Abstract
We describe a highly efficient use of lentiviral validation-based insertional mutagenesis (VBIM) to generate large populations of mammalian cells in which a strong promoter is inserted into many different genomic loci, causing greatly increased expression of downstream sequences. Many different selections or screens can follow, to isolate dominant mutant clones with a desired phenotypic change. The inserted promoter can be excised or silenced at will, to prove that the insertion caused the mutation. Cloning DNA flanking the insertion site identifies the locus precisely. VBIM virus particles are pseudotyped with VSV G protein, allowing efficient infection of most mammalian cell types, including non-dividing cells, and features are included that give high yields of stable virus stocks. In several different selections, useful mutants have been obtained at frequencies of approximately 10−6 or higher. We used the VBIM technique to isolate mutant human cells in which the F-box leucine-rich protein 11 (FBXL11), a histone H3K36 demethylase, is shown to be a negative regulator of NFκB. High levels of FBXL11 block the ability of NFκB to bind to DNA or activate gene expression, and siRNA-mediated reduction of FBXL11 expression has the opposite effects. The H212A mutation of FBXL11 abolishes both its histone H3K36 demethylase activity and its ability to inhibit NFκB. Thus, we have used a powerful tool for mutagenesis of mammalian cells to reveal an aspect of the complex regulation of NFκB-dependent signaling.
Keywords: Cre recombinase, lentivirus, promoter insertion
Genetic approaches to investigate signaling mechanisms fall into two broad categories: In forward genetics one creates random mutations in a population of cells, whereas in reverse genetics one manipulates a known gene. Both approaches have played critical roles in revealing the depth and complexity of mammalian signal transduction pathways. It is fair to say that only in recent years have we begun to comprehend the depth, breadth, and complexity of these pathways and of their interrelationships. Many of the intricate networks that provide sophisticated regulation of signaling pathways would have been very difficult to recognize or understand in the absence of powerful genetic techniques. Forward genetics seeks to associate a specific protein with a biological phenotype in a pathway of interest without the need to rely upon any previous knowledge. In a typical screen, one creates cell libraries containing millions of random mutations, applies selection or screening to isolate rare cells in which the targeted phenotype has been altered, identifies the mutated gene or gene product, and characterizes the function of the altered, over-expressed, or missing protein. In forward genetics, mutations can be induced by using chemical mutagens, insertional mutagens or through the delivery of diverse libraries containing, for example, cDNAs, shRNAs, or genetic suppressor elements, with each approach creating distinct genetic changes (1–4).
In one type of forward genetic screen, a defined DNA fragment is inserted into many different loci. If the inserted DNA includes a strong promoter, dominant mutants can be obtained by driving transcription into an adjacent gene, leading to the over-expression of an mRNA encoding a full-length or truncated protein, or an antisense RNA, depending the position and orientation of the inserted promoter. These dominant mutations can potentially identify either positive or negative regulators.
Previously, we described vectors for reversible promoter insertion, derived from murine leukemia virus (MLV), that were designed first to create promoter-dependent mutants and then to distinguish them from spontaneous mutants by removing the inserted promoter to prove that the mutant phenotype depends upon its function (5). However, this method does not allow the generation of high titers of viruses because the minimal CMV promoter is placed in an orientation opposite to that of the 5′ long terminal repeats (LTRs). Furthermore, target cells needed to be further modified if human cells were to be infected with mouse viruses. To improve the features of the previous reversible promoter insertional technique, we designed a set of lentiviral validation-based insertional mutagenesis (VBIM) vectors that extend applications to nearly any mammalian cell, even cells that are not dividing. The VBIM vectors allow one to generate high titers of viruses and allow the mutant phenotype to be reversed, thus validating that the insertion caused the mutation, either by removing the inserted promoter with Cre recombinase or by silencing it by expressing the Kruppel-associated box (KRAB) domain of the human Kox1 zinc finger protein. Following validation, each selected phenotype can be associated with a specific target gene after the insertion site has been cloned and sequenced.
We have used the VBIM system to identify regulators of nuclear factors κB (NFκB), central coordinators of innate and adaptive immune responses. There are five NFκB family members in mammals, RelA (p65), RelB, c-Rel, NFκB1 (p50), and NFκB2 (p52). The p65/p50 heterodimer functions most often in the so-called classical signaling pathway (6). Constitutive NFκB activation is frequently associated with a number of pathological conditions, including inflammation and cancer (7) and is well known to be involved in tumor angiogenesis and invasiveness (7). Loss of the normal regulation of NFκB is a major contributor to the deregulated growth, resistance to apoptosis and propensity to metastasize observed in many different cancers (8). Our data, showing NFκB activation in many cancer-derived cell lines (9, 10), further support the strong correlation between constitutive NFκB activation and cancer. In using VBIM vectors to identify positive and negative regulators of NFκB, we found that F-box leucine repeat rich protein 11 (FBXL11), identified in two independent mutant clones, is a potent negative regulator, vividly demonstrating the utility of the VBIM technique as a powerful tool for gene discovery.
Results
Creation and Characterization of the VBIM Vectors.
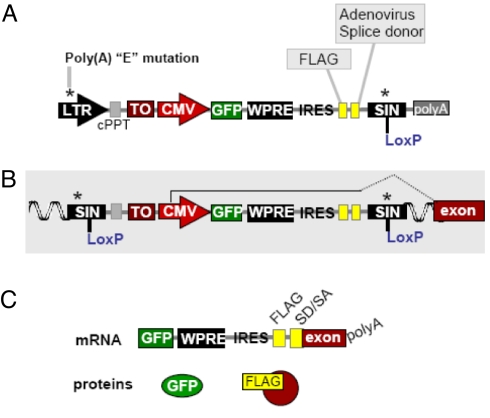
To improve the features of the MLV method (5), as shown in Fig. 1, we made four major changes, (1) replacing the MLV backbone with a lentiviral backbone, to enable the VBIM technique to be applied in any cell system, including non-dividing cells, (2) replacing the minimal CMV promoter with a much stronger full-length CMV promoter, (3) mutating the poly(A) sequence in the 3′ and 5′ LTRs, to allow the full length CMV promoter to be placed in a forward orientation, thus greatly increasing the virus titer, and (4) including a deletion to eliminate 3′ LTR promoter activity (self-inactivating, or SIN). Thus, the VBIM vectors have a number of elements to increase the efficiency of inducing and identifying dominant mutations. It is imperative to distinguish spontaneous mutants from those caused by targeted mutation. For this reason, a LoxP site in the 3′ LTR of the VBIM vectors permits excision of all but 238 nucleotides of inert proviral LTR sequence, allowing the promoter to be removed so that the dependence of the mutant phenotype on the presence of the promoter can be tested. The activity of the CMV promoter can be regulated by an upstream tet-operator (TO) element, following introduction of the TR-KRAB fusion protein, which binds to this element in a doxycycline-dependent manner to suppress the function of the adjacent promoter. An internal GFP reporter gene is under the control of the promoter and an internal ribosome entry site (IRES) downstream of GFP allows translation of the cellular protein encoded downstream of the insertion site. The protein product is tagged at its N terminus by a Flag epitope. A splice donor (SD) is provided and three different reading frames of Flag-SD are present in separate constructs, called VBIM-SD1, 2, and 3, so that each splice acceptor site in the genome can potentially function to allow the synthesis of a Flag-tagged fusion protein. Extensive data on the genesis and testing of the VBIM vectors are provided in SI Text. In Fig. S1, the structures of reverse and forward insertional mutagenesis vectors are compared, to demonstrate the benefit of forward orientation of the promoter. As shown in Fig. S2, a polyA mutation was successfully generated in the LTR of the lentiviral vector, and this vector can generate titers of virus similar to the high titers obtained with the wild-type LTR (Fig. S3). In addition, virus titers and infection efficiencies were compared for both the VBIM forward and reverse orientation promoter insertions, showing that the forward orientation vector can generate much higher titers of viruses and produced many more infected cells, as indicated by GFP expression (Fig. S4).
Fig. 1.
VBIM lentiviral vectors. (A) The VBIM vector backbone with all of the components of the promoter insertion complex. (B) Structure of the provirus following integration, including splicing into a neighboring exon. (C) Transcription from the CMV promoter produces a single mRNA encoding GFP and a FLAG/cellular fusion protein, if splicing is correct. Three separate constructs (SD1, -2, and -3) incorporate the three different reading frames. Abbreviations: CMV, cytomegalovirus promoter; cPPT, central polypurine tract; GFP, green fluorescent protein; IRES, internal ribosome entry sequence; Lox P, site for Cre-mediated recombination; LTR, long terminal repeat; SA, splice acceptor site; SD, splice donor site; SIN, self-inactivating LTR; TO, tetracycline operon; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element.
Use of VBIM Vectors To Obtain Mutants with Constitutive Activation of NFκB.
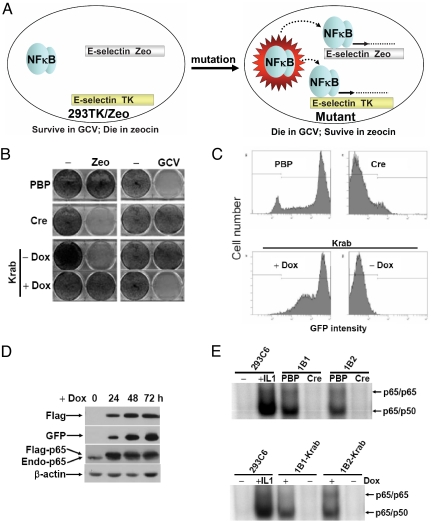
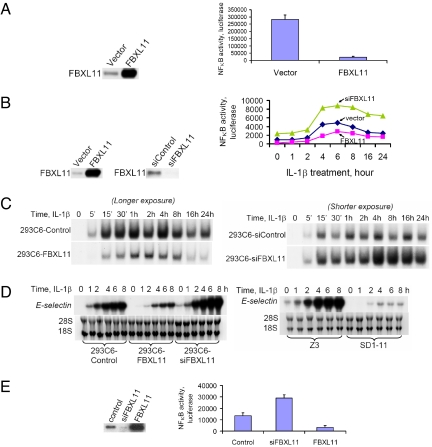
As a proof of principle, we performed a small scale of experiment to obtain mutants with constitutive activation NFκB from 293C6 cells. These cells have integrated NFκB-dependent promoters that drive the expression of thymidine kinase and a protein that degrades zeocin when NFκB is activated (11). Since the endogenous activity of NFκB is low in 293C6 cells, they are resistant to gancyclovir (GCV), which is a substrate of thymidine kinase, and sensitive to zeocin, i.e., the cells survive in GCV and die in zeocin (Fig. 2A, left panel). A mutation that drives constitutive activation of NFκB would switch these selections to GCV sensitivity and zeocin resistance (Fig. 2A, right panel). About 6 × 106 cells were infected with VBIM viruses generated from the SD1, –2, and –3 constructs. Three reversible clones were obtained at a frequency of approximately 5 × 10−7. In this experiment, about 90% of the 293C6 cells were positive for GFP expression, measured by FACS analysis. The selected reversible clones were further analyzed by the western method, demonstrating that Flag-tagged fusion proteins were produced abundantly and that all of the clones were GFP positive (Fig. 2 C and D). Following the expression of Cre, the promoter-dependent mutants were zeocin-sensitive and GCV-resistant and had no detectable κB DNA binding activity (Fig. 2A left panel, B, and E). GFP expression was also diminished to background levels (Fig. 2C). Southern analysis of control and Cre-expressing cells confirmed that the proviral DNA has been excised efficiently. To determine whether TR-Krab could fully repress the inserted promoters, cells were infected with pLV-CMV-TR-Krab and plated into medium with or without doxycycline, together with either GCV or zeocin. Cells expressing TR-Krab without doxycycline (in which TR-Krab is fully functional) behaved like wild-type 293C6 cells; they were resistant to GCV and lost κB-dependent DNA binding activity (Fig. 2 B and E). Additionally, expression of GFP and Flag/cellular fusion proteins (Fig. 2D) correlated with the functionality of TR-Krab (Fig. 2 C and D). Addition of doxycycline resulted in a time-dependent induction of Flag-tagged cellular fusion proteins and GFP (Fig. 2D), leading again to the mutant phenotype (Fig. 2 B and E, bottom panel). Validated mutants were obtained for each of the three SD reading frames, confirming that all of these vectors function as expected. In the mutants described in Fig. 2, the Flag-tagged fusion protein was identified as the NFκB subunit p65 (Fig. 2D). Therefore, we fully confirmed that the VBIM vectors and its Cre and doxycyclin/TR-Krab reversion system function as expected.
Fig. 2.
Use of VBIM forward vectors to generate mutants with constitutive activation of NFκB. (A) The principle of the 293TK/Zeo (293C6) cell screening system to screen for positive regulator of NFκB. (B) 293C6 cells infected with the three VBIM vectors were selected with zeocin. Mutant clones were infected subsequently with lentiviral vectors encoding Cre or TR-Krab, assayed with or without Dox, to demonstrate phenotypic reversion, shown by methylene blue staining for survival in Zeo or GCV. Cre, Cre recombinase; Dox, doxycycline; PBP, pBabepuro vector. (C) FACS analysis for GFP expression demonstrating that Cre or Krab excises or represses the promoter insertion cassette, resulting in the loss of GFP expression from the majority of the cells. For Krab-expressing cells, addition of Dox reversed the phenotype. (D) Western analysis of mutant 1B2, which has a promoter insertion in the gene encoding the p65 subunit of NFκB, expressing TR-Krab, showing loss of expression of both the Flag-tagged fusion protein and GFP in the absence of Dox, and induction after adding Dox for different times. (E) EMSA with an NFκB-specific probe, demonstrating reversion of NFκB DNA-binding activity by Cre or TR-Krab, with and without Dox. Mutants 1B1 and 1B2 both contain promoter insertions in the p65 gene.
SD1–11 Is a Reversible Mutant of Z3 Cells.
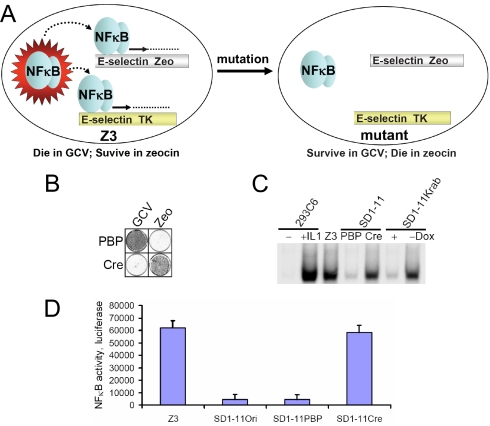
To screen for negative regulators of NFκB, we used Z3 cells, in which NFκB is constitutively active (12). These cells were obtained following chemical mutagenesis and selection for zeocin resistance of parental 293C6 cells (Fig. 2A, left panel). Z3 cells survive in zeocin and die in GCV (Fig. 3A, left panel). An additional mutation of Z3 cells that causes the constitutive NFκB activity to shut down can be selected for by requiring the cells to survive in GCV and die in zeocin (Fig. 3A, right panel). In total, about 3 × 106 Z3 cells were infected with the three VBIM viruses and selected in GCV for suppression of NFκB activity. Three reversible mutants were obtained, at a frequency of approximately 10−6. Mutant SD1–11 cells were infected with a control pBabepuro (PBP) vector or a vector encoding Cre. SD1–11PBP survived in GCV and died in zeocin (Fig. 3B), whereas SD1–11Cre died in GCV and survived in zeocin (Fig. 3B). This phenotype was confirmed in an EMSA assay (Fig. 3C) in which the ability of NFκB to bind to DNA was seen to be low in SD1–11PBP cells and high in SD1–11Cre cells. The reversibility of the SD1–11 phenotype was also confirmed by infection with TR-Krab, where the ability of NFκB to bind to DNA is low in the presence of doxycycline and high in its absence (Fig. 3C). A luciferase assay confirmed that SD1–11PBP has low NFκB activity and that SD1–11Cre has reverted to a level of NFκB similar to that of Z3 cells (Fig. 3D). Collectively, these experiments confirm that SD1–11 is a reversible mutant of Z3.
Fig. 3.
SD1–11 is a reversible mutant of Z3 cells. (A) The principle of the Z3 cell screening system to screen for negative regulator of NFκB. (B) Drug sensitivity test, showing that SD1–11 survives in GCV and dies in zeocin and that this phenotype is reversed by Cre. (C) EMSA assay to verify the reversibility of SD1–11, either by Cre or TR-Krab. Samples from 293C6 cells treated with IL-1β or Z3 cells were used as positive controls. (D) Luciferase assay of NFκB activity, showing that SD1–11Ori and SD1–11PBP have low NFκB activity, while SD1–11Cre has high NFκB activity. Ori, original. The results of triplicate luciferase assays are shown as means ± SD.
Identification of FBXL11 as a Negative Regulator of NFκB.
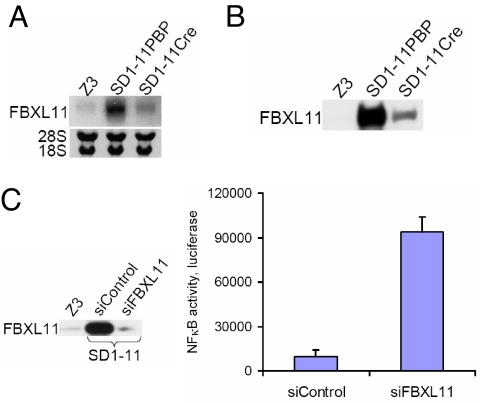
To identify the gene responsible for the phenotype of mutant SD1–11, a Southern experiment was carried out using a fragment from the VBIM vector as the probe, revealing a single band complementary to the VBIM vector which disappeared after introducing Cre. A two-step inverse PCR (iPCR) reaction to clone sequences flanking the single insertion site identified the gene as FBXL11. The insertion in the second intron of FBXL11 produces a Flag-tagged fusion protein lacking only 14 of 1,162 amino acid residues at its N terminus, leaving all of the functional domains intact. Analysis of total RNA (Fig. 4A) or protein (Fig. 4B) confirmed that FBXL11 is over-expressed in SD1–11 cells and that its high expression is reversed by Cre. Furthermore, using small interfering RNA (siRNA) to knock down the expression of FBXL11 (Fig. 4C, left panel) in SD1–11 cells increased NFκB activity (Fig. 4C, right panel). In the independent reversible mutant clone SD3–15-1, the insertion is also in the forward orientation in intron 2 of FBXL11, near the beginning of the intron. The third independent reversible mutant clone is still being characterized.
Fig. 4.
Identification of FBXL11 as a negative regulator of NFκB. (A) Northern assay showing that the high mRNA level of FBXL11 in SD1–11PBP control cells is decreased in SD1–11Cre cells. (B) Western analysis, showing that FBXL11 is over-expressed in SD1–11PBP, and decreased in SD1–11Cre. (C) Left panel, Western assay showing the expression levels of FBXL11 in SD1–11 cells and in cells in which its expression was knocked down with siRNA. Right panel, luciferase assay, showing that knocking the expression of FBXL11 down in SD1–11 cells increased NFκB activity. The results of triplicate luciferase assays are shown as means ± SD.
Confirmation of FBXL11 as a Negative Regulator of NFκB.
To confirm its function, we stably over-expressed full-length FBXL11 from a cDNA in a pool of Z3 cells (Fig. 5A, left panel). A κB-luciferase reporter was transfected transiently into either control or Z3-FBXL11 cells, and luciferase activity was determined and normalized to β-galactosidase activity. The data show that the over-expression of FBXL11 greatly decreased NFκB activity (Fig. 5A, right panel). Moreover, NFκB activity was affected as expected when FBXL11 was stably over-expressed, or down-regulated with siRNA, in 293C6 cells (Fig. 5B, left panels). As assayed by luciferase assay, upon treatment with IL-1β, NFκB activity was increased in siFBXL11 cells and decreased in cells over-expressing FBXL11 (Fig. 5B, right panel). Moreover, both EMSA and Northern assays indicated that the over-expression of FBXL11 decreased both the ability of NFκB to bind to DNA (Fig. 5C, left bottom panel) and the expression of the typical endogenous NFκB target gene E-selectin (Fig. 5D, left top panel). In siFBXL11 cells, the opposite effects were observed (Fig. 5C, right bottom panel, and D, left top panel). Similarly, in mutant SD1–11 cells, in which the expression of FBXL11 is high, the level of E-selectin is much lower than in Z3 cells (Fig. 5D, right top panel). Therefore, we have confirmed that FBXL11 is a negative regulator of NFκB in 293 cells. To test the generality of the effect of FBXL11 in different cells, we stably over-expressed it or knocked its expression down in human colon cancer HT29 cells (Fig. 5E, left panel), in which NFκB is constitutively activated (9), again finding that NFκB activity is decreased when FBXL11 expression is high and increased when it is low (Fig. 5E, right panel). Collectively, these data confirm that FBXL11 is a potent negative regulator of NFκB.
Fig. 5.
Confirmation of FBXL11 as a negative regulator of NFκB. (A) Increased expression of FBXL11 in Z3 cells inhibits NFκB activity. Left panel, Western analysis showing the stable over-expression of FBXL11 in Z3 cells. Right panel, reduced luciferase activity for NFκB in Z3 cells over-expressing FBXL11. (B) Effects of different FBXL11 levels on NFκB activity in 293C6 cells. Left panels, Western analyses showing over-expression or siRNA-mediated knock down of FBXL11 in 293C6 cells. Right panel, luciferase assay of NFκB activity in 293C6 cells treated with IL-1β for different times. Increased expression of FBXL11 inhibits NFκB and reduced expression increases NFκB activity. (C) EMSA assay, showing that increased stable expression of FBXL11 decreases the ability of NFκB to bind to DNA (left panel), and that reduced expression of FBXL11 increases NFκB DNA binding activity (right panel). (D) Northern analyses, showing the effect of FBXL11 on expression of endogenous E-selectin in response to IL-1β. (E) FBXL11 expression levels affect NFκB activity in colon cancer HT29 cells. Left panel, Western analysis showing increased or reduced expression of FBXL11. Right panel, luciferase assay of NFκB activity showing that increased expression of FBXL11 inhibits NFκB and that reduced expression increases NFκB activity. Results of triplicate luciferase assays are shown as means ± SD.
The Histone H3K36 Demethylase Activity Is Essential for the NFκB Inhibitory Function of FBXL11.
To test whether FBXL11 functions upstream or downstream of the IKK complex, we performed a western analysis, comparing 293C6 with the same cells stably over-expressing FBXL11 (Fig. S5). Upon treatment with IL-1β, the expression of FBXL11 did not affect either the phosphorylation of IκBα or its degradation and resynthesis significantly, indicating that FBXL11 functions downstream of IKK.
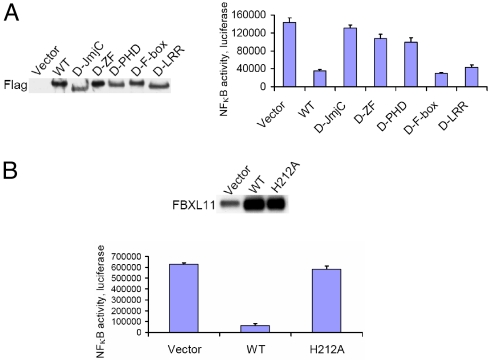
In addition to an F-box, FBXL11 contains a JmjC domain, a CxxC zinc finger, a PHD domain, and three leucine-rich repeats. FBXL11 demethylates histone H3K36, and the JmjC domain is required for this function (13). Expression of deletion constructs of FBXL11 in Z3 cells (Fig. 6A, left panel) revealed that the JmjC, zinc finger, and PHD domains are all required to suppress NFκB activation (Fig. 6A, right panel). These three domains are all necessary for the histone demethylase activity of FBXL11 (13). The H212A mutation of FBXL11 abolishes its histone demethylase activity completely (13). When we stably over-expressed the H212A mutant of FBXL11 in Z3 cells (Fig. 6B, top panel), its ability to inhibit NFκB was lost (Fig. 6B, bottom panel), demonstrating that the histone demethylase activity of FBXL11 is essential for this function.
Fig. 6.
The histone H3K36 demethylase activity is essential for the NFκB inhibitory function of FBXL11. (A) Domains required for NFκB negative regulatory function of FBXL11. Left panel, Western analysis of the expression of different Flag-tagged FBXL11 deletions in Z3 cells. Right panel, luciferase assays, showing the effects of different FBXL11 forms on NFκB activity. Abbreviation: D, deleted construct. (B) Histone H3K36 demethylase activity is essential for the NFκB inhibitory function of FBXL11. Top panel, Western analysis of the stable expression of wild-type (WT) FBXL11 or the H212A mutant in Z3 cells. Bottom panel, luciferase assay of NFκB activity, showing that the H212A mutant fails to inhibit NFκB activity in Z3 cells.
Discussion
The VBIM lentiviruses are designed to induce dominant changes in the expression of genomic sequences neighboring the insertion sites, with predicted changes that include the high-level expression of full-length or truncated proteins, microRNAs, or antisense RNAs. These diverse mutations can identify either positively or negatively acting factors from the same genetic screen, without the complications associated with technologies that use, for example, cDNA or shRNA libraries. The VBIM vectors use a lentiviral backbone with polyadenylation mutations in the 5′ and 3′ LTRs and a lox P site in the 3′ LTR. This design permits excision of all but 238 bp of inert proviral DNA, lacking both promoter activity and polyadenylation signals, following cleavage by Cre. The polyadenylation mutations also permit the CMV promoter to be placed so that it drives transcription in the same direction as transcription from the 5′ LTR during virus packaging. This important design feature allows in virus titers that are comparable to those obtained with standard lentiviral vectors (Fig. S3), eliminating promoter conflicts that occur with alternative designs (Fig. S4). Eliminating promoter interference also permits the use of a full-length CMV mutagenic promoter rather than a minimal tetracycline-regulated promoter, which requires the transactivating protein tTA to be present in the target cells (5). Thus, primary and even differentiated or senescent cells can be mutagenized without prior manipulation to express tTA or an ecotropic receptor (for adequate infection), as previously required for promoter insertion screens (5). Additionally, the VBIM vectors have a tetracycline-dependent operator upstream of the full-length CMV promoter, allowing tetracycline-regulated control of the mutant phenotype through the use of a TR-Krab fusion protein (14), providing an alternative means of validation.
While MLV-based insertional mutagenesis has a long history in gene discovery, we understand HIV-based insertion less well. Recent studies comparing MLV and HIV integration sites suggest that each virus does have an integration bias. For the purpose of promoter insertion in forward genetics, this difference raises the useful possibility that different targets may be identified from the same screen simply by using vectors derived from both viruses. MLV preferentially targets promoter regions, which may explain the tendency of MLV-based insertional mutagenesis to tag oncogenes rather than tumor suppressors, as the promoter insertion products created by MLV will favor over-expression of full-length or nearly full-length proteins (15, 16). In contrast, HIV integrations occur throughout coding regions, avoiding the CpG islands that are found in and near many promoters (15). This pattern of integration should increase the probability of generating truncated proteins and antisense transcripts, since insertion would occur more frequently downstream of initiation codons, with equal chances of integrating in either orientation. Integrations in both directions increase the likelihood of identifying genes that negatively affect the phenotype of interest. Our data from different screens has already yielded one revertible clone in which the insertion is in an antisense direction. Importantly, each mutation generated by promoter insertion will be dominant, since the product of promoter-driven expression is present in increased abundance, and thus the mutant phenotype will be evident in diploid cells. In addition, since the desired mutants can be identified by eliminating or silencing the inserted promoters, spontaneous mutants can be eliminated from consideration promptly, making the early steps of screening more efficient. In several additional different screens, including a screen for oncogenes that participate in the transformation of normal human mammary epithelial cells, we obtained one clone from 2.4 × 105 cells, with a frequency of validated mutants of 4 × 10−6, and three clones from another screen of 2.4 × 105 cells, at a frequency of 1.25 × 10−5. Additionally, in several different screens for over-expression of proteins that mediate drug resistance, we obtained one, two, or three reversible clones from a total 3 × 105 cells in each screen, with a frequency of validated mutants of 3.3 × 10−6 to 1 × 10−5. These high yields allow the selection of multiple validated mutants in experiments of reasonable scale. Therefore, the VBIM technique is a powerful tool for gene discovery that has broad applications in many different systems.
Constitutively active NFκB is a major factor in a number of diseases (8). Therefore, identification of regulators of NFκB is important for a fuller understanding of how it contributes to disease and of how it might be regulated in therapeutic approaches. There are many activators of NFκB but relatively few negative regulators have been reported to date. Most are associated with specific NFκB activation pathways. For example, CARMA1, a lymphocyte-specific member of the membrane-associated guanylate kinase family of scaffolding proteins, is a critical lipid raft-associated regulator of T cell receptor-induced NFκB activation (17). We have shown that SIGIRR (single Ig IL-1R-related molecule), a member of the Toll-like receptor-interleukin 1 receptor signaling (TLR-IL-1R) receptor superfamily, negatively modulates immune responses (18) by binding to TLR/IL-1R signaling components in a ligand-dependent manner. Over-expression of SODD (silencer of death domains) suppresses the ability of tumor necrosis factor (TNF) to activate NFκB-dependent reporter gene expression. SODD is preassociated with the intracellular domain of TNFR1, and TNF-induced aggregation of TNFR1 disrupts the TNFR1-SODD complex (19). Recently, the underlying mechanism by which A20 down-regulates NFκB activation in response to TNF has been described. A20 has two opposing activities: sequential de-ubiquitination and ubiquitination of the TNF receptor-interacting protein (RIP), targeting RIP for proteosomal degradation (20, 21). BCL3 (B cell leukemia-3) was identified as an essential negative regulator of TLR-dependent signaling that blocks the ubiquitination of the NFκB subunit p50, stabilizing a p50 complex that inhibits transcription (22). These examples reveal several different mechanisms of action, indicating the great complexity in the negative regulation of NFκB-dependent signaling.
Our data show that the negative regulatory function of FBXL11 is quite different from the functions of the factors discussed above. The JmjC domain (Fig. 6A), and especially the histone H3K36 demethylase activity that is encoded within it (Fig. 6B), are responsible for the negative regulative effect of FBXL11 on NFκB. A H212A point mutation in this domain abolishes the lysine demethylase activity (13) and also disables its negative regulative effect on NFκB. In contrast, the F-box domain does not play a key role in negative regulation of NFκB (Fig. 6A). These results open an avenue for future study. A detailed understanding of how FBXL11 negatively regulates NFκB through its lysine demethylase activity will provide further important insight into the complex regulation of NFκB-dependent signaling, and how this regulation may affect the function of NFκB in cancer and in other diseases that are driven by the constitutive NFκB activity.
Materials and Methods
Cell Culture and Plasmids.
All cells were cultured in Dulbecco's modified Eagle's medium with 10% FBS, and 1% penicillin and streptavidin. The cell lines used were: human 293C6 cells [293 cells previously transfected with constructs encoding IL-1R1 and accessory protein (11), and newly transfected with E-selectin-driven zeocin-resistance and thymidine kinase genes] and the Z3 mutant with constitutive NFκB generated by chemical mutagenesis, derived from 296C6 cells (12). All tumor cells were cultured in RPMI medium 1640 with 10% FBS and 1% penicillin and streptavidin. The FBXL11 expression plasmid was from American Tissue Culture Collection (ATCC). FBXL11 siRNA was from Open Biosystems. Full length and deletion constructs of Flag-tagged FBXL11 were described by Tsukada et al. (13).
Virus Production, Cell Infection and Selection.
The VBIM and pLV-tTR-KRAB-Red lentiviruses were packaged in 293T cells using second-generation packaging constructs pCMV-dR8.74 and pMD2G (both packaging plasmids and pLV-tTR-KRAB-Red were kind gifts of Dr. Didier Trono, University of Geneva, Switzerland) (23). Retroviruses encoding Cre recombinase or empty vector control were packaged in Phoenix-Ampho cells. Supernatant media containing virus, collected at 36–48 h, were supplemented with 4 μg/mL polybrene before being frozen in aliquots. To determine the titer of each VBIM virus, 293T cells were infected with serially diluted virus and analyzed for GFP expression by FACS. To perform selections in 293C6 cells and Z3 cells, infections were performed so that 70–90% of each population was GFP-positive before selection with drugs. Z3 cells, pretreated with 25 μg/mL zeocin for 7 days to remove any background mutants, were cultured at 1 × 105 cells/well, into 30 wells of 12-well plates. The next day, the cells were infected with the three different VBIM viruses (SD1, 2, and 3). The medium was replaced 24 h after infection and the cells from each well were split and transferred into a 15-cm plate 48 h later. After another 24 h, medium containing 0.1 μg/mL GCV was added and replaced every three days. Individual clones were picked after 2 weeks.
Western and Northern Analyses.
Cells were cultured to about 95% confluency and samples were collected and assayed by the western method as described by Lu et al. (9). Polyclonal anti-JHDM1a/FBXL11 (abcam) was used to detect FBXL11 and polyclonal anti-IκBα and anti-phospho-IκBα (Santa Cruz) were used to detect the corresponding proteins.
Northern analyses were carried out as described before (9). Cells were cultured to about 90% confluency, then treated with 10 ng/mL IL-1β (Peprotech) for different times. Total RNAs were purified and 20 μg of each was loaded into each well. Human cDNA probes for FBXL11 or E-selectin were made by reverse PCR from total RNA of 293C6 cells by using SuperScript III First-Strand Synthesis System (Invitrogen). The cDNA fragments were cloned into pCR8/GW/TOPOTA (Invitrogen) and the fragments were sequenced, excised with EcoRI, and used as probes.
Additional details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants CA95851 (to G.R.S.) and CA112586 and CA60730 (to A.V.G.) and National Cancer Institute Temin Award CA098176 (to E.S.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908560106/DCSupplemental.
References
- 1.Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 2.Gudkov AV, Kazarov AR, Thimmapaya R, Axenovich SA, Mazo IA, Roninson IB. Cloning mammalian genes by expression selection of genetic suppressor elements: Association of kinesin with drug resistance and cell immortalization. Proc Natl Acad Sci USA. 1994;91:3744–3748. doi: 10.1073/pnas.91.9.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kandel ES, Stark GR. Forward genetics in mammalian cells. In: Seghal PB, Levy DE, Hirano T, editors. Signal transducers and activators of transcription (STATs): Activation Biology. The Netherlands: Kluwer, Dordrecht; 2003. [Google Scholar]
- 4.Stark GR, Gudkov AV. Forward genetics in mammalian cells: Functional approaches to gene discovery. Hum Mol Genet. 1999;8:1925–1938. doi: 10.1093/hmg/8.10.1925. [DOI] [PubMed] [Google Scholar]
- 5.Kandel ES, Lu T, Wan Y, Agarwal MK, Jackson MW, Stark GR. Mutagenesis by reversible promoter insertion to study the activation of NFκB. Proc Natl Acad Sci USA. 2005;102:6425–6430. doi: 10.1073/pnas.0502463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karin M, Yamamoto Y, Wang QM. The IKK NFκB system: A treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 7.Karin M. Nuclear factor κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NFκB. J Clin Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu T, Sathe SS, Swiatkowski SM, Hampole CV, Stark GR. Secretion of cytokines and growth factors as a general cause of constitutive NFκB activation in cancer. Oncogene. 2004;23:2138–2145. doi: 10.1038/sj.onc.1207332. [DOI] [PubMed] [Google Scholar]
- 10.Lu T, et al. Secreted transforming growth factor β2 activates NFκB, blocks apoptosis, and is essential for the survival of some tumor cells. Proc Natl Acad Sci USA. 2004;101:7112–7117. doi: 10.1073/pnas.0402048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Commane M, Burns C, Vithalani K, Cao Z, Stark GR. Mutant cells that do not respond to interleukin-1 (IL-1) reveal a novel role for IL-1 receptor-associated kinase. Mol Cell Biol. 1999;19:4643–4652. doi: 10.1128/mcb.19.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sathe SS, et al. Mutant human cells with constitutive activation of NFκB. Proc Natl Acad Sci USA. 2004;101:192–197. doi: 10.1073/pnas.0306812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 14.Deuschle U, Meyer WK, Thiesen HJ. Tetracycline-reversible silencing of eukaryotic promoters. Mol Cell Biol. 1995;15:1907–1914. doi: 10.1128/mcb.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell RS, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 17.Gaide O, et al. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NFκB activation. Nat Immunol. 2002;3:836–843. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 18.Wald D, et al. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat Immunol. 2003;4:920–927. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Y, Woronicz JD, Liu W, Goeddel DV. Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science. 1999;283:543–546. doi: 10.1126/science.283.5401.543. [DOI] [PubMed] [Google Scholar]
- 20.Beyaert R, Heyninck K, Van Huffel S. A20 and A20-binding proteins as cellular inhibitors of nuclear factor κB-dependent gene expression and apoptosis. Biochem Pharmacol. 2000;60:1143–1151. doi: 10.1016/s0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 21.Heyninck K, Beyaert R. A20 inhibits NFκB activation by dual ubiquitin-editing functions. Trends Biochem Sci. 2005;30:1–4. doi: 10.1016/j.tibs.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH. Negative regulation of toll-like receptor signaling by NFκB p50 ubiquitination blockade. Science. 2007;317:675–678. doi: 10.1126/science.1142953. [DOI] [PubMed] [Google Scholar]
- 23.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: Lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.