Abstract
Inquiry-driven lab exercises require students to think carefully about a question, carry out an investigation of that question, and critically analyze the results of their investigation. Here, we describe the implementation and assessment of an inquiry-based laboratory exercise in which students obtain and analyze novel data that contribute to our understanding of macromolecular trafficking between the nucleus and cytoplasm in eukaryotic cells. Although many of the proteins involved in nucleocytoplasmic transport are known, the physical interactions between some of these polypeptides remain uncharacterized. In this cell and molecular biology lab exercise, students investigate novel protein–protein interactions between factors involved in nuclear RNA export. Using recombinant protein expression, protein extraction, affinity chromatography, SDS-polyacrylamide gel electrophoresis, and Western blotting, undergraduates in a sophomore-level lab course identified a previously unreported association between the soluble mRNA transport factor Mex67 and the C-terminal region of the yeast nuclear pore complex protein Nup1. This exercise immersed students in the process of investigative science, from proposing and performing experiments through analyzing data and reporting outcomes. On completion of this investigative lab sequence, students reported enhanced understanding of the scientific process, increased proficiency with cellular and molecular methods and content, greater understanding of data analysis and the importance of appropriate controls, an enhanced ability to communicate science effectively, and an increased enthusiasm for scientific research and for the lab component of the course. The modular nature of this exercise and its focus on asking novel questions about protein–protein interactions make it easily transferable to undergraduate lab courses performed in a wide variety of contexts.
INTRODUCTION
The process of “inquiry” has been defined most simply as “asking questions and finding answers” (French, 2005). For a biologist, scientific inquiry involves making an observation that leads to a hypothesis, then designing and carrying out an investigation that tests this hypothesis. Inquiry-based teaching and learning incorporates the process of inquiry into the classroom and facilitates learning by confronting students with questions for which they are expected to seek answers and by providing them with the opportunity to find answers for themselves (Prince and Felder, 2007). Well-designed inquiry-based methods of instruction improve student learning (Bransford et al., 2000; Schneider et al., 2002; Bryan, 2006; Lord et al., 2007), and the importance of inquiry-based experiences in science education has been emphasized repeatedly (Howard Hughes Medical Institute, 1996; National Science Foundation, 1996; National Research Council, 1996, 2000, 2003).
Although numerous methods exist for incorporating inquiry into science courses, laboratory-based experiences provide an excellent model for inquiry-based learning, because the questions that initiate many inquiry-based problems are often best addressed by experimental investigations (Prince and Felder, 2007). Investigative lab exercises that model the actual process of scientific inquiry result in improved student learning and retention, increased student understanding of and appreciation for the scientific research process, enhanced student investment in the course and increased interest in science in general, improved data analysis skills, and enhanced ability to integrate information learned in distinct contexts (National Science Teachers Association, 2001; Vallen, 2002; Gammie and Erdeniz, 2004; Glagovich and Swierczynski, 2004; Oliver-Hoyo et al., 2004; Howard and Miskowski, 2005; Bryan, 2006; Mitchell and Graziano, 2006; Goodman et al., 2007; Lord et al., 2007; Marshall, 2007). We also have used these exercises to teach scientific communication by requiring oral presentations, written reports that model primary journal articles, and mock grant proposals.
Although numerous exercises have been published describing inquiry-based learning exercises in the teaching lab, relatively few such labs are designed so that students collect truly novel data that add to the body of evidence in a particular field of research. Authors who do describe such exercises report enhanced enthusiasm and increased investment among student participants, better perceptions of how science is performed, and enhanced preparation for advanced work in research labs (Odom and Grossel, 2002; Gammie and Erdeniz, 2004; Howard and Miskowski, 2005; Marshall, 2007). Here, we describe an undergraduate lab exercise in which students generate and analyze novel data about protein–protein interactions that are important for cell function. This exercise was designed with a number of goals in mind: 1) To get early undergraduate students thinking about and involved in the investigative nature of science; 2) to have students actively apply the methods used by scientists to communicate with each other (reading and writing primary literature, grant proposal, lab notebook, journal club discussion); 3) to provide students with experience using modern techniques in cell and molecular biology; 4) to get students thinking carefully about the importance of proper controls in scientific experiments; 5) to have students analyze their own original data and carefully consider what to do next after those data are in hand; and 6) to facilitate student comprehension of concepts introduced in lecture. The exercise described here uses recombinant protein expression and affinity chromatography to investigate novel protein–protein interactions between proteins important for RNA transport in eukaryotes.
Yeast mRNA Export as an Experimental System
The yeast Saccharomyces cerevisiae provides an excellent model organism for undergraduate lab exercises and for gaining insight into eukaryotic cell function (Odom and Grossel, 2002; Marshall, 2007). Yeast cells are inexpensive to culture, grow rapidly, and are easily transformed with exogenous DNA. Many yeast strains are readily available that contain conditional and/or deletion mutations in genes encoding proteins that perform a diversity of cell functions. Basic cellular processes carried out in eukaryotes are performed in yeast, thus making them useful for investigating cell functions, including nucleocytoplasmic RNA transport.
In eukaryotic cells, the nucleus is separated from the cytoplasm by the two concentric membranes that form the nuclear envelope. For molecules to pass between the nuclear and cytoplasmic compartments, they must cross this double-membraned barrier. The sole conduits for transport of macromolecules across the nuclear envelope are large, multiprotein channels termed nuclear pore complexes (NPCs; for review, see Terry et al., 2007). Each NPC is composed of >30 different protein subunits, termed nucleoporins (Nups), most of which are present in eight to 32 copies per NPC (Rout et al., 2000; Cronshaw et al., 2002). Together, these Nups assemble to form NPCs, each of which is approximately 25 times larger than a ribosome. A typical eukaryotic nucleus may contain anywhere from 50 to several thousand NPCs. Each of the Nup subunits occupies a particular location and presumably carries out a specific function within the NPC. For example, Nup1 is a yeast nucleoporin localized to the nucleoplasmic face of the NPC that is important for both protein import and mRNA export across the nuclear envelope (Davis and Fink, 1990; Bogerd et al., 1994). Nup1 comprises three functional domains: an N terminus responsible for localization at the NPC, a central “repeats” domain that associates with protein import factors, and a C-terminal domain that is important for efficient protein import and RNA transport (Bogerd et al., 1994). In addition, methods similar to those described in this assay have revealed that a small region of the Nup1 C-terminus binds directly to the protein import factor Srp1/Kap-α (Belanger et al., 1994; Booth et al., 1999). In this laboratory exercise, students investigate the protein–protein interactions that may be important for Nup1 to participate in mRNA export from the nucleus.
mRNA is synthesized within the nucleus and needs to be exported to the cytoplasm to be translated at ribosomes. Mex67 is a yeast protein that associates with mRNA and is essential for mRNA transport from the nucleus to the cytoplasm through NPCs (Segref et al., 1997; for review, see Köhler and Hurt, 2007). The Mex67 protein binds directly to some Nups, suggesting that the physical interaction of Mex67 with the NPC is important for mRNA transport (Strässer et al., 2000; Strawn et al., 2001). Nup1 may provide one such binding site for Mex67 at the NPC (Fischer et al., 2002). However, the physical interaction between Mex67 and Nup1 has not been systematically investigated.
Using Affinity Chromatography to Test Protein–Protein Interactions
The functions of most proteins are dependent upon direct physical interactions with other polypeptides within a cell. Thus, to investigate the function of any given protein it is important to identify those macromolecules with which it interacts. Affinity chromatography provides one important method for identifying and characterizing these intermolecular interactions. Broadly defined, affinity chromatography is the use of one immobilized substrate to isolate a specific binding partner from a heterogeneous mixture of molecules based on the affinity of that partner for the substrate. When investigating protein–protein interactions, affinity chromatography typically involves linking one protein to an insoluble matrix and then incubating that matrix with a solution containing possible binding partners. This solution could be as simple as homogeneous solution containing a single recombinant protein or as complex as an entire cellular extract. After incubating the immobilized substrate protein with its potential binding partner(s) and washing away material associated nonspecifically, the binding partners are then eluted and detected by any of a variety of methods from chromatographic detection (the original source of the name “affinity chromatography”) to Western blotting to mass spectrometry.
In the exercises described here, students in undergraduate biology courses used affinity chromatography as a central technique in an in vitro investigation designed to identify Mex67-Nup1 interactions. The students in this course provide the first data suggesting a physical interaction between the C-terminal region of Nup1 and Mex67. In addition to learning important molecular biology and biochemistry methods, students also are exposed to reading, discussing, and writing in the primary literature and to the process of scientific discovery. Assessment of student outcomes reveals increased confidence in designing experiments and writing primary journal articles and an increased ability to understand primary literature, perform basic lab methods, and understand experimental controls. Students also report enthusiasm for the investigative nature of the project and perceive that it will benefit them in the future. Although the experiments described here use 5 wk of lab to investigate interactions between Nup1 and Mex67, the modular nature of the exercise and the methods used could be modified to examine interactions between any two proteins in an undergraduate laboratory course.
MATERIALS AND METHODS
Generation of Yeast Protein Extracts
Cells of yeast strains SWY518 (MATa ade2-1∷ADE2 ura3-1 his3-11,15 trp1-1 leu2-3112 can1-100) and SWY2154 (MATα MEX67-GFP:HIS5 ADE2 ura3-1 his3-11,15 trp1-1 leu2-3112 can1-100; Strawn et al., 2001) were grown to OD600 = 0.2–0.8 in 500 ml of SD −His media (Guthrie and Fink, 1991), with shaking at 30°C. Cells were harvested by centrifugation, washed with sorbitol wash buffer (300 mM sorbitol, 100 mM NaCl, 5 mM MgCl2, and 10 mM Tris, pH 7.5), and stored at −20°C until shortly before use. Students worked in pairs to generate extracts of soluble proteins from yeast. The cells were thawed on ice, and soluble proteins were extracted using glass bead lysis as described previously (Adams et al., 1997; see Supplemental Material for student protocol). All yeast and bacterial protein extracts were stored at −20°C until used in subsequent lab periods.
Expression and Extraction of Recombinant Proteins
Before the lab period, protease-deficient Escherichia coli (E. coli) strain BL21 (Miroux and Walker, 1996) was transformed with plasmid DNA expressing glutathione transferase (GST)-Nup1 domains and a GST control as appropriate for each experiment. (See Table 1 for plasmids used in various semesters). Before the lab, pellets containing cells expressing GST-Nups were prepared by growth in a 1 l culture of Luria broth + 50 μg/ml ampicillin. Expression of GST fusions was induced by adding isopropyl β-d-thiogalactoside to 1.0 mM, followed by incubation for 2.5 h at 37°C and centrifugation. The resulting samples were divided into four different tubes for purification by different lab groups. Each pair of students isolated two different GST-Nup fusions. E. coli were lysed, and GST-Nup1 protein extracts were isolated as described previously (Booth et al., 1999; see Supplemental Material).
Table 1.
Plasmids used in this study
| Plasmid name | Description | Source |
|---|---|---|
| pGEX-2TK | Vector for generating GST fusions | GE Healthcare |
| pSWB1/pLDB203 | GST-Nup1-N5-385 fusion in pGEX-2TK | Belanger et al. (1994) |
| pSWB5/pLDB209 | GST-Nup1-Rep432-816 in pGEX-2TK | Belanger et al. 1994 |
| pSWB6/pLDB205 | GST-Nup1-C778-1076 in pGEX-2TK | Belanger et al. (1994) |
| pLDB221 | GST-Nup1-C778-1041 in pGEX-2TK | This study |
| pKBB101/pLDB318 | GST-Nup1-C778-999 in pGEX-2TK | Booth et al. (1999) |
| pKBB106/pLDB321 | GST-Nup1-C1002-1076 in pGEX-2TK | This study |
Affinity Chromatography, Electrophoresis, and Western Blotting
Purification of GST-Nup1 and GST proteins from bacterial extracts by affinity chromatography was performed using glutathione-Sepharose beads (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom) as per Booth et al. (1999; also see Supplemental Material). Proteins bound to glutathione beads were solubilized in 50 μl of Laemmli buffer and separated using SDS-polyacrylamide gel electrophoresis (PAGE) on two identical 10% polyacrylamide gels. One gel was stained with Coomassie stain for total protein, and the other gel was transferred to nitrocellulose for Western blotting (see Supplemental Material). After protein transfer, the nitrocellulose filter was stained with Ponceau stain, blocked with 4% nonfat dry milk in Tris-buffered saline + 0.1% (vol/vol) Tween 20, and probed with anti-green fluorescent protein (GFP) antibody (Roche Diagnostics, Indianapolis, IN) at a 1:1000 dilution or with anti-Srp1 antibody (Booth et al., 1999) at 1:2500. Binding by primary antibodies was detected using horseradish peroxidase (HRP)-conjugated secondary antibodies and either colorimetric detection or Immun-Star HRP chemiluminescence as per manufacturer's instructions (Bio-Rad Laboratories, Hercules, CA). Primary and secondary antibody dilutions can be saved and stored at 4°C for up to a week for use in several lab periods.
Student Preparation
Before the beginning of the laboratory exercise, students completed a journal club discussion, generated a bibliography of articles examining nucleocytoplasmic protein transport and nuclear RNA export, discussed the experimental question they would be asking, and generated a grant proposal describing the experiments to be performed (see Results). Students received handouts providing background for each week's research question and experimental methods, as well as providing a protocol for completion of the week's lab activities. These handouts are provided in the Supplemental Material.
RESULTS
Student Background and Information
The investigative laboratory exercises described here have been used in cellular and molecular biology lab courses at two institutions over the course of six semesters. At Colgate University, these exercises were included in a sophomore-level Biol 212: Molecules, Cells, and Genes course that serves as an intensive introduction to basic biochemistry and molecular cell biology and is required of all biology, molecular biology, biochemistry, and environmental biology majors. In addition, students majoring in chemistry, physics, and neuroscience often complete this course as an elective, and all students who plan on attending medical school take the class as one of the required biology lab courses. Students taking Biol 212 have completed a year of general chemistry but are not required to have taken any biology; thus, they are most often inexperienced with molecular techniques, concepts, and instrumentation. A version of this exercise also was used at the University of Scranton for two semesters in a junior/senior level cell biology course. At Scranton, all students had taken an introductory biology sequence that included basic biochemistry, genetics, and cell biology, and many students also had taken upper-level courses in genetics, physiology, biochemistry, or a combination. At both institutions, students worked in pairs in lab sections of 14–18 students.
General Overview of the Exercise
Students investigated whether the yeast nucleoporin Nup1 physically interacts with soluble proteins involved in nucleocytoplasmic transport. Because one goal of this exercise is to have students perform novel research experiments, the lab exercise was slightly altered nearly every semester so that students were testing Nup1 for interaction with different soluble transport factors or were testing different domains of Nup1 for binding to RNA transporters. In the iterations reported here, students examined whether specific domains of Nup1 bind to the mRNA export factor Mex67. Although both Nup1 and Mex67 are important for efficient mRNA export (Bogerd et al., 1994, Segref et al., 1997), before the initial undertaking of this experiment a physical interaction between the two proteins had not been identified. Subsequent to our initial observations of Nup1-Mex67 binding in this lab and to the published report of a physical association between Nup1 and Mex67 (Fischer et al., 2002), students have investigated which domains of Nup1 are responsible for the interaction with Mex67.
To ask whether Nup1 associated with Mex67, students used several molecular and biochemical techniques over the 4 or 5 weeks of the exercise (Figure 1). During 1 wk, students expressed recombinant GST-Nup1 fusion proteins in E. coli, extracted soluble protein, precipitated the GST fusions, and examined the precipitated proteins by SDS-PAGE. In a second week, students generated soluble protein extracts from yeast expressing Mex67-GFP. During the final weeks of the exercise, students used their GST fusions to precipitate Nup1-binding proteins from the yeast extracts and then performed Western blotting to detect whether Mex67-GFP was among the precipitated proteins. In most semesters, this lab exercise was integrated with a journal club lab (see Prelaboratory Preparation), plus grant proposal and primary journal article writing assignments (see Supplemental Material).
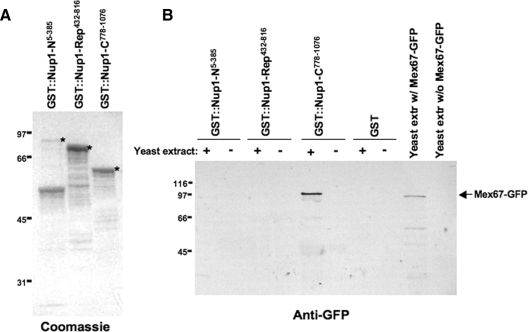
Figure 1.
Use of GST-Nup1 fusions to investigate protein–protein interactions with RNA transport factors. (A) Strategy for purification of proteins associated with Nup1 domains. Students expressed recombinant GST-Nup1 proteins in E. coli and purified the fusions using glutathione-Sepharose beads. After extracting soluble proteins (including a GFP-tagged form of Mex67, an RNA transport factor) from yeast, GST-Nup fusions associated with glutathione-Sepharose beads were used to isolate Nup1-binding proteins. Bound proteins were then run on polyacrylamide gels, and the presence or absence of Mex67-GFP was determined by probing a Western blot with anti-GFP antibodies. (B) GST-Nup1 fusions containing different domains of Nup1 were used to determine the region of Nup1 that associates with Mex67. Each group used two different GST-Nup1 constructs, each expressing a distinct region of Nup1, to determine the domain with which Mex67 associates in vitro. The cartoons labeled NUP1 and glutathione-S-transferase depict the full-length versions of each protein. The N-terminal region of Nup1 is shaded in gray, the “repeats” domain has vertical stripes, and the C terminus has dots. The GST-Nup1 fusions used in this study are depicted below, with the region of Nup1 included in each shown in cartoon form.
Prelaboratory Preparation
A primary goal of the courses in which this exercise has been used has been to integrate students into the process of doing science. To begin that integration, students participated in a “journal club” and generated a bibliography before undertaking the experiments described here. In the journal club, the students presented and discussed an article from Cell in which one transport factor for nuclear protein import was identified (Gorlich et al., 1994). This article provided important background for the exercise: it used some methods to be carried out by the students, incorporated some photomicroscopy that helped students visualize the questions being asked, included methods and results that are accessible to students with little experience with molecular cell biology, and it was well written and well cited. In this journal club, we discussed all sections of the article, from what contributions a researcher must make to be listed as an author and the significance of the order of the authors, to what should be included in a figure legend, to how to accurately cite the work of others. During the journal club period, the class was divided into groups of two to four students, and each group was responsible for describing the methods, outcomes, and implications of one figure from the article. Thus students gained experience analyzing the primary literature and exposure to some questions and methods important to cell and molecular biologists.
The students also spent part of a lab period generating a bibliography of citations relevant to NPC function, nucleoporins, RNA export, and Mex67 activity. In addition to receiving a brief lecture providing an overview of nucleocytoplasmic transport, students were introduced to several scientific journal databases and to the basics of the RefWorks citation management program. They then worked in pairs to contribute entries to a class bibliography that would be used for writing assignments later in the course.
Finally, every lab period opened with a 10- to 20-min discussion of the “big picture” being asked in our experiment and the relevance of that week's procedure to the overall experiment. Additional lectures and discussions were undertaken within each lab period as pauses in each protocol allowed.
Expression and Isolation of Recombinant GST-Nup1
For this experiment, we investigated whether Nup1 could associate with Mex67 in vitro and attempted to identify which specific region of Nup1 was responsible for this interaction (Figure 1). The exact experiment being performed changed each semester as we addressed new questions raised by the results obtained during the previous semester. This iterative process was emphasized to the students, reminding them that they were asking a novel research question and that the exact question they were asking was predicated on the findings of researchers (including other undergraduate students) who had preceded them. We also emphasized that the data they collected would be novel and would influence the future direction of research, in addition to being potentially publishable.
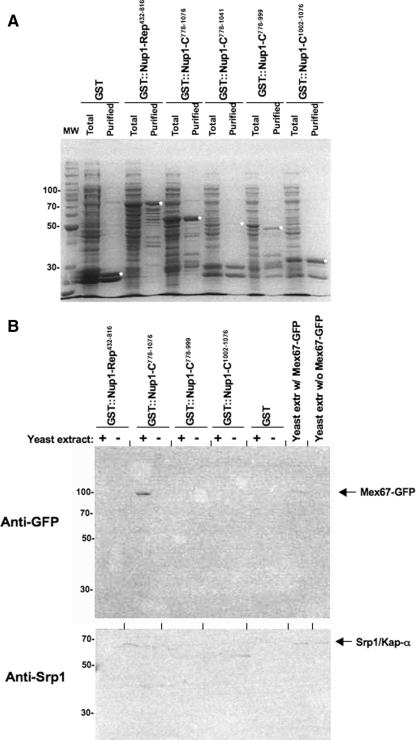
Each experiment began with students expressing GST-Nup1 fusion proteins to use for isolation of Nup1-binding proteins. To make an affinity matrix for isolating Nup1-binding proteins, we used plasmids containing GST-Nup1 gene fusions under control of an inducible promoter (Table 1 and Supplemental Material) and expressed the fusions in E. coli. GST alone was expressed as a control. Students extracted total protein from E. coli and affinity purified GST fusions from a portion of their extract by using glutathione-Sepharose beads. Proteins associated with the glutathione matrix were separated by SDS-PAGE, and gels were stained for total protein. Students observed a high level of protein purification with some apparent proteolytic degradation of the GST-Nup1 fusion proteins (Figures 2A and 3A). When the three major domains of Nup1 were purified, the majority of the repeats (GST-Nup1-Rep432-816) and C-terminal (GST-Nup1-C778-1076) fusions remained intact, whereas the N terminus was largely proteolyzed to a 50-kDa polypeptide (Figure 2A). When subunits of the Nup1 C-terminal domain were purified, the proteins remained largely intact except for GST-Nup1-C778-999, which retained less than half the protein undegraded, and GST-Nup1-C778-1041, for which intact protein was barely detectable (Figure 3A). These variations in protein content were consistent between lab groups. Although the amount of protein varied somewhat among groups, <10% of groups lacked detectable GST-Nup protein upon Coomassie staining. For those groups that did lack GST fusions, a visual check of their purified samples almost invariably revealed that they aspirated all of their glutathione-Sepharose beads from their tubes. The data obtained by every student group were discussed in each lab section, and all groups continued with the exercise the following week by using the two GST-Nup1 samples they had generated.
Figure 2.
Recombinant GST-Nup1 fusions are purified by students and used to identify the C terminus of Nup1 as a binding partner with Mex67. (A) GST-Nup1 fusion proteins were expressed in E. coli. Whole cell bacterial extracts containing GST-Nup1 proteins were incubated with glutathione-Sepharose beads and washed thoroughly (see Materials and Methods). Proteins remaining associated with the beads were separated by SDS-PAGE and stained with Coomassie Blue. Full-length GST-Nup1-N5-385, GST-Nup1-Rep432-816, and GST-Nup1-C778-1076 are indicated with an asterisk (*). (B) Affinity-purified GST-Nups bound to beads were incubated with yeast extracts containing Mex67-GFP (+) or with a mock solution lacking yeast proteins (−). After thorough washing, Western blotting using anti-GFP antibodies was used to detect the presence or absence of Mex67-GFP in each sample. Total yeast extracts containing or lacking Mex67-GFP were run as controls for detection of the GFP epitope. Bands representing Mex67-GFP are indicated with the labeled arrow. Locations of molecular weight standards in kilodaltons are shown to the left of each gel.
Figure 3.
Students did not identify a subdomain of GST-Nup1-C that associates with Mex67-GFP. (A) Students expressed GST-Nup1 fusion proteins in E. coli and purified the recombinant GST fusion proteins using glutathione-Sepharose beads. For each sample, a lane containing total bacterial protein extract (Total) and a lane containing purified GST-Nup1 protein (Purified) was separated by SDS-PAGE and stained with Coomassie Blue. Full-length purified proteins are indicated with a white asterisk (*). (B) Affinity-purified GST-Nups bound to beads were incubated with yeast extracts containing Mex67-GFP (+) or with a mock solution lacking yeast proteins (−). Students used Western blotting with anti-GFP (top) and anti-Srp1 (bottom) antibodies to detect the presence or absence of Mex67-GFP and Srp1/Kap-α in each sample. Total yeast extracts containing or lacking Mex67-GFP were run as controls for detection of the GFP epitope. Bands representing Mex67-GFP and Srp1/Kap-α are indicated with the labeled arrows. Molecular weight standards (in kilodaltons) are depicted to the left of each gel.
Detection of Mex67 Binding with Nup1
To investigate whether Nup1 domains associate with Mex67, students used recombinant Mex67 fused to GFP (Mex67-GFP) and expressed in yeast. The presence of the GFP epitope associated with Mex67 allowed for immunodetection of Mex67 protein during Western blotting later in the exercise. Students incubated the GST-Nup1 extracts isolated from E. coli the previous week with glutathione-Sepharose beads to generate a Nup1-containing affinity matrix. While the GST-Nup1 solution incubated, the students isolated total soluble protein extracts from yeast expressing Mex67-GFP. The students then mixed their fresh yeast extracts with GST-Nup1 bound to the glutathione-Sepharose beads to allow yeast proteins with affinity for Nup1 to bind to the recombinant Nup1 protein associated with the matrix. Each pair of students also performed one control “precipitation” for each GST-Nup1 fusion in which they added a mock solution instead of yeast extract to a tube containing Nup1 beads. After incubation, students thoroughly washed each sample with a buffered salt solution to remove proteins weakly associated with the Nup1 matrix. In subsequent weeks, each group separated the bound proteins by SDS-PAGE, transferred the proteins to nitrocellulose, and probed for the presence of Mex67-GFP on the filter by Western blotting using anti-GFP antibody (see Materials and Methods and Supplemental Material).
Experimental Outcomes: Nup1-C Associates with Mex67
In each semester, students tested specific regions of Nup1 for precipitation of Mex67-GFP (Figures 2 and 3) or other nuclear transport factors (data not shown) from their yeast protein extracts. In spring 2003, the students used GST fusions containing the N-terminal, FG-repeats, and C-terminal domains of Nup1 (Figure 1B). Although results between groups varied (see below), students most frequently identified Mex67-GFP associated with the GST∷Nup1-C region (Figure 2B). The band representing Mex67-GFP was found only in the GST-Nup-C sample that was incubated with yeast extract and not in the “mock” control to which yeast extracts were not added, helping confirm that the band detected was Mex67-GFP. In addition, a lane containing total yeast protein extracts from cells expressing Mex67-GFP and another lane containing yeast extract lacking Mex67-GFP were included on the Western blot. The appearance of a band at approximately 100 kDa in the lane containing extract with Mex67-GFP, but not in the lane lacking Mex67-GFP, confirmed the specificity of the antibody for Mex67-GFP. Although a slight majority of students observed this staining pattern, some students (approximately 30%) also observed bands of varying intensities in the GST-Nup1-N, GST-Nup1-Rep, and/or GST alone lanes (data not shown). In addition, approximately 10% of students observed a band only in the positive control lane (total yeast extract containing Mex67-GFP), and a smaller fraction did not observe bands in any lane at all (data not shown). To provide each lab group with an overview of their section's results, data from each lane of every student's blot were tabulated on the blackboard during the lab period so that students could see the pattern of Mex67-GFP precipitation specifically by Nup1-C. Pooled data from multiple lab sections confirmed the specificity of the Mex67–Nup1 interactions. These data and subsequent experiments by undergraduate research students (Belanger, Raclaw, and Walsh, unpublished data) provide the first strong evidence for the interaction of the C-terminal region of Nup1 with soluble Mex67 in vitro.
During spring 2006, students attempted to identify whether a smaller motif within the C terminus of Nup1 (Figure 1B) was responsible for the association with Mex67. Although several short regions from the C terminus of Nup1 were expressed and associated with glutathione-Sepharose beads (Figure 3A), students were only able to precipitate Mex67-GFP from yeast extracts using a GST-Nup1 fusion that contained the entire C-terminal domain from amino acids 778-1076 (Figure 3B), suggesting that a significant portion of this domain may be necessary for Mex67 binding. However, a control blot probing for the yeast karyopherin Srp1/Kap-α also revealed precipitation only by GST-Nup1-C778-1076, in disagreement with published work using the same GST-Nup1 constructs to identify a smaller Srp1/Kap-α binding region in Nup1-C (Booth et al., 1999). These observations provided an excellent focal point for discussion and helped reveal to students the importance of designing careful experimental controls and using those controls when interpreting data. Additional experiments will be required to determine whether specific subdomains within the C terminus of Nup1 are capable of binding Mex67 in vitro.
Student Assessment
Postlab assessments were completed by students after each semester in which an iteration of this lab was performed. During one semester (spring 2002), a prelab assessment also was performed that qualitatively inquired of students their self-perception of their ability to design an experiment asking an original research question in cell biology and to write a primary journal article based on data collected in a research lab. Fewer than half of these students (46.7%; n = 29) felt confident that they could design a cell biology experiment before this lab sequence, and 73.3% were confident they could write a good primary journal article (Table 2). After completing this lab exercise and its associated grant proposal and research article, these students were asked the same questions in a postlab written assessment. On completing the lab, 70.8% of the students were confident in being able to design an experiment and 87.5% felt they could write well in the format of the primary literature. These data suggest that a significant number of students perceive themselves to have improved in two of our targeted areas: scientific writing and the process of research.
Table 2.
Student perceptions of ability to design experiments and write primary journal articles as determined by pre- and postcourse surveysa
| % yes precourse | % yes postcourse | |
|---|---|---|
| I feel confident that I could design an experiment addressing a research question in cell biology. | 46.7 | 70.8 |
| I feel confident that I could write a good primary journal article based on results collected in a research lab. | 73.3 | 87.5 |
a Based on surveys from spring 2002 (n = 29).
After three of the semesters, quantitative postcourse assessments were performed. Fourteen questions investigating student perceptions of their gains in the scientific process, scientific communication, and content comprehension were asked (Table 3). Students reported strong gains in their understanding of the scientific process. Using a Likert scale (1–5, with 1, strongly disagree and 5, strongly agree), the mean student response to the first question inquiring about an increased understanding of lab methods used to address cell biological questions was 4.6, and additional questions on experimental design and the use of controls averaged 4.2 and 3.9, respectively. Similarly, student responses to statements about perceived gains in understanding the primary literature, writing a grant proposal, and explaining a research procedure to a lay audience all averaged over 4.0 on the Likert scale. Regarding specific scientific content in the lab, students reported that the exercise better helped them understand specifically how RNA molecules are transported in eukaryotic cells (mean = 4.3) but replied with lower Likert scores and a broader distribution of responses to questions 12 and 14, which addressed integration of concepts covered in lecture and lab.
Table 3.
Assessment of affinity chromatography lab
| Assessment statement | S05 (n = 31) | F05 (n = 44) | S06 (n = 49) | Total (n = 124) |
|---|---|---|---|---|
| Scientific process and lab methods | ||||
| 1. This lab helped me better understand lab techniques biologists use to address questions in cell and molecular biology. | 4.4 ± 0.3 | 4.6 ± 0.3 | 4.6 ± 0.5 | 4.6 ± 0.4 |
| 2. This lab helped me better understand the intellectual process biologists often follow when designing experiments to address novel biological questions. | 4.0 ± 0.7 | 4.1 ± 0.6 | 4.3 ± 0.6 | 4.2 ± 0.7 |
| 3. This exercise helped me think more carefully about the importance of appropriate controls in biology experiments. | 3.8 ± 0.6 | 4.1 ± 0.7 | 3.8 ± 1.0 | 3.9 ± 0.8 |
| Scientific communication | ||||
| 4. This lab helped me become better at reading and understanding primary journal articles. | 4.2 ± 0.6 | 4.1 ± 0.8 | 4.1 ± 0.9 | 4.1 ± 0.8 |
| 5. This lab helped me become better at identifying primary journal articles that are relevant to a specific topic. | 4.1 ± 0.6 | 4.0 ± 0.8 | 4.0 ± 0.9 | 4.0 ± 0.8 |
| 6. This lab enhanced my confidence in understanding primary literature. | 3.9 ± 0.5 | 3.9 ± 0.7 | 4.0 ± 0.7 | 4.0 ± 0.7 |
| 7. This exercise improved my ability to write a strong scientific article. | 4.0 ± 0.5 | 4.1 ± 0.7 | 4.2 ± 0.4 | 4.1 ± 0.5 |
| 8. The ″grant proposal″ assignment provided me with a strong understanding of the scientific question we were asking. | 4.4 ± 0.4 | 3.9 ± 1.0 | 3.9 ± 1.1 | 4.0 ± 0.9 |
| 9. The ″grant proposal″ assignment helped me better understand the goals and format of a typical research grant proposal. | 4.5 ± 0.4 | 4.1 ± 0.8 | 4.3 ± 0.5 | 4.3 ± 0.6 |
| 10. I could now explain to my roommate or my mother how to perform a Western blot. | 4.3 ± 0.4 | 4.2 ± 0.4 | 4.2 ± 1.0 | 4.2 ± 0.7 |
| 11. I could now explain to my roommate or my mother why someone would choose to perform a Western blot: | 4.1 ±0.7 | 4.2 ± 0.7 | 4.1 ± 0.9 | 4.1 ± 0.8 |
| Content | ||||
| 12. This lab exercise helped me better understand topics discussed in lecture relating to protein/protein interactions. | 4.0 ± 0.5 | 3.4 ± 1.0 | 3.8 ± 0.7 | 3.7 ± 0.8 |
| 13. This lab exercise helped me better understand how proteins and RNAs are transported inside cells. | 4.4 ± 0.7 | 4.3 ± 1.0 | 4.4 ± 0.5 | 4.3 ± 0.7 |
| 14. This lab exercise built on concepts covered in class. | 3.7 ± 0.5 | 3.2 ± 1.1 | 3.7 ± 0.7 | 3.6 ± 0.8 |
| Overall lab perceptions | ||||
| 15. This lab exercise was frustrating. | 2.6 ± 1.0 | 2.7 ± 1.4 | 2.7 ± 1.4 | 2.7 ± 1.3 |
| 16. This lab exercise was interesting. | 4.0 ± 0.3 | 4.1 ± 0.5 | 3.9 ± 0.7 | 4.0 ± 0.6 |
| 17. I became invested in this project. | 3.5 ± 0.6 | 3.6 ± 0.7 | 3.4 ± 1.1 | 3.5 ± 0.9 |
| 18. This exercise was too intellectually difficult. | 2.0 ± 0.6 | 2.0 ± 0.6 | 1.9 ± 0.7 | 2.0 ± 0.6 |
| 19. This exercise was too technically difficult. | 1.8 ± 0.8 | 1.8 ± 0.6 | 1.8 ± 0.7 | 1.8 ± 0.7 |
| 20. This exercise will help me in the future. | 4.0 ± 0.5 | 4.1 ± 0.6 | 3.9 ± 1.0 | 4.0 ± 0.8 |
Values are means ± SD.
Students also were asked to provide a general perception of the Nup1 lab in questions 15–20 (Table 3). Students in general did not perceive that the lab was too technically difficult (mean = 1.8) or too intellectually difficult (2.0), whereas the level of frustration with the lab seemed to vary (mean = 2.7 ± 1.3). A higher fraction of students reported becoming invested in the project (3.5), and students generally agreed with statements indicating that the exercise was interesting (4.0) and will help them in the future (4.0).
In every semester in which the various iterations of this lab were used, students were provided with a qualitative assessment tool that asked open-ended questions about aspects of the lab they perceived as being positive or negative and about what changes they would suggest making to the exercise. Student responses to these open-ended questions were overwhelmingly positive and reflected strong enthusiasm for the lab from a majority of the student participants. Particularly notable were the number of comments on how students appreciated being a part of the process of discovery and how they gained an understanding both of how scientific questions are asked and how accessible these questions can be to them. A sampling of the quotes from students that emphasized the process of science includes the following:
I really like the fact that we were actually researching something for which we didn't know what the “right” answer would be. This put more of a focus on understanding how the experiment was designed and what it was meant to do.
I enjoyed seeing actual biological laboratory techniques. So much of what we talk about in class I have to accept without understanding the experimentation. This allowed me to do a novel experiment and see what scientists really do and how they do it. I really like that we didn't know what the results were going to be, it made me feel like I was doing something worthwhile rather than cookbook lab assignments.
It engaged my thirst for more such projects and I can't wait to get involved in future grant proposals and projects.
The original experiment at the end was more interesting than cookie cutter textbook labs and made it much easier to take an interest in, making my understanding of the lab better and writing of the article easier.
I think it is useful for students to experience a lab exercise that might not work. Too often, labs are very monotonous because the result is known and works exactly as planned. I have found that I learn better from labs that yield unexpected results.
The fact that we were truly after new data and not repeating an experiment previously performed and reported on. That made it exciting and gave a reason to be vested in really doing it right.
I liked how each week built on the last. It allowed increased understanding as opposed to separate labs each week.
I think there should be an original experiment every semester like the one we did. It's a lot more fun when you don't know what is supposed to happen.
Doing an original experiment was GREAT!! A good lesson in what science is really like!
Students also indicated that the lab enhanced their ability to read and write scientific literature:
I enjoyed finally being able to understand primary journals. It was like learning a secret code.
It really taught me a lot about the process of answering research questions, not only by performing the experiment but by reading the primary literature as well.
I really enjoyed turning in a ‘draft’ form of the results, figures, and materials/methods. The comments were helpful and it took some stress out of writing the primary journal article.
I probably learned the most through actually writing the discussion of the research paper, as it forces you to look at all the implications of your results instead of simply saying: Mex67 associates with the C-terminal region of Nup1.
I really enjoyed the process of writing the final report. It brought a lot of satisfaction to think through each and every step of the experiment and identify the possible sources of error and the ways of changing the experimental design to improve the significance of the final results.
Not all of the student comments were positive. The majority of the constructive criticisms noted in response to an assessment question asking for ways to improve the lab inquired about whether the length of some of the incubation periods during the affinity chromatography and Western blotting labs could be shortened. A few students also indicated feeling confused during the lab and expressed frustration at the lack of concrete results for their group at the end of the exercise. The majority of the remaining comments reflected the challenging nature of the grant proposal and research article writing assignments that accompanied the lab. The students offered the following constructive criticisms in response to the request, “Please indicate what you liked least about the Nup1 lab exercise and accompanying assignments:”
Some of the labs were really long.
Is there any way to cut out some of the sitting around?
The time it took!!!
Long incubation periods.
I had trouble understanding the topic when I wrote the grant proposal because I hadn't done the experiment.
It was extremely difficult to write the grant proposal without doing any of the prior research.
Researching past journal articles was very time-consuming and often confusing to put everything together.
I found writing the paper extremely difficult and time consuming.
I only wish I knew what I was doing earlier in the assignment.
It was incredibly intense.
I didn't like having to keep everything straight for 5 or so weeks.
The samples were so small and we had to be very precise.
It didn't really work out for everyone.
I didn't like that our data didn't work. It was frustrating to put so much effort into a project and have to use others' results instead of our own.
I wish we'd spent more time on the NPC in class.
I didn't really enjoy reading the primary journal articles because there were a lot of concepts I did not understand, but after I figured them out it really did help me understand the lab.
These criticisms not only provide constructive feedback to take into account as we design future labs but also provide additional data that we are achieving some of our goals, including getting students into the primary literature, making them aware of the process of science, and enhancing their science writing skills.
DISCUSSION
Here, we report the outcomes of a multiweek lab experience for students in a sophomore- to junior-level molecular biology course. Students investigated whether physical interactions occur in vitro between the yeast nucleoporin Nup1 and the mRNA transport factor Mex67 and subsequently sought to determine which region of Nup1 is important for the association. The experiments reported in detail here describe three of the six semesters this exercise has been used, with different protein–protein interactions tested during most every iteration of the course. Students obtained evidence that the C terminus of Nup1 is sufficient to mediate a physical interaction with Mex67. Student assessments provide strong evidence of student gains in experimental design, writing, data analysis, and enthusiasm for science.
The modular nature of this multiweek lab exercise makes it potentially useful in a breadth of cell biology, molecular biology, or biochemistry courses. First, the methods described can be used to test for physical interactions between any two or more proteins, allowing instructors to select experiments that examine novel questions in their own area of expertise to use plasmids and strains easily available to them. Second, the set of techniques used can be altered from course to course. The exercise described here has been taught as a 4- to 6-wk lab sequence emphasizing affinity chromatography and Western blotting techniques. However, one could redesign the lab to emphasize only recombinant protein expression and purification. Or one could develop several additional weeks of lab before this exercise in which GST-Nup1 (or a recombinant protein of choice) encoding plasmids are isolated, confirmed by restriction enzyme digestion and electrophoresis, and transformed into E. coli. One also could follow this lab with fluorescent microscopy experiments using GFP-tagged proteins and yeast mutants to investigate changes in protein localization in cells lacking functional transport factors or other proteins.
The goals for this exercise included helping students experience the process of scientific exploration, enhancing scientific communication skills, exposing students to specific research techniques, making clear the importance of appropriate controls in experimental design, gaining confidence and competence in data analysis, and aiding in the understanding and retention of concepts in cellular and molecular biology. Formal and informal assessments suggest that we have accomplished each of these goals to an extent. However, some alterations to the exercise may enhance the value of this experience for some students.
To provide students with a realistic representation of the process of science, this exercise was designed to test a novel question in cell and molecular biology for which the result had not been determined, but for which previous experiments led to a reasonable hypothesis. In most semesters, we had students develop a bibliography and write a grant proposal to stimulate them to explore the primary literature and investigate published information about Nup1 and Mex67. After this exploration, they were required to synthesize what they had read to generate a hypothesis and to design an experiment to test that hypothesis. Some students verbally expressed frustration while carrying out this exercise, indicating that it was too difficult to write a proposal on a question about which they knew very little in advance and to depend entirely on the primary literature for information. However, surprisingly few students commented on the difficulty of the grant proposal on their postcourse assessments, even when specifically prompted for anonymous constructive criticism. This may be due to the length of time (3 or 4 wk) between the proposal due date and the assessment, but informal comments from several students indicated that this was because they saw the value of the proposal once they began carrying out the experiments and they appreciated the insight the proposal provided for them when it came time to write their final research reports. Through the combination of the literature review, grant proposal, experimentation, and research article, students gained insight into the process of science and the importance of effectively reading and writing scientific literature.
Students also obtained their own novel research results. Many students reported feeling pride in collecting their own data and an increased level of investment when asking a novel research question that could potentially contribute to a scientific publication. Students clearly understood that they were not undertaking a “cookbook” lab exercise and responded with enhanced enthusiasm and investment. However, some students expressed disappointment at the outcome of their 4 or 5 weeks of lab and were frustrated by their apparent lack of results. Students have often become accustomed to labs that have a “right” answer at the end and thus are confused by differences in results between different lab groups. We sought to use this variation in outcomes as a learning tool to provide insight to the students about the process of science. We spent a significant portion of the last lab period of this exercise discussing the data obtained by every lab group and thinking carefully about the conclusions the data allowed us to draw about the experiment. Were all groups in agreement about the outcome? (They rarely were). Were there any patterns in the data? (Usually, but not always). When do we consider a piece of data to be an outlier and what do we do with those data? How many replicates are enough to be convinced of an outcome? How do our controls provide insight into the possible reasons we see bands in each of our experimental lanes? Are there any other controls or any other experiments we should perform that would provide further evidence in support of conclusions we think can be made from these data? This time spent analyzing and discussing the data may have been the aspect of the lab exercise that did the most toward achieving our goals, because it forced students to think about the methods and controls they used; required them to analyze their data carefully and critically; asked them to think about the biological questions they were attempting to address; required them to communicate orally about their results and the implications of those results; and, most importantly, helped them gain important insight into the process of science.
The following three comments are representative of many of the written responses received on assessment forms over the six semesters this lab was performed:
I truly felt like a scientist after I handed in my final paper, and I felt as though I contributed to science.
It was definitely the most interesting lab experience ever!
This lab was great. I learned a lot and even had fun. It is the first lab ever that I have not dreaded going to!
These provide some additional insight into one outcome I had not necessarily predicted when designing the lab. Many students seemed to come out of the lab not only with an increased understanding of the process of science but also with an increased enthusiasm for this process and a greater appreciation of what they can accomplish even as undergraduates in a laboratory-based course.
In summary, we have described an investigative lab exercise in which students collect novel, potentially publishable data about protein–protein interactions involved in nucleocytoplasmic RNA transport. Students in introductory cell and molecular biology courses at Colgate University and the University of Scranton have identified a region of the C terminus of the nucleoporin Nup1 as being sufficient for association with the mRNA transport factor Mex67 in vitro. The procedures outlined here can be used by students to examine physical interactions between virtually any two soluble proteins. It is important that this lab allowed students to gain experience collecting and analyzing data, learn new laboratory techniques, gain deeper insight into course content, communicate scientific information orally and in writing, and experience the process of science through the collection and reporting of original research results.
Supplementary Material
ACKNOWLEDGMENTS
I acknowledge the Colgate University Biology 212 students and University of Scranton Biology 350 students for efforts in carrying out this lab exercise. Thanks also to Biology faculty from Colgate University and the University of Scranton (J. Voltzow, T. Sweeney, M. Kainz, N. Pruitt, B. Hoopes, K. G. Belanger, and S. Geier) for work implementing this exercise and for helpful constructive criticisms; to S. Wente (Vanderbilt University) for sharing yeast strains and reagents; and to N. Pruitt and K. G. Belanger for critically reading this manuscript. Special thanks to Kathleen Baier for many hours of technical assistance and insight in the Biol 212 lab. This work was supported by funding from Colgate University and National Institutes of Health grant R05 GM-65107 (to K.D.B.).
REFERENCES
- Adams A., Gottschling D. E., Kaiser C. E., Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Belanger K. D., Kenna M. A., Wei S., Davis L. I. Genetic and physical interactions between Srp1p and nuclear pore complex proteins Nup1p and Nup2p. J. Cell Biol. 1994;126:619–630. doi: 10.1083/jcb.126.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd A. M, Hoffman J. A., Amberg D. C., Fink G. R., Davis L. I. nup1 mutants exhibit pleiotropic defects in nuclear pore complex function. J. Cell Biol. 1994;127:319–332. doi: 10.1083/jcb.127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth J. W., Belanger K. D., Sannella M. I., Davis L. I. The yeast nucleoporin Nup2p is involved in nuclear export of importin alpha/Srp1p. J. Biol. Chem. 1999;274:32360–32367. doi: 10.1074/jbc.274.45.32360. [DOI] [PubMed] [Google Scholar]
- Bransford J. D., Brown A. L., Cocking R. R. How People Learn: Brain, Mind, Experience, and School: Expanded Edition. Washington, DC: National Academies Press; 2000. [Google Scholar]
- Bryan J. A. Assessment results following inquiry and traditional physics laboratory activities. J. Coll. Sci. Teach. 2006;35:56–61. [Google Scholar]
- Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L. I., Fink G. R. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell. 1990;61:965–978. doi: 10.1016/0092-8674(90)90062-j. [DOI] [PubMed] [Google Scholar]
- Fischer T., Strässer K., Rácz A., Rodriguez-Navarro S., Oppizzi M., Ihrig P., Lechner J., Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French D. P. Was “inquiry” a mistake? It's all in the name. J. Coll. Sci. Teach. 2005;35:60. [Google Scholar]
- Gammie A. E., Erdeniz N. Characterization of pathogenic human MSH2 missense mutations using yeast as a model system: a laboratory course in molecular biology. Cell Biol. Educ. 2004;3:31–48. doi: 10.1187/cbe.03-08-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. Transformation of yeast by the LiAc/SS carrier DNA/PEG method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Glagovich N., Swierczynski A. Teaching failure in the laboratory. J. Coll. Sci. Teach. 2004;33:45–47. [Google Scholar]
- Goodman B. E., Eisenhart M., DeHaan R. L., Kemm R. E., Rodenbaugh D. W., Pelaez N. Scientific principles of education research: experimental biology 2007. Adv. Physiol. Educ. 2007;31:374–376. doi: 10.1152/advan.00056.2007. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Prehn S., Laskey R. A., Hartmann G. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R., editors. Methods in Enzymology. Vol. 194. San Diego, CA: Academic Press; 1991. Guide to Yeast Genetics and Molecular Biology. [PubMed] [Google Scholar]
- Howard Hughes Medical Institute. Beyond Bio 101, The Transformation of Undergraduate Biology Education. Chevy Chase, MD: Howard Hughes Medical Institute; 1996. [Google Scholar]
- Howard D. R., Miskowski J. A. Using a module-based laboratory to incorporate inquiry into a large cell biology course. Cell Biol. Educ. 2005;4:249–260. doi: 10.1187/cbe.04-09-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Lord T., Chad S., Zimmerman R. Putting inquiry teaching to the test: enhancing learning in college botany. J. Coll. Sci. Teach. 2007;36:62–65. [Google Scholar]
- Marshall P. A. Using Saccharomyces cerevisiae to test the mutagenicity of household compounds: an open ended hypothesis-driven teaching lab. CBE Life Sci. Educ. 2007;6:307–315. doi: 10.1187/cbe.06-12-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B., Walker J. E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Mitchell B. F., Graziano M. R. From organelle to protein gel: a 6-wk laboratory project on flagellar proteins. CBE Life Sci. Educ. 2006;5:239–246. doi: 10.1187/cbe.05-07-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (NRC) National Science Education Standards. Washington, DC: National Academies Press; 1996. [Google Scholar]
- NRC. Inquiry and the National Science Education Standards: A Guide for Teaching and Learning. Washington, DC: National Academies Press; 2000. [Google Scholar]
- NRC. BIO 2010, Transforming Undergraduate Education for Future Research Biologists. Committee on Undergraduate Biology Education to Prepare Research Scientists for the 21st Century; Washington, DC: National Academies Press; 2003. [PubMed] [Google Scholar]
- National Science Foundation. Shaping the Future. Washington, DC: National Science Foundation; 1996. [Google Scholar]
- National Science Teachers Association. Practicing Science: The Investigative Approach in College Science Teaching. Arlington, VA: National Science Teachers Association Press; 2001. [Google Scholar]
- Odom D. P., Grossel M. J. Using the two-hybrid screen in the classroom laboratory. CBE Life Sci. Educ. 2002;1:43–62. doi: 10.1187/cbe.02-02-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Hoyo M. T., Allen D., Anderson M. A. Implementing inquiry-guided instruction: practical issues. J. Coll. Sci. Teach. 2004;33:20–24. [Google Scholar]
- Prince M., Felder R. M. The many faces of inductive teaching and learning. J. Coll. Sci. Teach. 2007;36:14–20. [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R. M., Krajcik J., Marx R. W., Soloway E. Performance of students in project-based science classrooms on a national measure of science achievement. J Res. Sci. Teach. 2002;39:410–422. [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., Lührmann R., Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K., Bassler J., Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L. A., Shen T., Wente S. R. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J. Biol. Chem. 2001;276:6445–6452. doi: 10.1074/jbc.M008311200. [DOI] [PubMed] [Google Scholar]
- Terry L. J., Shows E. B., Wente S. R. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Vallen E. Analysis of protein localization and secretory pathway function using the yeast Saccharomyces cerevisiae. Cell Biol. Educ. 2002;1:173–192. doi: 10.1187/cbe.02-08-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.