Abstract
Three-dimensional (3D) strain maps of the myocardium provide a coordinate-system-independent quantification of myocardial deformation and kinematics. We combine two MRI techniques, displacement encoding with stimulated echoes (DENSE) and strain encoding (SENC), to fully formulate a 3D strain map in a single slice of myocardium. The method utilizes two-dimensional DENSE in-plane displacement measurements in two adjacent slices in conjunction with a single SENC through-plane strain measure to calculate the 3D strain tensor. Six volunteers were imaged and the technique demonstrated 3D strain measures in all volunteers that are consistent with those reported in the literature from 3D tagging. The mean peak strain (+/− standard deviation) for six healthy volunteers for the first, second and third principal strains are 0.42 +/−0.11, −0.10 +/−0.03, and −0.21 +/−0.02, respectively. These results show that this technique is capable of reliably quantifying 3D cardiac strain.
Keywords: 3D, myocardium, cardiac, strain, SENC, DENSE
Introduction
MRI provides unique mechanisms for imaging functional cardiac kinematics with high spatial and temporal resolution. In this article we present a technique that combines two of these techniques, displacement encoding with stimulated echoes (DENSE) [1,2] and strain encoding (SENC) [3], to calculate a fully descriptive three-dimensional (3D) strain map of a single slice of myocardium. Myocardial strain describes contraction and orientation, and also gives insight into functional properties such as myofiber orientation and electrical activation patterns. Three-dimensional strain is independent of the image plane and is ideal to fully quantify these myocardial properties.
Several techniques have been used to calculate myocardial strain in one or two dimensions (1D or 2D). These include single-plane myocardial tagging (1D and 2D) [4,5,6], DENSE [1,2,7], SENC, and velocity encoded phase-contrast imaging [8,9]. Three-dimensional strain tensor calculations, to date, have been demonstrated using multi-plane acquisitions with myocardial tagging [10,11,12,13,14], phase-contrast velocity encoding [15], stimulated-echo techniques [16] and a combination of the stimulated-echo and tagging techniques called zHARP [17].
Myocardial tagging has the advantage that strain (1D, 2D or 3D) can be measured with high accuracy. Measuring 3D strain using tagging typically requires the registration of various image sets consisting of multiple parallel slices imaged twice with orthogonal in-plane tag lines, and a third set of images perpendicular to these with tags orthogonal to the first two tag orientations [10,11,12,13,14]. Strain is calculated using the differential displacements of adjacent intersections between three tag planes. The main disadvantages of myocardial tagging are its low spatial resolution limited by tag spacing and the time consuming identification of tag lines in post processing. Harmonic phase (HARP) analysis [18] has been introduced to automate the post processing and has been used to track the myocardial displacement in 3D [19]. This could be extended to a 3D strain measurement.
Phase-contrast velocity encoding measures instantaneous strain rate using the differential velocity between neighbouring voxels. By imaging two contiguous image planes in four encoding directions the full 3D strain rate tensor can be calculated [15, 16]. A measure of strain is obtained by integrating the strain rate over the cardiac cycle, taking care to ensure that the integral traverses the same material points in the myocardium [15]. This procedure is not a trivial operation and is confounded by movement of the myocardium out of the image plane.
Stimulated-echo techniques have been used to measure 3D strain by encoding two contiguous slices in a single shot [16]. This method uses displacement encoding in four or more directions across two adjacent contiguous slices to measure the differential displacement, between and along the Cartesian axes. This 3D differential displacement trivially formulates a full 3D strain tensor. The method eliminates registration errors that may occur during multiple breathhold acquisitions, but introduces artefacts as a result of the increased readout duration associated with measuring two echoes from two slices in a single readout.
zHARP [17] is a technique that encodes through-plane displacement onto the phase of tagged images thus quantifying both in-plane and through-plane motion simultaneously, using HARP analysis and stimulated-echoes, respectively. Each zHARP image requires two breathholds to acquire. The technique calculates 3D strain for pairs of adjacent slices so that four breathholds are required to image a single plane of 3D strain. zHARP provides a technique whereby 3D strain can be calculated without the need to image and co-register orthogonal data sets. The technique uses slice-followed two-point phase cycling which allows one to follow the same voxel of myocardium through the cardiac cycle.
SENC and harmonic phase analysis of tagged images have been combined to calculate an incomplete 3D strain tensor, yielding three in-plane components and one normal through-plane strain [20]. This cannot be used to analyse the principal strains and is thus limited to imaging within the standard cardiac coordinate image planes (radial-circumferential or longitudinal).
SENC derives through-plane strain by estimating the frequency of multiple tag planes applied parallel to and within an image plane. It performs a centre-of-mass estimate in the slice encoding direction of the kz slice profile of the stimulated-echo by acquiring two image planes at different slice encodings about the centre of the stimulated-echo. SENC is limited to 1D strain, which in a Lagrangian frame of reference is perpendicular to the image plane, or in the Eulerian frame of reference is along the direction determined by the spatial gradient of phase within one of the SENC encoded phase images. A limitation of this technique is that the range of measurable strain values is constrained by the width of the slice profile in k-space. Finally, SENC requires prior knowledge of the range of strain values expected and the k-space slice profile.
DENSE calculates the Eulerian displacement by encoding tissue displacement onto the MRI phase data. These displacement fields can be used to calculate 2D strain of a single image plane at a pixel resolution [1,2,7] and the calculation thereof requires little user interaction. Spottiswoode et al [21] have demonstrated the ability to track the myocardium and thus calculate Lagrangian displacement using DENSE.
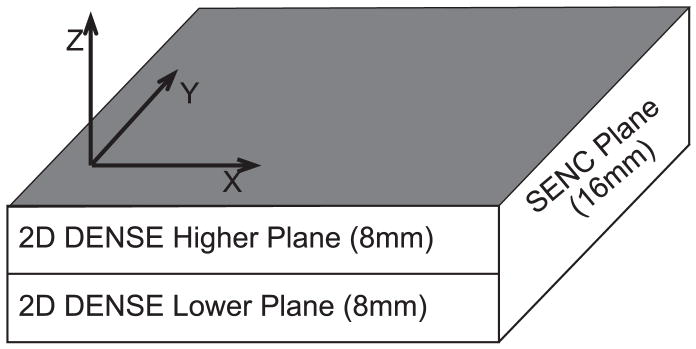
The method presented here acquires a time series of cine DENSE images of two adjacent 8mm thick slices, repeated for encodings applied in each of two orthogonal in-plane directions, and a cine SENC acquisition with a 16 mm slice thickness that encompasses both the cine DENSE planes as illustrated in Figure 1. These data are combined to yield uniplanar 3D cine strain tensor fields at a high spatial resolution. This method combines the advantages inherent in DENSE, such as high spatial resolution and convenient post processing, with the key advantage of SENC, which is its ability to image through-plane deformation in a single acquisition. We present here the modifications made to an existing cine DENSE [2] sequence to perform cine SENC, the computation of 3D strain, and a comparison of our results with values that have been reported previously.
Figure 1.
A single 16mm cine SENC slice encompasses two 8mm slices imaged with cine DENSE.
Methods
The cine DENSE sequence developed by Kim et al [2] was adapted to perform both cine DENSE and cine SENC imaging. This cine DENSE sequence uses a segmented EPI readout trajectory and performs two-point phase cycling for T1 relaxation echo suppression as described in [2]. The sequence acquires three cine image groups per breathhold, a phase reference series, and two corresponding groups for two-point phase cycling. Due to short repetition times (TR) and the small RF excitation angle, a dummy scan with duration equal to a single cardiac cycle is performed at the start of each of the three image acquisitions to achieve a steady state that serves to reduce artefacts.
The sequence in [2] applies an encoding kernel at end diastole for each heart beat which is triggered by the QRS complex of the ECG signal. Following a slice selective RF excitation pulse a decoding gradient is applied.
The sequence in [2] was modified to perform, upon operator selection, either cine SENC or cine DENSE. For a cine SENC selection, Figure 2a depicts the sequence timing in which a series of three cine image groups are acquired in a single breathhold, in this case a phase reference image series and two SENC encoded image groups. For each SENC encoded cine image series spatial encoding was applied through the image plane in the same manner as with through-plane cine DENSE. For cine SENC the applied decoding gradient had a different magnitude for each of the two cine SENC image groups. The cine SENC imaging sequence is shown in Figure 2b.
Figure 2.
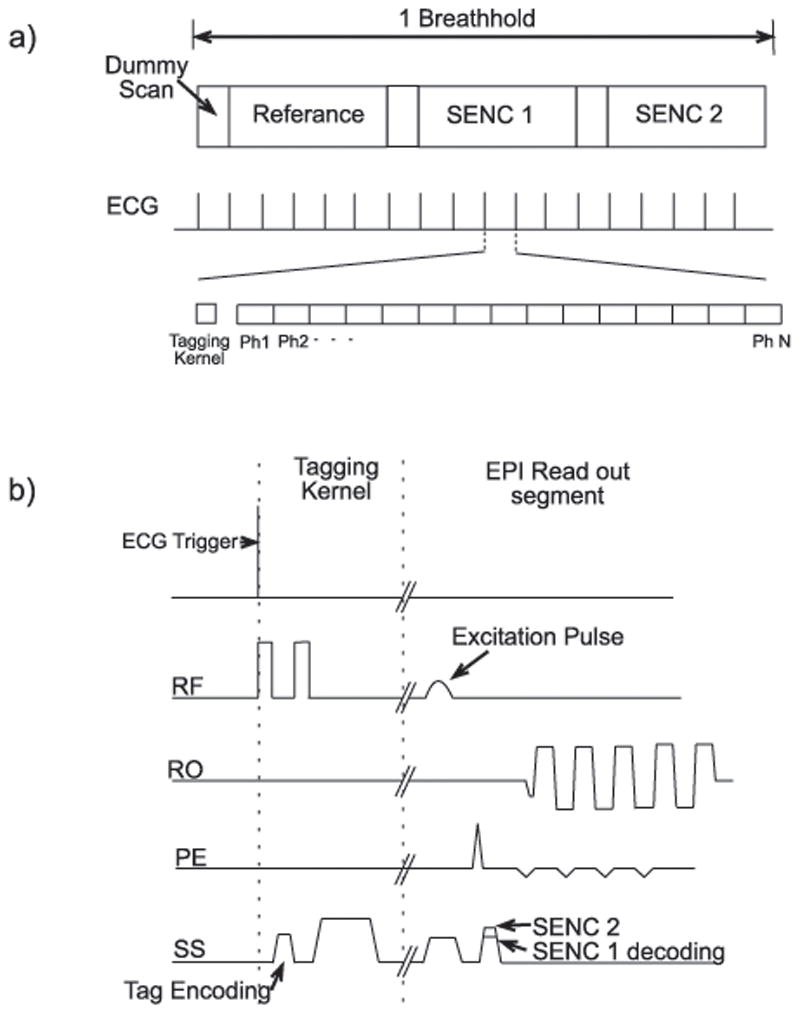
a) Image timing demonstrating acquisition of three imaging groups in one breathhold. b) Sequence timing diagram for cine SENC imaging
The optimal SENC encoding value was found by using data from a healthy volunteer. Images were acquired with encoding values ranging from 0.1 cyc/mm to 0.4 cyc/mm in steps of 0.01 cyc/mm (with matching decoding gradients). The appropriate encoding was found by determining which image possessed the greatest magnitude variation over time with the myocardium never completely fading. The optimal value was found to be 0.15 cyc/mm.
The decoding value of the first cine SENC acquisition was selected to be equal to the SENC encoding gradient to allow imaging of strain from its onset in early systole. The optimal value for the second decoding gradient was found to be 0.185 cyc/mm by imaging the same volunteer with decoding values ranging from 0.150 cyc/mm to 0.200 cyc/mm in steps of 0.005 cyc/mm. The optimal value was determined by plotting the results of this experiment at end systole, thus presenting the k-space slice profile, and selecting the centre (or peak) of the profile at end systole as demonstrated by Osman et al in [3].
The MRI measurements we obtained to formulate 3D strain consist of two adjacent 2D (in-plane) cine DENSE datasets and one cine SENC acquisition encompassing both cine DENSE slices (Figure 1).
Images were acquired in five breathholds of less than 20 s each: four for cine DENSE (i.e. two images encoded in X and Y, respectively, for each of two adjacent image planes) and one for cine SENC (refer to Figure 1). The protocol used for imaging included an echo train length of 9, 18 lines per heart beat, 18 cardiac phases, a flip angle of 15°, a TR of 22 ms and a matrix size of 128×96. The parameters specific to cine DENSE were FOV of 360 mm, a slice thickness of 8 mm and an encoding and decoding frequency of 0.1 cyc/mm. The cine SENC specific parameters were FOV of 480 mm, a slice thickness of 16 mm, an encoding frequency of 0.15 cyc/mm and decoding frequencies of 0.15 cyc/mm and 0.185 cyc/mm. A larger voxel size (resulting in a larger FOV) was used for SENC in order to increase the signal-to-noise ratio (SNR). Due to the difference in FOV between cine SENC and cine DENSE a bilinear interpolation was used to match the pixel size of cine SENC to that of cine DENSE. Images were manually contoured to aid the post processing and the visualisation of the data.
This method was used to image six volunteers, three at the University of Virginia on a Siemens 1.5T Magnetom Avanto scanner and three at Groote Schuur Hospital on a Siemens 1.5T Magnetom Symphony Quantum. All subjects imaged in this study provided informed consent and were studied in accordance with research protocols approved by the Human Investigations Committee of the respective institutions. All volunteers were imaged along a mid-ventricular short-axis slice.
Three post-processing steps were performed on the cine DENSE data sets, of which the first two could be executed online. The first step removed the T1 relaxation echo using two-point phase cycling. The cine DENSE image was then phase corrected by subtracting the phase of the phase reference image from the phase of the encoded image. The third step, which was performed offline using MATLAB (The Mathworks Inc, Natick, MA, USA), involved spatio-temporal phase unwrapping and quantification of absolute displacement for each voxel [21]. Typically, this phase unwrapping is not essential for 2D DENSE strain analysis [1], however, as strain was to be calculated from two separate cine-DENSE image planes (higher and lower) this procedure was necessary to ensure that there was no displacement offset between the two planes.
For cine SENC, two post-processing steps were performed offline in MATLAB. Firstly, phase correction was done on each of the two cine SENC images by subtracting the phase of the reference images from that of the cine SENC images. Secondly, magnitude and phase data at each myocardial voxel was used to calculate a vector describing the frequency and orientation of its cine SENC tag planes. The in-plane and through-plane components of the cine SENC tag vector were calculated using phase and magnitude data, respectively.
The frequency of the tag planes perpendicular to the image plane, known as the SENC vector v=v ẑ, was calculated as follows. Let P1x,y and P2x,y be the complex pixel values from the SENC images with decoding values of W1 = 0.15 cyc/mm and W2 = 0.185 cyc/mm, respectively. The through-plane component of the vector is calculated using a centre of mass estimate as follows:
| (1) |
This centre of mass estimate (u) of the through-plane tag frequency contains a linear error (as reported by Osman et al [3]) which was corrected using
| (2) |
in which optimal results were found for values of a = 0.96 and b = 0.004 using the data from all of our volunteer data sets. The tilt of the tag plane out of the image plane (θ) was calculated using the spatial gradient of phase [3]. The spatial gradient of phase is the phase difference between neighbouring voxels in one SENC image along the X- and Y- directions. The spatial gradient of phase is calculated using the SENC image in which the voxel in question has greatest magnitude, noting that a change in phase between neighbouring voxels is directly proportional to the difference in voxel displacement. As v is the tag frequency perpendicular to the image plane and not the tag planes, the frequency of the tag planes, fSENC = v/sin(θ), where θ is the angle between the image plane and the tag plane.
Lagrangian strain is calculated as follows:
| (3) |
where I is the identity matrix. F is the deformation gradient tensor which is given by
| (4) |
where x, y and z represent components at the measurement time and X, Y and Z represent mutually orthogonal components at the reference time, noting that in our coordinate system x and y are in the DENSE image plane and z is through the image plane. The deformation gradient tensor depicts the change in each axis of a unit vector from the reference time to the time of measurement.
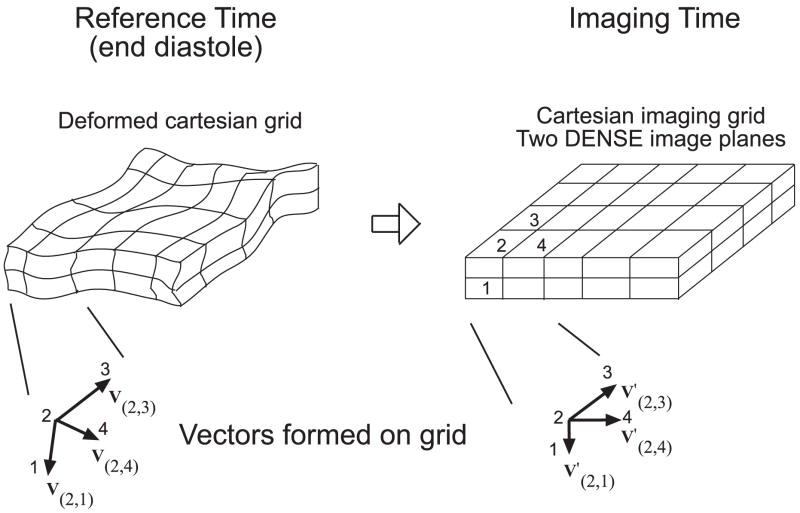
The paradigm used to formulate F is described in Figure 3 where a Cartesian image grid is shown at the imaging time and deformed at the reference time. Vectors are formed between the centres of imaged neighbouring voxels at the reference time as well as at the imaging time where they are aligned to a Cartesian grid. Consider the numbered voxels 1, 2, 3 and 4 in Figure 3 and the vectors formed between them at the imaging time (V′(2,1), V′(2,3), V′(2,4)) and at the reference time (V(2,1), V(2,3), V(2,4)), respectively. The deformation gradient F is calculated as follows:
| (5) |
where the/operator is a matrix right division.
Figure 3.
Depiction of how a segment of myocardium deforms into the Cartesian imaging grid of two contiguous DENSE imaged slices. Vectors are formed between these imaged voxels: V(2,1), V(2,3), V(2,4). The change of these vectors from the reference time to the imaging time are described by F. All vector X and Y components are determined from DENSE. The Z component of V(2,3), V(2,4) is determined from the SENC tag plane tilt (spatial gradient of phase), and the Z component of V(2,1) from using fenc and fSENC.
The X and Y components of all V vectors are trivially derived from the known DENSE displacement of each voxel from its Cartesian position at the imaging time. The tag plane tilt, as determined from the cine SENC spatial gradient of phase, is used to measure the Z components of the in-plane vectors (V(2,3) and V(2,4)). The fSENC measure is then used to calculate the Z component of the through-plane vector (V(2,1)), as follows:
| (6) |
where V(2,1)Z is the z element of vector V(2,1), h is the offset between the two cine DENSE image planes (8mm), and fenc is the SENC SPAMM encoding frequency (0.150cyc/mm).
In summary, the 3D strain calculation is formulated from F using the 2D cine DENSE data acquired in two adjacent image planes to measure in-plane differential displacements. The through-plane differential displacements (∂z/∂X, ∂z/∂Y, ∂z/∂Z) are measured using the cine SENC acquisition in a plane that encompasses both of the cine DENSE acquisitions.
This right-angled three vector kernel (as demonstrated in Figure 3) can be rotated through up to eight different orientations about the point of strain calculation covering all the voxels demonstrated in Figure 4. The average deformation gradient of all the orientations of the kernel was found while taking care to exclude kernels which fell outside of the myocardial boundaries. The Lagrangian strain for each voxel was then found using this averaged deformation gradient and Eq. 3.
Figure 4.
A cluster of voxels through which a 3 element kernel is rotated to form an average deformation gradient for the central voxel.
In order to understand the errors introduced in the strain tensor due to inter-breathhold myocardial position variability, a two-step experiment was performed. In the first step, we measured the inter-breathhold myocardial misalignment in a healthy volunteer and, in the second, we estimated what effect DENSE and SENC data acquired at randomly offset myocardial positions would have on strain measurements. Ten series of long-axis gradient recalled echo cine EPI images were acquired in ten separate breathholds for a healthy volunteer. Five of these series were acquired in the two chamber long-axis view and the other five with a four chamber long-axis view. These images were contoured and four measurements were obtained from each to measure myocardial tilt, shift in the long axis, and shift in the mid-ventricular short axis.
In the second step we considered twelve typical end-systolic strain tensors with known deformation gradients and mapped them to the Radial, Circumferential and Longitudinal (R, C and L) coordinate system for ease of comparison. We then proceeded to calculate the effect on our measurements of a random translation and rotation applied to each acquisition (four DENSE and one SENC). The random translations and rotations were normally distributed with a standard deviation equal to the peak translation and rotation measured in the volunteer data.
This model of through-plane slice variability was repeated 5000 times for each of 12 typical end-systolic strain tensors and deformation gradients. These 5000 simulations were separated into 10 groups of 500 each, for which the deformation gradient was scaled resulting in a linear scaling of strain tensor values from 0.1 to 1 in increments of 0.1. The scaling was performed to illuminate the effect of the strain magnitude on the error. For each of the 10 groups of 500 the RMS strain error was calculated for each of the six components of the strain tensor.
Results
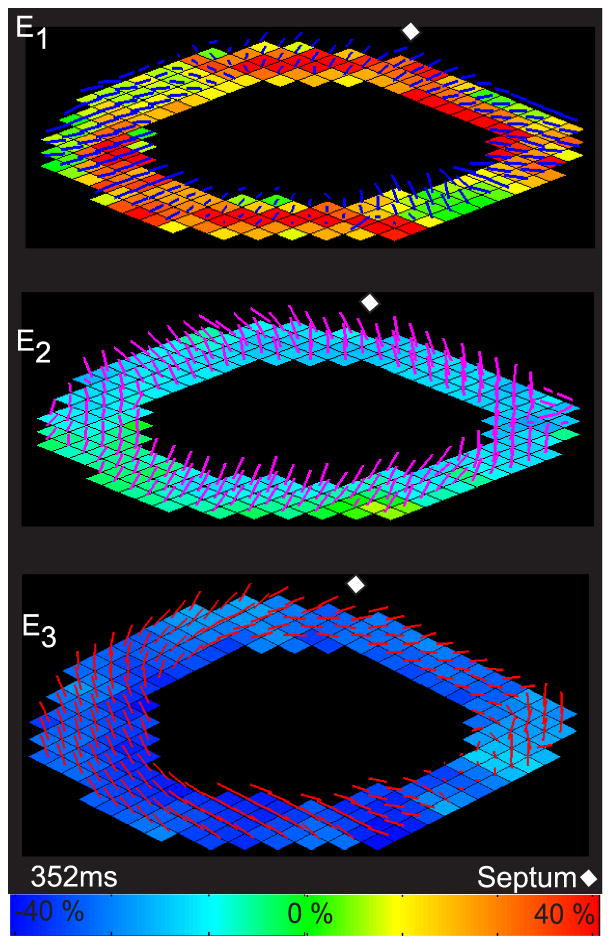
Figure 5 shows magnitude and direction maps of the principal strains for a single volunteer. The principal strains were sorted from largest positive to largest negative. The principal strain orientations were found to be consistent among all six volunteers with the first principal strain (E1 most positive) oriented radially within the image plane. The second principal strain (E2) is oriented in the long axis with a significant circumferential component and the third principal strain (E3 most negative) is oriented circumferentially with a significant longitudinal component. E3 forms a left-handed, clockwise, spiral from the apex to the base, as viewed from the base in all six volunteers compared to E2 which is counter-clockwise. There are a few locations where the direction of the E2 and E3 principal strains are switched, but this occurs when their values are close to one another and thus can not be distinguished by magnitude.
Figure 5.
End systolic principal strains for one volunteer depicted on a colour map overlaid with the vector orientation
The principal strain values were segmented into six standard segments, anteroseptal, inferoseptal, inferior, inferolateral, anterolateral and anterior. Outliers with strain greater than 0.8 or less than −0.8 were excluded from the datasets. No data set had more than five outliers in the myocardium at any one time frame. Figure 6 shows all three principal strains averaged across volunteers in each of the six segments. The peak average principal strains and the time at which they occurred are listed in Table 1.
Figure 6.
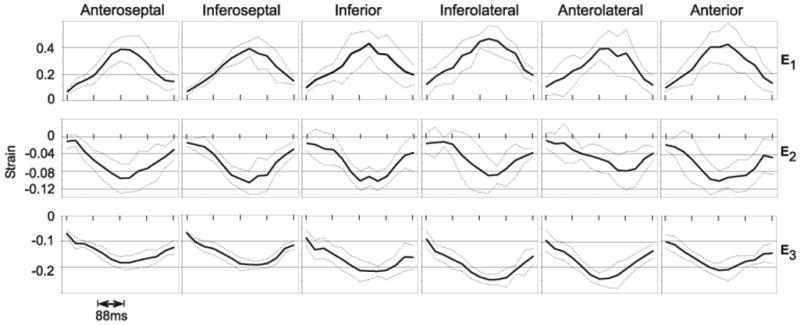
Principal mid-ventricular strain averaged across subjects. The temporal resolution is 44ms with a total of 572ms displayed. The dotted lines above and below the average strain are plotted at +/− 1 standard deviation.
Table 1.
Peak average principal strains for six standard cardiac segments of a mid-ventricular short-axis slice.
| E1 | E2 | E3 | ||||
|---|---|---|---|---|---|---|
| Time (ms) | Strain | Time (ms) | Strain | Time (ms) | Strain | |
| Anteroseptal | 310 | 0.39±0.09 | 310 | −0.10±0.03 | 350 | −0.18±0.02 |
| Inferoseptal | 350 | 0.39±0.05 | 350 | −0.11±0.03 | 400 | −0.19±0.02 |
| Inferior | 350 | 0.43±0.10 | 400 | −0.10±0.02 | 350 | −0.21±0.02 |
| Inferolateral | 350 | 0.47±0.10 | 350 | −0.09±0.04 | 350 | −0.25±0.02 |
| Anterolateral | 350 | 0.39±0.13 | 440 | −0.08±0.04 | 310 | −0.24±0.02 |
| Anterior | 350 | 0.43±0.16 | 310 | −0.10±0.02 | 310 | −0.21±0.03 |
When measuring the inter-breathhold myocardial position variability in the four chamber view, the maximum separation of the contours in the long-axis direction at the base and apex is 3.4mm and 2.3mm respectively. The contours in this image group have a maximum mid-ventricular separation of 3.4mm. The angle of the long-axis to the horizontal axis for each image in the group is 51.45°, 50.98°, 51.22°, 48.58° and 48.99°, respectively. In the two chamber long-axis view the maximum separation of the contours at the base and apex is 3.7mm. The maximum separation of the contours in the short-axis plane is 2.7mm and the five angles of the long-axis to the horizontal are 34.67°, 35.36°, 35.71°, 35.71°, and 35.71°, respectively.
In Figure 7 the strain error is plotted as a function of strain on a separate graph for each component of the strain tensor that is aligned to the RCL coordinate system. Each line corresponds to a different input strain. The largest RMS errors occur in the radial longitudinal shear strain (Erl) with maximum amplitude 1.5% strain. The next largest RMS errors are in the circumferential longitudinal (Ecl) shear components with a maximum of 0.82% strain. The RMS errors of the remaining components of the strain tensor are all negligible with values less than 0.5% strain.
Figure 7.
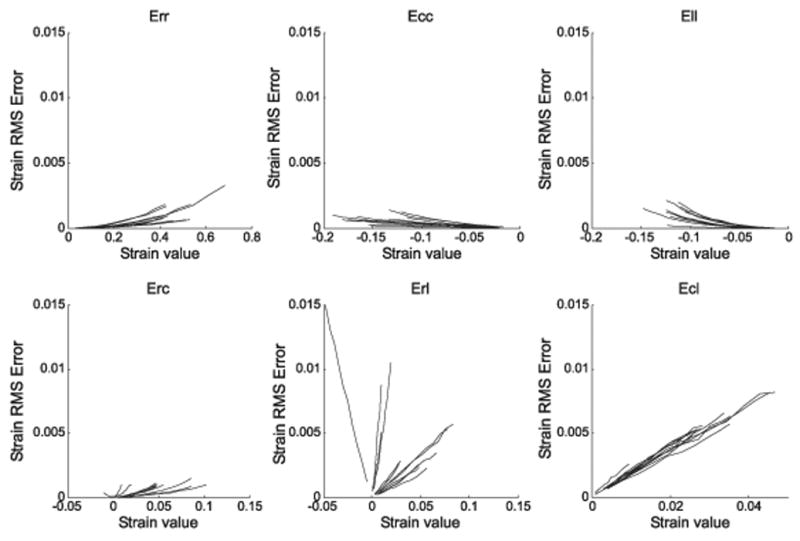
RMS error plotted as a function of strain for the simulation that explores the effects of variability in the myocardial position between breathholds. The error for each of the 12 input strain values is plotted on a separate line for each strain component in the RCL system. Err is the radial normal strain, Ecc the circumferential, Ell the Longitudinal, Erc the radial circumferential shear strain, Erl the radial longitudinal shear strain, and Ecl is the circumferential longitudinal shear strain.
Discussion
The directions of the principal strain vectors agree with those reported by Moore et al [11]. The results consistently demonstrate a single positive principal strain and two negative principal strains corresponding to radial thickening and circumferential/longitudinal shortening, respectively.
The mean strains observed agree to within two standard deviations (SD) with the values reported by Moore et al with the exception of the mean peak strain of E2 in the anterolateral and anterior segments and E3 in the anterior segment which all fall minimally (0.02) below this 2SD range. However, it was noted that the strains observed, for E2 in particular, are consistently lower than those reported by Moore et al [11]. A possible reason for a reduced E2 could be that the parameters chosen for Eq. 2 may not be optimal. Equation 2 results in a direct scaling of the magnitude of the observed strain and is the primary contributor to E2.
As expected E1, which is in a radial direction, is significantly larger than E2 and E3. As can be observed in Figure 5, E1 is not transmurally and circumferentially uniform, however this variance is shown in Figure 6 to be in proportion to the magnitude of the strain. It should be noted that an increase in the positive Lagrangian strain results in an increased variance or error.
A Lagrangian frame of reference was chosen for this work to be consistent with values reported in the literature, a Lagrangian calculation uses as the point of reference the time of initial encoding. An Eulerian calculation uses the measurement time as the point of reference and infers kinematic information about the time of encoding. Thus an Eulerian description is synonymous with our measurement techniques and the resultant data. The differences between the strain values from these two approaches are that a positive Lagrangian strain (such as E1) is larger than its Eulerian counterpart and vice versa for negative strains (such as circumferential or longitudinal). This is simply explained with reference to a 1D strain formula of change in length divided by length (E = Δl/l). For Lagrangian strain, the referenced l would be at its shortest (before stretching) compared to Eulerian strain for which the referenced l would be at its longest (after stretching) with Δl remaining the same. This further implicates and explains the magnification of measured noise in Lagrangian E1, where the error primarily lies in Δl, noting that E2 and E3 would suffer in the same way in an Eulerian calculation. The Lagrangian strains depicted in Figure 6 are plotted over time. However, as no tissue tracking was performed, the plots do not represent strain from exactly the same tissue over time as would be the case in a true Lagrangian frame of reference.
The spatial resolution of strain is determined by the size of the strain kernel and the resolution of the underlying measurements, the base kernel used here spans 5.6 mm x 5.6 mm × 16 mm. The DENSE data is sampled at an optimal resolution (2.8 mm × 2.8 mm × 8 mm) for this kernel as two DENSE samples are required in each of X, Y and Z. The SENC frequency measure is over-sampled at 3.75 mm × 3.75 mm × 16 mm as it is only used once per strain kernel. However, the SENC tag plane tilt measure (spatial gradient of phase) is under-sampled as two samples are required within the kernel in both X and Y resulting in the real resolution of this measure being 7.5 mm × 7.5mm × 16 mm, which is larger than the strain kernel. This under-sampling was not taken into consideration when adjusting the SENC FOV and reduces the resolution of the through-plane shear components of the strain.
The use of cine SENC to measure the through-plane differential (∂z/∂Z) is faster than imaging two separate slices using through-plane encoded cine DENSE which would have required two separate acquisitions and increasingly suffer from inaccuracies due to heart translation along the Z axis between breathholds.
The technique described here requires five breathholds, which may be too demanding for clinical application. This is, however, comparable to other techniques like zHARP which requires four breathholds to compute 3D strain for a single imaging plane. With tagging it is not possible to compute strain for a single slice and the entire heart needs to be imaged with tagging applied in orthogonal directions, which requires 18 breathholds [11]. Both our technique and zHARP have the advantage that strain can be calculated in a single image plane at a reduced acquisition time, namely five and four breathholds, respectively.
The simulations of the effect of inter-breathhold myocardial position variability on the strain calculation demonstrates that the largest error relates to the through-plane shear strains. The magnitude of this error is, however, relatively small. It is noteworthy how insensitive the through-plane normal strain is to this kind of error. In a parallel simulation we investigated how a 3D tensor calculated with two planes of 3D DENSE is affected by the same inter-breathhold myocardial position variability and found a significant increase in the through-plane normal strain error (RMS error of 0.04 strain). More information is needed to fully quantify the order of this error as our estimation of myocardial position variability between breathholds was based on data obtained from a single volunteer in a single scanning session.
In order to encompass the two independent cine DENSE slices the strain was imaged over a large 16mm slice. This may disguise pathology that does not span the entire slice due to partial voluming, particularly at the apex and base. This large measurement slice also reduces the definition of the myocardial boundaries so that sub-endocardial and sub-epicardial strains cannot be clearly defined. This slice thickness could be reduced but at the expense of ∂x/∂Z and ∂y/∂Z, the longitudinal shear parameters of the deformation gradient (F), which would increasingly suffer from inaccuracies due to differing heart positions between imaging breathholds. It should be noted, however, that the myocardium was segmented on the 8mm DENSE images in order to exclude SENC data for which the myocardium does not span the entire 16mm.
For a reduced slice separation the problem of inaccuracies in the longitudinal shear components of F could be addressed by acquiring cine DENSE data for a single encoding direction for two adjacent slices simultaneously and in the same breathhold. Reese et al [16] proposed a method where they encoded two adjacent slices of myocardium in a single shot. Alternatively, this could be done by acquiring cine DENSE data without two-point phase cycling for the two adjacent slices with encoding in a single direction in a single breathhold. This would require only two phase reference and two encoded images (one for each slice) to be acquired in a single breathhold. However, this could only be achieved by sacrificing temporal resolution.
Conclusion
A method has been demonstrated to measure the complete 3D strain tensor field in a single slice of myocardium over the cardiac cycle using a combination of breathhold cine DENSE and cine SENC measurement techniques. It has been validated in six healthy volunteers. The strain results agree to within experimental accuracy with values reported in literature. These strain results demonstrate that this technique is capable of reliably quantifying 3D cardiac strain at a high spatial resolution for a single slice.
Acknowledgments
This project was supported by NIH Research Grant R03 TW07633 funded by the Fogarty International Center and the National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH/NIBIB grant R01 EB001763, Focus Area grant FA2005040800023 of the National Research Foundation of South Africa, and an international student travel grant from the University of Cape Town. We thank Galiema Adams, Sharon Heyne, Petronella Samuels, Kamillah Salie, and Nadia Solomon for their contributions to this research.
References
- 1.Aletras AH, Ding S, Balaban RS, Wen H. DENSE: Displacement Encoding with Stimulated Echoes in Cardiac Functional MRI. Journal of Magnetic Resonance. 1999;137:247–252. doi: 10.1006/jmre.1998.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim D, Gilson WD, Kramer CM, Epstein FH. Myocardial Tissue Tracking with Two-dimensional Cine Displacement-encoded MR Imaging: development and Initial Evaluation. Radiology. 2004;230:862–871. doi: 10.1148/radiol.2303021213. [DOI] [PubMed] [Google Scholar]
- 3.Osman NF, Sampath S, Atalar E, Prince JL. Imaging Longitudinal Cardiac Strain on Short-Axis Images Using Strain-Encoded MRI. Magnetic Resonance in Medicine. 2001;46:324–334. doi: 10.1002/mrm.1195. [DOI] [PubMed] [Google Scholar]
- 4.Zerhouni EA, Parish DM, Rogers WJ, Yang A, Shapiro EP. Human Heart: Tagging with MR Imaging – A new Method for Noninvasive Assement of Myocardial Motion. Cardiac Radiology. 1988;169:59–63. doi: 10.1148/radiology.169.1.3420283. [DOI] [PubMed] [Google Scholar]
- 5.Axel L, Dougherty L. MR Imaging of Motion with Spatial Modulation of Magnetization. Radiology. 1989;171:841–845. doi: 10.1148/radiology.171.3.2717762. [DOI] [PubMed] [Google Scholar]
- 6.McVeigh ER, Zerhouni EA. Noninvasive Measurement of Transmural Gradients in Cardiac Strain with MR Imaging. Radiology. 1991;180:677–683. doi: 10.1148/radiology.180.3.1871278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aletras AH, Balaban RS, Wen H. High-Resolution Strain Analysis of the Human Heart with Fast-DENSE. Journal of Magnetic Resonance. 1999;140:41–57. doi: 10.1006/jmre.1999.1821. [DOI] [PubMed] [Google Scholar]
- 8.Van Dijk P. Direct cardiac NMR imaging of heart wall and blood flow velocity. Journal of Computer Assisted Tomography. 1984;8:429–436. doi: 10.1097/00004728-198406000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Bryant D, Firmin D, Longmore D. Measurement of flow with NMR imaging using gradient pulse and phase difference technique. Journal of Computer Assisted Tomography. 1984;8:588–593. doi: 10.1097/00004728-198408000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Young AA, Axel L. Three-Dimensional Motion and Deformation of the Heart Wall: Estimation with Spatial Modulation of Magnetization – a Model-based Approach. Radiology. 1992;185:241–247. doi: 10.1148/radiology.185.1.1523316. [DOI] [PubMed] [Google Scholar]
- 11.Moore CC, Lugo-Olivieri CH, McVeigh ER, Zerhouni EA. Three-dimensional systolic strain patterns in the normal human left ventricle: characterization with tagged MR imaging. Radiology. 2000;214:453–466. doi: 10.1148/radiology.214.2.r00fe17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young AA, Kramer CM, Ferrari VA, Axel L, Reichek N. Three-dimensional left ventricular deformation in hypertrophic cardiomyopathy. Circulation. 1994;90:854–867. doi: 10.1161/01.cir.90.2.854. [DOI] [PubMed] [Google Scholar]
- 13.Azhari H, Weiss JL, Rogers WJ, Siu CO, Zerhouni EA, Shapiro EP. Noninvasive quantification of principal strains in normal canine hearts using tagged MRI images in 3-D. The American Journal of Physiology. 1993;264:H205–H216. doi: 10.1152/ajpheart.1993.264.1.H205. [DOI] [PubMed] [Google Scholar]
- 14.Kuijer JPA, Marcus JT, Gotte MJW, van Rossum AC, Heethaar RM. Three-dimensional myocardial strain analysis based on short- and long- axis magnetic resonance tagged images using a 1D displacement field. Magnetic Resonance imaging. 2000;18:553–564. doi: 10.1016/s0730-725x(00)00146-6. [DOI] [PubMed] [Google Scholar]
- 15.Tseng WY, Reese TG, Weisskoff RM, Brady TJ, Wedeen VJ. Myocardial fiber shortening in humans: initial results of MR imaging. Radiology. 2000;216:128–139. doi: 10.1148/radiology.216.1.r00jn39128. [DOI] [PubMed] [Google Scholar]
- 16.Reese T, Feinberg D, Dou J, Wedeen V. Phase Contrast MRI of Myocardial 3D strain by Encoding contiguous Slices in a Single Shot. Magnetic Resonance in Medicine. 2002;47:665–676. doi: 10.1002/mrm.10111. [DOI] [PubMed] [Google Scholar]
- 17.Abd-Elmoniem KZ, Stuber M, Prince JL. Direct three-dimensional myocardial strain tensor quantification and tracking using zHARP. Medical Image Analysis. 2008;12(6):778–786. doi: 10.1016/j.media.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osman NF, McVeigh ER, Prince JL. Imaging heart motion using harmonic phase MRI. IEEE Transactions on Medical Imaging. 2000;19:186–202. doi: 10.1109/42.845177. [DOI] [PubMed] [Google Scholar]
- 19.Pan L, Lima JAC, Osman NF. Fast Tracking of Cardiac Motion Using 3D-HARP. IEEE Transactions on Biomedical Engineering. 2005;52:1425–1435. doi: 10.1109/TBME.2005.851490. [DOI] [PubMed] [Google Scholar]
- 20.Sampath S, Osman N, Prince J. Imaging the evolution of 3-D myocardial strains using a combined HARP-SENC MRI pulse sequence. Proc Intl Soc Mag Res Med. 2005:13. [Google Scholar]
- 21.Spottiswoode BS, Zhong X, Hess AT, Kramer CM, Meintjes EM, Mayosi BM, Epstein FH. Tracking Myocardial Motion From Cine DENSE Images Using Spatiotemporal Phase Unwrapping and Temporal Fitting. IEEE Transactions on medical imaging. 2007;26:15–30. doi: 10.1109/TMI.2006.884215. [DOI] [PubMed] [Google Scholar]