Abstract
Background and Aims
Actin-myosin II motor converts chemical energy into force/motion in muscle and non-muscle cells. The phosphorylation of regulatory light chain (MLC20) is critical to the cytoplasmic functions of these motors. We do not know whether myosin II and actins in the nucleus function as motors to generate relative motion, such as that between RNA polymerase II holoenzyme and DNA, for assembly of the preinitiation complex.
Methods
The experiments were performed on primary cultures of human colonic circular smooth muscle cells (HCCSMCs) and rat colonic circular muscle strips.
Results
We show that myosin II and α- and β-actins are present in the nuclei of colonic smooth muscle cells. The nuclear myosin II is tethered to recognition sequence AGCTCC (−39/−34) in the ICAM-1 core promoter region. The actins are known to complex with RNA polymerase II and they are tethered to the nucleoskeleton. The dephosphorylation of MLC20 increases the transcription of ICAM-1, whereas its phosphorylation decreases it. Colonic inflammation suppresses nuclear MLCK, which increases the unphosphorylated form of nuclear MLC20, resulting in enhanced transcription of ICAM-1.
Conclusions
1) Myosin II is a core transcription factor; 2) the phosphorylation/dephosphorylation of nuclear MLC20 results in the sliding of myosin and actin molecules past each other producing relative motion between the DNA bound to the myosin II and RNA polymerase II holoenzyme bound to actins and nucleoskeleton.
Keywords: Inflammation, smooth muscle, RNA polymerase II, actin, preinitiation complex
Gene transcription is a tightly regulated multi-step process. These steps include the binding of a gene specific transcription factor to its recognition sequence on the proximal promoter, chromatin remodeling, assembly of the preinitiation complex (PIC) at the core promoter region, elongation, and RNA splicing. Some of these steps, such as chromatin remodeling, assembly of PIC and elongation, require the generation of force resulting in motion. The mechanisms of generation of motion within the nucleus are not known. In the cytoplasm, actin-based myosin motors transduce chemical energy stored in ATP into mechanical energy resulting in generation of force/motion for functions, such as cell contraction, cytokinesis, cell polarization and cell motility.1,2 The motion in these processes results from the sliding of actin and myosin filaments past each other following the phosphorylation of the 20 kDa regulatory myosin light chain (MLC20). Previous studies have established that β-actin and myosin I are present also in the nucleus.3–10 These studies also found that each of these proteins is involved separately in chromatin remodeling and PIC assembly, which requires generation of force and motion in the nucleus. However, we do not know whether the nuclear myosins and actins function as molecular motors to accomplish these processes.
The studies to date have focused primarily on the regulation of ribosomal RNA genes by nuclear myosin I and β-actin.5 RNA polymerase I (RNAP I) mediates transcription of these genes. In differentiated cells, the ribosomal genes are expressed at a relatively steady rate. However, the majority of genes regulate the expression of proteins. The expressions of protein-coding genes are acutely susceptible to micro-environmental stimuli, such as hormones, inflammatory mediators and neurotransmitters.11,12 Consequently, the protein-coding genes require mechanisms that can fine-tune their rate of expression. In this study, we investigated whether an actin (both α-actin and β-actin)-based myosin II (both smooth muscle and non-muscle) motor regulates transcription of the ICAM-1 gene. Previous studies show that an increase in the transcription of ICAM-1 gene plays a critical role in smooth muscle dysfunction in colonic inflammation.13
Our findings show that both types of myosin II as well as α-actin and β-actin are present in the nuclei of human colonic circular smooth muscle cells. Nuclear myosin II is tethered to its recognition sequence in the core promoter region of the ICAM-1 gene. The actins are known to form complexes with RNAP II and the nucleoskeleton.7,8,14,15 Dephosphorylation of the nuclear MLC20 increases transcription of ICAM-1 gene, whereas its phosphorylation decreases it. We established the translational relevance of our cellular findings by showing that dephosphorylation of nuclear MLC20 in colonic inflammation enhances gene expression of ICAM-1 in the muscularis externa of the colon in intact rats.
Materials and Methods
Primary cultures of human colonic circular smooth muscle cells (HCCSMCs)
Primary cultures of HCCSMCs in passages three to five were used in all experiments.12
Animals and induction of colonic inflammation
Sprague Dawley rats were anaesthetized with 2% isoflurane for intraperitoneal injections of ML-7 (1 mg/kg) and microcystin-LR (MCLR) (150 μg/kg) or intracolonic administration of 2,4,6-Trinitrobenzene sulfonic acid (TNBS). The colon was cleansed with oral Colyte® and rats were fasted 24 hours before induction of inflammation with 68 mg/kg TNBS (dissolved in 40% ethanol (v/v) and 250 μL injected 8 cm into the colon via a catheter). Animals were kept in a head-down position for one minute to prevent leakage of TNBS. Rats in the control group received infusion of physiological saline. All animal procedures were approved by the IACUC at the University of Texas Medical branch.
Construction of plasmids
Human ICAM-1 promoter-luciferase reporter construct was engineered by cloning PCR fragment of ICAM-1 promoter (nt −1939 ~ +12) between Kpn I and Mlu I sites of the reporter luciferase vector pGL3-Basic (Promega, Madison, WI). All ICAM-1 promoter constructs with mutant binding sites of myosin II were generated by using GeneTailor™ Site-Directed Mutagenesis System (Invitrogen, Carlsbad, CA). pcDNA3.1(+)-smMLC20 was generated by subcloning the corresponding full-length human smooth muscle MLC20 cDNA into pcDNA3.1(+) (Invitrogen). pCMV6-nmMLC20 was purchased from ORIGENE (Rockville, MN). All constructs were confirmed by sequencing in both directions.
Chromatin fractionation
Chromatin fractions were prepared as described by Carriere et al.16 Briefly, HCCSMCs were washed in PBS, resuspended in 2 mL of chromatin fractionation buffer (0.15 M NaCl/10 mM MgCl2/10 mM CaCl2/1 mM PMSF/15 mM Tris, pH 7.5/0.1% Tween 20), and ruptured by using Ultra-Turrax (Labortechnik, Staufen, Germany) in the presence of 0.1% NP-10. After centrifugation at 800 × g (10 min at 4°C), nuclei were digested with DNase I (0.2 μg/L for 10 min at 30°C) and pelleted by brief centrifugation. Chromatin fractions were prepared by adding NaCl, to a final concentration of 400 mM, to the nuclear pellets resuspended in chromatin fractionation buffer. After 30 min at 4°C, the nuclei were centrifuged at 21,000 × g for 10 min, and the supernatant was saved as chromatin fraction 0.4 M. Chromatin fraction 0.8 M was similarly prepared by adding NaCl to a final concentration of 0.8 M NaCl. The final pellet was saved as residual pellet.
Transfection of MLC20 RNAi in HCCSMCs
MLC20-specific RNAi and scrambled control RNAi were purchased from Dharmacon (Chicago, IL). Cells (5 × 104 in 1 mL growth medium without antibiotics) were plated into each well of a 12-well culture plate one day before transfection. For each well, 40 pmol RNAi and 4.0 μL Lipofectamine 2000 (Invitrogen) were diluted in 100 μL Opti-MEM I Reduced Serum Medium, separately. After 5-minute incubation, diluted RNAi and Lipofectamine 2000 were combined and incubated for 20 minutes at room temperature. The complexes were then added to each well containing cells and medium in a drop-wise manner.
Chromatin immunoprecipitation (ChIP) assay
For ChIP assay, ChIP-IT™ Express Enzymatic Kit (Active Motif, Carlsbad, CA) was used. Histones and transcription factors were cross-linked to DNA by adding formaldehyde to culture medium to a final concentration of 1% and incubating for 10 minutes at room temperature. After washing, cells were collected, pelleted by centrifugation for 10 min at 720 × g at 4°C, and resuspended in 1 mL ice-sold lysis buffer supplemented with 5 μL Protease Inhibitor Cocktail and 5 μL PMSF. The nuclei were pelleted and then resuspended in 0.5 mL shearing buffer. The DNA was sheared with enzymatic shearing cocktail for 12 min at 37°C. After centrifugation at 12,500 rpm and 4°C for 10 min, the supernatant containing the sheared chromatin was collected. Magnetic beads and antibodies were used to capture chromatin. Immunoprecipitates were eluted with 50 μL Elution Buffer AM2. Eluates were heated at 94°C for 15 min to reverse formaldehyde cross-linking (Input sample as well), followed by proteanase K digestion at 37°C for 1 hour. For PCR, 5 μL of the eluted DNA and 36 cycles of amplification were used with five sets of ICAM-1 promoter-specific primers covering different regions (nt −245~−6, −474~−328, −725~−573, −1103~−871, and −1590~−1373) of the promoter. PCR products were subjected to electrophoresis on 2% agarose gels, stained with ethidium bromide.
Trasient transfection and reporter assay
Transfection was performed using Lipofectamine™ 2000 (Invitrogen) or FuGENE 6 (Roche, Indianapolis, IN), and cells were harvested 48 h post-transfection. β-galactosidase and luciferase assays were performed.17
Please see supplemental materials for the following methods
Immunostaining and confocal microscopy, Immunoblotting and co-immunoprecipitation, Detection of mRNA by RT-PCR or real-time RT-PCR and Agarose-oligonucleotide pulldown assay.
Statistics
All data are expressed as mean ± SE and analyzed by two-tailed Student’s t-test, considering p < 0.05 as significant.
Results
Nuclear myosin II in colonic circular smooth muscle cells
Immunostaining and confocal microscopy showed the presence of smooth muscle-specific α-actin, β-actin and MLC20 in the nuclei of HCCSMCs (Figures 1A to H). Immunoblotting with nuclear and cytoplasmic extracts showed that MLC20, MLC17 and MHC are present in both the cytoplasmic and the nuclear fractions, while MLC23 is present only in the cytoplasmic fraction (Figure 1l), indicating that myosin II is also a nuclear protein in HCCSMCs. Both α-actin and β-actin are expressed in the cytoplasm and the nucleus (Figure 1l). α-Tubulin, a cytosolic marker, was detected only in the cytoplasmic fractions and histone H1, a nuclear marker, only in the nuclear fractions, indicating clean separation of the cytoplasmic and nuclear fractions. The specificities of the antibodies were established by immunostaining with IgG (see supplementary material) instead of the primary antibody and western blotting.
Figure 1.
Myosin II and actin exist in the nuclei of HCCSMCs. (A to H) Immunostaining and confocal microscopy of α-actin (green), β-actin (green) and pT18/S19MLC20 (red) in HCCSMCs. TO PRO 3 (blue) was used for counterstaining of nuclei. (I) Western blot analyses of myosin heavy chain, light chains, α-actin and β-actin in cytoplasmic and nuclear fractions from HCCSMCs. α-Tubulin and histone H1 were used as cytoplasmic and nuclear markers, respectively. Cyt, cytoplasmic fraction; Nuc, nuclear extracts. (J) MHC, MLC20, MLC17 and α-actin are associated with the chromatins in HCCSMCs. Chromatin fractions were analyzed by immunoblotting with antibodies indicated. 1, chromatin fraction 0.4M; 2, chromatin fraction 0.8M; 3, chromatin residual pellet. Ctr, control.
We then investigated whether myosin II associates with chromatin. Chromatin fractionation followed by western blotting showed that α-Tubulin is undetectable in chromatin fractions, as expected, but MLC17 and α-actin are present in all three chromatin fractions (Figure 1J). MHC, smooth muscle MLC20 (smMLC20) and nonmuscle MLC20 (nmMLC20) were detected in the residual pellet (Figure. 1J), indicating that myosin II interacts with chromatin in the nucleus. MLC23 was not found in chromatin fractions.
MLC20 induces ICAM-1 gene expression
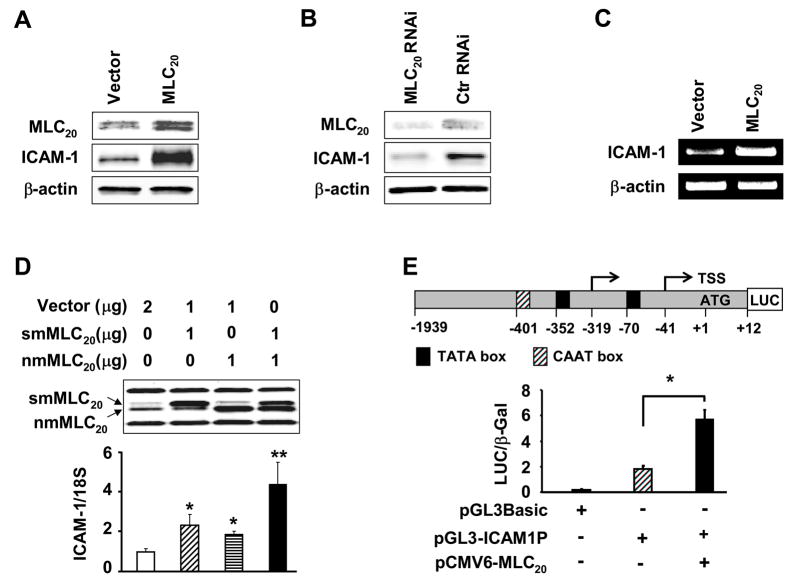
Myosin is associated with numerous motion-based functions in the cytoplasm.18–23 Since this is the first finding that myosin II is also a chromatin-associated nuclear protein (Figure 1J), we hypothesized that nuclear myosin II may function as an actin-based nuclear motor to regulate gene transcription. We tested this hypothesis by over-expressing MLC20, the regulatory light chain of myosin II, in HCCSMCs, and examining whether it affects the expression of ICAM-1, a protein expressed by these cells in response to inflammation.13,24 The over-expression of MLC20 enhanced expression of ICAM-1 by about three-fold (Figure 2A). On the other hand, the knock down of MLC20 by its RNAi markedly decreased the expression ICAM-1 protein (Figure 2B). RT-PCR analysis showed more than two-fold increase of ICAM-1 mRNA by the over-expression of MLC20 (Figure 2C), suggesting that MLC20 regulates ICAM-1 gene transcription. We then investigated whether smMLC20, nmMLC20 or both induce ICAM-1 gene transcription. We over-expressed smMLC20 alone, nmMLC20 alone, or both together in HCCSMCs and analyzed ICAM-1 mRNA by real-time RT-PCR. We found that both smMLC20 and nmMLC20 significantly increase the transcription of ICAM-1 gene (Figure 2D). The effect is additive when both proteins are expressed together.
Figure 2.
MLC20 induces ICAM-1 protein and mRNA expressions, and ICAM-1 promoter activity in HCCSMCs. (A) Overepression of MLC20 increases ICAM-1 protein expression. HCCSMCs were transfected with pCMV6-nmMLC20 and pcDNA-smMLC20, or vector control (Ctr). (B) RNAi-mediated knockdown of MLC20 inhibits ICAM-1 protein expression. Scrambled RNAi (Ctr RNAi) were used as controls. MLC20 depletion and ICAM-1 reduction were monitored 96 hours after transfection. (C) MLC20 over-expression stimulates ICAM-1 mRNA. Total RNA was isolated from cells transfected with pCMV6-nmMLC20 and pcDNA-smMLC20, or vector control. (D) smMLC20, nmMLC20 or their combination induces ICAM-1 mRNA expression, measured using real-time RT-PCR (n=3, *p<0.05 vs control, **p<0.05 vs smMLC20 and nmMLC20). smMLC20, smooth muscle MLC20; nmMLC20, nonmuscle MLC20. (E) MLC20 increases ICAM-1 promoter activity in HCCSMCs. Cells were extracted using 1 X Reporter Lysis Buffer, 48 hr post-transfection (n=3, *p<0.05). Values were normalized for transfection efficiency using pSV-β-Gal 17.
We generated a reporter construct (pGL3-ICAM1P) with luciferase gene driven by the human ICAM-1 promoter (−1939 – +12 nt).25 The co-transfection of this reporter construct and MLC20 in HCCSMCs significantly increased the ICAM-1 promoter activity (5.7 ± 0.7 vs 1.8 ± 0.3) (Figure 2E).
Smooth muscle myosin II associates with the RNA polymerase II (RNAP II) transcription machinery
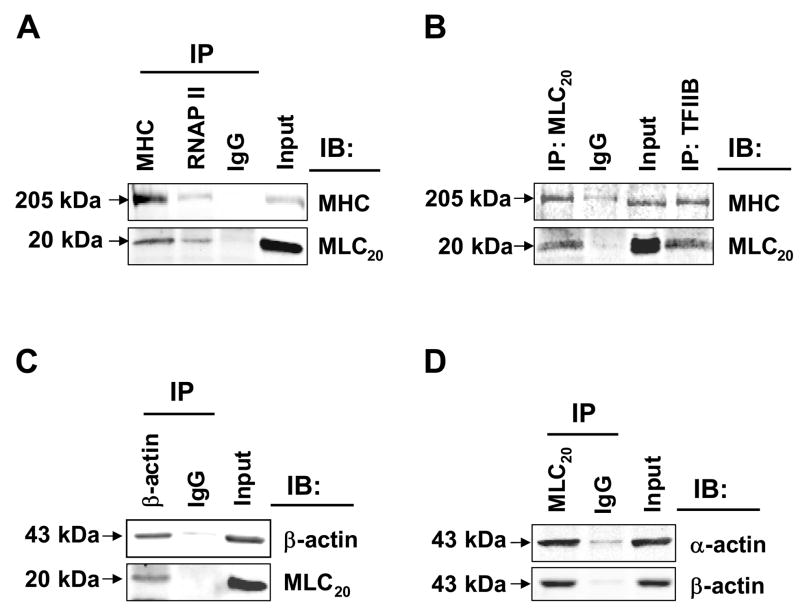
We tested the hypothesis that myosin II is a part of the RNAP II transcription complex. We used co-immunoprecipitation assays followed by immunoblotting to investigate whether nuclear myosin II interacts with RNAP II, TFIIB and actins. We found that RNAP II antibody pulled down MHC and MLC20; MHC antibody pulled down MLC20, as expected (Figure 3A). TFIIB antibody immunoprecipitated MHC and MLC20, whereas MLC20 antibody immunoprecipated MHC (Figure 3B). β-actin antibody precipitated MLC20 (Figure 3C), and MLC20 antibody pulled down both α-actin and β-actin (Figure 3D). In all cases immunoprecipitation with IgG was used as negative control and input was used as positive control. These data support our hypothesis that myosin II is part of the transcriptional machinery.
Figure 3.
Myosin II is associated with RNAP II, TFIIB, α-actin and β-actin in the nucleus. Nuclear extracts were immunoprecipitated (IP) with indicated antibodies, followed by immunoblotting (IB). Rabbit IgG was used as negative control. (A) RNAP II co-precipitated with MHC and MLC20. MHC co-precipitated MLC20. (B) TFIIB antibody pulled down MHC and MLC20 and MLC20 pulled down MHC. (C) β-actin co-precipitated with MLC20. (D) Anti-MLC20 antibody precipitated both α-actin and β-actin.
Smooth muscle myosin II binds to a specific cis-element on ICAM-1 core promoter
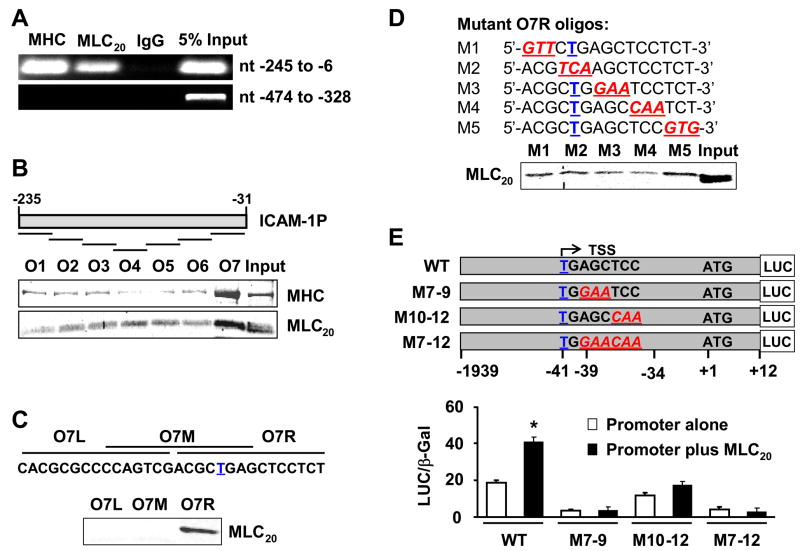
Next, we performed ChIP assay with anti-MHC and anti-MLC20 antibodies. The immunoprecipitated DNA was analyzed by PCR using five sets of primers that amplify the 5′-flanking region on ICAM-1 gene (nt −1590 ~ −6) (Figure 2E). Our data show that myosin II occupies the −245 ~ −6 nt region of the ICAM-1 promoter in which potential myosin II binding motif may be located (Figure 4A). This region covers the reported dominant downstream transcription start site of ICAM-1 in several cell lines26 (Figure 2E). All other primers showed negative signals (data not shown).
Figure 4.
Myosin II binds to the AGCTCC sequence downstream of the transcription start site. (A) MHC and MLC20 bind to the ICAM-1 core promoter. Lack of MLC20 binding to other regions shown only for nt −474/−328. (B) Oligo O7 pulled down MHC and MLC20 from nuclear extracts of HCCSMCs. Bound myosin heavy chain and light chains were detected by immunoblotting. (C) Only the Oligo O7R interacted with myosin II. (D) Mutations of nucleotides 7–9 or 10–12 attenuated O7R binding to MLC20. (E) MLC20 induction of ICAM-1 promoter activity is abrogated by mutation of identified myosin II binding motif (AGCTCC). Upper panel, schematic presentation of reporter constructs with wild type (WT) or mutant myosin II binding sites. Lower panel, mutations nearly abolished basal ICAM-1 promoter activity and its induction by MLC20. Luciferase and β-Gal (internal control) activities of cell lysates were measured 48 hrs later (n=3, *p<0.05 vs control). Underlined bolded T, transcription start site (TSS). Underlined bold and italicized, mutated nucleotides.
We identified the myosin II binding motif within the human ICAM-1 promoter by agarose-oligonucleotide pulldown assays using nuclear extracts of HCCSMCs and biotinylated oligonucleotides. Among the seven overlapping oligos (Figure 4B) used in the first-round screening, oligo O7 (5′-CAC GCG CCC CAG TCG ACG CTG AGC TCC TCT-3′) pulled down MHC and MLC20 (Figure 4B), suggesting that oligo O7 with the transcription start site (T nucleotide) contains the myosin II binding motif. Second round screening with three smaller overlapping oligos showed that only O7R pulls down MLC20 (Figure 4C), indicating myosin II binding motif is in the right half of this oligonucleotide. To identify the myosin II binding motif more precisely, three-point mutations (shown by underlined italics) were introduced into oligo O7R (Figure 4D); each mutant oligo was used for pulldown assays. Less MLC20 was pulled down by mutations of nucleotides 7 to 9 from AGC to GAA, and mutations of nucleotides 10 to 12 from TCC to CAA (Figure 4D). These data suggest that nucleotides 7 to 12 (AGCTCC) in O7R are critical for myosin II/DNA binding.
We used promoter-reporter assays to investigate whether the above mutations abrogate the induction of ICAM-1 by over-expression of MLC20. We mutated nucleotides AGC to GAA (M7–9), TCC to CAA (M10–12), or AGCTCC to GAACAA (M7–12) in plasmid pGL3-ICAM1P (Figure 4E) by using GeneTailor™ Site-Directed Mutagenesis System (Invitrogen). We examined the promoter activities with and without the over-expression of MLC20 in HCCSMCs transfected with these mutations. ICAM-1 promoter with each mutation exhibited significantly less promoter activity, compared with the wild type promoter, suggesting that myosin II binding site (AGCTCC −39/−34) is critical to the constitutive expression and MLC20-induced transcription of the ICAM-1 promoter (Figure 4E).
Phosphrylation/dephosphorylation of MLC20 regulates ICAM-1 promoter activity and protein expression
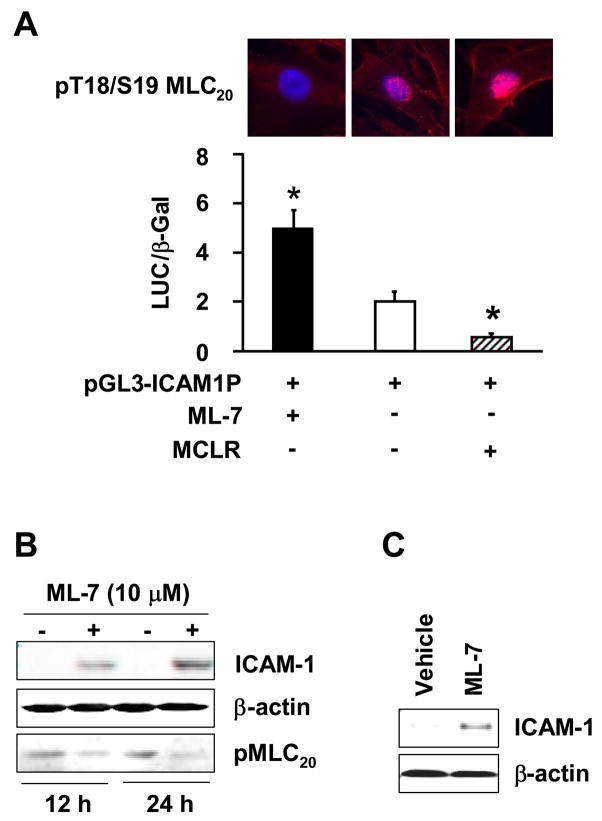
The cytosolic smooth muscle myosin II functions by phosphorylation of its regulatory 20 kDa light chains; myosin II with MLC20 phosphorylation has a 500-fold greater actin-activated ATPase activity.27,28 The myosin-actin interaction results in cross-bridge cycling and relative motion between myosin and actin associated structures. We hypothesized that myosin II with unphosphorylated MLC20 is the active form for transcriptional activity. We tested this hypothesis by examining the regulation of ICAM-1 promoter activity by myosin II in the presence or absence of myosin light chain kinase (MLCK) inhibitor, ML-7, or myosin light chain phosphatase (MLCP) inhibitor, MCLR. ML-7 significantly enhanced the ICAM-1 promoter activity (Figure 5A). On the other hand, MLCP inhibitor MCLR decreased ICAM-1 promoter activity (Figure 5A). In agreement with the effects of phosphorylation/dephosphorylation of MLC20 on ICAM-1 promoter activity, the incubation of fresh rat colonic muscularis externa tissue with ML-7 time-dependently increased the expression of ICAM-1 (Figure 5B). Note that ML-7 decreased pMLC20, whereas MCLR increased it (Figure 5 A and B). In addition, intraperitoneal injection of ML-7 in intact rats induced the expression of ICAM-1 protein in colonic muscularis externa twenty-four hours later (Figure 5C).
Figure 5.
Phosphorylation status of MLC20 affects myosin II transcriptional activity on ICAM-1. (A) ML-7 (5 μM) induced and MCLR (5 nM) suppressed ICAM-1 promoter activity (n=3), *p<0.05 vs control. pT18/S19MLC20 was monitered by immunostaining (top panel). (B) Twenty-four hour incubation with ML-7 increased ICAM-1 protein expression in muscularis externa of rat distal colon. (C) Intraperitoneal administration of ML-7(1 mg/kg) in naïve intact rats induced ICAM-1 expression in the muscularis externa of the distal colon after 24 hours.
Nuclear MLC20 phosphorylation/dephosphorylation regulates ICAM-1 gene expression in colonic inflammation
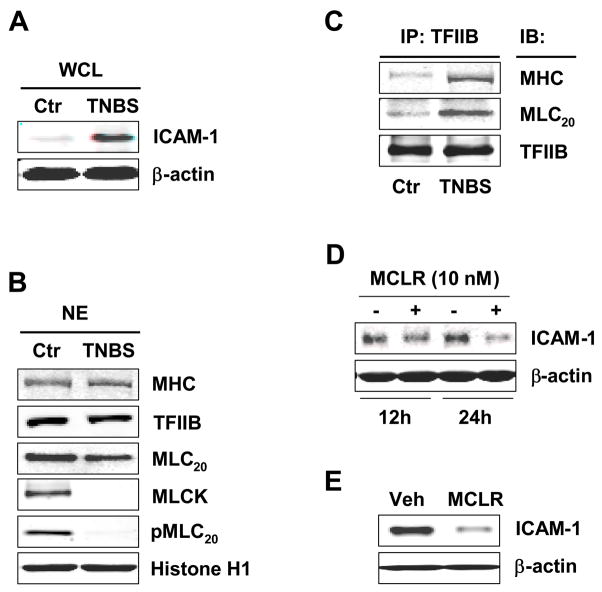
We investigated whether the cellular findings in cultured cells and nuclei translate to physiological/pathological processes in intact animals. We induced colonic inflammation in intact rats by the well established method of intraluminal administration of TNBS.29 Inflammation induced by TNBS mimics key features of the human Crohn’s disease.30 The inflammatory response enhanced expression of ICAM-1 in the muscularis externa of the colon, 24 hours after the induction of inflammation (Figure 6A). The total MHC, MLC20 and TFIIB protein levels in the nuclei from the colonic muscularis externae of the vehicle-treated and TNBS-treated rats were not significantly different from each other (Figure 6B). However, MLCK and pMLC20(Thr18/Ser19) proteins were significantly reduced in the nuclei obtained from the colonic muscularis externa of the inflamed colon, when compared with those obtained from the normal colon (Figure 6B). In addition, significantly greater levels of MHC and MLC20 were immunoprecipitated by anti-TFIIB antibody from the nuclear extracts of the muscularis externae of the TNBS rats than those from the vehicle-treated control rats, indicating increase in the association of myosin II with the preinitiation complex (Figure 6C). The total TFIIB was not different between the two groups of the nuclear extracts.
Figure 6.
TFIIB/myosin II association is augmented in the muscularis externa from distal colon of TNBS rats. (A) ICAM-1 increased in whole cell lysates (WCL) from muscularis externa of TNBS rats. (B) Significantly less MLC20 phosphorylation in the nucleus was observed in TNBS rats. (C) TFIIB/myosin II association in the nucleus is greatly enhanced in TNBS rats. (D) ICAM-1 in inflamed muscularis externa was reduced by MCLR treatment in vitro. (E) Intraperitoneal administration of 150 μg/kg MCLR one-hour before TNBS administration reduced the expression of ICAM-1 24 hours later, when compared with TNBS controls.
The above findings suggest that TFIIB/nuclear myosin II interaction might be a key step in the regulation of ICAM-1 gene in inflammation. Therefore, we investigated whether MLC20 phosphorylation/dephosphorylation could serve as a potential target to minimizing the adverse effects of inflammation on smooth muscle function.13,24 We incubated the strips of colonic muscularis externae, obtained twenty-four hours after the induction of inflammation, with MLCP inhibitor MCLR for twelve or twenty-four hours. Incubation with MCLR time-dependently reduced the enhanced expression of ICAM-1 (Figure 6D). In addition, we found that intraperitoneal injection of MCLR significantly decreased ICAM-1 protein expression in the rat colonic muscularis externa twenty-four hours after the induction of inflammation, when compared with vehicle treated controls (Figure 6E).
Discussion
The presence of myosin I and β-actin in the nuclei of several cell-types, and their associations with RNA polymerases is widely accepted.4,5,31’3,6–10 Loss-of-function (using antibodies and siRNAs) and gain-of-function (overexpression) paradigms show that the nuclear myosin I and actins are involved in the transcription of ribosomal RNA genes. However, we do not know whether myosin I and -actin interact with each other to regulate the transcription of rRNA genes.
Very little is known about the presence or the function of myosin II in the nucleus. A previous study identified a myosin heavy chain of size identical to that of muscle myosin heavy chain in the nuclear pore complexes32 and embryonic myosin heavy chain was identified in myoblast nuclei.33 However, another study ruled out a role of myosin II in the nucleus because its antibodies failed to block the transcription of rRNA genes mediated by RNAP I.5 These investigators did not examine the potential role of myosin II in mediating the transcription of protein-coding genes by RNAP II. Our immunohistochemical and immunoblotting data show that both smooth muscle and non-muscle myosin II are present in the nuclei of the human colonic circular smooth muscle cells. The nuclear myosin II contains all the essential components, such as MLC20, MLC17, and MHC, to function as an actin-based motor. The nuclei of these cells also express α-actin and β-actin. Previous studies focused only on β-actin because they used non-muscle cells.
Both smooth muscle and non-muscle myosin IIs in the nucleus are associated with chromatin, which suggests they might regulate transcription. We examined this possibility by investigating the regulation of ICAM-1 gene expression in HCCSMCs. These cells express ICAM-I at a low level in the resting state. Colonic inflammation enhances the expression of ICAM-1 gene by transcriptional upregulation.13 We found that overexpression of smMLC20 or nmMLC20 increases the protein and mRNA expressions of ICAM-I in HCCSMCs. On the other hand, the suppression of MLC20 by its RNAi decreases the expression of ICAM-I. The transcriptional effects of smMLC20 and nmMLC20 are additive, suggesting that they may act independent of each other. Therefore, the non-muscle myosin II alone may regulate gene transcription in cell-types that do not express the smooth muscle-specific myosin II.
The presence of actins and myosin II in the nucleus raised the possibility that they function as a nuclear motor. The classic motor action of actin-based myosin II motor depends on the phosphorylation of MLC20. The phosphorylation of MLC20 primarily at Ser19 by kinases, such as MLCK or Rho-kinase,34–38 increases myosin’s affinity for actin.27, 28 The binding of actin results in the hydrolysis of ATP, assembly of myosin II into filaments, and the sliding of myosin and actin filaments past each other to generate motion. The dephosphorylation of MLC20 by MLCP, reduces the myosin-actin interaction to reverse motion-induced effects.39
We tested the above hypothesis by investigating whether the phosphorylation/dephosphorylation of MLC20 alters the transcription of ICAM-1. We found that the dephosphorylation of MLC20 by MLCK inhibitor ML-7 time-dependently enhances the luciferase activity of the ICAM-1/Luc construct as well as the expression of ICAM-1 protein. On the other hand, the phosphorylation of MLC20 by MLCP inhibitor MCLR time-dependently suppresses the activity of the promoter-reporter construct and the protein expression of ICAM-1. This suggests that unphosphorylated MLC20 is the active form to induce transcription. The motion produced by the phosphorylation of MLC20 may distance the core promoter region and RNAP II holoenzyme attached to actins, while its dephosphorylation brings them together.
ChIP and agarose nucleotide pulldown assays showed that myosin II is tethered to the cis-element AGCTCC (−39~−34). The general understanding is that the negatively charged tail of myosin (MHC) binds to the cargo-requiring motion, in this case the positively charged DNA,4 while the myosin heads walk-along the actin polymers/filaments to produce motion. In the nucleus, the actins bind to the RNA polymerases as well as to the nucleoskeleton.4,8,14,15 This configuration suggests that an interaction between the actins bound to the nucleoskeleton and RNAPs, and myosin II bound to the DNA may produce relative motion between them to bring together the RNAP II and its associated general transcription factors (GTFs) to the core promoter region of the DNA to form the PIC. The immunoprecipitation experiments showing that myosin II associates with RNAP II, TFIIB, α-actin and β-actin support this suggestion.
The ratio of actin monomers to polymers (about 4:1) in the nucleus is about the same as that in the cytoplasm.40 The major difference in the two compartments is that the nuclear polymers do not bundle up to form stress fibers. The formation of stress fibers in the cytoplasm may allow the generation of larger forces required for deformations of the cytoskeleton along more or less fixed directions. On the other hand, the actin oligomers in the nucleus may allow more flexibility to the nuclear myosin in moving its cargo in different directions required for the concurrent transcriptional regulation of various genes along the DNA.31,41 The force required to bend or move a small segment of DNA is expected to be much less than that required deforming the stiffer cytoskeleton, therefore, the formation of stress fibers may not be necessary in the nucleus.
The actin-based myosin motor is a ubiquitous transducer of chemical energy stored in ATP to mechanical energy. Our study is the first to show that the phosphorylation/dephosphorylation of MLC20 in the nucleus regulates gene transcription. Some of the other prominent nuclear functions that require motion are chromatin-remodeling, elongation and nucleocytoplasmic trafficking. ATP-hydrolysis and the presence of both actins and myosin I have been associated with these functions also.42,43 Our findings suggest that actin-myosin II motors may also regulate these nuclear functions requiring the generation of force and motion.
We established the translational relevance of cellular findings by showing that dephosphorylation of MLC20 is associated with enhanced transcription of ICAM-1 gene in the muscularis externa during colonic inflammation. The inflammatory response suppressed the expression of MLC20 in the cytoplasm and in the nucleus. It also suppressed the expression of MLCK in the cytoplasm and almost obliterated it in the nucleus. As a result, the phospho-MLC20 was almost absent in the nucleus, but unphosphorylated MLC20 was increased, which is equivalent to the blockade of MLCK by ML-7. The absence of phosphorylated MLC20 resulted in an increase in the association of TFIIB with myosin II and increase in the expression of ICAM-1. On the other hand, when we treated the strips of muscularis externa taken from the animals with inflamed colons with MCLR, the expression of ICAM-1 in these tissues decreased time-dependently. MCLR increases the phosphorylation of MLC20 by inhibiting MLCP. In vivo treatment of rats with MCLR prior to the induction of inflammation also reduced the expression of ICAM-1.
We conclude that the phosphorylation/dephosphorylation of nuclear MLC20 regulates the expression of ICAM-1 gene in vitro and in intact animals. Myosin II is a core transcription factor. It binds to its recognition sequence in the core promoter region. MHC may tether to this cis-element. On the other hand, α-actin and β-actin are associated with the RNAP II holoenzyme containing TFIIB and other GTFs. The phosphorylation of MLC20 initiates motor action to distance DNA from the RNAP II and its associated GTFs and decreases transcription. The dephosphorylation of MLC20 reverses the motor action to bring DNA closer to the RNAP II and enhances transcription.
Materials and Methods
Immunostaining and confocal microscopy
HCCSMCs cultured on coverslips were fixed in cooled methanol for 10 min at −20°C, permeabilized with cooled acetone for 1 min at −20°C, blocked with 5% normal donkey serum/PBS for 1 hr at room temperature and incubated overnight at 4 °C with primary antibodies diluted in 1.5% normal donkey serum/PBS. After washing three times 15 min each with 1X PBS, cells were incubated for 1 hr at room temperature with ALEXA-conjugated antibody (Invitrogen) diluted 1:400 in PBS, followed by incubation for 10 min at room temperature with TO PRO 3/PBS (1:500 dilution) for staining of nuclei. Coverslips were mounted to the slides in FluorSave Reagent (Calbiochem) and viewed under confocal microscope (Zeiss LSM 510 UV META Laser Scanning Confocal Microscope). Primary antibodies were: mouse anti-α-actin (Abcam), mouse antiβ-actin (Sigma) and goat anti-pMLC (Thr18/Ser19, Santa Cruz). Secondary antibodies were: donkey anti-goat IgG (594) and donkey anti-mouse IgG (488).
Immunoblotting and co-immunoprecipitation
Whole cell lysates were prepared using IP Lysis Buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerolphosphate, 1 mM Na3VO4, 1 μg/mL leupeptin) and the protein concentration was determined, whereas nuclear extracts were obtained using NE-PER Extraction Reagents (Pierce Biotechnology, Rockford, IL). Proteins were separated on 8–16% or 8% tris-glycine gels (Invitrogen), they were transferred to nitrocellulose membranes (Invitrogen), and after incubation with primary antibody followed by secondary antibody IRDye 800 conjugated anti-mouse IgG (ROCKLAND) or Alexa Fluor 680 goat anti-rabbit IgG (Invitrogen), detection was by ODYSSEY® Infrared Imaging System (LI-COR Biosciences, Lincoln, Nebraska). Antibodies were as follows: mouse anti-myosin light chain monoclonal (Sigma), mouse anti-myosin II monoclonal (Abcam), mouse anti-β-actin monoclonal (AC-15, Sigma), rabbit anti-histone H1 polyclonal (FL-219, Santa Cruz), rabbit anti-α-tubulin polyclonal (H-300, Santa Cruz), rabbit anti-TFIIB polyclonal (C-18, Santa Cruz), sheep anti-MLCK polyclonal (COVANCE), and mouse anti-ICAM-1 monoclonal (G-5, Santa Cruz). Although β-actin served as a loading control in most experiments, we routinely confirmed the clean separation of nuclear and cytoplasmic fractions by immunoblotting with anti-α-tubulin antibody and anti-histone H1 antibody. For co-immunoprecipitation, either whole cell lysates or nuclear extracts were used. Supernatants were precleared with protein G sepharose for 1.5 hr at 4°C. Immunoprecipitation was conducted overnight at 4°C with 2 μg of the indicated antibody. Immunocomplexes were collected by adding 100 μL of 50% protein G sepharose slurry and incubating for 1.5 hr at 4°C. The beads were washed 5 times with lysis buffer, resuspended in 35 μL SDS sample buffer, heated to 90°C for 10 min, and subjected to Western blotting analysis with corresponding antibodies.
Detection of mRNA by RT-PCR or real-time RT-PCR
Total RNA was extracted using RNeasy Mini Kit (QIAGEN, Valencia, CA). One μg of total RNA was reverse-transcribed using SuperScript™ III First-Strand Synthesis System for RT-PCR (Invitrogen). Primers 5′-CTT CTC CTG CTC TGC AAC CC-3′ (Forward) and 5′-GGG AGA GCA CAT TCA CGG TC-3′ (Reverse) were used for ICAM-1 amplification. Primers 5′-ATG GAT GAT GAT ATC GCC GC-3′ (Forward) and 5′-TTA ATG TCA CGC ACG ATT TC-3′ (Reverse) were used for β-actin. Reactions were conducted in a thermal cycler (GeneAmp PCR system 9700, Perkin-Elmer, Foster City, CA), initiated by hot start at 94°C for 2 min. Cycling conditions were: 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s, with 35 cycles for ICAM-1 and 25 cycles for β-actin. All reactions ended with a 5 min elongation step at 72°C. PCR products were subjected to electrophoresis on 2% agarose gels, stained with ethidium bromide. The amount of amplified product was confirmed by this method to be in linear range with respect to the input RNA for both ICAM-1 and β-actin primers. Quantification of ICAM-1 gene expression by real-time PCR was performed with a ABI Prism 7000 Thermal Cycler, and ICAM-1 was amplified in the presence of Taqman probe and primers specific to human ICAM-1. 18S expression was also quantified as an internal control for the amount and quanlity of cDNA. All samples were assayed in triplicate in an Optical 96-well reaction plate with Optical Adhesive Covers (Applied Biosystems) in a 20-μL volume containing 5 μL (2 μL for 18S) diluted cDNA (1:10 dilution with MQ water).
Agarose-oligonucleotide pulldown assay
DNA affinity purification was performed as described previously, with modifications11. The oligonucleotides from human ICAM-1 promoter and their complementary strands were synthesized by Sigma Genosys, and biotinylated by using Biotin 3′ End DNA Labeling Kit (Pierce, Rockford, IL). After annealing of the two single-strand oligonucleotides, the double-stranded oligonucleotide was incubated with streptavidin-conjugated agarose beads (Pierce) for 1 hour at 4°C and washed twice with IP lysis buffer. Nuclear extract (50 μg) suspended in 300 μL of IP lysis buffer was precleared with agarose beads for 1 hour at 4°C to remove any nonspecific binding to the beads. The lysates were then incubated with oligo/streptavidin-conjugated beads overnight at 4°C. The beads were washed 5 times with IP lysis buffer, and the affinity-adsorbed protein was eluted by boiling in SDS sample buffer for 5 minutes and subjected to Western blotting.
Supplementary Material
Acknowledgments
Supported in part by NIDDK Grants DK 032346 and DK 072414 (SKS)
Footnotes
No conflict of interest exists
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–8. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 2.Bahler M. Are class III and class IX myosins motorized signalling molecules? Biochim Biophys Acta. 2000;1496:52–9. doi: 10.1016/s0167-4889(00)00008-2. [DOI] [PubMed] [Google Scholar]
- 3.Nowak G, Pestic-Dragovich L, Hozak P, Philimonenko A, Simerly C, Schatten G, de Lanerolle P. Evidence for the presence of myosin I in the nucleus. J Biol Chem. 1997;272:17176–81. doi: 10.1074/jbc.272.27.17176. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann WA, Johnson T, Klapczynski M, Fan JL, de Lanerolle P. From transcription to transport: emerging roles for nuclear myosin I. Biochem Cell Biol. 2006;84:418–26. doi: 10.1139/o06-069. [DOI] [PubMed] [Google Scholar]
- 5.Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozak P, Grummt I. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6:1165–72. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 6.Kysela K, Philimonenko AA, Philimonenko VV, Janacek J, Kahle M, Hozak P. Nuclear distribution of actin and myosin I depends on transcriptional activity of the cell. Histochem Cell Biol. 2005;124:347–58. doi: 10.1007/s00418-005-0042-8. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, Hozak P, de Lanerolle P. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 8.Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 2004;18:3010–5. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheer U, Hinssen H, Franke WW, Jockusch BM. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 1984;39:111–22. doi: 10.1016/0092-8674(84)90196-x. [DOI] [PubMed] [Google Scholar]
- 10.Pestic-Dragovich L, Stojiljkovic L, Philimonenko AA, Nowak G, Ke Y, Settlage RE, Shabanowitz J, Hunt DF, Hozak P, de Lanerolle P. A myosin I isoform in the nucleus. Science. 2000;290:337–41. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 11.Shi XZ, Pazdrak K, Saada N, Dai B, Palade P, Sarna SK. Negative transcriptional regulation of human colonic smooth muscle Cav1. 2 channels by p50 and p65 subunits of nuclear factor-kappaB. Gastroenterology. 2005;129:1518–32. doi: 10.1053/j.gastro.2005.07.058. [DOI] [PubMed] [Google Scholar]
- 12.Shi XZ, Choudhury BK, Pasricha PJ, Sarna SK. A novel role of VIP in colonic motility function: induction of excitation-transcription coupling in smooth muscle cells. Gastroenterology. 2007;132:1388–400. doi: 10.1053/j.gastro.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Pazdrak K, Shi XZ, Sarna SK. TNFalpha suppresses human colonic circular smooth muscle cell contractility by SP1- and NF-kappaB-mediated induction of ICAM-1. Gastroenterology. 2004;127:1096–109. doi: 10.1053/j.gastro.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Wasser M, Chia W. The EAST protein of drosophila controls an expandable nuclear endoskeleton. Nat Cell Biol. 2000;2:268–75. doi: 10.1038/35010535. [DOI] [PubMed] [Google Scholar]
- 15.Clark TG, Merriam RW. Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell. 1977;12:883–91. doi: 10.1016/0092-8674(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 16.Carriere V, Roussel L, Ortega N, Lacorre DA, Americh L, Aguilar L, Bouche G, Girard JP. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci U S A. 2007;104:282–7. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Dashwood RH. Activator protein 2alpha associates with adenomatous polyposis coli/beta-catenin and Inhibits beta-catenin/T-cell factor transcriptional activity in colorectal cancer cells. J Biol Chem. 2004;279:45669–75. doi: 10.1074/jbc.M405025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray RM, Guo H, Patel M, Jin S, Bhattacharya S, Johnson LR. Role of myosin regulatory light chain and Rac1 in the migration of polyamine-depleted intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G983–95. doi: 10.1152/ajpgi.00356.2006. [DOI] [PubMed] [Google Scholar]
- 19.Cai S, Pestic-Dragovich L, O’Donnell ME, Wang N, Ingber D, Elson E, De Lanerolle P. Regulation of cytoskeletal mechanics and cell growth by myosin light chain phosphorylation. Am J Physiol. 1998;275:C1349–56. doi: 10.1152/ajpcell.1998.275.5.C1349. [DOI] [PubMed] [Google Scholar]
- 20.Yamakita Y, Yamashiro S, Matsumura F. In vivo phosphorylation of regulatory light chain of myosin II during mitosis of cultured cells. J Cell Biol. 1994;124:129–37. doi: 10.1083/jcb.124.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ihnatovych I, Hu W, Martin JL, Fazleabas AT, de Lanerolle P, Strakova Z. Increased phosphorylation of myosin light chain prevents in vitro decidualization. Endocrinology. 2007;148:3176–84. doi: 10.1210/en.2006-1673. [DOI] [PubMed] [Google Scholar]
- 22.Moss RL, Fitzsimons DP. Myosin light chain 2 into the mainstream of cardiac development and contractility. Circ Res. 2006;99:225–7. doi: 10.1161/01.RES.0000236793.88131.dc. [DOI] [PubMed] [Google Scholar]
- 23.Rottbauer W, Wessels G, Dahme T, Just S, Trano N, Hassel D, Burns CG, Katus HA, Fishman MC. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ Res. 2006;99:323–31. doi: 10.1161/01.RES.0000234807.16034.fe. [DOI] [PubMed] [Google Scholar]
- 24.Shi XZ, Sarna SK. Transcriptional Regulation Of Inflammatory Mediators Secreted By Human Colonic Circular Smooth Muscle Cells. Am J Physiol Gastrointest Liver Physiol. 2005 doi: 10.1152/ajpgi.00512.2004. [DOI] [PubMed] [Google Scholar]
- 25.Voraberger G, Schafer R, Stratowa C. Cloning of the human gene for intercellular adhesion molecule 1 and analysis of its 5′-regulatory region. Induction by cytokines and phorbol ester. J Immunol. 1991;147:2777–86. [PubMed] [Google Scholar]
- 26.Wawryk SO, Cockerill PN, Wicks IP, Boyd AW. Isolation and characterization of the promoter region of the human intercellular adhesion molecule-1 gene. Int Immunol. 1991;3:83–93. doi: 10.1093/intimm/3.1.83. [DOI] [PubMed] [Google Scholar]
- 27.Cross RA. What is 10S myosin for? J Muscle Res Cell Motil. 1988;9:108–10. doi: 10.1007/BF01682153. [DOI] [PubMed] [Google Scholar]
- 28.Morano I. Tuning smooth muscle contraction by molecular motors. J Mol Med. 2003;81:481–7. doi: 10.1007/s00109-003-0451-x. [DOI] [PubMed] [Google Scholar]
- 29.Morris GP, Beck PL, Herridge MS, Depew WT, Szewczuk MR, Wallace JL. Hapten-induced model of chronic inflammation and ulceration in the rat colon. Gastroenterology. 1989;96:795–803. [PubMed] [Google Scholar]
- 30.Elson CO, Sartor RB, Tennyson GS, Riddell RH. Experimental models of inflammatory bowel disease. Gastroenterology. 1995;109:1344–67. doi: 10.1016/0016-5085(95)90599-5. [DOI] [PubMed] [Google Scholar]
- 31.Fomproix N, Percipalle P. An actin-myosin complex on actively transcribing genes. Exp Cell Res. 2004;294:140–8. doi: 10.1016/j.yexcr.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 32.Berrios M, Fisher PA. A myosin heavy-chain-like polypeptide is associated with the nuclear envelope in higher eukaryotic cells. J Cell Biol. 1986;103:711–24. doi: 10.1083/jcb.103.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodgers BD. Insulin-like growth factor-I downregulates embryonic myosin heavy chain (eMyHC) in myoblast nuclei. Growth Horm IGF Res. 2005;15:377–83. doi: 10.1016/j.ghir.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Gallagher PJ, Herring BP, Griffin SA, Stull JT. Molecular characterization of a mammalian smooth muscle myosin light chain kinase. J Biol Chem. 1991;266:23936–44. [PMC free article] [PubMed] [Google Scholar]
- 35.Hai CM, Murphy RA. Ca2+, crossbridge phosphorylation, and contraction. Annu Rev Physiol. 1989;51:285–98. doi: 10.1146/annurev.ph.51.030189.001441. [DOI] [PubMed] [Google Scholar]
- 36.Filenko AM, Danilova VM, Sobieszek A. Smooth muscle myosin light chain kinase, supramolecular organization, modulation of activity, and related conformational changes. Biophys J. 1997;73:1593–606. doi: 10.1016/S0006-3495(97)78191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikebe M, Hartshorne DJ. Phosphorylation of smooth muscle myosin at two distinct sites by myosin light chain kinase. J Biol Chem. 1985;260:10027–31. [PubMed] [Google Scholar]
- 38.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–9. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 39.Somlyo AP, Somlyo AV. Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiol Rev. 2003;83:1325–58. doi: 10.1152/physrev.00023.2003. [DOI] [PubMed] [Google Scholar]
- 40.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–52. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldman MA. The epigenetics of the cell. Genome Biol. 2003;4:309. doi: 10.1186/gb-2003-4-3-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–81. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 43.Percipalle P, Fomproix N, Cavellan E, Voit R, Reimer G, Kruger T, Thyberg J, Scheer U, Grummt I, Farrants AK. The chromatin remodelling complex WSTF-SNF2h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep. 2006;7:525–30. doi: 10.1038/sj.embor.7400657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.