose gels, vacuum transferred to nylon membranes (Hybond-N; Amersham Biosciences), and fixed to the membrane by baking for 30 min at 120°C using standard protocols (Sambrook and Russell, 2001). The probes were amplified using PCR (Expand High Fidelity Plus PCR System; Roche Applied Science) and simultaneously labeled using a digoxigenin DNA labeling mix (Roche Applied Science). Plasmids containing the genomic DNA of VChR1 and VChR2 were used as templates. The VChR1 probe (probe 1) was amplified using the primers 5′-TGGTTACAAAGCGAAAGCTAACAC-3′ and 5′-ACAGAGTCGCAGGTGAATCG-3′ and resulted in an 867-bp fragment (nucleotide positions 846–1,712). The VChR2 probe (probe 2) was amplified using the primers 5′-GAAAACGGTTCACCAGCACAAG-3′ and 5′-GCATAGTAGATGAGGATTATGACC-3′ and resulted in a 741-bp fragment (nucleotide positions 794–1,534). For the DNA gel blots, prehybridization, hybridization, and washing steps were carried out using the standard solutions (Roche Applied Science); the prehybridization and hybridization temperature was 52°C, and the washing procedure was as described (Sambrook and Russell, 2001). The hybridizing bands were detected using an anti-digoxigenin-alkaline phosphatase conjugate (1:7,500 dilution) and the chemiluminescent substrate CDP Star, in accordance with the instructions of the supplier of the chemiluminescence reagent (Roche Applied Science). Chemiluminescence-sensitive films (Retina XBA; Fotochemische Werke) were subsequently exposed to the membranes for 2 to 15 min.
Separation of Cell Types
Ten-liter cultures of synchronously grown V. carteri spheroids were harvested by filtration on a 100-μm mesh nylon screen shortly before the onset of cell cleavage. The mother spheroids were disrupted using a 50-mL Dounce homogenizer with a tight-fitting pestle (B. Braun), and the gonidia and somatic cell sheets were separated as described previously (Hallmann et al., 2001; Nematollahi et al., 2006).
Isolation of Total RNA
Total RNA was extracted from 1 g of frozen gonidia or somatic cells using 10 mL of the phenol-based TRI Reagent (Sigma-Aldrich) and 3 mL of trichloromethane. The RNA was precipitated from the aqueous phase with isopropanol. The RNA pellets were washed twice with 75% ethanol, air dried, and dissolved in RNase-free distilled water. The quantity and purity of the RNA were checked using agarose-formaldehyde gel electrophoresis and by measuring the absorption at 260 and 280 nm using the Ultrospec 2100 pro UV/Visible Spectrophotometer (GE Healthcare).
Cloning of VChR1 and VChR2 Fragments
cDNA fragments of VChR1 and VChR2 were obtained by RT-PCR using total RNA as a template. The oligonucleotide primer pairs 5′-AGTCACGAGATGCATACTTGGC-3′ (antisense) and 5′-TGGTTACAAAGCGAAAGCTAACAC-3′ (sense) and 5′-GGAAACCAGACATGCTCAGAC-3′ (antisense) and 5′-CGACTTTGTGCTGATGCACC-3′ (sense) produced VChR1 cDNA fragments of lengths 1,004 and 1,238 bp, respectively. The oligonucleotide primer pair 5′-GTGGACTTCTTGACGGTGTC-3′ (antisense) and 5′-GAAAACGGTTCACCAGCACAAG-3′ (sense) produced a VChR2 cDNA fragment 943 bp long. The cDNAs were cloned and sequenced.
Quantitative Real-Time RT-PCR
The SensiMix One-Step Kit (Quantace) and a DNA Engine Opticon Continuous Fluorescence Detection System (MJ Research) were used for real-time RNA quantification as described previously (Nematollahi et al., 2006). All real-time RT-PCR experiments were carried out in triplicate together with controls lacking RT or template. The final products of all real-time RT-PCRs were visualized using agarose gel electrophoresis to ensure amplification of a single product of the correct size. The specific primers were as follows: 5′-CAATTGTTTTCTCAATGCTGGTACC-3′ (antisense) and 5′-TCCACTATGGATTATCCCGTTGC-3′ (sense) for VChR1 (expected cDNA length, 145 bp) and 5′-CTTCAGCGCTACAGGTGAACC-3′ (antisense) and 5′-GAAAACGGTTCACCAGCACAAG-3′ (sense) for VChR2 (expected, 158 bp). The Volvox actin gene (Cresnar et al., 1990) was used as a control; the actin-specific primers were 5′-TGAGAAGACGTACGAGCTGC-3′ and 5′-CCTCCATGCCGATTAGGCTA-3′ (expected, 104 bp).
Analysis of Gene Expression Using the 2−ΔΔCt Method
The expression levels of VChR1 and VChR2 were analyzed using real-time RT-PCR and the 2−ΔΔCt method (Bustin, 2000; Pfaffl, 2001). The Volvox actin gene, which is expressed at a constant level in both cell types throughout the life cycle (Cresnar et al., 1990; Amon et al., 1998; Nematollahi et al., 2006), was used as an internal control in all real-time RT-PCR experiments. The Ct, ΔCt, and ΔΔCt values were calculated as described previously (Nematollahi et al., 2006).
Database Sequence Search
BLASTn and tBLASTx algorithms (Altschul et al., 1990) were used to search for opsin-related sequences in the databases of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), V. carteri whole-genome shotgun reads at the Chlamydomonas reinhardtii Web site (version 3.0) of the JGI (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), and later, in genomic sequences at the V. carteri Web site (JGI Volvox genome portal 1.0; http://genome.jgi-psf.org/Volca1/Volca1.home.html), which was established in 2007.
Phylogenetic Analysis
The protein sequences were aligned using the Multiple Sequence Comparison by Log-Expectation program (MUSCLE; Edgar, 2004). Minor manual optimization of the alignments, trimming, and management of multialigned data were done using BioEdit version 7.0.9 (Hall, 1999). The alignments were illustrated using GeneDoc 2.6 (Nicholas et al., 1997). The unrooted tree was calculated using the PHYLogeny Inference Package (PHYLIP; Felsenstein, 1989). In these calculations, 10,000 bootstrap resamplings of multiply aligned sequences were generated using Seqboot. Distance matrices (Dayhoff's point-accepted mutation matrix) were computed with Protdist, trees were constructed using the neighbor-joining method (Saitou and Nei, 1987) as implemented in Neighbor, and finally, a consensus tree was built using Consense. Phylogenetic trees were drawn with TreeView (Page, 1996).
VChR1 and VChR2 Expression in COS-1 Cells
For expression in COS-1 cells (CRL-1650; American Type Culture Collection), human codon-optimized synthetic VChR DNA fragments (corresponding to amino acids 1–317 of VChR1 and 1–317 VChR2 of the complete polypeptide sequences; accession nos. EU285658 and EU285660) and a C-terminal ETSQVAPA sequence (1D4 epitope; Molday and MacKenzie, 1983) were designed by and purchased from GeneArt. The synthetic VChR DNAs were inserted between the EcoRI-NotI sites of the expression vector pMT4. Tissue culture, transient transfection with the resulting ChR2-pMT4 vector, reconstitution with the chromophore, and the subsequent purification of ChR2 were all carried out essentially as described for bovine rhodopsin (Meyer et al., 2000). Three days after transfection, the cells were harvested and reconstituted with all-trans-retinal (final concentration, 30 μm). The VChRs were solubilized using dodecyl maltoside and purified by immunoaffinity adsorption using the rho 1D4 antibody coupled to cyanogen bromide-activated Sepharose 4 Fast Flow resin (GE Healthcare). The VChRs were eluted using 100 μm of an 18-mer peptide corresponding to the C-terminal rhodopsin sequence dissolved in 0.03% (w/v) dodecyl maltoside, 10 mm 1,3-bis-[tris(hydroxymethyl)-methylamino]propane, pH 6.0. The eluates were concentrated using Centricon YM-10 (Millipore) concentrators and stored at −80°C.
UV/Visible Spectroscopy
For absorption spectroscopy and measuring the slow kinetics at 20°C, a Cary 50 Bio spectrophotometer (Varian) was used at a spectral resolution of 2 nm. The samples were illuminated for 5 s using a blue Luxeon light-emitting diode (Philips Lumileds) with a wavelength of 540 nm (VChR1) or 456 nm (VChR2) at 90 mW cm−2 and 2 × 1021 photons m−2 s−1. Transient spectroscopy was performed using an LKS.60 flash photolysis system (Applied Photophysics) at 22°C. Excitation pulses (10 ns, 540 or 456 nm) were provided by a tunable Rainbow OPO/ Nd:YAG laser system. The laser energy was adjusted to 5 mJ per shot. The instrument used a Xe-Lamp (150 W) as the monitoring light source, which was pulsed during short-time experiments. Monochromators placed before and after the sample were set to a 2-nm spectral resolution. A 1P28 photomultiplier (Hamamatsu Photonics) was used for detection, and the signal was recorded with an Infinium Oscilloscope (Agilent Technologies). In total, 32,000 data points were recorded in each measurement and compressed by LKS.60 software to files of 500 data points. Fifty of these points were recorded before laser excitation, and 450 points were recorded after laser excitation. To avoid artifacts and scatter, only data points 60 to 500 were used in the analysis. The data were analyzed using Matlab 7.01 (MathWorks). Singular value decomposition of representative data sets was carried out to identify significant components that were used for reconstruction of the three-dimensional spectra. Time constants were obtained by fitting exponential functions to the data. Freshly prepared VChRs were used for the measurements at pH 6, and freshly prepared VChRs were diluted with buffer containing 0.03% dodecyl maltoside for the experiments at pH 8 and 4.5.
Sequence data of Volvox channelrhodopsins are available under GenBank accession numbers EU285658 (VChR1, complete mRNA), EU285659 (VChR1, complete genomic DNA), EU285660 (VChR2, complete mRNA), and EU285661 (VChR2, complete genomic DNA). Other sequence data from this article can be found under accession numbers EU622855 (VChR1, mRNA fragment, synthetic construct), DQ094781 (chimeric mRNA/genomic DNA of VChR1 and VChR2), AF508965 (C. reinhardtii channelrhodopsin-1, ChR1), AF508966 (C. reinhardtii channelrhodopsin-2, ChR2), NP_923144 (Gloeobacter violaceus bacterioopsin, Br), BAB74864 (Nostoc species bacteriorhodopsin, Br), ABA08437 (Guillardia theta opsin-1, Ops1), ABA08438 (Guillardia theta opsin-2, Ops2), ABA08439 (Cryptomonas species opsin, Ops), AF135863 (Neurospora crassa opsin-1, Nop-1), YP_446872 (Salinibacter ruber halorhodopsin, Hop), P94853 (Haloarcula vallismortis cruxhalorhodopsin-3, ChoP3), YP_136278 (Haloarcula marismortui halorhodopsin, Hop), P15647 (Natronomonas pharaonis halorhodopsin, Hop), CAA49773 (Halobacterium species halorhodopsin, Hop), O93742 (Halorubrum sodomense halorhodopsin, Hop), O93741 (Haloterrigena species halorhodopsin, Hopa), BAA75201 (Haloterrigena species halorhodopsin, Hopb), P16102 (Halobacterium salinarum halorhodopsin, Hop), YP_658762 (Haloquadratum walsbyi halorhodopsin, Hop), YP_135281 (Haloarcula marismortui opsin, Xop2), P33743 (Halobacterium species sensory rhodopsin-1, Sop1), O93743 (Halorubrum sodomense rhodopsin, Sop), P25964 (Halobacterium salinarum sensory rhodopsin-1, Sop1), Q48334 (Haloarcula vallismortis bacterial rhodopsin, Csr3), YP_446609 (Salinibacter ruber sensory rhodopsin a, Sria), YP_446677 (Salinibacter ruber sensory rhodopsin b, Srib), P71411 (Halobacterium salinarum sensory rhodopsin-2, Sop2), BAB86796 (Halobacterium species phoborhodopsin, Pr), P42197 (Haloarcula vallismortis sensory rhodopsin-2, Sop2), P42196 (Natronomonas pharaonis sensory rhodopsin-2, Sop2), YP_656804 (Haloquadratum walsbyi bacteriorhodopsin II, BopII), YP_656801 (Haloquadratum walsbyi bacteriorhodopsin I, BopI), YP_136594 (Haloarcula marismortui bacteriorhodopsin, Xop1), P02945 (Halobacterium salinarum bacteriorhodopsin, Bop), P29563 (Halobacterium species archaerhodopsin-2, Ar2), P96787 (Halorubrum sodomense archaerhodopsin-2, Ar3), J05165 (Halobacterium species archaerhodopsin-1, Ar1), AAS15567 (Halorubrum xinjiangense archaerhodopsin, Aop), AAU04564 (Halobiforma lacisalsi bacteriorhodopsin, Br), O93740 (Haloterrigena species bacteriorhodopsin, Bop), AAB32951 (Haloarcula species cruxrhodopsin-2, Cop2), YP_137573 (Haloarcula marismortui bacteriorhodopsin, Bop), BAA81816 (Haloarcula japonica cruxrhodopsin, Cop), and P94854 (Haloarcula vallismortis cruxrhodopsin-3, Cop3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide sequence alignment of the VChR1 and VChR2 coding sequences.
Supplemental Figure S2. Nucleotide sequence alignment of the VChR1 and Chlamydomonas ChR1 coding sequences.
Supplemental Figure S3. Nucleotide sequence alignment of the VChR2 and Chlamydomonas ChR2 coding sequences.
Supplemental Figure S4. Aligned coding sequences of VChR1, VChR2, ChR1, and ChR2 with highlighted intron positions.
Supplemental Figure S5. Protein sequence alignment of the con1, con2, and con3 regions of VChR1 and VChR2.
Supplemental Figure S6. Trimmed alignment of 45 rhodopsin-related proteins from green algae, fungi, cyanobacteria, cryptomonads, and halobacteria.
Supplemental Figure S7. Multiple alignment of the con1, con2, and con3 regions of the channelrhodopsins from Volvox and Chlamydomonas.
Supplemental Table S1. Sequence comparison of VChR1 and VChR2 to related rhodopsins.
Supplemental Data Set S1. Trimmed sequences of 45 rhodopsin-related proteins from green algae, fungi, cyanobacteria, cryptomonads, and halobacteria corresponding to the alignment in Supplemental Figure S6.
Supplementary Material
Acknowledgments
We thank the JGI for providing Volvox and Chlamydomonas sequence information on its Web sites. We also thank T. Schmiedel for support with the DNA gel blots, K. Puls for biological technical assistance, and Dr. Rolf Hagedorn and Christina Mrosek for assistance with the laser system and data evaluation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Armin Hallmann (armin.hallmann@gmx.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Adams CR, Stamer KA, Miller JK, McNally JG, Kirk MM, Kirk DL (1990) Patterns of organellar and nuclear inheritance among progeny of two geographically isolated strains of Volvox carteri. Curr Genet 18 141–153 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Amon P, Haas E, Sumper M (1998) The sex-inducing pheromone and wounding trigger the same set of genes in the multicellular green alga Volvox. Plant Cell 10 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann M, Hegemann P (1991) In vitro identification of rhodopsin in the green alga Chlamydomonas. Biochemistry 30 3692–3697 [DOI] [PubMed] [Google Scholar]
- Berthold P, Tsunoda SP, Ernst OP, Mages W, Gradmann D, Hegemann P (2008) Channelrhodopsin-1 initiates phototaxis and photophobic responses in Chlamydomonas by immediate light-induced depolarization. Plant Cell 20 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BJ, Anderson DM (1986) Green light-mediated photomorphogenesis in a dinoflagellate resting cyst. Nature 322 659–661 [Google Scholar]
- Bogomolni RA, Spudich JL (1982) Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc Natl Acad Sci USA 79 6250–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun FJ, Hegemann P (1999) Two light-activated conductances in the eye of the green alga Volvox carteri. Biophys J 76 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LS (2004) Fungal rhodopsins and opsin-related proteins: eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem Photobiol Sci 3 555–565 [DOI] [PubMed] [Google Scholar]
- Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25 169–193 [DOI] [PubMed] [Google Scholar]
- Cresnar B, Mages W, Müller K, Salbaum JM, Schmitt R (1990) Structure and expression of a single actin gene in Volvox carteri. Curr Genet 18 337–346 [DOI] [PubMed] [Google Scholar]
- Deininger W, Kröger P, Hegemann U, Lottspeich F, Hegemann P (1995) Chlamyrhodopsin represents a new type of sensory photoreceptor. EMBO J 14 5849–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann CL (2003) Eyespot placement and assembly in the green alga Chlamydomonas. Bioessays 25 410–416 [DOI] [PubMed] [Google Scholar]
- Ebnet E, Fischer M, Deininger W, Hegemann P (1999) Volvoxrhodopsin, a light-regulated sensory photoreceptor of the spheroidal green alga Volvox carteri. Plant Cell 11 1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender F, Hallmann A, Amon P, Sumper M (1999) Response to the sexual pheromone and wounding in the green alga Volvox: induction of an extracellular glycoprotein consisting almost exclusively of hydroxyproline. J Biol Chem 274 35023–35028 [DOI] [PubMed] [Google Scholar]
- Ernst OP, Murcia PA, Daldrop P, Tsunoda SP, Kateriya S, Hegemann P (2008) Photoactivation of channelrhodopsin. J Biol Chem 283 1637–1643 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1989) Phylip: Phylogeny Inference Package (version 3.2). Cladistics 5 164–166 [Google Scholar]
- Foster KW, Smyth RD (1980) Light antennas in phototactic algae. Microbiol Rev 44 572–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Deininger W, Kateriya S, Hegemann P (2003) Rhodopsin-related proteins, cop1, cop2 and chop1 in Chlamydomonas reinhardtii. In A Batschauer, ed, Photoreceptors and Light Signalling. Comprehensive Series in Photochemical and Photobiological Sciences. Royal Society of Chemistry, Cambridge, UK, pp 124–135
- Fuhrmann M, Stahlberg A, Govorunova E, Rank S, Hegemann P (2001) The abundant retinal protein of the Chlamydomonas eye is not the photoreceptor for phototaxis and photophobic responses. J Cell Sci 114 3857–3863 [DOI] [PubMed] [Google Scholar]
- Gilles R, Gilles C, Jaenicke L (1984) Pheromone-binding and matrix-mediated events in sexual induction of Volvox carteri. Z Naturforsch C 39c 584–592 [Google Scholar]
- Gloeckner G, Beck CF (1995) Genes involved in light control of sexual differentiation in Chlamydomonas reinhardtii. Genetics 141 937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Jung KH, Sineshchekov OA, Spudich JL (2004) Chlamydomonas sensory rhodopsins A and B: cellular content and role in photophobic responses. Biophys J 86 2342–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR, Lohr M, Im CS (2004) Chlamydomonas reinhardtii in the landscape of pigments. Annu Rev Genet 38 119–173 [DOI] [PubMed] [Google Scholar]
- Guschina IA, Harwood JL (2006) Lipids and lipid metabolism in eukaryotic algae. Prog Lipid Res 45 160–186 [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41 95–98 [Google Scholar]
- Hallmann A (2006) The pherophorins: common, versatile building blocks in the evolution of extracellular matrix architecture in Volvocales. Plant J 45 292–307 [DOI] [PubMed] [Google Scholar]
- Hallmann A (2007) A small cysteine-rich extracellular protein, VCRP, is inducible by the sex-inducer of Volvox carteri and by wounding. Planta 226 719–727 [DOI] [PubMed] [Google Scholar]
- Hallmann A, Amon P, Godl K, Heitzer M, Sumper M (2001) Transcriptional activation by the sexual pheromone and wounding: a new gene family from Volvox encoding modular proteins with (hydroxy)proline-rich and metalloproteinase homology domains. Plant J 26 583–593 [DOI] [PubMed] [Google Scholar]
- Hand WG, Haupt W (1971) Flagellar activity of the colony members of Volvox aureus Ehrbg. during light stimulation. J Protozool 18 361–364 [Google Scholar]
- Harz H, Hegemann P (1991) Rhodopsin-regulated calcium currents in Chlamydomonas. Nature 351 489–491 [Google Scholar]
- Harz H, Nonnengässer C, Hegemann P (1992) The photoreceptor current of the green alga Chlamydomonas. Philos Trans R Soc Lond B Biol Sci 338 39–52 [Google Scholar]
- Hegemann P (2008) Algal sensory photoreceptors. Annu Rev Plant Biol 59 167–189 [DOI] [PubMed] [Google Scholar]
- Hegemann P, Ehlenbeck S, Gradmann D (2005) Multiple photocycles of channelrhodopsin. Biophys J 89 3911–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann P, Fuhrmann M, Kateriya S (2001) Algal sensory photoreceptors. J Phycol 37 668–676 [Google Scholar]
- Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH (1990) Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol 213 899–929 [DOI] [PubMed] [Google Scholar]
- Holland EM, Harz H, Uhl R, Hegemann P (1997) Control of phobic behavioral responses by rhodopsin-induced photocurrents in Chlamydomonas. Biophys J 73 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops HJ (1984) Somatic cell flagellar apparatuses in two species of Volvox (Chlorophyceae). J Phycol 20 20–27 [Google Scholar]
- Hoops HJ (1993) Flagellar, cellular and organismal polarity in Volvox carteri. J Cell Sci 104 105–117 [Google Scholar]
- Hoops HJ (1997) Motility in the colonial and multicellular Volvocales: structure, function, and evolution. Protoplasma 199 99–112 [Google Scholar]
- Ishizuka T, Kakuda M, Araki R, Yawo H (2006) Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res 54 85–94 [DOI] [PubMed] [Google Scholar]
- Jung KH (2007) The distinct signaling mechanisms of microbial sensory rhodopsins in Archaea, Eubacteria and Eukarya. Photochem Photobiol 83 63–69 [DOI] [PubMed] [Google Scholar]
- Kavaliers M, Macvean C (1980) Effect of temperature and lunarphase on the phototactic responses of larvae of the wax moth, Galleria mellonella (Lepidoptera, Pyralidae). Entomol Exp Appl 28 222–228 [Google Scholar]
- Kirk DL (1998) Volvox: Molecular-Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge University Press, Cambridge, UK
- Kirk DL, Kirk MM (1986) Heat shock elicits production of sexual inducer in Volvox. Science 231 51–54 [DOI] [PubMed] [Google Scholar]
- Kirk MM, Kirk DL (1985) Translational regulation of protein synthesis, in response to light, at a critical stage of Volvox development. Cell 41 419–428 [DOI] [PubMed] [Google Scholar]
- Klare JP, Chizhov I, Engelhard M (2008) Microbial rhodopsins: scaffolds for ion pumps, channels, and sensors. Results Probl Cell Differ 45 73–122 [DOI] [PubMed] [Google Scholar]
- Kreimer G (1994) Cell biology of phototaxis in flagellate algae. Int Rev Cytol 148 229–310 [Google Scholar]
- Kreimer G (2009) The green algal eyespot apparatus: a primordial visual system and more? Curr Genet 55 19–43 [DOI] [PubMed] [Google Scholar]
- Kreimer G, Melkonian M (1990) Reflection confocal laser scanning microscopy of eyespots in flagellated green algae. Eur J Cell Biol 53 101–111 [PubMed] [Google Scholar]
- Litvin FF, Sineshchekov OA, Sineshchekov VA (1978) Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis. Nature 271 476–478 [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK (1999) Structure of bacteriorhodopsin at 1.55 A resolution. J Mol Biol 291 899–911 [DOI] [PubMed] [Google Scholar]
- Mages H-W, Tschochner H, Sumper M (1988) The sexual inducer of Volvox carteri: primary structure deduced from cDNA sequence. FEBS Lett 234 407–410 [DOI] [PubMed] [Google Scholar]
- Matsuno-Yagi A, Mukohata Y (1977) Two possible roles of bacteriorhodopsin: a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem Biophys Res Commun 78 237–243 [DOI] [PubMed] [Google Scholar]
- Melkonian M, Robenek H (1984) The eyespot apparatus of green algae: a critical review. Prog Phycol Res 3 193–268 [Google Scholar]
- Meyer CK, Böhme M, Ockenfels A, Gartner W, Hofmann KP, Ernst OP (2000) Signaling states of rhodopsin: Retinal provides a scaffold for activating proton transfer switches. J Biol Chem 275 19713–19718 [DOI] [PubMed] [Google Scholar]
- Miller G (2006) Optogenetics: shining new light on neural circuits. Science 314 1674–1676 [DOI] [PubMed] [Google Scholar]
- Molday RS, MacKenzie D (1983) Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry 22 653–660 [DOI] [PubMed] [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P (2002) Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296 2395–2398 [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA 100 13940–13945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Bray DF, Costerton JW, Wagenaar EB (1973) The eyespot of Chlamydomonas eugametos: a freeze-etch study. Can J Bot 51 817–819 [Google Scholar]
- Nematollahi G, Kianianmomeni A, Hallmann A (2006) Quantitative analysis of cell-type specific gene expression in the green alga Volvox carteri. BMC Genomics 7 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Deerfield DW (1997) GeneDoc: analysis and visualization of genetic variation. EMBnet.news 4 14 [Google Scholar]
- Oesterhelt D, Stoeckenius W (1973) Functions of a new photoreceptor membrane. Proc Natl Acad Sci USA 70 2853–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto OK, Hastings JW (2003) Genome-wide analysis of redox-regulated genes in a dinoflagellate. Gene 321 73–81 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 357–358 [DOI] [PubMed] [Google Scholar]
- Pan JM, Haring MA, Beck CF (1997) Characterization of blue light signal transduction chains that control development and maintenance of sexual competence in Chlamydomonas reinhardtii. Plant Physiol 115 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Bercoff Å, Koch J, Burglin TR (2006) LogoBar: bar graph visualization of protein logos with gaps. Bioinformatics 22 112–114 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provasoli L, Pintner IJ (1959) Artificial media for freshwater algae: problems and suggestions. In CA Tyron, RT Hartman, eds, The Ecology of Alga. Pymatuning Laboratory of Field Biology, University of Pittsburgh, Pittsburgh, pp 84–96
- Ritter E, Stehfest K, Berndt A, Hegemann P, Bartl FJ (2008) Monitoring light-induced structural changes of channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J Biol Chem 283 35033–35041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 [DOI] [PubMed] [Google Scholar]
- Sakaguchi H, Iwasa K (1979) Two photophobic responses in Volvox carteri. Plant Cell Physiol 20 909–916 [Google Scholar]
- Sakaguchi H, Tawada K (1977) Temperature effect on the photo-accumulation and phobic response of Volvox aureus. J Protozool 24 284–288 [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaller K, Uhl R (1997) A microspectrophotometric study of the shielding properties of eyespot and cell body in Chlamydomonas. Biophys J 73 1573–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schletz K (1976) Phototaxis bei Volvox: Pigmentsysteme der Lichtrichtungsperzeption. Z Pflanzenphysiol 77 189–211 [Google Scholar]
- Schmidt JA, Eckert R (1976) Calcium couples flagellar reversal to photostimulation in Chlamydomonas reinhardtii. Nature 262 713–715 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, Kaminski M, Geimer S, Eitzinger N, Reissenweber T, Voytsekh O, et al (2006) Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18 1908–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert B, Lanyi JK (1982) Halorhodopsin is a light-driven chloride pump. J Biol Chem 257 10306–10313 [PubMed] [Google Scholar]
- Sharma AK, Spudich JL, Doolittle WF (2006) Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol 14 463–469 [DOI] [PubMed] [Google Scholar]
- Shimono K, Furutani Y, Kandori H, Kamo N (2002) A pharaonis phoborhodopsin mutant with the same retinal binding site residues as in bacteriorhodopsin. Biochemistry 41 6504–6509 [DOI] [PubMed] [Google Scholar]
- Sineshchekov OA, Govorunova EG, Jung KH, Zauner S, Maier UG, Spudich JL (2005) Rhodopsin-mediated photoreception in cryptophyte flagellates. Biophys J 89 4310–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineshchekov OA, Jung KH, Spudich JL (2002) Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 99 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JL (2006) The multitalented microbial sensory rhodopsins. Trends Microbiol 14 480–487 [DOI] [PubMed] [Google Scholar]
- Starr RC (1969) Structure, reproduction and differentiation in Volvox carteri f. nagariensis Iyengar, strains HK9 & 10. Arch Protistenkd 111 204–222 [Google Scholar]
- Starr RC (1970) Control of differentiation in Volvox. Dev Biol (Suppl) 4 59–100 [DOI] [PubMed] [Google Scholar]
- Starr RC, Jaenicke L (1974) Purification and characterization of the hormone initiating sexual morphogenesis in Volvox carteri f. nagariensis Iyengar. Proc Natl Acad Sci USA 71 1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr RC, O'Neil RM, Miller CE (1980) L-Glutamic acid as a mediator of sexual differentiation in Volvox capensis. Proc Natl Acad Sci USA 77 1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Yamasaki K, Fujita S, Oda K, Iseki M, Yoshida K, Watanabe M, Daiyasu H, Toh H, Asamizu E, et al (2003) Archaeal-type rhodopsins in Chlamydomonas: model structure and intracellular localization. Biochem Biophys Res Commun 301 711–717 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Mochizuki Y, Kamo N, Kobatake Y (1985) Evidence that the long-lifetime photointermediate of s-rhodopsin is a receptor for negative phototaxis in Halobacterium halobium. Biochem Biophys Res Commun 127 99–105 [DOI] [PubMed] [Google Scholar]
- Tam LW, Kirk DL (1991) Identification of cell-type-specific genes of Volvox carteri and characterization of their expression during the asexual life cycle. Dev Biol 145 51–66 [DOI] [PubMed] [Google Scholar]
- Tschochner H, Lottspeich F, Sumper M (1987) The sexual inducer of Volvox carteri: purification, chemical characterization and identification of its gene. EMBO J 6 2203–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda SP, Ewers D, Gazzarrini S, Moroni A, Gradmann D, Hegemann P (2006) H+-pumping rhodopsin from the marine alga Acetabularia. Biophys J 91 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda SP, Hegemann P (2009) Glu 87 of Channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem Photobiol 85 564–569 [DOI] [PubMed] [Google Scholar]
- Van Uytvanck J, De Meester L (1990) Phototaxis in Daphnia magna: the influence of temperature and acidity on the phototactic behaviour of Daphnia genotypes. J Plankton Res 12 1089–1097 [Google Scholar]
- Zhang F, Prigge M, Beyriere F, Tsunoda SP, Mattis J, Yizhar O, Hegemann P, Deisseroth K (2008) Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat Neurosci 11 631–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.