Abstract
RADICAL-INDUCED CELL DEATH1 (RCD1) and SIMILAR TO RCD ONE1 (SRO1) are the only two proteins encoded in the Arabidopsis (Arabidopsis thaliana) genome containing both a putative poly(ADP-ribose) polymerase catalytic domain and a WWE protein-protein interaction domain, although similar proteins have been found in other eukaryotes. Poly(ADP-ribose) polymerases mediate the attachment of ADP-ribose units from donor NAD+ molecules to target proteins and have been implicated in a number of processes, including DNA repair, apoptosis, transcription, and chromatin remodeling. We have isolated mutants in both RCD1 and SRO1, rcd1-3 and sro1-1, respectively. rcd1-3 plants display phenotypic defects as reported for previously isolated alleles, most notably reduced stature. In addition, rcd1-3 mutants display a number of additional developmental defects in root architecture and maintenance of reproductive development. While single mutant sro1-1 plants are relatively normal, loss of a single dose of SRO1 in the rcd1-3 background increases the severity of several developmental defects, implying that these genes do share some functions. However, rcd1-3 and sro1-1 mutants behave differently in several developmental events and abiotic stress responses, suggesting that they also have distinct functions. Remarkably, rcd1-3; sro1-1 double mutants display severe defects in embryogenesis and postembryonic development. This study shows that RCD1 and SRO1 are at least partially redundant and that they are essential genes for plant development.
Poly(ADP-ribose) polymerases (PARPs) are a class of enzymes that posttranslationally add negatively charged ADP-Rib (PAR) polymers synthesized from NAD+ to Lys residues on target proteins (Altmeyer et al., 2009). Depending on the specific PARP involved, one to hundreds of ADP-Rib units can be attached to the target (Kim et al., 2005). PARPs are found in all groups of eukaryotes and are characterized by the catalytic site, a β-α-loop-B-α NAD+ fold, also called the PARP signature (Ruf et al., 1996; Oliver et al., 2004). This family has been best characterized in humans, where there are 18 family members with diverse functional domains outside of the PARP signature (Ame et al., 2004; Schreiber et al., 2006; Hassa and Hottiger, 2008). It is postulated that different PARPs participate in diverse events through these domains. However, it is unclear if all proteins with PARP signatures actually function as enzymes. For example, while TCDD-inducible PARP, PARP-9, and PARP-10 have replaced an important catalytic residue (Glu) with nonconserved residues (Aguiar et al., 2000; Ma et al., 2001; Yu et al., 2005), PARP-9 is enzymatically inactive (Aguiar et al., 2005), while TCDD-inducible PARP is enzymatically active (Ma et al., 2001) and PARP-10 has transferase activity rather than polymerase activity, adding one ADP-Rib subunit to target proteins (Yu et al., 2005; Chou et al., 2006; Kleine et al., 2008).
PARPs have been implicated to be involved in DNA damage repair, cell death pathways, transcription, and chromatin modification/remodeling (for review, see Kim et al., 2005; Schreiber et al., 2006; Hassa and Hottiger, 2008). Human PARPs have been placed into five classes according to their functions: DNA-dependent PARPs, tankyrases, CCCH-type PARPs, macroPARPs, and a diverse orphan group with no known functions (Schreiber et al., 2006). The original PARPs identified, PARP-1 and PARP-2, are DNA-dependent PARPs that function in DNA damage repair and are characterized by DNA-binding, WGR, and PARP regulatory domains N terminal to their catalytic sites (Satoh and Lindahl, 1992; Trucco et al., 1998; Schreiber et al., 2002). Recent work has also implicated PARP-1 in other DNA-associated functions such as transcriptional control, DNA methylation, and modulation of promoter chromatin state by binding nucleosomes (Hassa et al., 2003; Carrillo et al., 2004; Fossati et al., 2006; Ju and Rosenfeld, 2006; Cohen-Armon et al., 2007; Ishiguro et al., 2007; Choi et al., 2008; Guastafierro et al., 2008; Krishnakumar et al., 2008; Caiafa et al., 2009). The third member of the DNA-dependent PARPs, PARP-3, is similar to PARP-1 and PARP-2 except that it is missing a known DNA-binding domain (Johansson, 1999). PARP-3 has been shown to associate with the centrosome, polycomb group proteins, and DNA repair machinery (Augustin et al., 2003; Rouleau et al., 2007). A recent study in astrocytes suggests that all three DNA-dependent PARPs can act cooperatively during activation of this cell type (Phulwani and Kielian, 2008).
The other groups of PARPs have not been as extensively studied. The tankyrases are involved in telomere length control (Smith et al., 1998; Smith and de Lange, 2000; Cook et al., 2002), while CCCH-type PARPs, characterized by CCCH zinc fingers and a WWE domain in addition to the PARP catalytic domain, bind RNA, particularly viral RNA, through their zinc fingers and target the RNA for degradation (Gao et al., 2002; Guo et al., 2004, 2007; Kerns et al., 2008). The macroPARPs contain macro domains (Aguiar et al., 2005), recently shown to bind ADP-Rib and PAR (Karras et al., 2005; Egloff et al., 2006); members of this family have been implicated in regulation of gene expression through interactions with transcriptional cofactors (Goenka and Boothby, 2006; Cho et al., 2009) and in protecting cells from DNA damage-induced apoptosis (Cho et al., 2009).
PARPs and the role of poly(ADP-ribosyl)ation have not been as well studied in plants as in animal systems. PARP inhibitor studies have demonstrated the involvement of PARPs in abiotic stress (Amor et al., 1998; De Block et al., 2005) and defense responses (Berglund et al., 1996; Adams-Phillips et al., 2008). A role for PARP activity in seeds to protect against genotoxic stress has also been inferred (Hunt and Gray, 2009).
Arabidopsis (Arabidopsis thaliana) encodes nine putative PARP-encoding genes (Supplemental Fig. S1A). Orthologs of PARP-1 and PARP-2, the DNA-dependent PARPs, AtPARP1 (At4g02390) and AtPARP2 (At2g31320), respectively, have been identified (Lepiniec et al., 1995; Babiychuk et al., 1998). These genes have been implicated in DNA repair (Doucet-Chabeaud et al., 2001; De Block et al., 2005) and have been shown to be associated with chromosomes through their N-terminal zinc fingers (Babiychuk et al., 2001). AtPARP1 and AtPARP2 are up-regulated during geminivirus infection, most likely in response to accumulation of nicked viral DNA forms (Ascencio-Ibanez et al., 2008), and also accumulate under conditions of DNA stress (Culligan et al., 2006). The finding that two putative transcriptional coactivators can bind to the zinc fingers of AtPARP1 suggests that, like mammalian PARP-1, this protein may also be involved in regulation of gene expression in addition to its role in DNA repair (Storozhenko et al., 2001). Another gene, At5g22470, which is similar to the other two DNA-dependent PARPs, is also found in Arabidopsis. No functional data on this gene have been published; however, it is highly expressed during seed development (Becerra et al., 2006). As expected, these three genes group together in a phylogenetic tree of flowering plant PARPs (Supplemental Fig. S1A). Down-regulation of AtPARP1 and AtPARP2 causes resistance to a number of abiotic stresses, including heat, cold, and drought (Amor et al., 1998; De Block et al., 2005; Vanderauwera et al., 2007), as does application of PARP inhibitors to plants (De Block et al., 2005). This effect may be mediated directly through PARP targets or could be a consequence of altered NAD metabolism in the plants (Hashida et al., 2009).
No orthologs of the three other functional groups of PARPs known in humans have been identified. However, a group of four genes encoding relatively short proteins with the PARP signature but no other known functional domain(s) has been found (SRO2–SRO5; Belles-Boix et al., 2000; Ahlfors et al., 2004). This family consists of two gene pairs: SRO2/SRO3 and SRO4/SRO5 (Supplemental Fig. S1A). These pairs likely arose from the relatively recent genome duplication in Arabidopsis evolutionary history (Henry et al., 2006); the gene pairs are found in regions of synteny within the Arabidopsis genome (Plant Genome Duplication Database, http://chibba.agtec.uga.edu/duplication/index/home; Tang et al., 2008). These genes may be involved in stress signaling; SRO5 is necessary for response to both salt and oxidative stress (Borsani et al., 2005), and SRO2 is up-regulated in chloroplastic ascorbic peroxidase mutants (Kangasjarvi et al., 2008).
Arabidopsis has two other genes, RADICAL-INDUCED CELL DEATH1 (RCD1) and SIMILAR TO RCD ONE1 (SRO1), that encode putative PARPs with WWE domains N terminal to the PARP signature (Belles-Boix et al., 2000; Ahlfors et al., 2004). This gene pair likely also arose from a genome duplication, as the two genes fall into two syntenic chromosomal regions (Supplemental Fig. S1A; Plant Genome Duplication Database, http://chibba.agtec.uga.edu/duplication/index/home; Tang et al., 2008). WWE domains are postulated to be protein-protein interaction domains and are found in proteins involved in the ubiquitin/proteosome pathway or in PARPs (Aravind, 2001), for example, in the animal CCCH-type PARPs (Katoh, 2003). Proteins with a similar domain structure to RCD1/SRO1 have been found in other plants, including crop species such as rice (Oryza sativa; Supplemental Fig. S1A; Ahlfors et al., 2004), suggesting that these proteins may play a conserved and vital role. In addition, these proteins are similar to a PARP family member of unknown function from human, PARP11 (Supplemental Fig. S2; Hakme et al., 2008).
RCD1 was originally identified as a stress response gene (Overmyer et al., 2000). It is involved in the response to several abiotic stresses, including ozone. All rcd1 alleles confer dominant ozone hypersensitivity, although they seem to be recessive for other phenotypes (Ahlfors et al., 2004; Fujibe et al., 2004). rcd1 mutants display accumulation of both ethylene and salicylic acid and altered expression of ethylene- and abscisic acid-responsive genes (Ahlfors et al., 2004). In addition, rcd1 plants are hypersensitive to ozone (Overmyer et al., 2005) and conversely are resistant to UV-B light (Fujibe et al., 2004). The phenotypic analysis suggests that RCD1 may be a convergence point for several hormone signaling pathways involved in the stress response and act both negatively and positively in stress responses (Ahlfors et al., 2004). rcd1 mutants also display pleiotropic developmental defects, including reduced stature, malformed leaves, and early flowering, consistent with the fact that the gene is expressed in all tissues examined. These developmental defects do not seem to be associated with the defects in the hormonal signaling pathways examined to date (Ahlfors et al., 2004). Recently, phenotypes of RCD1 overexpression transgenic lines have been reported. The overexpression caused dwarfing, normal response to an inducer of free radicals, methyl viologen, and hypersensitivity to ozone. Intriguingly, the 35S∷RCD1 transgene was able to complement all tested rcd1-2 phenotypes (Fujibe et al., 2006). A third RCD1 allele, rcd1-3, has been identified (Katiyar-Agarwal et al., 2006) and displays similar phenotypes to rcd1-1 and rcd1-2, although it has not been fully characterized. Taken together, the available data suggest that there is an optimal level of RCD1 activity/level for normal development and stress response.
The developmental phenotypes of rcd1 alleles have not been extensively analyzed, and despite the significant sequence similarity, almost nothing is known about SRO1 function. This study presents data indicating that SRO1 possesses both unique and overlapping functions with RCD1 and that RCD1 and SRO1 function in previously unidentified developmental pathways, including embryogenesis.
RESULTS
RCD1 and SRO1 Are Paralogous Genes with Similar Expression Patterns
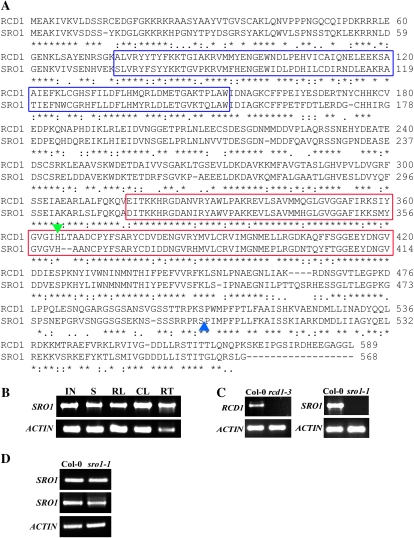
RCD1 and SRO1 encode similar proteins that have 76% similarity throughout their entire length (Fig. 1A; Belles-Boix et al., 2000; Ahlfors et al., 2004). Phylogenetic analysis indicates that these two genes are paralogs (Supplemental Fig. S1A), likely arising from a genome duplication (Plant Genome Duplication Database, http://chibba.agtec.uga.edu/duplication/index/home; Tang et al., 2008). This suggests that these two genes may be partially redundant. Functional redundancy would require that the two genes have similar expression patterns. RCD1 is expressed in all plant parts (Ahlfors et al., 2004). SRO1 is also expressed in all plant parts examined (Fig. 1B). SRO1 protein, like RCD1 (Fujibe et al., 2006), is localized to the plant nucleus (data not shown).
Figure 1.
RCD1 and SRO1 are similar proteins. A, Predicted amino acid sequences of the RCD1 and SRO1 proteins. SRO1 is 76% similar to RCD1. Asterisks, colons, and periods indicate identical, similar, and semiconserved amino acid residues, respectively. Hyphens correspond to gaps introduced to improve the alignment. The blue boxes mark the WWE domain, and the red boxes indicate the putative PARP catalytic domain. The insertion sites in rcd1-3 and sro1-1 are indicated by green (inverted) and blue (upright) triangles, respectively. B, SRO1 is expressed in all plant parts tested. RT-PCR was done using primers SRO1-F and SRO1-R to amplify SRO1 and Actin-F and Actin-R to amplify the actin control gene (Supplemental Table S1). IN, Inflorescence; S, 7-d-old seedlings; RL, rosette leaves; CL, cauline leaves; RT, roots. C, The T-DNA insertions in the mutant alleles disrupt gene expression. rcd1-3 and sro1-1 do not accumulate any detectable full-length transcript. RT-PCR was done using primers RCD1-F/RCD1-R and SRO1-F/SRO1-R, respectively. Col-0, Columbia. D, Transcription upstream and downstream of the T-DNA insertion site is seen in sro1-1. RT-PCR upstream (top panel; using primers SRO1-150F and SRO1-1360R) and downstream (middle panel; primers SRO1-1600F and SRO1-R) of the T-DNA insertion produces products.
In order to compare the expression patterns of RCD1 and SRO1 during development in more detail, we utilized the AtGenExpress atlas (www.weigelworld.org/resources/microarray/AtGenExpress/; Schmid et al., 2005). This atlas compares the expression profiles of 22,746 probe sets on the Affymetrix ATH1 microarray from almost 80 diverse developmental samples. RCD1 and SRO1 were expressed in all samples; this was expected from our reverse transcription (RT)-PCR data and previous data (Fig. 1B; Ahlfors et al., 2004). For the most part, the pattern of transcript accumulation was similar between the two paralogous genes; however, SRO1 is consistently expressed at a lower level than RCD1 (Supplemental Fig. S3A). This similarity of expression pattern is reinforced by examining the correlation coefficients for expression of all gene expression vectors compared with that of SRO1 using Expression Angler on the Botany Array Resource data set (http://bar.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi; Toufighi et al., 2005). The data from the Botany Array Resource consist of 93 samples, with plant age, experiment type, tissue type, and treatment information appended. Using this tool, RCD1 is among the top three genes with the most similar expression pattern to that of SRO1 based on the Pearson correlation coefficient (r value of at least 0.92; Supplemental Fig. S3C). Another of the top genes (At1g61420) has been implicated in Arabidopsis innate immunity (Qutob et al., 2006), consistent with a function in stress response similar to RCD1 and SRO1.
Since RCD1 has been implicated in abiotic stress response, RCD1 and SRO1 expression profiles in Arabidopsis samples challenged with abiotic stresses were compared, again using the AtGenExpress data (www.weigelworld.org/resources/microarray/AtGenExpress/; Kilian et al., 2007). As shown in Supplemental Figure S3B, the two expression patterns are similar, with the level of SRO1 transcript lower than that of RCD1. SRO1 expression level varies less in response to abiotic stress than RCD1, but neither gene shows large changes in expression in response to any of the abiotic stresses tested by microarray. However, RCD1 transcript has been shown to accumulate under high light stress (Bechtold et al., 2008); SRO1 transcript has not been reported to do so. This suggests that transcription or transcript stability of RCD1 may be regulated during the response to specific abiotic stresses. The similarity in expression pattern between RCD1 and SRO1 is consistent with the hypothesis that these genes have at least partially redundant functions.
Identification of Mutations in RCD1 and SRO1
We independently isolated a T-DNA insertion allele of RCD1, rcd1-3, which has previously been described (Katiyar-Agarwal et al., 2006). This allele can be complemented by a 35S∷RCD1 transgene (see Fig. 5C below), as reported previously for other rcd1 alleles (Ahlfors et al., 2004; Fujibe et al., 2006). We have identified a T-DNA insertion allele in SRO1, sro1-1. Both rcd1-3 and sro1-1 do not accumulate any detectable full-length transcript (Fig. 1C). However, sro1-1 appears to have partial transcripts arising both upstream and downstream of the T-DNA insertion site (Fig. 1D), while rcd1-3 also accumulates transcript 5′ to the insertion (data not shown), suggesting that these alleles may be partial loss of function. However, an allele of RCD1, rcd1-4, an RNA null, is indistinguishable from rcd1-3 (J. Kangasjarvi, personal communication). Unlike rcd1-3 mutants, sro1-1 mutants cannot be complemented by overexpressing SRO1 (data not shown). Complementation of sro1-1 mutants was achieved by transforming mutants with a 5-kb genomic fragment including SRO1. This fragment was introduced into the rcd1-3; sro1-1 double mutant background, where the restoration of SRO1 function restores plants to an rcd1-3 mutant phenotype (see Fig. 5F below). Complementation was done in this background because sro1-1 has very mild phenotypes as a single mutant (see below).
Figure 5.
rcd1-3 and sro1-1 display pleiotropic developmental defects. A, Double mutant seedlings are small and have malformed, light green leaves. Fifteen-day-old seedlings are shown: rcd1-3; sro1-1 (top), rcd1-3 (bottom left), sro1-1 (bottom center), and Columbia (Col-0; bottom right). B, RCD1 controls plant height. Plants shown are 45 d old. C, Complementation of rcd1-3 mutants by 35S∷RCD1. Four independent transgenic lines are shown. D, sro1-1 is dominant in an rcd1-3 mutant background, causing further reduction in plant height. In each pot, rcd1-3; +/+ plants are at left and rcd1-3; sro1-1/+ plants are at right. E, The rcd1-3; sro1-1 plants are dwarf (mature height averages about two inches; see ruler) and have a bushy habit. The plant shown is 55 d old. F, Complementation of sro1-1 mutants by a 5-kb genomic fragment covering the SRO1 gene. Complementation is shown in the rcd1-3 background, where restoration of SRO1 function confers an rcd1-3 mutant phenotype. Three independent lines are shown.
RCD1 and SRO1 Both Function in Abiotic Stress Response
It has previously been reported that rcd1 is involved in response to a number of abiotic stresses (Overmyer et al., 2000, 2005; Ahlfors et al., 2004; Fujibe et al., 2004, 2006; Katiyar-Agarwal et al., 2006). The rcd1-3 allele behaves similarly to the previously described alleles by displaying increased resistance to chloroplastic reactive oxygen species (ROS) induced by paraquat (Table I). In contrast, rcd1-3 mutant plants are more sensitive to apoplastic ROS (hydrogen peroxide [H2O2]; Table I). sro1-1 plants are also resistant to chloroplastic ROS (Table I), although to a lesser degree. Surprisingly, sro1-1 plants are resistant to apoplastic ROS (Table I), in contrast to the sensitivity of rcd1-3 plants. This suggests that these two genes are not always redundant in function but may have independent functions under certain stress conditions. Similarly, rcd1-3 and sro1-1 display opposite salt stress phenotypes (Table II); rcd1 plants are salt sensitive while sro1 plants are resistant.
Table I.
rcd1-3 and sro1-1 plants display different responses to oxidative stress
Seeds were plated on MS medium supplemented with the indicated amounts of H2O2 or paraquat. Seed germination was assessed as emergence of cotyledons after 5 d of light exposure. Values are mean percentages ± se of germinated seeds using data from three independent experiments. Values of mock treatment were set to 100.
| Conditions | Columbia | rcd1-3 | sro1-1 |
|---|---|---|---|
| Mock | 100 | 100 | 100 |
| 2.5 mm H2O2 | 83.6 ± 3.8 | 73.5 ± 3.6 | 94.1 ± 1.9 |
| 0.25 μm paraquat | 22.5 ± 1.3 | 77.9 ± 3.3a | 38.9 ± 2.2a |
Values significantly different from the wild type at P < 0.05.
Table II.
rcd1-3 and sro1-1 plants display different responses to salt stress
Seeds were plated on MS medium supplemented with 80 mm NaCl. Seed germination was assessed as emergence of cotyledons after 5 d of light exposure. Values are mean percentages ± se of germinated seeds using data from three independent experiments. Values of mock treatment were set to 100.
| Conditions | Columbia | rcd1-3 | sro1-1 |
|---|---|---|---|
| Mock | 100 | 100 | 100 |
| 80 mm NaCl | 85.8 ± 1.3 | 71.1 ± 7.0 | 92.5 ± 0.7a |
Values significantly different from the wild type at P < 0.05.
In contrast to the opposing roles in oxidative and salt stress, RCD1 and SRO1 appear to act similarly in response to osmotic stress. rcd1-1 plants have been reported to display increased resistance to Glc (Ahlfors et al., 2004). In our hands, rcd1-3 plants display more resistance to both Glc and mannitol (Table III), suggesting that this gene is involved in the response to osmotic stress, rather than being specifically involved in Glc signaling in the plant. Loss of SRO1 confers resistance to osmotic stress, as does loss of RCD1 (Table III). The individual roles of RCD1 and SRO1 during stress response, therefore, appears to be complex.
Table III.
rcd1-3 and sro1-1 mutants are resistant to osmotic stress
Seeds were plated on MS medium supplemented with the indicated amounts of Glc or mannitol. Seed germination was assessed as emergence of cotyledons after 4 d of light exposure. Values are mean percentages ± se of germinated seeds using data from three independent experiments. Values of mock treatment were set to 100.
| Conditions | Columbia | rcd1-3 | sro1-1 |
|---|---|---|---|
| Mock | 100 | 100 | 100 |
| 2% Glc | 92.9 ± 0.6 | 91.1 ± 0.4 | 93.8 ± 1.9 |
| 4% Glc | 78.7 ± 4.1 | 80.1 ± 1.8 | 81.7 ± 0.6 |
| 6% Glc | 36.5 ± 3.5 | 54.7 ± 5.6a | 64.5 ± 2.0a |
| 2% mannitol | 97.1 ± 1.2 | 95.4 ± 1.8 | 99.2 ± 0.3 |
| 4% mannitol | 87.7 ± 1.2 | 96.1 ± 1.6a | 96.1 ± 0.8a |
| 6% mannitol | 58.2 ± 2.2 | 73.9 ± 0.6a | 93.0 ± 0.5a |
Values significantly different from the wild type at P < 0.05.
Analysis of Single Mutant Plants Indicates That RCD1 Has a Larger Developmental Role Than SRO1
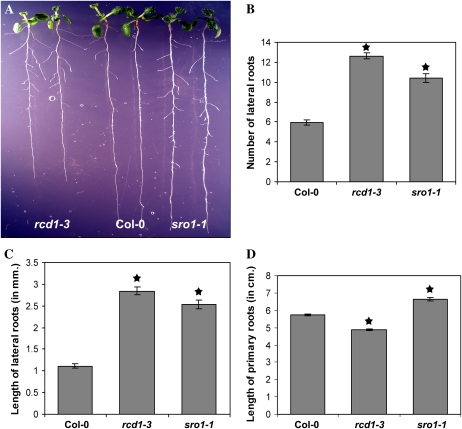
The developmental defects of rcd1-3 and sro1-1 single mutants are of very different magnitudes. rcd1-3 plants display similar phenotypic defects to those reported for previously isolated alleles, rcd1-1 and rcd1-2 (Ahlfors et al., 2004; Fujibe et al., 2004, 2006), and rcd1-3 itself (Katiyar-Agarwal et al., 2006). These phenotypes include abnormally shaped leaves and mild early bolting (Fig. 2). Additional developmental defects were noted in rcd1-3 mutant plants, including small petals and abnormal hypocotyl elongation in the dark (data not shown) as well as changes in root architecture (Fig. 3). rcd1-3 mutants have shorter primary roots, while lateral root number and length are increased. We observed the same trends in root growth in 6-, 16-, and 21-d-old seedlings (data not shown).
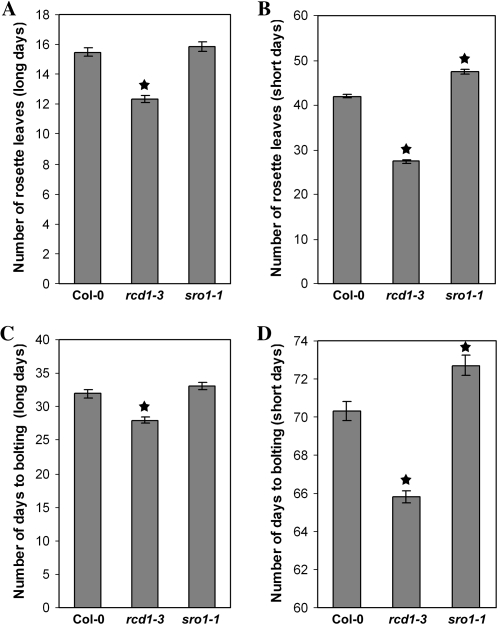
Figure 2.
RCD1 and SRO1 have opposite roles in control of bolting. rcd1-3 plants bolt early in both long days and short days, as measured by number of rosette leaves at bolting (A and B) and days to bolting (C and D). sro1-1 plants bolt late in both long and short days. Stars indicate values significantly different from the wild type at P < 0.01. Error bars indicate se. Col-0, Columbia.
Figure 3.
Root length and architecture are controlled by RCD1 and SRO1 function. A, The length of the primary root is shortened in rcd1-3 seedlings, while sro1-1 seedlings have longer primary roots. Eleven-day-old seedlings are shown. B, rcd1-3 and sro1-1 seedlings have extra lateral roots. C, In both mutant backgrounds, lateral roots are longer. D, The lengths of the primary roots in both mutants differ from that in the wild type. rcd1-3 primary roots are shorter, while sro1-1 primary roots are longer. Stars indicate values significantly different from the wild type at P < 0.01. Error bars indicate se. Col-0, Columbia. [See online article for color version of this figure.]
In contrast to rcd1 mutant plants, sro1-1 plants display only minor developmental defects, some of which are shared by rcd1 plants and some of which are not. Plant height, leaf shape, and floral architecture are normal. Root architecture seems to be controlled by both RCD1 and SRO1. In contrast to the rcd1-3 primary roots, those of sro1-1 are longer (Fig. 3); however, sro1-1 mutant seedlings have both an increased number and length of lateral roots, similar to rcd1-3. Like rcd1-3, sro1-1 seedlings are wild type in appearance when germinated in the light but have a longer hypocotyl when germinated in the dark (data not shown). SRO1 does not always act in the same direction as RCD1. For example, sro1-1 plants are mildly late flowering, although this is only significant in short days (Fig. 2), in contrast to rcd1-3, which is early bolting. Taken together, our analysis of the single mutant phenotypes suggests that RCD1 has a larger developmental role than does SRO1 but that SRO1 is important for specific developmental events, such as root development.
RCD1 Has a Role in Maintaining Reproductive Fate
Arabidopsis is a facultative long-day plant. As such, it takes much longer to bolt and flower under short-day conditions than under long-day conditions. rcd1-3 plants bolt slightly early compared with wild-type Columbia in both daylength conditions as measured by both number of rosette leaves and days to bolting (Fig. 2). However, there is a defect in the transition to flowering after bolting that is particularly strong under noninducing short-day conditions. Upon bolting, wild-type plants make three to four (long days) or five to six (short days) cauline leaves before producing solitary flowers. In both daylength conditions used in this study, rcd1-3 plants form aerial rosettes instead (Fig. 4, A–E; data not shown), while wild-type plants form none. Only one to two such rosettes are formed by rcd1-3 plants grown in long days before the formation of cauline leaves with associated branches and then solitary flowers. In short days, up to four to five aerial rosettes and the same number of aerial half-rosettes were formed before the formation of cauline leaves. Only some rcd1-3 plants ever form flowers under short-day conditions; however, these flowers never fully mature and do not open or form seed. In contrast, once sro1-1 flowers are formed in short days, they are normal and fertile (Fig. 4B).
Figure 4.
Loss of RCD1 function causes formation of aerial rosettes. A, Under short-day conditions, rcd1-3 plants form many aerial rosettes (arrowhead) before the formation of cauline leaves with axillary branches (arrow). B, sro1-1 plants do not form aerial rosettes and form fertile flowers (stars). C to E, Examples of aerial rosettes formed on rcd1-3 plants. F and G, Examples of extra leaves formed in the axils of cauline leaves of sro1-1 (F) and Columbia (Col-0; G) plants in short-day conditions. H, FLC expression is misregulated in rcd1-3 plants. RT-PCR was done using aerial rosettes (rcd1-3) or cauline leaves and associated structures (sro1-1 and Col-0). The number of amplification cycles is indicated.
The aerial rosettes formed on rcd1-3 plants are of two types: some completely encircle the circumference of the stem (Fig. 4C), suggesting that they arise from the shoot apical meristem and not from an axillary meristem, while others are formed in the axils of leaves, either cauline-like leaves (Fig. 4D) or aerial rosette leaves (Fig. 4E). At a low frequency, both wild-type and sro1-1 plants grown in short days have extra leaves forming in association with cauline leaves (Fig. 4, F and G). However, these structures never fully surround the stem and do not resemble aerial rosettes; instead, they appear to arise from suppression of internode elongation.
The flowering repressor FLOWERING LOCUS C (FLC) is strongly expressed during vegetative development but decreases upon reproductive induction and is not detected in inflorescences (Michaels and Amasino, 1999). Overexpression of FLC can cause formation of aerial rosettes (Wang et al., 2007). Consistent with the aerial rosettes formed by rcd1-3 plants in short days being caused by a reversion to vegetative growth, these structures express FLC at a high level compared with wild-type cauline leaves associated with extra leaves (Fig. 4H). FLC expression is also misregulated in sro1-1 cauline leaves associated with extra leaves, but not to as high a level as in rcd1-3 (Fig. 4H). This may contribute to the mild late flowering of sro1-1 plants.
RCD1 and SRO1 Control Plant Height and Vegetative Development
rcd1-3 plants, like other characterized alleles, are significantly shorter than wild-type plants; on the other hand, sro1-1 plants do not differ from wild-type plants in height (Fig. 5B; Table IV). As single sro1-1 mutants have only mild developmental defects, it is possible that the function of SRO1 is mostly complemented by the intact RCD1 locus but becomes necessary when RCD1 is absent or reduced. Therefore, reducing the dose of SRO1 may enhance the phenotype of rcd1. To test this, rcd1-3 was crossed to sro1-1 to construct rcd1-3; sro1-1 double mutants. In the F2 generation, plants with two novel phenotypes were observed. One of the novel phenotypes corresponded to the double mutant (see below), and one was determined to be rcd1-3/rcd1-3; sro1-1/+ by PCR genotyping. Specifically, the plants that were homozygous for rcd1 and heterozygous for sro1 were significantly shorter than rcd1-3 single mutants (Fig. 5D; Table IV). Other developmental defects seen in rcd1-3 single mutants were also enhanced, including flower size and lateral root number (data not shown). In contrast, rcd1-3/+; sro1-1/sro1-1 plants did not differ significantly from sro1-1 single mutants (data not shown).
Table IV.
RCD1 and SRO1 control plant height
Values are means ± se.
| Genotype | Height |
|---|---|
| cm | |
| Columbiaa | 47.13 ± 0.841 |
| sro1-1a | 45.4 ± 0.81 |
| rcd1-3a | 23.2 ± 0.58d |
| rcd1-3; sro1-1a | 3.35 ± 0.49d |
| 35S∷RCD1; rcd1-3 (5)b | 43.6 ± 1.04 |
| 35S∷RCD1; rcd1-3 (7)b | 43.4 ± 0.97 |
| 35S∷RCD1; rcd1-3 (8)b | 46.0 ± 0.93 |
| Columbiab | 42.7 ± 1.01 |
| rcd1-3b | 22.16 ± 0.57d |
| rcd1-3;+/+c | 21.67 ± 0.54 |
| rcd1-3; sro1-1/+c | 14.94 ± 0.68d |
Plants grown in respective identical conditions. The numbers 5, 7, and 8 in parentheses represent three independent transgenic lines.
Values significantly different from the wild type at P < 0.01.
The rcd1-3; sro1-1 double mutant plants were severely defective. On soil, only a very few double mutant plants were recovered, even though they should occur at a frequency of one in 16. Even on growth medium, the double mutant seeds fail to germinate effectively, with only 39.5% ± 2.5% of mutant seeds germinating; this germination defect was not significantly improved by application of gibberellic acid (data not shown). The seedlings that form are small, mostly pale green, and malformed. In particular, the leaves are very small and sessile (Fig. 5A). Without supplementation of the growth medium with Suc, the seedlings will not progress beyond the cotyledon stage. Even with sugar supplementation, only a few seedlings will progress to adulthood, and those plants have severe defects.
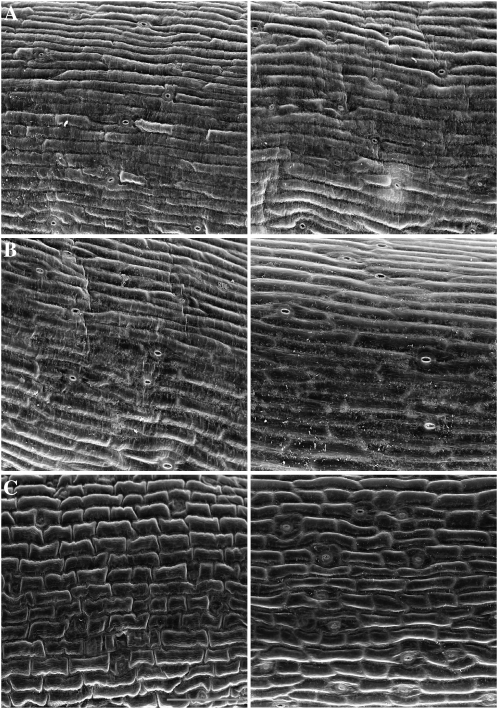
The adult double mutant plants are extremely dwarfed, typically only reaching approximately one to two inches in height, have a bushy growth habit, and have very small malformed leaves (Fig. 5E; Table IV). In addition, they have small flowers and produce few seeds. To determine if the extreme dwarfness of rcd1-3; sro1-1 plants was caused by defects in cell size and/or cell number, inflorescence stems were examined by scanning electron microscopy. While the cells of rcd1-3 and wild-type plants were not significantly different, the cells of the double mutant were smaller, with less elongation along the axis of the stem; cell size in the double mutant was also much more variable than that seen in the wild type (Fig. 6). When cell size was measured, rcd1-3; sro1-1 cells were only 50% as large as the wild type on average (10,536 ± 4,622 versus 20,148 ± 5,887 pixels). rcd1-3 cells are on average slightly larger than Columbia cells (22,147 ± 5,025). In addition to the cell elongation problem, the margins of the cells appear less smooth in rcd1-3; sro1-1 than in the wild type and rcd1-3 (Fig. 6), and the rcd1-3; sro1-1 stems contain fewer cells than the wild type (data not shown).
Figure 6.
Scanning electron microscopy images of epidermal cells of the inflorescence stems of wild-type (A), rcd1-3 (B), and rcd1-3; sro1-1 (C) plants. The double mutant stems have less cell elongation. The middle portion of two representative stems from each genotype is shown. Bar = 100 μm.
Double mutant plants are abnormal from germination. Close examination of rcd1-3; sro1-1 seedlings showed that the hypocotyls are much shortened or missing (Fig. 7A). In addition, little to no hypocotyl lengthening can be observed when the double mutant seedlings are exposed to exogenous gibberellin, suggesting that these structures are missing in the mutants (Fig. 7B). The phenotype of the rcd1-3; sro1-1 double mutant seedlings superficially resembles that of mutations in components of the brassinosteroid biosynthetic pathway, such as det2 mutants (Noguchi et al., 1999); however, the double mutant seedlings do expand their leaves, in a manner similar to the wild type, when exposed to exogenous brassinosteroid (Fig. 7D). Consistent with rcd1-3; sro1-1 leaves lacking petioles rather than having very short petioles, no visible lengthening of petioles could be seen in the rcd1-3; sro1-1 seedlings exposed to brassinosteroid, unlike in the wild type or the rcd1-3 or sro1-1 single mutant seedlings (Fig. 7D).
Figure 7.
rcd1-3; sro1-1 seedlings respond to exogenous gibberellic acid (GA3) and brassinosteroid (BR). A, Mock treatment. B, Treatment with 10 μm GA3. The hypocotyls of the GA3-deficient mutant (ga1-2; Sun and Kamiya, 1994) and the wild type elongate on application of GA3, while only some rcd1-3; sro1-1 seedlings respond due to the fact that these seedlings have short or absent hypocotyls. C, Mock treatment. D, Treatment with 0.1 μm BR. The leaves of rcd1-3; sro1-1 seedlings expand in response to BR, similar to the wild type and a BR-deficient mutant (det-2). However, petioles are absent on rcd1-3; sro1-1 leaves. Plants shown in A and B are 1 week old, and those shown in C and D are 3 weeks old.
RCD1 and SRO1 Are Necessary for Embryo and Seed Development
The presence of only a fraction of the expected double mutant plants among the F2 progeny of crosses between rcd1-3 and sro1-1 plants suggested that most rcd1-3; sro1-1 mutants aborted prematurely. This proved to be the case. Although rcd1-3; sro1-1 plants produce seeds, a large fraction of those seeds are not normal and none of the double mutant embryos or seedlings are normal. Seeds were classified into three classes: normal (Fig. 8A), class I (misshapen; Fig. 8B), and class II (misshapen and shrunken; Fig. 8C). More than 80% of seeds from a wild-type plant are normally shaped. Similarly, rcd1-3 and sro1-1 single mutants produce at least 75% normal seeds (Table V). sro1-1 plants produce a slightly lower number of normal seeds than the wild type and rcd1-3. rcd1-3; sro1-1 plants produce less than 10% normally shaped seeds. The seeds of the double mutants also are darker in color (Fig. 8, B and C).
Figure 8.
RCD1 and SRO1 are necessary for embryogenesis and seed development. A, Wild-type seeds. B, Class I rcd1-3; sro1-1 seeds are oddly shaped but of approximately wild-type size. C, Class II rcd1-3; sro1-1 seeds are both oddly shaped and shrunken. D, Mature wild-type embryos. E and F, Mature rcd1-3; sro1-1 embryos. These embryos are abnormally shaped and sized. The roots and hypocotyls are most affected. Pink arrowheads, Absence of hypocotyls; red arrowheads, short hypocotyls; green arrowheads, abnormal shape and structure of cotyledons; yellow arrowhead, arrest of embryo development at approximately the globular stage. G and H, Siliques of rcd1-3; sro1-1 plants. G, A young rcd1-3; sro1-1 silique in which seeds appear to be of normal size and shape. H, A mature rcd1-3; sro1-1 silique. By this stage in fruit development, the seeds have become small and shrunken.
Table V.
rcd1-3; sro1-1 seeds are abnormal
Classes are defined in the text. Values are mean percentages ± se from two independent seed stocks.
| Genotype | Wild Type | Class I | Class II |
|---|---|---|---|
| Columbia | 83.8 ± 10.3 | 10.5 ± 6.0 | 5.16 ± 4.2 |
| rcd1-3 | 82.6 ± 5.25 | 11.9 ± 5.37 | 5.4 ± 0.19 |
| sro1-1 | 75.2 ± 1.9 | 18.5 ± 1.04 | 6.15 ± 2.95 |
| rcd1-3; sro1-1 | 7.4 ± 2.6 | 59.6 ± 1.85 | 33.0 ± 0.75 |
To determine the origin of the misshapen seeds produced by the double mutant, both ovule development and embryogenesis in the double mutant were examined. rcd1-3; sro1-1 ovules appear smaller than wild-type ovules and are not normally shaped (Fig. 9B). The misshapen integuments likely contribute most to the shape changes seen in class I seeds. This conclusion is supported when seeds produced by rcd1-3; sro1-1 plants fertilized by wild-type pollen are examined. Nearly all seeds produced, although the embryos are heterozygous at both loci, are similar in appearance to class I seeds (data not shown). However, the ovule defects alone do not explain the seed defects.
Figure 9.
RCD1 and SRO1 control ovule morphogenesis and embryogenesis. A and B, Ovules 4 d after emasculation. A, The wild type. B, rcd1-3; sro1-1 mutant ovule is misshapen and small. C to E, Early globular stage embryos. C, The wild type. D and E, rcd1-3; sro1-1 globular embryos are normal. F to H, Heart-stage embryos. F, The wild type. G and H, rcd1-3; sro1-1 heart-stage embryos are broader than the wild type. I to N, Mature embryos. I, The wild type. J to N, rcd1-3; sro1-1 mature embryos display a range of phenotypes and do not fill seed. Bars = 50 μm. Each row has one bar that applies to all images in that row.
Embryo development in rcd1-3; sro1-1 mutants was examined next. Double mutant embryos from mature, but still green, seeds all have abnormal shapes and sizes, but there is considerable heterogeneity in the severity of the phenotype (Fig. 8, E and F). Some embryos appear not to have progressed much past the globular stage. Most embryos developed further but have abnormal shapes; the basal portions of the embryo (root and hypocotyl) are most affected. The root was often short and sometimes had no clearly differentiated root cap. The hypocotyls were most affected, being shortened or missing. This is consistent with the abnormal hypocotyls seen in germinated rcd1-3; sro1-1 seedlings (Fig. 7B). The shape and structure of the cotyledons was also not fully wild type. Overall, the rcd1-3; sro1-1 embryos were smaller than wild-type embryos and did not bend as normal. In contrast, the embryos of the rcd1-3 and sro1-1 single mutants are wild type in appearance (data not shown). Consistent with small embryos that do not fill the seed, developing seeds in young double mutant siliques appear to be relatively normal, although there is variation in size and shape (Fig. 8G), but by fruit maturity, the seeds have become very abnormal, most likely due to collapse of the seed around small embryos during desiccation (Fig. 8H).
To determine at what stage or stages developmental defects appear in rcd1-3; sro1-1, earlier embryonic stages were examined. Embryo development in the double mutant appears normal until the globular stage (Fig. 9D). From this stage on, variable defects are seen. By the heart stage, embryos have become broader than the wild type, with misoriented cell divisions in the region of the developing hypocotyl (Fig. 9, G and H). At maturity, the most normal rcd1-3; sro1-1 mutants have short hypocotyls and rounded cotyledons (Fig. 9J), and none of the mutant embryos fill the seed (Fig. 9, J–N). In addition, there was often asymmetric growth of the cotyledons (Fig. 9L), and some embryos do not appear to have differentiated into recognizable structures (Fig. 9M).
Differences in Function between RCD1 and SRO1 Are Encoded in Both the Promoter and Coding Sequences of the Two Genes
As mentioned above, differences in the ability of the 35S promoter to complement mutations in RCD1 and SRO1 suggest that there are differences in transcription level and/or pattern, transcript stability, and/or protein stability between these two paralogs. However, expression analysis indicates that both RCD1 and SRO1 have broadly similar expression patterns (Supplemental Fig. S3) and, as demonstrated above, the two genes do share some functions. To access possible differences between RCD1 and SRO1, we generated chimeric constructs between the genes and examined the ability of these constructs to complement both rcd1-3 and sro1-1 as well as examining the 35S constructs more closely (Fig. 10). 35S∷SRO1 failed to complement rcd1-3, similar to its inability to replace SRO1 function (data not shown). pRCD1∷SRO1g and pSRO1∷RCD1 both complemented the rcd1-3 single mutant; however, the level of complementation by pRCD1∷SRO1g was more variable between independent transgenic lines, as visualized by plant height, with fewer lines conferring wild-type height (Fig. 10B). Both of these chimeric constructs ameliorated the rcd1-3; sro1-1 phenotype, with pSRO1∷RCD1 again having a higher activity, as is the case in the rcd1-3 background (Fig. 10A). Although pSRO1∷SRO1g can fully complement sro1-1 (Fig. 5F), this construct cannot complement rcd1-3 either as a single mutant (data not shown) or in the double mutant background (Figs. 5F and 10B). Together, these results reinforce the idea that, although RCD1 and SRO1 are paralogs, some differences in regulation or function have differentiated the two loci.
Figure 10.
Both the regulatory regions and the coding regions of RCD1 and SRO1 vary in activity. A, Complementation of rcd1-3 mutants by pRCD1∷SRO1g and pSRO1∷RCD1. Three independent transgenic lines are shown for each transgenic construct to illustrate variation in complementation. B, Complementation of rcd1-3; sro1-1 mutants by pRCD1∷SRO1g and pSRO1∷RCD1. Five independent transgenic lines are shown for pRCD1∷SRO1g (center) and two independent lines for pSRO1∷RCD1 (extreme right). The ability of pRCD1∷SRO1g to rescue the double mutant is variable. Plants shown are 45 d old.
DISCUSSION
RCD1 and SRO1 Exhibit Partially Redundant But Not Identical Functions
In this study, we investigated the functions of RCD1 and its paralog SRO1 in Arabidopsis. We discovered that these two genes play redundant roles during several aspects of development including embryogenesis, revealing a previously unknown role of RCD1 during embryonic development. SRO1 seems to have a more minor role compared with that of RCD1; for example, the single sro1-1 mutation does not affect plant height, while rcd1-3 does, but sro1-1/+ can enhance the dwarf phenotype of rcd1-3. Also, we found that RCD1 is involved in stabilization of reproductive development.
RCD1 and SRO1 have very similar expression patterns; both genes are expressed in all plant organs at all times tested (Fig. 1B; Supplemental Fig. S3A) and show relatively little change in expression under various abiotic stresses (Supplemental Fig. S3B). Mutant analysis has demonstrated that both RCD1 and SRO1 are necessary for normal stress response. Since the transcripts of these genes do not change drastically in response to such stresses, it is more likely that protein level, activity, and/or localization are likely to be altered. For example, it has been shown that RCD1 can traffic into the cytoplasm under salt stress (Katiyar-Agarwal et al., 2006).
The ability of various transgenes to complement the rcd1-3 and sro1-1 mutants suggests that RCD1 and SRO1 genes are not equivalent. Expression data show that the RCD1 promoter drives a higher level of expression (Supplemental Fig. S3); this higher expression level presumably allows the pRCD1∷SRO1g transgene to complement rcd1-3 and rcd1-3; sro1-1 plants, while pSRO1∷SRO1g can only complement sro1-1. However, the SRO1 promoter can complement rcd1-3 when driving RCD1 but not SRO1. This suggests that the coding regions of the two genes are not equivalent. This could be due to transcript instability conferred by the SRO1 coding region, lower translation ability of the SRO1 message, or differences in the stability or activity between RCD1 and SRO1 proteins. Similar variability in the complementation ability of chimeric transgenes between RCD1 and SRO1 has been seen by others (J. Kangasjarvi, personal communication).
Control of Reproductive Growth by RCD1 and SRO1
Although RCD1 and SRO1 share many functions, the single mutants do not show identical phenotypes. In particular, the functions of the two genes in control of the transition to reproductive growth in Arabidopsis are different. sro1-1 plants flower slightly late compared with wild-type plants under both long-day and short-day conditions (Fig. 2); once the transition to reproductive growth has been made in these plants, it proceeds normally (Fig. 4). rcd1-3 plants, on the other hand, bolt early under both daylength conditions (Fig. 3). They do not retain reproductive identity correctly, however. Rather, rcd1-3 plants produce aerial rosettes, especially under short-day conditions (Fig. 4). rcd1-3; sro1-1 plants do not bolt under short-day conditions, even after 5 months (data not shown), suggesting that RCD1/SRO1 function is essential for the reproductive transition under noninducing conditions. Furthermore, this suggests that SRO1 function in rcd1-3 mutants allows bolting in short days.
The Arabidopsis ecotype Sy-0 has a distinct shoot morphology that includes formation of aerial rosettes (Poduska et al., 2003). rcd1-3's short-day phenotype is somewhat similar to the morphology of this accession. It has been determined that the HUA2/ART1 allele present in the Sy-0 background causes enhanced expression of the floral repressor FLC, contributing to the formation of aerial rosettes. 35S∷FLC plants phenocopy the Sy-0 shoot morphology (Wang et al., 2007). The aerial rosettes of rcd1-3 do express FLC at a higher level compared with wild-type cauline leaves (Fig. 4H), suggesting that misexpression of this gene may contribute to the formation of aerial rosettes seen in rcd1-3 plants. However, Sy-0 and 35S∷FLC plants both produce many vegetative leaves before bolting and formation of aerial rosettes, in contrast to the mild reduction of rosette leaf number seen in rcd1-3 plants (Fig. 3), suggesting that there may be additional factors involved. The Sy-0 accession has an active FRIGIDA allele, which Columbia does not (Poduska et al., 2003). Crossing the rcd1-3 allele into a background with an active FRIGIDA allele may lead to late bolting as well as aerial rosette formation.
The genetically separable bolting versus flowering seen in rcd1-3 mutants suggests a two-phase transition during flowering: first an inflorescence-producing phase, then a flower-producing phase. Loss of RCD1 function would interfere with the transition from inflorescence to flowering. Hempel and Feldman (1994) provided evidence for a single-phase transition in Arabidopsis in which existing leaf primordia could be transformed to flowers upon floral induction (acropetal development) whereas paraclades were formed from later-arising primordia. However, later work using Columbia grown under short-day conditions supported a two-step phase transition under noninducing conditions (i.e. paraclades arise first and flowers second) and further suggested that flowering time mutations have different effects on phase transition during flowering under different regimes (Suh et al., 2003). Our data support such a two-phase transition and suggest that RCD1 functions in the transition from inflorescence production to flower production, perhaps through control of FLC expression. FLC expression is controlled on many levels, including epigenetic modification of chromatin (Dennis and Peacock, 2007), transcriptional activation (Kim et al., 2006), and mRNA processing (Liu et al., 2007; Xing et al., 2008). Poly(ADP-ribosyl)ation has been implicated in all three of these processes in other systems (Hakme et al., 2008; Ji and Tulin, 2009; Quenet et al., 2009); therefore, it is difficult to assign RCD1 a specific function in the control of FLC expression without further experimentation.
RCD1 and SRO1 Are Essential for Normal Embryogenesis
RCD1 mutants were originally identified on the basis of their response to the abiotic stress ozone (Ahlfors et al., 2004). While rcd1 mutants have a number of developmental defects, most noticeably in stature, there was no lethality associated with loss of RCD1 function. Similarly, we have shown that sro1-1 mutants have only subtle developmental defects. However, rcd1-3; sro1-1 plants do not all proceed successfully through embryogenesis, demonstrating that these genes are essential for embryogenesis and act redundantly during this developmental stage.
The defects seen in rcd1-3; sro1-1 mutant embryos are completely expressive, as all have some abnormality, but they are heterogeneous in severity, ranging from an apparent arrest at the globular stage to formation of mature embryos that can germinate and survive (Figs. 8 and 9), although the plants formed are abnormal. This heterogeneity in phenotype could be due to the nature of the sro1-1 allele itself. This allele is not an RNA null mutation (Fig. 1D), and there likely remains some level of gene function present. The amount of function present may vary in individual embryos, leading to defects at various steps in embryogenesis. If a null allele in the SRO1 locus is identified, it may have a more severe phenotype, both as a single mutation and in combination with rcd1-3. It is not surprising that both members of a pair of paralogous genes need to be mutated before a function in embryogenesis is seen. At least 35 gene pairs that do not have clear embryonic lethality as single mutants do encode essential functions, as indicated by the lethality of double mutant combinations (http://www.seedgenes.org/7R_Double_Mutant_List.html; Tzafrir et al., 2003).
The heterogeneous nature of the embryonic defects in rcd1-3; sro1-1 plants makes determining the process or processes these two genes are involved with more difficult to determine. However, since embryogenesis proceeds normally through the globular stage (Fig. 9D), it is likely that delineation of the basic axes of the embryo is normal, as this is completed by the end of the globular stage (Jenik et al., 2007). In general, the basal region of the rcd1-3; sro1-1 embryos is more severely disrupted than the apical region, where the two cotyledons are usually relatively normal (Figs. 8 and 9). The hypocotyls of the mutant embryos, in particular, are shortened, as are the roots to a lesser extent. Most of the embryos that have progressed past the globular stage do have both root and shoot apical meristems, again suggesting that the apical-basal polarity of the embryo is intact. This suggests that the function(s) of RCD1/SRO1 may be most necessary for hypocotyl formation. This conclusion is further supported by the appearance of the rcd1-3; sro1-1 seedlings that do germinate. These seedlings have no hypocotyls or very shortened hypocotyls (Fig. 7). In general, these defects suggest that there may be a deficit in auxin signaling or transport, as this plant hormone is essential for hypocotyl formation (Jenik and Barton, 2005). This hypothesis is currently being tested.
Possible Mechanisms of RCD1 and SRO1 Action
RCD1 has been found to bind a number of Arabidopsis transcription factors by yeast two-hybrid assay (Belles-Boix et al., 2000; Ahlfors et al., 2004). We propose that RCD1 and SRO1 act by binding specific transcription factors and associating with chromatin through them. RCD1 is normally localized in the nucleus (Fujibe et al., 2006), supporting such a role. Once there, they poly(ADP-ribosyl)ate target protein(s) to regulate transcription. The PARP catalytic domains of RCD1 and SRO1 retain several important conserved residues that are present in the human, murine, chicken, maize (Zea mays), and Arabidopsis PARPs (Ruf et al., 1996, 1998a, 1998b; Ame et al., 2004; Kinoshita et al., 2004; Oliver et al., 2004; Bellocchi et al., 2005). Importantly, the residues necessary to form the donor site (Gly-347, Leu-348, Ser-375, and Tyr-378) are present in the two proteins, as is the residue Tyr-378, necessary for forming the acceptor site (Ruf et al., 1996; Oliver et al., 2004). The presence of these residues suggests that RCD1 and SRO1 function as PARPs, although this will need to be tested experimentally. Target proteins are likely to include RCD1 and SRO1 themselves, as most active PARPs automodify themselves (Mendoza-Alvarez and Alvarez-Gonzalez, 1993, 1999; Lindahl et al., 1995). Substrates could also include the transcription factors that RCD1 and SRO1 bind to, histones, transcriptional coactivators, and/or chromatin-remodeling factors. Examples of such activity by PARPs in other systems are abundant in the literature (Goenka et al., 2007; Krishnakumar et al., 2008; Sala et al., 2008). Given the pleiotropic nature of the rcd1-3; sro1-1 mutant phenotype, it is likely that the RCD1 and SRO1 proteins have multiple targets that vary both temporally and spatially.
The transcription factors that bind to RCD1 in yeast two-hybrid assays have demonstrated roles in stress response. For example, RCD1 can bind to STO, a transcription factor involved in salt stress response (Belles-Boix et al., 2000). This correlates well with the salt sensitivity of rcd1 mutants (Table II; Ahlfors et al., 2004; Katiyar-Agarwal et al., 2006). However, it was also found that RCD1 interacts with the plasma membrane Na+/H+ antiporter SOS1 under salt stress conditions (Katiyar-Agarwal et al., 2006); under this stress, some of the protein appears to accumulate in the cytoplasm, where it interacts with the cytoplasmic tail of SOS1. This suggests that RCD1 may also function outside of the nucleus.
Surprisingly, the region of RCD1 shown to bind to the transcription factors in yeast is the very C-terminal region, consisting of the end of the PARP catalytic domain and sequences beyond that (Belles-Boix et al., 2000) and not the WWE domain, although this domain is known to mediate protein-protein interactions. This suggests that the WWE domain brings other protein(s) into complex with RCD1/SRO1 and possible transcription factor-binding partners. These other proteins might be the actual targets of ADP-ribosylation by RCD1/SRO1. While association of RCD1 with transcription factors that mediate stress responses may explain the stress phenotypes seen in rcd1 mutants, to date none of the identified binding partners can explain the developmental defects, especially those of rcd1-3; sro1-1 embryos. This suggests that other binding partners need to be identified. Recent unpublished work has identified RCD1 binding to transcription factors implicated in development (J. Kangasjarvi, personal communication); of particular interest, RCD1 can bind to IAA11, an auxin-responsive transcription factor that is expressed in embryos (Genevestigator; Zimmermann et al., 2005). Given that the embryonic defects seen in rcd1-3; sro1-1 embryos suggest auxin defects, interaction with this transcription factor may help explain some of the developmental defects.
Consistent with a role of RCD1 in abiotic stress response, microarray expression analysis of the rcd1-1 mutant identified genes involved in such responses as having changed levels of expression in the mutant (Ahlfors et al., 2004). Up-regulated genes include At3g22370, which encodes ALTERNATIVE OXIDASE1a, shown to be necessary for normal response to light and drought stress (Giraud et al., 2008), and COR47, encoding a dehydrin also implicated in drought stress (Mouillon et al., 2008). The genes down-regulated in an rcd1 background included three other genes encoding dehydrins, ATDI8 (Puhakainen et al., 2004), ERD10 (Kovacs et al., 2008), and RAB18 (Lang and Palva, 1992). All of the proteins encoded by these genes are involved in the response to problems in either redox balance in the cell and/or problems in protein folding under stress conditions. Changes of these transcripts can help explain changes in abiotic stress response seen in the rcd1-3 mutants, but not necessarily the developmental defects. It is also difficult to determine if changes in the expression of stress response genes are a direct effect of the loss of RCD1 function or a consequence of altered redox metabolism in mutant plants, since loss of PARP function can lead to accumulation of NAD and oxidative stress (Formentini et al., 2009; Hashida et al., 2009). Therefore, more work will need to be done before the mechanism of action of RCD1 and SRO1 can be determined.
CONCLUSION
The putative PARPs RCD1 and SRO1 represent an important class of regulatory molecules with roles in embryogenesis, vegetative and reproductive development, and abiotic stress responses in Arabidopsis. Among the nine putative PARPs identified in the Arabidopsis genome, RCD1 and SRO1 are most similar to each other, and we show that they exhibit both redundant and divergent functions during development and stress response. Our study provides important insights into the complexity in the relationship between two highly similar paralogous genes. In addition, orthologs of RCD1 and SRO1 are present throughout the flowering plants, and similar proteins are found all the way to humans. Therefore, it is likely that information generated on the molecular mechanism of action of these genes in Arabidopsis will be applicable to other systems as well.
MATERIALS AND METHODS
Phylogenetic Analysis
Sequences of the Arabidopsis (Arabidopsis thaliana) PARP catalytic domain-containing proteins were retrieved from the National Center for Biotechnology Information (http://www.ncbi.nim.nih.gov/Tools/), and the PARP catalytic regions were identified using Pfam version 23.0 (http://pfam.sanger.ac.uk/; Coggill et al., 2008; Finn et al., 2008). Sequences from angiosperm species were identified and retrieved using National Center for Biotechnology Information BLAST searches. The PARP catalytic regions within these proteins were identified using Pfam. These regions were isolated using Perl scripts from the “Wildcat Toolbox” (http://proteomics.arizona.edu/toolbox.html; Haynes et al., 2006). ClustalX (http://www.clustal.org; Larkin et al., 2007) was used to generate alignments of the PARP catalytic regions. Phylogenetic trees were generated using ClustalX and the neighbor-joining method excluding gaps. Bootstrap values are included in the tree (Supplemental Fig. S1A). The tree was plotted using NJplot software (http://pbil.univ-lyon1.fr/software/njplot.html; Perriere and Gouy, 1996).
Plant Growth Conditions, Mutant Identification, and Genetic Crosses
The rcd1-3 allele has been described previously (Katiyar-Agarwal et al., 2006) and was obtained from the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org/abrc/). The sro1-1 allele was also obtained from the ABRC and is a T-DNA insertion from the SALK collection (SALK_126383; http://signal.salk.edu/; Alonso et al., 2003). rcd1-3; sro1-1 double mutant lines were created by crossing the respective homozygous mutants. F1 plants were confirmed by PCR genotyping (see below) and allowed to self-pollinate. Double mutant lines were identified from the F2 population by segregation of a novel phenotype and confirmed by PCR genotyping. All genotypes were confirmed in the F3 generation.
Arabidopsis seeds were vernalized for 3 to 5 d and were grown on Fafard-2 Mix soil (55% peat, perlite, and vermiculite) with subirrigation at 22°C with 50% relative humidity under long-day (16 h, 80 μmol m−2 s−1) irradiance in controlled growth chambers (Enconair Ecological Chambers) or growth rooms under similar conditions. Short-day conditions varied only in that the illumination was limited to 8 h of approximately 45 μmol m−2 s−1. Starting 2 weeks after planting, flats were regularly watered with fertilizer water (Peters Professional 20-10-20 Peat-Lite special fertilizer; Scotts) with a final concentration of 180 μL L−1. Plants studied for height and other phenotypes were grown side by side under identical conditions.
Seeds used in germination and root growth assays were sterilized with 70% ethanol followed by 10% (v/v) hypochlorite (bleach) and placed on Murashige and Skoog (MS) medium (RPI) agar plates with the indicated amounts of Suc (see below), incubated in the dark for 3 d at 4°C, and then grown under long-day conditions at 22°C in a CU-36L Plant Growth Chamber (Percival Scientific).
Genotyping and Cloning
For genotyping, genomic DNA was extracted from the seedlings or leaves by crushing plant material in liquid nitrogen and extracting with urea extraction buffer (7.0 m urea, 0.31 m NaCl, 0.05 m Tris-Cl, pH 8, 0.02 m EDTA, pH 8, and 1% [w/v] sarcosine) followed by phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v) extraction. The genomic DNA was used as template in PCR; the primers used to detect the presence of the T-DNA insertion in rcd1-3 were S383-LP and LBb1, and for the sro1-1 insertion, S432-RP and LBb1 (Supplemental Table S1). The wild-type locus was amplified with primers S383-LP/S383-RP and S432-LP/S432-RP, respectively. The sequence of primer LBb1 was obtained from the SALK Web site (http://signal.salk.edu/; Supplemental Table S1). PCR was done using Biolase Red DNA Polymerase (Bioline) on a conventional PCR machine (Bio-Rad icycler Thermal Cycler).
Cloning and Arabidopsis Transformation
RCD1 cDNA (stock no. U11347) was obtained from the ABRC. Amplification of the RCD1 coding sequence was done using the cDNA as template and the primers RCD1-F and RCD1-R (Supplemental Table S1). PCR was done using Platinum Pfx DNA Polymerase (Invitrogen). The PCR product was introduced into the pENTR-D Gateway entry vector (Invitrogen) to form pENTRD-RCD1 and sequenced at the Plant-Microbe Genomics Facility at Ohio State University. pENTRD-RCD1 was then recombined with pGWB2 (Gateway destination vector; a gift from T. Nakagawa, Shimane University, Matsue, Japan) to form p35S∷RCD1, which was then introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. rcd1-3 plants were transformed using the floral dip method (Clough and Bent, 1998). Transformants were selected on MS agar plates with hygromycin (30 μg mL−1). The heights of three independent transgenic lines were compared with those of the wild type and the untransformed rcd1-3 mutant to determine if the transgene complemented the mutant (Table IV).
Complementation of sro1-1 mutants was achieved by transforming sro1-1 mutants with a 5-kb genomic fragment that includes SRO1. This fragment was cloned by PCR using Columbia genomic DNA (prepared as above) as template and the primers SRO1-221-F and SRO1-221-R (Supplemental Table S1). PCR was performed using Pfu DNA polymerase (Stratagene). The PCR product was then recombined with the Gateway entry vector pDONR221 (Invitrogen) to form pDONR-gSRO1 and sequenced as above. pDONR-SRO1g was recombined with pGWB1 Gateway destination vector and introduced into Agrobacterium as above to form pSRO1∷SRO1g. Transformation and selection of transgenic plants was done as with RCD1. Complementation of sro1-1 was achieved in the rcd1-3 background.
Chimeric constructs of pRCD1∷SRO1g and pSRO1∷RCD1 were generated using the MultiSite Gateway Pro 2.0 Kit (Invitrogen). The promoters of RCD1 (pRCD1) and SRO1 (pSRO1) were amplified using Pfu Ultra DNA polymerase (Stratagene) and the primers pRCD1-F/pRCD1-R and pSRO1-F/pSRO1-R, respectively (Supplemental Table S1). The PCR products were then recombined with the pDONR P1-P5r Gateway vector (Invitrogen) to form entry clones pDONR-pRCD1 and pDONR-pSRO1, respectively. RCD1 coding sequence (RCD1) and SRO1 gene (SRO1g) were amplified using Platinum Pfx DNA Polymerase (Invitrogen) and the primers RCD1c-F/RCD1c-R and SRO1g-F/SRO1g-R, respectively (Supplemental Table S1). The PCR products were then recombined with the pDONR P5-P2 Gateway vector (Invitrogen) to form entry clones pDONR-RCD1 and pDONR-SRO1g. pDONR-pRCD1 with pDONR-SRO1g and pDONR-pSRO1 with pDONR-RCD1 were recombined separately with pGWB1 Gateway destination vector to form pRCD1∷SRO1g and pSRO1∷RCD1. Agrobacterium and plant transformation was done as described above. The chimeric constructs were used to complement both rcd1-3 and rcd1-3; sro1-1.
RNA Isolation and RT-PCR
RNA isolation was done from various plant tissues as described using Trizol according to the manufacturer's instructions (Invitrogen). cDNA was prepared from total RNA according to the manufacturer's instructions using the Transcriptor First Strand cDNA Synthesis Kit (Roche). PCR was done on cDNA using Biolase Red DNA Polymerase and primers as mentioned above and in Supplemental Table S1. PCR to determine the expression of SRO1 in different tissues was done for 45 cycles. FLC expression was assayed using cDNA made from material collected from plants grown under short-day growth conditions. rcd1-3 cDNA was made from young aerial rosettes, including the apical meristem, while wild-type and sro1-1 cDNA was made from young nodes where cauline leaves were associated with several leaves. RT-PCR was done as above. Primers used to amplify FLC were as described (Wang et al., 2007).
Phenotypic Analysis of the Mutants
Root phenotypes and flowering time were analyzed in the wild type (Columbia), rcd1-3, and sro1-1; all other phenotypes were observed in the wild type, rcd1-3, sro1-1, and rcd1-3; sro1-1. Wherever indicated in the text, significant difference between the phenotypes of the mutants and the wild type was calculated, at P < 0.01 or 0.05, by Student's t test.
Plant height was measured when the plants reached maturity and the flowers of the primary inflorescence had formed siliques; length of the primary inflorescence stem was measured from three independent biological replicates of each genotype. Each replicate consisted of 10 plants of each genotype. In total, the height of 30 plants of every genotype was measured, and the average height and se were calculated.
The roots of 11-d-old seedlings were analyzed from three independent biological replicates of each genotype. For each replicate, at least 40 seedlings of every genotype were examined. In total, 130 seedlings were analyzed. In order to determine the length of the lateral roots, the five longest lateral roots from each seedling were measured, for a total of at least 650 lateral roots per genotype.
Flowering time was analyzed using 30 plants of each genotype in three independent biological replicates (10 plants per replicate) grown either in long-day or short-day conditions as described above. Both days from germination to bolting and number of rosette leaves when the bolt length was approximately 5 cm were recorded.
In order to determine if seeds produced by the different genotypes were normal, mature dry seeds were examined with a dissecting microscope. Two independent seed stocks of each genotype with at least 100 seeds per replicate were analyzed. Based on physical appearance, seeds were placed into three classes: normal, class I, and class II (for more detail, see “Results”). In addition, mature embryos from green, fully formed seeds were excised and observed with the microscope. Differential interference contrast techniques on a Nikon Eclipse E600 microscope were used to observed mature ovules and embryonic development in wild-type and rcd1-3; sro1-1 plants. Ovules were dissected from siliques collected 4 d after emasculation of mature flower buds, while embryos were observed after dissection of fertilized siliques at various developmental stages. Dissected siliques were cleared with a chloral hydrate:glycerol:water solution (8:1:2, w/v/v) without prior fixation.
Stress Response and Hormone Assays
To assay germination, seeds of Columbia, rcd1-3, and sro1-1 were sown on MS plates containing 0.7% (w/v) agar and 3% (w/v) Suc supplemented with NaCl, H2O2, or paraquat as described (Katiyar-Agarwal et al., 2006). Seed germination was defined as the emergence of cotyledons and assayed after 5 d. Three biological replicates were analyzed for each treatment, and for each replicate approximately 150 seeds of each genotype were used. Evaluation of the results of these assays involved setting germination percentage of the mock treatments for each genotype to 100%, and the respective stress conditions are presented as the percentage of this value.
Osmotic stress assays were done by sowing seeds on MS medium with 0%, 2%, 4%, or 6% Glc or mannitol (w/v), vernalizing for 5 d at 4°C, and incubating as above for 4 d as described (Ahlfors et al., 2004). Germination percentage was analyzed as above.
Hormone response assays were done by plating seeds on MS + 1% Suc supplemented with 10 μm gibberellic acid or 0.1 μm brassinosteroid. The seeds were vernalized for 5 d, and seedlings were analyzed as indicated above.
Scanning Electron Microscopy
Five-week-old plants were fixed in 3% (v/v) glutaraldehyde + 2% (v/v) paraformaldehyde (Electron Microscopy Sciences) in 0.1 m phosphate buffer, pH 7.2, by vacuum infiltration and then overnight at 4°C. Fixed samples were washed three times for 15 min each with 0.1 m potassium phosphate buffer, pH 7.2. Samples were then postfixed in 1% (v/v) osmium tetroxide (Sigma-Aldrich) for 1 h and dehydrated through an ethanol series of 25%, 50%, 75%, 95%, and 100% (three times). Tissues were coated with platinum and examined with a scanning electron microscope (Hitachi S-3500N) at the Molecular and Cellular Imaging Center, Ohio Agricultural Research and Development Center, Ohio State University.
Photography
Photographs of adult plants were taken on an Olympus digital camera (C-5500). Photographs of seedlings, flowers, seeds, and embryos were taken on a Nikon Digital Sight DS-5M camera on a Nikon SMZ800 dissecting microscope. All photographs were taken with equal magnification for each plant part studied. All images of equal magnification were put into equal-sized canvases of the same resolution to make a composite figure with Adobe Photoshop version 7.0.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of the PARP family in angiosperms.
Supplemental Figure S2. RCD1 and SRO1 are similar to PARP11.
Supplemental Figure S3. Comparison of the expression patterns of RCD1 and SRO1.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Tea Meulia and the Molecular and Cellular Imaging Center, Ohio Agricultural Research and Development Center, Ohio State University, for scanning electron microscopy and differential interference contrast assistance, Matteo Citarelli for generating the phylogenetic tree, the Arabidopsis Biological Resource Center for rcd1-3 and sro1-1 seeds, Dr. J.C. Jang for help with hormone experiments, Dr. Jang and Dr. Iris Meier for critical reading of the manuscript, Dr. T. Nakagawa for the gift of the pGWB destination vectors, and Alyssa LaRue for assistance with media preparation, plant care, and general laboratory services.
This work was supported by the Ohio Plant Biotechnology Consortium (grant to R.S.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rebecca S. Lamb (lamb.129@osu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams-Phillips L, Wan J, Tan X, Dunning FM, Meyers BC, Michelmore RW, Bent AF (2008) Discovery of ADP-ribosylation and other plant defense pathway elements through expression profiling of four different Arabidopsis-Pseudomonas R-avr interactions. Mol Plant Microbe Interact 21 646–657 [DOI] [PubMed] [Google Scholar]
- Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA (2005) B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J Biol Chem 280 33756–33765 [DOI] [PubMed] [Google Scholar]
- Aguiar RC, Yakushijin Y, Kharbanda S, Salgia R, Fletcher JA, Shipp MA (2000) BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood 96 4328–4334 [PubMed] [Google Scholar]
- Ahlfors R, Lang S, Overmyer K, Jaspers P, Brosche M, Tauriainen A, Kollist H, Tuominen H, Belles-Boix E, Piippo M, et al (2004) Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 16 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO (2009) Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res 37 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G (2004) The PARP superfamily. Bioessays 26 882–893 [DOI] [PubMed] [Google Scholar]
- Amor Y, Babiychuk E, Inze D, Levine A (1998) The involvement of poly(ADP-ribose) polymerase in the oxidative stress responses in plants. FEBS Lett 440 1–7 [DOI] [PubMed] [Google Scholar]
- Aravind L (2001) The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci 26 273–275 [DOI] [PubMed] [Google Scholar]
- Ascencio-Ibanez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin A, Spenlehauer C, Dumond H, Menissier-De Murcia J, Piel M, Schmit AC, Apiou F, Vonesch JL, Kock M, Bornens M, et al (2003) PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci 116 1551–1562 [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Cottrill PB, Storozhenko S, Fuangthong M, Chen Y, O'Farrell MK, Van Montagu M, Inze D, Kushnir S (1998) Higher plants possess two structurally different poly(ADP-ribose) polymerases. Plant J 15 635–645 [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Van Montagu M, Kushnir S (2001) N-terminal domains of plant poly(ADP-ribose) polymerases define their association with mitotic chromosomes. Plant J 28 245–255 [DOI] [PubMed] [Google Scholar]
- Becerra C, Puigdomenech P, Vicient CM (2006) Computational and experimental analysis identifies Arabidopsis genes specifically expressed during early seed development. BMC Genomics 7 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Richard O, Zamboni A, Gapper C, Geisler M, Pogson B, Karpinski S, Mullineaux PM (2008) Impact of chloroplastic- and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. J Exp Bot 59 121–133 [DOI] [PubMed] [Google Scholar]
- Belles-Boix E, Babiychuk E, Van Montagu M, Inze D, Kushnir S (2000) CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett 482 19–24 [DOI] [PubMed] [Google Scholar]
- Bellocchi D, Macchiarulo A, Costantino G, Pellicciari R (2005) Docking studies on PARP-1 inhibitors: insights into the role of a binding pocket water molecule. Bioorg Med Chem 13 1151–1157 [DOI] [PubMed] [Google Scholar]
- Berglund T, Kalbin G, Strid A, Rydstrom J, Ohlsson AB (1996) UV-B- and oxidative stress-induced increase in nicotinamide and trigonelline and inhibition of defensive metabolism induction by poly(ADP-ribose)polymerase inhibitor in plant tissue. FEBS Lett 380 188–193 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiafa P, Guastafierro T, Zampieri M (2009) Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J 23 672–678 [DOI] [PubMed] [Google Scholar]
- Carrillo A, Monreal Y, Ramirez P, Marin L, Parrilla P, Oliver FJ, Yelamos J (2004) Transcription regulation of TNF-alpha-early response genes by poly(ADP-ribose) polymerase-1 in murine heart endothelial cells. Nucleic Acids Res 32 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Goenka S, Henttinen T, Gudapati P, Reinikainen A, Eischen CM, Lahesmaa R, Boothby M (2009) PARP-14, a member of the B aggressive lymphoma family, transduces survival signals in primary B cells. Blood 113 2416–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Hwang CK, Kim CS, Song KY, Law PY, Loh HH, Wei LN (2008) Transcriptional regulation of mouse mu opioid receptor gene in neuronal cells by poly(ADP-ribose) polymerase-1. J Cell Mol Med 12 2319–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HY, Chou HT, Lee SC (2006) CDK-dependent activation of poly(ADP-ribose) polymerase member 10 (PARP10). J Biol Chem 281 15201–15207 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coggill P, Finn RD, Bateman A (2008) Identifying protein domains with the Pfam database. Curr Protoc Bioinformatics Chapter 2: Unit 2.5 [DOI] [PubMed]
- Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R (2007) DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell 25 297–308 [DOI] [PubMed] [Google Scholar]
- Cook BD, Dynek JN, Chang W, Shostak G, Smith S (2002) Role for the related poly(ADP-ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol 22 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB (2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48 947–961 [DOI] [PubMed] [Google Scholar]
- De Block M, Verduyn C, De Brouwer D, Cornelissen M (2005) Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J 41 95–106 [DOI] [PubMed] [Google Scholar]
- Dennis ES, Peacock WJ (2007) Epigenetic regulation of flowering. Curr Opin Plant Biol 10 520–527 [DOI] [PubMed] [Google Scholar]
- Doucet-Chabeaud G, Godon C, Brutesco C, de Murcia G, Kazmaier M (2001) Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol Genet Genomics 265 954–963 [DOI] [PubMed] [Google Scholar]
- Egloff MP, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, Gruez A, Campanacci V, Cambillau C, Ziebuhr J, et al (2006) Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80 8493–8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al (2008) The Pfam protein families database. Nucleic Acids Res 36 D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formentini L, Macchiarulo A, Cipriani G, Camaioni E, Rapizzi E, Pellicciari R, Moroni F, Chiarugi A (2009) Poly(ADP-ribose) catabolism triggers AMP-dependent mitochondrial energy failure. J Biol Chem 284 17668–17676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati S, Formentini L, Wang ZQ, Moroni F, Chiarugi A (2006) Poly(ADP-ribosyl)ation regulates heat shock factor-1 activity and the heat shock response in murine fibroblasts. Biochem Cell Biol 84 703–712 [DOI] [PubMed] [Google Scholar]
- Fujibe T, Saji H, Arakawa K, Yabe N, Takeuchi Y, Yamamoto KT (2004) A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol 134 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibe T, Saji H, Watahiki MK, Yamamoto KT (2006) Overexpression of the RADICAL-INDUCED CELL DEATH1 (RCD1) gene of Arabidopsis causes weak rcd1 phenotype with compromised oxidative-stress responses. Biosci Biotechnol Biochem 70 1827–1831 [DOI] [PubMed] [Google Scholar]
- Gao G, Guo X, Goff SP (2002) Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297 1703–1706 [DOI] [PubMed] [Google Scholar]
- Giraud E, Ho LH, Clifton R, Carroll A, Estavillo G, Tan YF, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka S, Boothby M (2006) Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc Natl Acad Sci USA 103 4210–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka S, Cho SH, Boothby M (2007) Collaborator of Stat6 (CoaSt6)-associated poly(ADP-ribose) polymerase activity modulates Stat6-dependent gene transcription. J Biol Chem 282 18732–18739 [DOI] [PubMed] [Google Scholar]
- Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, Zupi G, Calabrese L, Caiafa P (2008) CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem 283 21873–21880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G (2004) The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78 12781–12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ma J, Sun J, Gao G (2007) The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA 104 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakme A, Wong HK, Dantzer F, Schreiber V (2008) The expanding field of poly(ADP-ribosyl)ation reactions. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep 9 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida SN, Takahashi H, Uchimiya H (2009) The role of NAD biosynthesis in plant development and stress responses. Ann Bot (Lond) 103 819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO (2003) Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J Biol Chem 278 45145–45153 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO (2008) The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 13 3046–3082 [DOI] [PubMed] [Google Scholar]
- Haynes PA, Miller S, Radabaugh T, Galligan M, Breci L, Rohrbough J, Hickman F, Merchant N (2006) The wildcat toolbox: a set of perl script utilities for use in peptide mass spectral database searching and proteomics experiments. J Biomol Tech 17 97–102 [PMC free article] [PubMed] [Google Scholar]
- Hempel F, Feldman LJ (1994) Bi-directional inflorescence development in Arabidopsis thaliana: acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192 276–286 [Google Scholar]
- Henry Y, Bedhomme M, Blanc G (2006) History, protohistory and prehistory of the Arabidopsis thaliana chromosome complement. Trends Plant Sci 11 267–273 [DOI] [PubMed] [Google Scholar]
- Hunt L, Gray JE (2009) The relationship between pyridine nucleotides and seed dormancy. New Phytol 181 62–70 [DOI] [PubMed] [Google Scholar]
- Ishiguro A, Ideta M, Mikoshiba K, Chen DJ, Aruga J (2007) Zic2-dependent transcriptional regulation is mediated by DNA-dependent protein kinase, poly-ADP ribose polymerase, and RNA helicase A. J Biol Chem 282 9983–9995 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Barton MK (2005) Surge and destroy: the role of auxin in plant embryogenesis. Development 132 3577–3585 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W (2007) Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23 207–236 [DOI] [PubMed] [Google Scholar]
- Ji Y, Tulin AV (2009) Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res 37 3501–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M (1999) A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologues. Genomics 57 442–445 [DOI] [PubMed] [Google Scholar]
- Ju BG, Rosenfeld MG (2006) A breaking strategy for topoisomerase IIbeta/PARP-1-dependent regulated transcription. Cell Cycle 5 2557–2560 [DOI] [PubMed] [Google Scholar]
- Kangasjarvi S, Lepisto A, Hannikainen K, Piippo M, Luomala EM, Aro EM, Rintamaki E (2008) Diverse roles for chloroplast stromal and thylakoid-bound ascorbate peroxidases in plant stress responses. Biochem J 412 275–285 [DOI] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG (2005) The macro domain is an ADP-ribose binding module. EMBO J 24 1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, Zhu JK (2006) The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 103 18816–18821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M (2003) Identification and characterization of human TIPARP gene within the CCNL amplicon at human chromosome 3q25.31. Int J Oncol 23 541–547 [PubMed] [Google Scholar]
- Kerns JA, Emerman M, Malik HS (2008) Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet 4 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 347–363 [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL (2005) Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 19 1951–1967 [DOI] [PubMed] [Google Scholar]
- Kim S, Choi K, Park C, Hwang HJ, Lee I (2006) SUPPRESSOR OF FRIGIDA4, encoding a C2H2-type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell 18 2985–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Nakanishi I, Warizaya M, Iwashita A, Kido Y, Hattori K, Fujii T (2004) Inhibitor-induced structural change of the active site of human poly(ADP-ribose) polymerase. FEBS Lett 556 43–46 [DOI] [PubMed] [Google Scholar]
- Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell 32 57–69 [DOI] [PubMed] [Google Scholar]
- Kovacs D, Kalmar E, Torok Z, Tompa P (2008) Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol 147 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL (2008) Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319 819–821 [DOI] [PubMed] [Google Scholar]
- Lang V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20 951–962 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Babiychuk E, Kushnir S, Van Montagu M, Inze D (1995) Characterization of an Arabidopsis thaliana cDNA homologue to animal poly(ADP-ribose) polymerase. FEBS Lett 364 103–108 [DOI] [PubMed] [Google Scholar]
- Lindahl T, Satoh MS, Poirier GG, Klungland A (1995) Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci 20 405–411 [DOI] [PubMed] [Google Scholar]
- Liu F, Quesada V, Crevillen P, Baurle I, Swiezewski S, Dean C (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28 398–407 [DOI] [PubMed] [Google Scholar]
- Ma Q, Baldwin KT, Renzelli AJ, McDaniel A, Dong L (2001) TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem Biophys Res Commun 289 499–506 [DOI] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R (1993) Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem 268 22575–22580 [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R (1999) Biochemical characterization of mono(ADP-ribosyl)ated poly(ADP-ribose) polymerase. Biochemistry 38 3948–3953 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillon JM, Eriksson SK, Harryson P (2008) Mimicking the plant cell interior under water stress by macromolecular crowding: disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol 148 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J (1999) Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol 120 833–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver AW, Ame JC, Roe SM, Good V, de Murcia G, Pearl LH (2004) Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res 32 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Brosche M, Pellinen R, Kuittinen T, Tuominen H, Ahlfors R, Keinanen M, Saarma M, Scheel D, Kangasjarvi J (2005) Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death1 mutant. Plant Physiol 137 1092–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H Jr, Kangasjarvi J (2000) Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12 1849–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriere G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78 364–369 [DOI] [PubMed] [Google Scholar]
- Phulwani NK, Kielian T (2008) Poly (ADP-ribose) polymerases (PARPs) 1-3 regulate astrocyte activation. J Neurochem 106 578–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduska B, Humphrey T, Redweik A, Grbic V (2003) The synergistic activation of FLOWERING LOCUS C by FRIGIDA and a new flowering gene AERIAL ROSETTE 1 underlies a novel morphology in Arabidopsis. Genetics 163 1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhakainen T, Hess MW, Makela P, Svensson J, Heino P, Palva ET (2004) Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol Biol 54 743–753 [DOI] [PubMed] [Google Scholar]
- Quenet D, El Ramy R, Schreiber V, Dantzer F (2009) The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol 41 60–65 [DOI] [PubMed] [Google Scholar]
- Qutob D, Kemmerling B, Brunner F, Kufner I, Engelhardt S, Gust AA, Luberacki B, Seitz HU, Stahl D, Rauhut T, et al (2006) Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18 3721–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, McDonald D, Gagne P, Ouellet ME, Droit A, Hunter JM, Dutertre S, Prigent C, Hendzel MJ, Poirier GG (2007) PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J Cell Biochem 100 385–401 [DOI] [PubMed] [Google Scholar]
- Ruf A, de Murcia G, Schulz GE (1998. a) Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry 37 3893–3900 [DOI] [PubMed] [Google Scholar]
- Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE (1996) Structure of the catalytic fragment of poly(ADP-ribose) polymerase from chicken. Proc Natl Acad Sci USA 93 7481–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf A, Rolli V, de Murcia G, Schulz GE (1998. b) The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J Mol Biol 278 57–65 [DOI] [PubMed] [Google Scholar]
- Sala A, La Rocca G, Burgio G, Kotova E, Di Gesu D, Collesano M, Ingrassia AM, Tulin AV, Corona DF (2008) The nucleosome-remodeling ATPase ISWI is regulated by poly-ADP-ribosylation. PLoS Biol 6 e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh MS, Lindahl T (1992) Role of poly(ADP-ribose) formation in DNA repair. Nature 356 356–358 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem 277 23028–23036 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7 517–528 [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T (2000) Tankyrase promotes telomere elongation in human cells. Curr Biol 10 1299–1302 [DOI] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282 1484–1487 [DOI] [PubMed] [Google Scholar]
- Storozhenko S, Inze D, Van Montagu M, Kushnir S (2001) Arabidopsis coactivator ALY-like proteins, DIP1 and DIP2, interact physically with the DNA-binding domain of the Zn-finger poly(ADP-ribose) polymerase. J Exp Bot 52 1375–1380 [PubMed] [Google Scholar]
- Suh SS, Choi KR, Lee I (2003) Revisiting phase transition during flowering in Arabidopsis. Plant Cell Physiol 44 836–843 [DOI] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and collinearity in plant genomes. Science 320 486–488 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43 153–163 [DOI] [PubMed] [Google Scholar]
- Trucco C, Oliver FJ, de Murcia G, Menissier-de Murcia J (1998) DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res 26 2644–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, De Block M, Van de Steene N, van de Cotte B, Metzlaff M, Van Breusegem F (2007) Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci USA 104 15150–15155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sajja U, Rosloski S, Humphrey T, Kim MC, Bomblies K, Weigel D, Grbic V (2007) HUA2 caused natural variation in shoot morphology of A. thaliana. Curr Biol 17 1513–1519 [DOI] [PubMed] [Google Scholar]
- Xing D, Zhao H, Xu R, Li QQ (2008) Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J 54 899–910 [DOI] [PubMed] [Google Scholar]
- Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, Vervoorts J, Walsemann G, Grotzinger J, Kremmer E, et al (2005) PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene 24 1982–1993 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hennig L, Gruissem W (2005) Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci 10 407–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.