Abstract
Introduction
The Cu-PTSM (pyruvaldehyde bis(N4-methylthiosemicarbazonato)copper(II)) and Cu-ATSM (diacetyl bis(N4-methylthiosemicarbazonato)copper(II)) radiopharmaceuticals exhibit strong, species-dependent binding to the IIA site of human serum albumin (HSA), while the related Cu-ETS (ethylglyoxal bis(thiosemicarbazonato)copper(II)) radiopharmaceutical appears to only exhibit non-specific binding to human and animal serum albumins.
Methods
To further probe the structural basis for the species-dependence of this albumin binding interaction, protein binding of these three radiopharmaceuticals was examined in solutions of albumin and/or serum from a broader array of mammalian species (rat, sheep, donkey, rabbit, cow, pig, dog, baboon, mouse, cat, elephant). We also evaluated the albumin binding of several copper(II) bis(thiosemicarbazone) chelates offering more diverse substitution of the ligand backbone.
Results
Cu-PTSM and Cu-ATSM exhibit a strong interaction with HSA that is not apparent with the albumins of other species, while the binding of Cu-ETS to albumin is much less species-dependent. The strong interaction of Cu-PTSM with HSA does not appear to simply correlate with variation, relative to the animal albumins, of a single amino acid lining HSA's IIA site. Those agents that selectively interact with HSA share the common feature of only methyl or hydrogen substitution at the carbon atoms of the diimine fragment of the ligand backbone.
Conclusions
The interspecies variations in albumin binding of Cu-PTSM and Cu-ATSM are not simply explained by unique amino acid substitutions in the IIA binding pocket of the serum albumins. However, the specific affinity for this region of HSA is disrupted when substituents bulkier than a methyl group appear on the imine carbons of the copper bis(thiosemicarbazone) chelate.
Keywords: Serum albumin binding, Cu-PTSM, Cu-ATSM, Cu-ETS, Copper radiopharmaceutical, PET radiopharmaceuticals
Introduction
Copper(II) bis(thiosemicarbazone) radiopharmaceuticals for quantitatively mapping regional perfusion with positron emission tomography (PET) can exhibit highly species-dependent binding to serum albumin [1-5]. The pyruvaldehyde bis(N4-methylthiosemicarbazonato)copper(II) (Cu-PTSM) perfusion tracer, and the diacetyl bis(N4-methylthiosemicarbazonato)copper(II) (Cu-ATSM) hypoxia tracer, are both much more extensively protein bound in the presence of human serum albumin (HSA) than in the presence of various animal albumins [1,2]. This is in sharp contrast with the behavior of the related ethylglyoxal bis(thiosemicarbazonato)copper(II) (Cu-ETS) chelate, which seems to exhibit only non-specific binding to HSA and animal albumins [1,2] (Figure 1). These uncharged square-planar copper(II) chelates share rigid π-conjugated ligand backbones, and differ only in various peripheral substitutions at the imine carbon, and terminal amine nitrogen, locations (Figure 2 and Tables 1 and 2). The high-affinity HSA binding of Cu-PTSM and Cu-ATSM appears [3] to occur in the well characterized IIA subdomain that is also responsible for binding the drugs warfarin and furosemide, among others [6-10].
Figure 1.
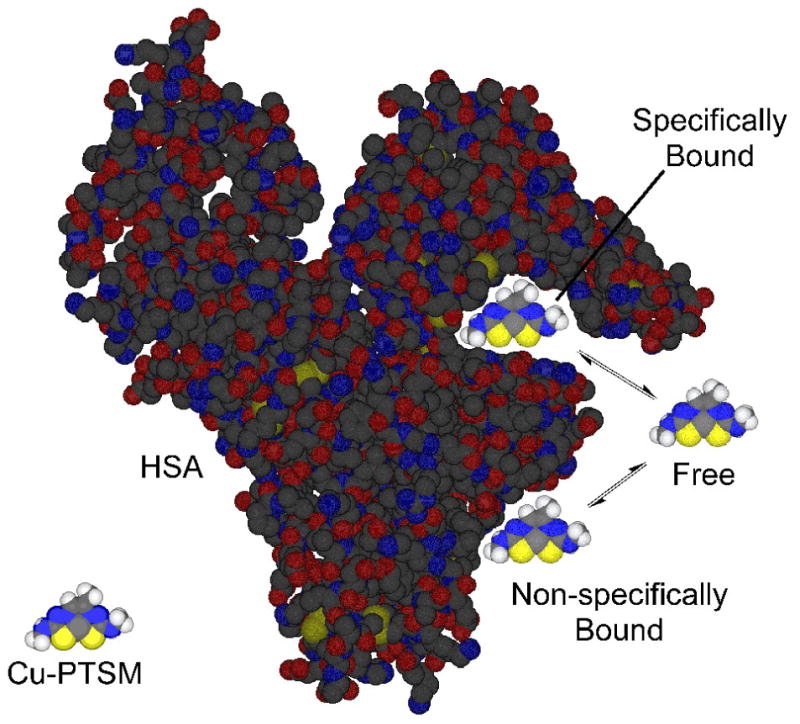
Illustration of the equilibria involved in the HSA-binding of Cu-PTSM. The specific HSA binding of Cu-PTSM (and Cu-ATSM) occurs in the IIA site of HSA known to be responsible for binding the drugs warfarin and phenylbutazone [3].
Figure 2.
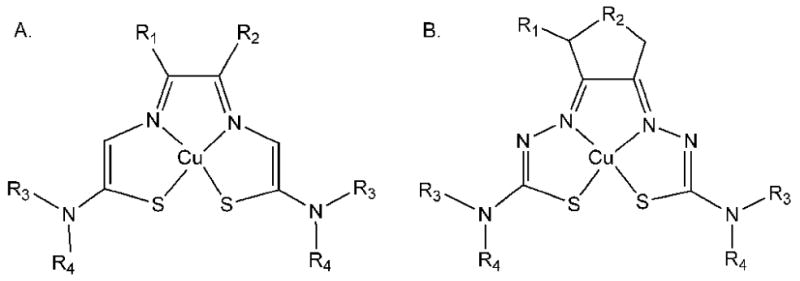
General structural formulae for the copper(II) bis(thiosemicarbazone) chelates studied. A. Bis(thiosemicarbazones derived from α-ketoaldehydes and diacetyl. B. Bis(thiosemicarbazones) derived from 3-methyl-1,2-cyclopentanedione and 1,2-cyclohexanedione.
Table 1.
Identity and characteristics of the principle bis(thiosemicarbazonato)copper(II) chelates studied. The substituents R1, R2, R3, and R4 are designated as shown in Figure 2A.
| % Free ± SD (n ≥ 3)b | ||||||||
|---|---|---|---|---|---|---|---|---|
| [64/67Cu]Cu-L | R1 | R2 | R3 | R4 | MW | Log Pa | HSA | CSA |
| Cu-PTSM | -CH3 | -H | -CH3 | -H | 308 | 1.92 ± 0.04 | 4.0 ± 0.1 | 42.0 ± 1.3 |
| Cu-PTSM2 | -CH3 | -H | -CH3 | -CH3 | 336 | 3.18 ± 0.05 | 4.0 ± 0.3 | 15.8 ± 0.7 |
| Cu-PTSE | -CH3 | -H | -CH2CH3 | -H | 336 | 2.76 ± 0.05 | 5.9 ± 0.2 | 27.0 ± 1.6 |
| Cu-ATSM | -CH3 | -CH3 | -CH3 | -H | 322 | 2.20 ± 0.12 | 5.9 ± 0.4 | 36.3 ± 1.5 |
| Cu-2,3-PnTSM | -CH2CH3 | -CH3 | -CH3 | -H | 337 | 2.80 ± 0.1 | 6.6 ± 0.3 | 9.6 ± 0.4 |
| Cu-PTS | -CH3 | -H | -H | -H | 280 | 0.76 ± 0.04 | 10.6 ± 0.2 | 50.1 ± 1.0 |
| Cu-ATS | -CH3 | -CH3 | -H | -H | 294 | 1.04 ±0.13 | 13.2 ± 0.3 | 37.6 ± 1.8 |
| Cu-ETSM | -CH2CH3 | -H | -CH3 | -H | 322 | 2.65 ± 0.07 | 21.3 ± 0.6 | 20.3 ± 0.4 |
| Cu-KTSM | -CH3(CH2CH3O)CH | -H | -CH3 | -H | 366 | 2.64 ± 0.11 | 30.4 ± 1.6 | 23.4 ± 0.6 |
| Cu-ETS | -CH2CH3 | -H | -H | -H | 294 | 1.35 ± 0.02 | 40.6 ± 1.1 | 44.2 ± 1.7 |
| Cu-KTS | -CH3(CH2CH3O)CH | -H | -H | -H | 338 | 1.71 ± 0.04 | 46.1 ± 0.6 | 55.2 ± 0.5 |
With the exception of Cu-ATS, the tabulated log P values were either previously reported in the literature, or experimentally measured in this study using the literature technique [16]. The log P value for Cu-ATS was calculated using: log PCu-ATS = log PCu-PTS + [log PCu-ATSM – log PCu-PTSM], according to the relationship of alkyl substitution and lipophilicity reported by Hansch [26].
Serum albumin concentrations were 40 mg/mL. The reported values are corrected for non-specific binding to the ultrafiltration membrane. These saline control values reflect significant, but reproducible, levels of radiopharmaceutical binding to the ultrafiltration device (mean % free ± SD, n≥3): Cu-PTSM (63.5 ± 3.9), Cu-PTSM2 (21.3 ± 0.9), Cu-PTSE (48.1 ± 2.9), Cu-ATSM (65.3 ± 3.7), Cu-2,3-PnTSM (35.1 ± 1.8), Cu-PTS (70.8 ± 1.1), Cu-ATS (63.7 ± 1.6), Cu-ETSM (48.5 ± 1.6), Cu-KTSM (72.6 ± 3.3), Cu-ETS (70.6 ± 2.5), Cu-KTS (46.1 ± 0.6).
Table 2.
Identity and characteristics of the bis(thiosemicarbazonato)copper(II) chelates studied with ligands derived from 3-methyl-1,2-cyclopentanedione and 1,2-cyclohexanedione. The substituents R1, R2, R3, and R4 are designated as shown in Figure 2B.
| % Free ± SD (n ≥ 3)b | ||||||||
|---|---|---|---|---|---|---|---|---|
| [64/67Cu]Cu-L | R1 | R2 | R3 | R4 | MW | Log Pa | HSA | CSA |
| Cu-1,2-CyHexTSM | -H | -C2H4- | -H | -CH3 | 348 | 2.90 ± 0.05 | 9.2 ± 0.1 | 6.9 ± 0.6 |
| Cu-3Me-1,2-CyPentTSM | -CH3 | -CH2- | -H | -CH3 | 348 | 2.06 ± 0.08 | 16.0 ± 0.1 | 15.2 ±0.4 |
| Cu-1,2-CyHexTS | -H | -C2H4- | -H | -H | 320 | 1.64 ± 0.07 | 21.0 ± 0.5 | 12.4 ±0.7 |
| Cu-3Me-1,2-CyPentTS | -CH3 | -CH2- | -H | -H | 320 | 1.34 ± 0.03 | 31.0 ± 1.0 | 28.5 ± 0.7 |
The tabulated log P values were experimentally measured in this study using the literature technique [16].
Serum albumin concentrations were 40 mg/mL. The reported values are corrected for non-specific binding to the ultrafiltration membrane. These saline control values reflect significant, but reproducible, levels of radiopharmaceutical binding to the ultrafiltration device (mean % free ± SD, n≥3): Cu-1,2-CyHexTSM (35.9 ± 1.0), Cu-3-Me-1,2-CyPentTSM (48.7 ± 0.7), Cu-1,2-CyHexTS (50.1 ± 0.3), Cu-3-Me-1,2-CyPentTS (52.3 ± 1.6).
HSA is the most abundant human plasma protein (46 mg/mL), and is responsible for transporting a number of endogenous compounds and drugs [11]. Serum albumins are nearly ubiquitous throughout the animal kingdom, and albumins from phenotypically diverse species share a high degree of similarity. Serum albumins from nearly every species share: three flexible domains; consistent molecular weights (65–69 kDa); similar disulfide linkage patterns; and a limited number of tryptophan residues (typically one or two) in highly conserved locations [12]. However, for a number of drugs the extent of binding to serum albumin is known to be species-dependent, with affinity for nonhuman serum albumins not reliably predicted by drug affinity for HSA [13-15].
To determine whether the observed interspecies variations in albumin binding of Cu-PTSM and Cu-ATSM could be attributed to simple differences in the primary structures of various serum albumins, we have evaluated the albumin binding of 64Cu-PTSM, 64Cu-ATSM, and 64Cu-ETS with the commercially available albumins of eight mammalian species for which the amino acid sequences are known. Also, we have extended our previous studies of related copper(II) bis(thiosemicarbazone) radiopharmaceuticals, identifying additional structural derivatives of these chelates that do, and do not, selectively interact with HSA.
Materials and Methods
Radiochemical Synthesis
Copper-64 was obtained as no-carrier-added 64Cu2+ in dilute HCl from the Research Resource for Radionuclide Applications at Washington University via Isotrace, Inc. (St. Louis, MO). No-carrier-added copper-67 was obtained as 67Cu2+ in 0.1 N HCl (0.05 mL) from Trace Radiopharmaceuticals (Denton, TX). Diacetyl bis(thiosemicarbazone) (H2ATS), pyruvaldehyde bis(N4-methylthiosemicarbazone) (H2PTSM), pyruvaldehyde bis(thiosemicarbazone) (H2PTS), ethylglyoxal bis(thiosemicarbazone) (H2ETS), 2-keto-3-ethoxybutyraldehyde bis(thiosemicarbazone) (H2KTS), diacetyl bis(N4-methylthiosemicarbazone) (H2ATSM), ethylglyoxal bis(N4-methylthiosemicabazone) (H2ETSM), 2-3-keto-ethoxybutyraldehyde bis(N4-methylthiosemicarbazone) (H2KTSM), pyruvaldehyde bis(N4-dimethylthiosemicarbazone) (H2PTSM2), pyruvaldehyde bis(N4-ethylthiosemicarbazone) (H2PTSE), 1,2-cyclohexyl bis(thiosemicarbazone) (H2-1,2-CyHexTS), 1,2-cyclohexyl bis(N4-methylthiosemicarbazone) (H2-1,2-CyHexTSM), 3-methyl-1,2-cyclopentyl bis(thiosemicarbazone) (H2-3-Me-1,2-CyPentTS), 3-methyl-1,2-cyclopentyl bis(N4-methylthiosemicarbazone) (H2-3-Me-1,2-CyPentTSM), and 2,3-pentane bis(N4-methylthisoemicarbazone) (H2-2,3-PnTSM) were prepared as described previously [16]. The [64/67Cu] radiopharmaceuticals were prepared as described previously, and purified using C18 SepPak Light (Waters, Milford, MA) solid-phase extraction cartridges to remove any residual ionic radiocopper [2]. The radiochemical purity of each radiopharmaceutical was determined by thin layer chromatography using silica gel plates with 100 % EtOH as the mobile phase, and always exceeded 98%. The lipophilicity of each chelate was either available from the literature, or measured as described previously [16]. For the log P values reported previously, and in the present study, the octanol phases were repartitioned into fresh aqueous solutions to reduce the effect of any ionic radiocopper contamination of the original aqueous phase.
Protein Binding
Ultrafiltration binding assays were used to quantify the binding of these 15 radiocopper chelates in 40 mg/mL solutions of human and canine serum albumins. In addition, protein binding of the Cu-PTSM, Cu-ATSM, and Cu-ETS chelates was measured in 40 mg/mL solutions of commercial sheep, donkey, rabbit, and cow serum albumins (Sigma-Aldrich, St. Louis, MO), as well as in serum from sheep, donkey, rabbit, cow, cat, and elephant. The donkey, sheep, and canine serum albumin (CSA) were ethanol fraction V powders. The rabbit and bovine serum albumin materials were purified by the vendor, with claim of >99% purity for each protein. The HSA was essentially globulin-free, and at least 99.5% fatty acid free. Sheep, donkey, rabbit, and cow serums were also purchased from Sigma-Aldrich, and stored frozen until being thawed immediately prior to use. Cat serum was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA) and reconstituted per the vendor instructions prior to use in 0.9% normal saline. The elephant serum was a generous gift from the Elephant Sanctuary (Hohenwald, TN) [17]. Total protein concentrations, as reported in the vendor certificates of analysis for sheep, rabbit, bovine, and donkey serums, were 62, 53, 76, and 68 mg/mL, respectively. For sheep, rabbit, and bovine sera, the vendor also reported serum albumin concentrations of 37.4, 28.1, and 35.8 mg/mL, respectively.
Centrifree (Millipore, Bedford, MA) ultrafiltration devices (30,000-Da nominal molecular weight limit methylcellulose micropartition membranes) were used to mechanically separate the free fraction of each radiochelate from the protein-bound fraction. Typically, a 4 μL aliquot of each radiotracer was added to 1 mL of serum albumin or serum solution and vortex mixed for 10 s at room temperature. Then, 300 μL aliquots of each tracer + protein solution were loaded into the ultrafiltration devices and centrifuged, within 5 min. of being mixed, in a fixed angle rotor for 10 min at 2000×G in a centrifuge maintained at 20°C. To account for the non-specific ultrafiltration membrane binding of each chelate, individual correction factors were calculated by subjecting saline solutions of each [64/67Cu]Cu-bis(thiosemicarbazone) to the ultrafiltration process. Equal volume samples of each ultrafiltrate and loading solution were counted in a Packard Autogamma 5530 automatic gamma counter to quantify radiocopper levels. “Mean corrected free fractions” were calculated as the ratios of “free fraction in protein solution” to “apparent free fraction in protein-free saline solution;” the associated variance was calculated as described previously using the Delta method [2,18].
Serum Albumin Sequence Homology
Serum albumin amino acid sequences from 17 mammal species were analyzed using various proteomic tools available through the UniProt database [19]. SSEARCH and ClustalW (version 1.82) were used for pairwise and multiple alignments, respectively [20,21]. These alignment tools calculate a “% Identity” value, which is a simple ratio of matched residues compared to the total number of residues. In addition to global comparisons of entire serum albumin sequences (each ∼585 residues), two SSEARCH sequence alignments were used to evaluate sequence homology between species in locations analogous to the IIA drug binding site of HSA. One narrow-scope alignment was restricted to 22 amino acids that directly delineate the void of the IIA site; Tyr-150, Lys-195, Lys-199, Phe-211, Trp-214, Ala-215, Arg-218, Leu-219, Arg-222, Phe-223, Leu-234, Leu-238, Val-241, His-242, Arg-257, Leu-260, Ala-261, Ile-264, Ser-287, Ile-290, Ala-291, and Glu-292 (Figure 3) [22-24]. A second comparison with a slightly expanded scope used 107 total residues in two sequence segments near the IIA site, resides 148 – 152, and 193 – 294. This comparison included 22 amino acids shown to directly line the IIA site, as well as 85 additional residues sequentially within +/- 2 residues of these 22 key amino acids. This criteria traces a roughly spherical shape with an approximate radius of 20Å originating in the center of the IIA site (Figure 2A).
Figure 3.
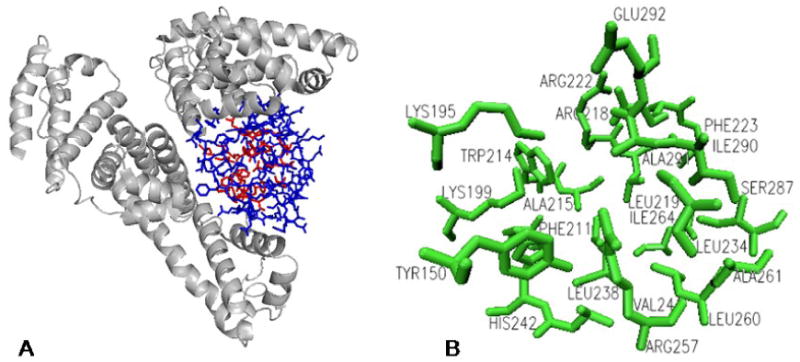
A. The structure of HSA and the location of the IIA drug binding site. The 22 amino acids adjacent to the void created by the IIA binding site are shown in red. An 85 additional residues in the vicinity of this site, used for the expanded comparisons, are shown in blue. B. Detailed view of 22 amino acids lining the IIA site. The Supplementary Material includes stereoviews of the IIA binding site, and an illustration with the superimposed positions of bound warfarin and phenylbutazone, based HSA-drug adduct crystal structures 2bxc and 2bxd from the Protein Data Bank [24].
Results and Discussion
Species Dependence of Albumin Binding
Table 3 presents the measured protein binding of Cu-PTSM, Cu-ATSM, and Cu-ETS in solutions of the commercially available mammalian serum albumins and sera. The species- and compound-dependence of the level of protein binding of these radiopharmaceuticals has been reported, but the present study extends those data to six additional species [2]. In the absence of a crystal structure defining the nature of the strong interaction of Cu-PTSM and Cu-ATSM with HSA, we hoped the protein binding data for these additional species might help identify specific amino acids lining the warfarin binding pocket whose variation might account for the strong species-dependence of the albumin binding of Cu-PTSM and Cu-ATSM.
Table 3.
Binding of [64Cu]Cu-PTSM, [64Cu]Cu-ATSM, and [64Cu]Cu-ETS to the Serum Albumins and Sera of Various Species
| Complex | Species | %Freea | |||
|---|---|---|---|---|---|
| Serum Albuminb | n | Serum | n | ||
| Cu-PTSM | Humanc | 4.0 ± 0.1 | 8 | 4.3 ± 0.2 | 5 |
| Ratc | 14.1 ± 0.3 | 6 | 14.7 ± 1.2 | 6 | |
| Sheep | 15.1 ± 0.8 | 5 | 13.9 ± 0.5 | 5 | |
| Donkey | 15.4 ± 0.5 | 2 | 17.5 ± 0.9 | 5 | |
| Rabbit | 25.2 ± 0.9 | 5 | 15.3 ± 0.9 | 5 | |
| Cow | 28.8 ± 1.4 | 5 | 15.2 ± 1.4 | 5 | |
| Pigc | 39.9 ± 0.9 | 3 | 26.3 ± 1.2 | 6 | |
| Dogc | 42.0 ± 1.3 | 4 | — | — | |
| Baboonc | 12.6 ± 0.4 | 4 | — | — | |
| Mousec | — | — | 14.6 ± 0.6 | 5 | |
| Cat | — | — | 18.3 ± 0.5 | 5 | |
| Elephant | — | — | 22.7 ± 2.2 | 3 | |
|
| |||||
| Cu-ATSM | Human | 5.9 ± 0.4 | 9 | 6.2 ± 0.4 | 5 |
| Ratc | 6.3 ± 0.2 | 6 | 5.7 ± 0.2 | 6 | |
| Sheep | 3.9 ± 0.3 | 5 | 2.4 ± 0.1 | 5 | |
| Donkey | 9.9 ± 0.2 | 2 | 10.1 ± 0.5 | 5 | |
| Rabbit | — | — | 9.9 ± 0.4 | 5 | |
| Cow | 10.0 ± 0.6 | 5 | 5.4 ± 0.2 | 4 | |
| Pigc | 16.8 ± 0.7 | 7 | 11.2 ± 0.3 | 6 | |
| Dogc | 36.3 ± 1.5 | 6 | — | — | |
| Baboonc | 24.6 ± 0.6 | 4 | — | — | |
| Mousec | — | — | 6.9 ± 0.2 | 5 | |
| Cat | — | — | 10.1 ± 0.5 | 4 | |
| Elephant | — | — | 20.8 ± 2.4 | 3 | |
|
| |||||
| Cu-ETS | Human | 40.6 ± 1.1 | 14 | 34.4 ± 2.3 | 4 |
| Ratc | 19.3 ± 0.2 | 3 | 11.7 ± 1.5 | 4 | |
| Sheep | 21.1 ± 0.5 | 5 | 17.7 ± 0.8 | 4 | |
| Donkey | 23.5 ± 1.5 | 2 | 28.1 ± 2.0 | 5 | |
| Rabbit | 38.7 ± 1.1 | 5 | 21.1 ± 0.5 | 5 | |
| Cow | 30.5 ± 0.5 | 5 | 22.5 ± 1.0 | 5 | |
| Pigc | 42.5 ± 1.1 | 5 | 28.7 ± 1.0 | 6 | |
| Dogc | 44.2 ± 1.7 | 4 | — | — | |
| Baboon | 36.4 ± 0.4 | 3 | — | — | |
| Mousec | — | — | 14.9 ± 1.0 | 5 | |
| Cat | — | — | 12.7 ± 0.4 | 5 | |
| Elephant | — | — | 30.6 ± 2.8 | 3 | |
Corrected for nonspecific radiotracer association with the ultrafiltration membrane.
Serum albumin concentration of 40 mg/mL.
From reference 2.
Multiple Tukey standardized range tests with α = 0.05 were applied using ANOVA models generated by SAS software (SAS Institute, Cary, NC) to compare the mean free fractions [25]. [64Cu]Cu-PTSM binding to rat, baboon, sheep, and donkey serum albumins is statistically indistinguishable. In addition, the canine and porcine albumin binding values are comparable to one another, and significantly below the highest binding observed with HSA. For [64Cu]Cu-ATSM, binding did not significantly differ between rat, human, and sheep albumins, but the binding variations relative to the canine, porcine, and baboon albumins were statistically significant. Levels of unbound [64Cu]Cu-ATSM in donkey and bovine albumin solutions were similar, and slightly higher, than free tracer levels in the group that included rat, human and sheep albumins.
Overall, [64Cu]Cu-ETS binding to the albumins of these 12 species appears more uniform than the binding observed with [64Cu]Cu-PTSM and [64Cu]Cu-ATSM, but there is still evidence of subtle species-dependent binding variations. [64Cu]Cu-ETS binding was similar for canine, porcine, human, or rabbit serum albumins, and a second statistical grouping the included donkey, sheep, and rat serum albumins.
Drugs may bind to other plasma proteins in addition to albumin; therefore, measuring protein binding in serum may yield further insights for predicting drug bioavailability and its variations between species. The measured [64Cu]Cu-PTSM, [64Cu]Cu-ATSM, and [64Cu]Cu-ETS binding in human, sheep, and donkey sera indicates that binding to albumin dominates the overall protein binding in serum (Table 3). Rat serum and rat albumin also provided similar results for both [64Cu]Cu-PTSM and [64Cu]Cu-ATSM, while [64Cu]Cu-ETS binding in rat serum was slightly higher than in purified albumin solutions.
Conversely, our data (Table 3) indicate additional proteins may be present in sera of the rabbit, cow, and pig that exhibit significant additional affinity for binding these agents, manifested as higher protein binding in serum than in pure serum albumin solutions. For example, [64Cu]Cu-PTSM was 28.8 ± 1.4%-free in 40 mg/mL bovine serum albumin solution, but 50% lower (15.2 ± 1.4%-free) in serum (containing 35.8 mg/mL albumin). These findings are consistent with our previous study which showed the pig serum free fraction was ∼65% of the free fraction observed in albumin solution at equal concentration [2]. The commercial availability of porcine IgG (PIgG), allowed us to show that [64Cu]Cu-PTSM, [64Cu]Cu-ATSM, and [64Cu]Cu-ETS bind PIgG at significant, but comparable, levels [2]. Unfortunately, purified serum proteins other than albumin were not commercially available for the rabbit and cow to allow further examination of the specific protein(s) that account for the observed differences in protein binding between the serum and purified albumin solutions.
Serum Albumin Sequence Homology
To evaluate whether specific amino acid substitutions between HSA and the other mammalian albumins might explain the observed variations in albumin binding between species, we analyzed albumin amino acid sequences in the eight species for which we have 64Cu-bis(thiosemicarbazone) radiopharmaceutical binding data, as well as nine additional species for which the sequences are available. This analysis (tabulated data available in the Supplementary Material) included comparison of the proteins as a whole, and perhaps the more germane subset of amino acids comprising the IIA binding site of HSA (Figure 3) [3]. Baboon serum albumin was the only protein directly tested in this study without a completed protein sequence. The mean “% Identity Value” for all the complete sequence comparisons was 75.1 ± 5.5% (n = 136 comparisons). Considering the diversity of these species, the relatively small range of % Identity (67.7% for elephant vs. guinea pig to 100.0% identity for human vs. orangutan) demonstrates significant overall conservation of structural elements.
Of the eight serum albumins directly assayed in this project, rat serum albumin has the lowest level of overall sequence homology (73.4%) with HSA. Surprisingly, [64Cu]Cu-ATSM binding to rat serum albumin and HSA was almost identical, 5.9 ± 0.4%-free, and 6.3 ± 0.2%-free, respectively, indicating [64Cu]Cu-ATSM binding to be insensitive to sequence differences between these two species. Conversely, [64Cu]Cu-PTSM and [64Cu]Cu-ATSM binding to HSA and canine serum albumin (CSA) were the most dramatic examples of interspecies variation in this study; yet, overall sequence homology between HSA and CSA (80.1%) is among the highest values for any non-primate. The disparities between [64Cu]Cu-PTSM (4.0 ± 0.1%-free vs. 42.0 ± 1.3%-free) and [64Cu]Cu-ATSM (5.9 ± 0.4%-free vs. 36.3 ± 1.5%-free) binding to HSA and CSA, respectively, clearly indicate significant species-dependence that is not explained by comparisons of entire proteins.
Unfortunately, restricting the analysis to the amino acids associated with the IIA drug binding site (Table 4 and Supplementary Material) also does not point to a clear cause for the stronger interaction of Cu-PTSM and Cu-ATSM with HSA compared to the other albumins. Sequence conservation in the IIA site was highest for pig serum albumin, increasing incrementally from 76% to 91% as the focus of the comparison narrows to only residues lining the IIA site. No single amino acid lining the IIA site of HSA appears to account for the increased ability of HSA to bind Cu-PTSM and Cu-ATSM relative to the other albumins studied, as there are no residues in this region that are unique to HSA (Table 4).
Table 4.
Sequence homology within 22 amino acids that line the void of the IIA drug binding site of serum albumin. The amino acids that differ from those found in HSA are shown in bold typeface.
| Residue | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 150 | 195 | 199 | 211 | 214 | 215 | 218 | 219 | 222 | 223 | 234 | 238 | 241 | 242 | 257 | 260 | 261 | 264 | 287 | 290 | 291 | 292 | |
| Human | Y | K | K | F | W | A | R | L | R | F | L | L | V | H | R | L | A | I | S | I | A | E |
| Rat | Y | R | K | F | W | A | R | M | R | F | L | V | I | N | R | L | A | M | S | L | A | E |
| Sheep | Y | R | R | L | W | S | R | L | K | F | I | L | V | H | R | L | A | I | S | I | A | E |
| Donkey | Y | K | K | F | W | S | R | L | K | F | I | L | V | H | R | L | T | I | S | I | A | E |
| Rabbit | Y | Q | R | Y | W | A | R | L | R | F | I | L | V | H | R | L | A | M | A | I | Y | G |
| Cow | Y | R | R | L | W | S | R | L | K | F | L | L | V | H | R | L | A | I | S | I | A | E |
| Pig | Y | K | K | F | W | S | R | L | R | F | I | L | V | H | R | L | A | I | S | I | A | E |
| Dog | Y | K | K | F | W | S | R | L | R | F | V | L | V | H | R | L | A | M | S | L | A | E |
| Mouse | Y | R | K | F | W | A | R | L | T | F | L | L | V | N | R | L | A | M | A | L | S | E |
| Cat | Y | K | K | F | W | S | R | L | K | F | L | L | I | H | R | L | A | I | S | I | S | E |
| Elephant | Y | R | K | F | W | A | H | L | R | F | L | L | V | Y | R | L | A | I | S | I | A | E |
| Gerbil | Y | R | K | F | W | A | R | M | T | F | L | L | V | T | R | L | A | M | S | L | A | E |
| Vole | Y | K | K | F | W | A | R | M | K | F | L | L | V | T | R | L | A | M | A | L | A | E |
| Guinea Pig | Y | Q | K | F | W | S | R | L | K | F | I | L | V | T | R | L | A | M | A | I | L | E |
| Orangutan | Y | K | K | F | W | A | R | L | R | F | L | L | V | H | R | L | A | I | S | I | A | E |
| Rhesus Monkey | Y | K | K | F | W | A | R | L | K | F | L | L | V | H | R | L | A | M | S | L | A | E |
| Horse | Y | K | K | V | W | S | R | L | K | F | I | L | V | H | R | L | A | I | S | I | A | E |
Comparing the serum albumins having the highest and lowest observed [64Cu]Cu-PTSM and [64Cu]Cu-ATSM binding, these tracers consistently had the highest affinity for HSA, and the lowest affinity for pig and dog albumins (Table 3). Yet, canine and porcine serum albumins are most similar to HSA, with combined substitutions at only 4 residues in the IIA binding site. The Ile→Met substitution at residue 264, and the Ile→Leu substitution at residue 292 are unique to canine serum albumin (CSA), and these residues in pig serum albumin and HSA are identical. In HSA, Ile-292 is located in the deepest part of the binding pocket that receives the coumarin moiety of warfarin, and the replacement with structurally similar Leu in CSA would seem not likely to be solely responsible for the dramatic difference in tracer binding [23]. The Ile→Met substitution between HSA and CSA at residue 264 is slightly more dramatic. In HSA, hydrophobic Ile-264 bifurcates a sub-chamber from the main cavity. The Met-264 substitution in CSA is a less bulky residue, but comparable in terms of polarity and hydrophobicity. Given the similarly low tracer binding (especially for [64Cu]Cu-PTSM) to both pig serum albumin and CSA, an isolated substitution in only this location does not explain the variation in tracer binding to HSA.
Compared to the IIA binding site of HSA, pig serum albumin presents only two substitutions; residue 215 has an Ala→Ser substitution, and there is a Leu→Ile substitution at residue 234 (Table 4). [64Cu]Cu-PTSM and [64Cu]Cu-ATSM both have a higher affinity for HSA (only 4% and 6% free) than pig serum albumin (roughly 40% and 17% free). The Leu→Ile substitution at residue 234 is very conservative, and is not expected to greatly alter any physical properties in this region of the IIA site. At least in the warfarin-HSA adduct crystal structures (1h9z or 1ha2), Leu-234 is not actively involved in drug binding [23]. Ile is slightly more flexible, hydrophobic, and polar than Leu, but any differences are expected to be negligible. With similar hydrocarbon structures, these residues are identical in terms of sidechain bulk.
The Ala→Ser substitution common to both pig serum albumin and CSA at residue 215 (Table 4) is attractive as a single variation that might, at least in part, explain the disparity in binding relative to HSA. Crystal structures of HSA-drug adducts reveal that non-polar Ala-215 is adjacent to the lone Trp-214 in a sub-pocket of the IIA site that accepts substituent groups from several drugs [24]. Additional residues within two residues of residue 215 (Ala-213 and Ala-217) are also conserved between pig serum albumin, HSA, and CSA. The hydroxyl group at residue 215 from a polar serine may change the micro-environment in this immediate location, but there is no obvious physical mechanism involving this single substitution that explains the dramatic disruption in binding to CSA and porcine serum albumins.
The high affinity of both [64Cu]Cu-PTSM and [64Cu]Cu-ATSM for HSA appears to be unique among the species tested, but the observed interspecies variations do not appear to be readily explained by a common pattern of substitutions in the IIA site. By contrast, the albumin binding of 64Cu-ETS is relatively species-independent (Table 3).
Relationship of Chelate Structure to Albumin Binding
To further probe how chelate structure impacts albumin binding, the protein binding of 12 additional copper(II) bis(thiosemicarbazone) chelates (Figure 2 and Tables 1 and 2) was evaluated with both human and canine serum albumin. These include additional derivatives that retain the methyl substitution of the diimine fragment, as in Cu-PTSM and Cu-ATSM, as well as a variety of α-ketoaldehyde and 1,2-diketone derivatives that, like Cu-ETS, provide an increase in steric bulk surrounding the diimine carbons of the bis(thiosemicarbazone) chelates.
The previously observed discrepancies between Cu-PTSM/Cu-ATSM and Cu-ETS binding to HSA are representative of the large overall range observed for the HSA binding within this broader set of chelates. HSA binding ranged from 4% free (Cu-PTSM) to 46% free (Cu-KTS) (Tables 1 and 2). CSA binding of the same 16 chelates spans a similar range, 10% free for Cu-2,3-PnTSM to 55% free for Cu-KTS. The data in Table 1 are organized from highest HSA binding (Cu-PTSM) to lowest (Cu-KTS). Ordering the compounds based on their affinity for HSA suggests that these complexes can be grouped into two categories. One group includes Cu-PTS, Cu-PTSM, Cu-PTSM2, Cu-PTSE, Cu-ATSM, and Cu-ATS. These chelates have one or two methyl substituent groups on the diimine backbone, and show high affinities for HSA that are not apparent in the binding to canine serum albumin. The second group includes the chelates with larger substituents (-CH2CH3) or (-CH3(CH2CH3O)CH) on the diimine fragment, which do not bind with as high an affinity for HSA and do not show the great disparities in binding affinity between HSA and canine albumin. Thus, additional steric bulk at the diimine backbone appears to disrupt the specific HSA binding seen with Cu-PTSM and Cu-ATSM. Interestingly, similarly increased steric bulk in the amine functions at the other end of the planar chelate (Cu-PTSE) does not disrupt the specific binding to HSA.
Comparing some of the isomeric chelates perhaps best illustrates the finding that HSA binding is quite sensitive to steric bulk in the diimine backbone. Cu-ATS and Cu-ETS, derived from 2,3-butanedione and 1,2-butanedione, respectively, differ dramatically in their binding to HSA (13.2% free vs. 40.6% free, respectively). Differences in non-specific binding do not explain this dramatic difference, given the results for canine albumin, where we presume the interactions to be largely nonspecific. The binding of the isomeric Cu-ATSM and Cu-ETSM yields a similar conclusion; Cu-ETSM is more lipophilic than Cu-ATSM (log P = 2.7 vs. 2.2), but has a much lower affinity for HSA (21.3% free vs. 5.9% free). [64Cu]Cu-ETSM shows similar binding to HSA and CSA (21.3 ± 0.6%, and 20.3 ± 0.4% free), while [64Cu]Cu-ATSM binds to HSA much more strongly than it binds CSA (5.9 ± 0.4%, and 36.3 ± 1.5% free). Strong chelate association with HSA, but not CSA, was also observed in two previous studies of “mixed” N4-methylated bis(thiosemicarbazone) complexes containing dissimilar amine substituents, but retaining only methyl substitution about the diimine fragment [4,5].
In the four chelates derived from cyclic 1,2-diketones (Figure 2B and Table 2), steric bulk at the diimine carbon atoms would exceed that of Cu-ETS, and presumably similarly disrupt binding in the IIA site of HSA. This does seem to be the case, as none of these agents interacts more strongly with HSA than canine albumin (Table 2). However, the canine albumin binding of these four chelates, and Cu-2,3-PnTSM (Table 1), does become stronger than seen with many of the other derivatives, we believe reflecting lipophilicity-dependent non-specific binding to the protein.
To a first approximation the binding of these radiopharmaceuticals to CSA appears to be strongly influenced by lipophilicity (Figure 4A), as expected for nonspecific binding. This relationship also appears to hold for chelate binding to HSA (Figure 4B), if one excludes from consideration the subset of chelates that bind the IIA site of HSA. The measured binding to CSA is largely predictive of the measured binding to HSA, as might be expected for non-specific binding, if one excludes from the analysis the subset of agents appearing to also specifically bind in the IIA site of HSA (Figure 5). However, analysis as in Figure 5 alone does not prospectively allow distinction between the agents that exhibit high binding to HSA via the IIA site, and those that simply exhibit high, largely lipophilicity-dependent, non-specific binding to both HSA and CSA.
Figure 4.
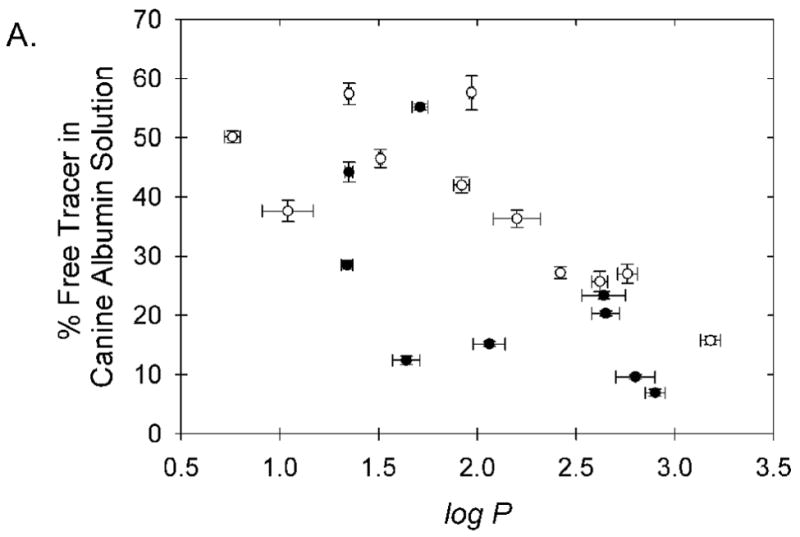
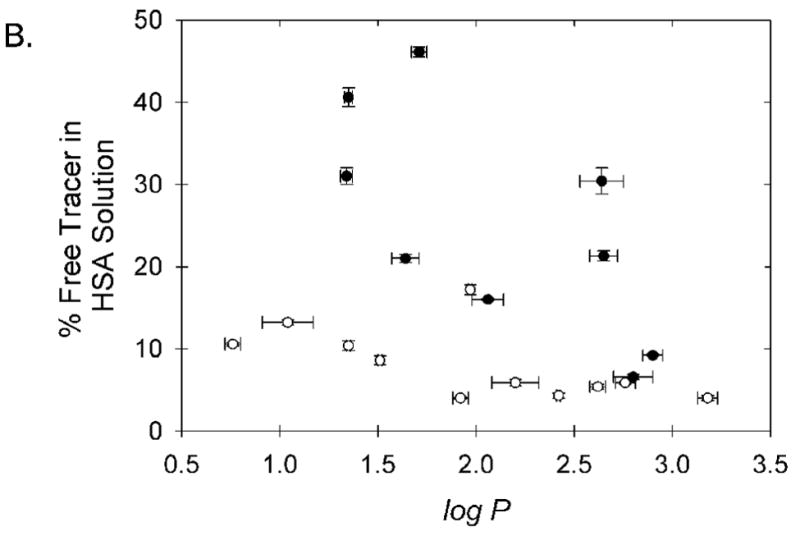
Relationship between chelate lipophilicity and observed binding to (A) canine serum albumin and (B) human serum albumin. The six chelates believed to bind in the high affinity IIA site of HSA; Cu-PTS, Cu-ATS, Cu-ATSM, Cu-PTSM, Cu-PTSE, and Cu-PTSM2, are shown with open symbols. The displayed error bars represent ±1 standard deviation about the mean measured values.
Figure 5.
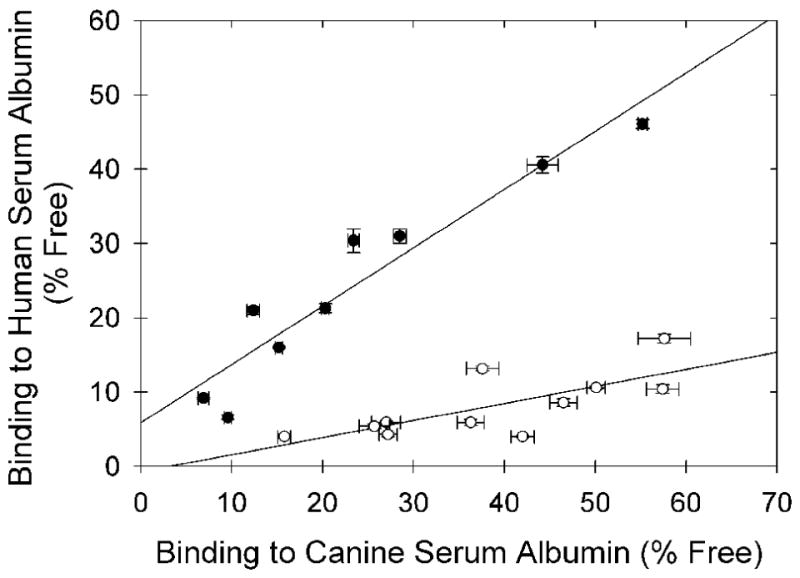
Correlation between the binding to human and canine serum albumin for the 15 copper(II) bis(thiosemicarbazone) chelates shown in Tables 1 and 2, and five additional mixed bis(thiosemicarbazone) chelates from references 4 and 5. The open dots are the data for the subset of eleven chelates believed to exhibit high affinity for the IIA site of HSA, and sharing the common structural feature of presenting only methyl (-CH3) or -H substituents at the carbon atoms of the diimine backbone. These data are fit by the line: y = 0.23x - 0.72; r2 = 0.54 The solid dots are the data for the remaining chelates, containing bulkier substituents on at least one imine carbon atom of the ligand backbone, and are fit by the line: y = 0.79x + 5.9; r2 = 0.91.
Assuming the association of these chelates with canine albumin to largely represent non-specific binding, the HSA binding data can be normalized to potentially remove the contributions of lipophilicity-dependent, non-specific binding through calculation of the ratio of free tracer in HSA solution to free tracer in canine albumin solution (Figure 6). The subset of copper(II) bis(thiosemicarbazone) tracers exhibiting high and specific affinity for HSA, attributed to binding in the IIA site, are readily and consistently identified in this plot (Figure 6), and share the common structural feature of no substituent larger than a methyl group on the imine carbon atoms.
Figure 6.
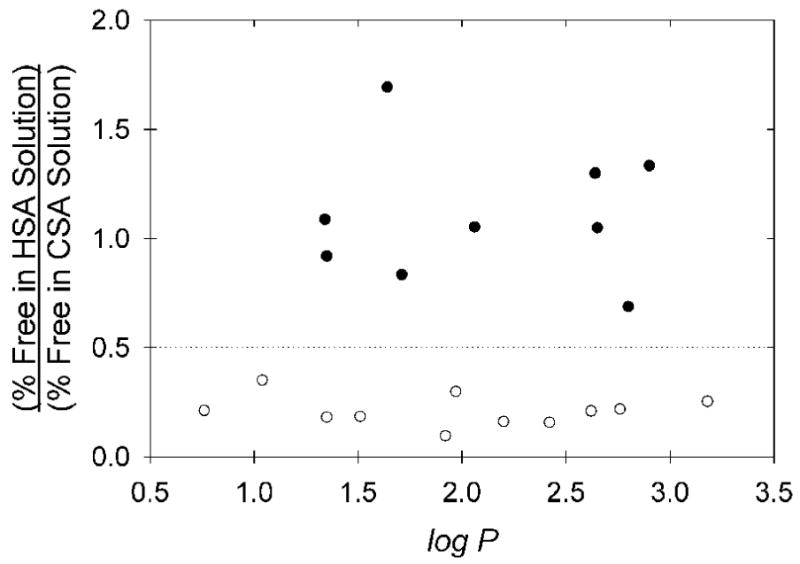
Ratio of measured HSA binding to CSA binding for the various copper(II) bis(thiosemicarbazone) radiopharmaceuticals, including values for the five pyruvaldehyde-derived “mixed” bis(thiosemicarbazone) copper(II) radiopharmaceuticals previously reported [4,5]. The dashed line (HSA/CSA Ratio = 0.5) is an apparent threshold above which binding to HSA is believed to be entirely non-specific, and largely approximated by the binding to CSA. The common structural feature of the eleven chelates below the dotted line, all appearing to bind with high affinity in the IIA site of HSA, is the presence of only methyl (-CH3) or -H substituents at the carbon atoms of the diimine backbone. The HSA/CSA binding ratios for these eleven chelates are less than from those of the nine chelates with bulkier substituents with P = 0.0001 (Mann-Whitney test).
Qualitative inspection of the fit of Cu-PTSM vs. Cu-ETS into the crystallographically-defined IIA site of HSA does not reveal an obvious structural basis for the observed selectivity of HSA in binding the pyruvaldehyde and diacetyl-derived chelates, as adequate volume seemingly exists to accommodate both agents. Quantitative analysis the physicochemical forces impacting protein binding of these chelates in the IIA site is not possible, at present, as the tools for modeling the solvent accessible surface of the copper chelates are not available as they are for organic molecules. Crystallographic characterization of the HSA-Cu-PTSM adduct can be expected to provide a clear definition of the molecular nature of the Cu-PTSM/Cu-ATSM interaction with the IIA site of HSA; efforts are underway to obtain suitable protein crystals to allow such an analysis.
From this in vitro study it is not clear whether extensive non-specific protein binding, intrinsic to the highly lipophilic tracers, will alter tracer distribution in vivo. In humans, the first-pass tissue extraction of Cu-PTSM in high-flow tissue is clearly affected by the uniquely high affinity of the tracer for HSA. However, high binding that is restricted to non-specific interactions may be less significant, if the rate of dissociation is significantly greater than the rate of dissociation from the specific IIA binding site. The reported similarity in rat brain uptake of (R)TSM and (R)TSM2 chelates argues that lipophilicity-dependent non-specific binding does not appreciably alter first pass extraction in the fashion seen when agents bind the IIA site of HSA [16].
Conclusions
The species-dependent specific binding of the Cu-PTSM and Cu-ATSM radiopharmaceuticals to human serum albumin does not appear to be readily explained by variations in a discrete amino acid associated with the IIA drug binding site of albumin. A molecular understanding of the nature of the strongly compound-dependent interaction of these chelates with the IIA binding site will seemingly need to await crystallographic characterization of the HSA adducts of these drugs. However, strong copper bis(thiosemicarbazone) interaction with the IIA site of HSA does appear to have some clear structural requirements, with strong specific HSA binding appearing to be disrupted when substituents bulkier than a methyl group appear on the imine carbons of the ligand backbone. Chelates such as Cu-ETSM and Cu-KTSM may merit further investigation for use as radiopharmaceuticals for perfusion imaging, including application for evaluation of cerebral blood flow, since these agents do not selectively bind HSA and readily penetrate the blood-brain barrier [16].
Supplementary Material
Acknowledgments
This work was supported by a research grant from the Purdue Research Foundation, and R01-CA092403. Development of the production of Cu-64 at Washington University School of Medicine was supported by the NCI grant R24 CA86307. Special thanks to Dr. Susan Mikota of Elephant Care International for the elephant serum.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mathias CJ, Bergmann SR, Green MA. Species-dependent binding of copper(II) bis(thiosemicarbazone) radiopharmaceuticals to serum albumin. J Nucl Med. 1995;36:1451–5. [PubMed] [Google Scholar]
- 2.Basken NE, Mathias CJ, Lipka AE, Green MA. Species dependence of [64Cu]Cu-bis(thiosemicarbazone) radiopharmaceutical binding to serum albumins. Nucl Med Biol. 2008;35:281–6. doi: 10.1016/j.nucmedbio.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basken NE, Mathias CJ, Green MA. Elucidation of the human serum albumin (HSA) binding site for the Cu-PTSM and Cu-ATSM radiopharmaceuticals. J Pharm Sci. doi: 10.1002/jps.21570. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackerman LJ, West DX, Mathias CJ, Green MA. Synthesis and evaluation of copper radiopharmaceuticals with mixed bis(thiosemicarbazone) ligands. Nucl Med Biol. 1999;26:551–4. doi: 10.1016/s0969-8051(99)00020-7. [DOI] [PubMed] [Google Scholar]
- 5.Kim JL, Mathias CJ, Green MA. Mixed bis(thiosemicarbazone) ligands for the preparation of copper radiopharmaceuticals; synthesis and evaluation of tetradentate ligands containing two dissimilar thiosemicarbazone functions. J Med Chem. 1997;40:132–6. doi: 10.1021/jm9605703. [DOI] [PubMed] [Google Scholar]
- 6.Sudlow G, Birkett DJ, Wade DN. The characterization of two specific drug binding sites on human serum albumin. Mol Pharmacol. 1975;11:824–32. [PubMed] [Google Scholar]
- 7.Sudlow G, Birkett DJ, Wade DN. Further characterization of two specific drug binding sites on human serum albumin. Mol Pharmacol. 1976;12:1052–61. [PubMed] [Google Scholar]
- 8.Kragh-Hansen U, Chuang VTG, Otagiri M. Practical aspects of the ligand-binding and enzymatic properties of human serum albumin. Biol Pharm Bull. 2002;25(6):695–704. doi: 10.1248/bpb.25.695. [DOI] [PubMed] [Google Scholar]
- 9.Tillement JP, Houin G, Zini R, Urien S, Albengres E, Barré J, Lecomte M, D'Athis P, Sebille B. Advances in Drug Research. Vol. 13. London: Academic Press; 1984. The binding of drugs to blood plasma macromolecules: Recent advances and therapeutic significance; pp. 59–94. [Google Scholar]
- 10.Sjöholm I, Ekman B, Kober A, Ljungstedt-Pahlman I, Seiving B, Sjödin T. Binding of drugs to serum albumin XI: The specificity of three binding sites as studied with albumin immobilized in microparticles. Mol Pharmacol. 1979;16:767–77. [PubMed] [Google Scholar]
- 11.Lenter C, editor. Geigy Scientific Tables. CIBA GEIGY; 1984. pp. 136–9. [Google Scholar]
- 12.Peters T. All about Albumin. San Diego (Calif): Academic Press; 1994. [Google Scholar]
- 13.Panjehshahin MR, Yates MS, Bowmer CJ. A comparison of drug binding sites on mammalian serum albumins. Biochem Pharmacol. 1992;44(5):873–9. doi: 10.1016/0006-2952(92)90118-3. [DOI] [PubMed] [Google Scholar]
- 14.Robertson A, Karp W, Broderson R. Comparison of the binding characteristics of serum albumins from various animal species. Dev Pharmacol Ther. 1990;15:106–111. doi: 10.1159/000457629. [DOI] [PubMed] [Google Scholar]
- 15.Kosa T, Maruyama T, Otagiri M. Species differences of serum albumins I: drug binding sites. Pharm Res. 1997;14(11):1607–12. doi: 10.1023/a:1012138604016. [DOI] [PubMed] [Google Scholar]
- 16.John EK, Green MA. Structure-activity relationships for metal-labeled blood flow tracers: comparison of keto aldehyde bis(thiosemicarbazone)copper(II) derivatives. J Med Chem. 1990;33:1764–70. doi: 10.1021/jm00168a035. [DOI] [PubMed] [Google Scholar]
- 17.http://www.elephants.com
- 18.Casella G, Berger R. Statistical Inference. Pacific Grove (Calif): Duxbury; 2002. pp. 240–5. [Google Scholar]
- 19.The UniProt Consortium. The universal protein resource. Nucleic Acids Res. 2007;35:D193–7. doi: 10.1093/nar/gkl929. http://www.uniprot.org. [DOI] [PMC free article] [PubMed]
- 20.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larkin MA, Blackshields G, Brown NP, et al. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 22.Curry S, Mandelkow H, Bricks P, Franks N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat Struct Mol Biol. 1998;5:827–35. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- 23.Petitpas I, Bhattacharya AA, Twine S, East M, Curry S. Crystal structure analysis of warfarin binding to human serum albumin. J Biol Chem. 2001;276:22804–9. doi: 10.1074/jbc.M100575200. [DOI] [PubMed] [Google Scholar]
- 24.Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, Curry S. Structural basis of the drug-binding specificity of human serum albumin. J Mol Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 25.SAS, version 9.1.3. Cary (NC): SAS Institute Inc; 20022003. [Google Scholar]
- 26.Fujita T, Iwasa J, Hansch C. A new substituent constant, Π, derived from partition coefficients. J Am Chem Soc. 1964;86:5175–80. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


