Abstract
Mycobacterium ulcerans is the causative agent of Buruli ulcer, the third most common mycobacterial disease after tuberculosis and leprosy. It is an emerging infectious disease that afflicts mainly children and youths in West Africa. Little is known about the evolution and transmission mode of M. ulcerans, partially due to the lack of known genetic polymorphisms among isolates, limiting the application of genetic epidemiology. To systematically profile single nucleotide polymorphisms (SNPs), we sequenced the genomes of three M. ulcerans strains using 454 and Solexa technologies. Comparison with the reference genome of the Ghanaian classical lineage isolate Agy99 revealed 26,564 SNPs in a Japanese strain representing the ancestral lineage. Only 173 SNPs were found when comparing Agy99 with two other Ghanaian isolates, which belong to the two other types previously distinguished in Ghana by variable number tandem repeat typing. We further analyzed a collection of Ghanaian strains using the SNPs discovered. With 68 SNP loci, we were able to differentiate 54 strains into 13 distinct SNP haplotypes. The average SNP nucleotide diversity was low (average 0.06–0.09 across 68 SNP loci), and 96% of the SNP locus pairs were in complete linkage disequilibrium. We estimated that the divergence of the M. ulcerans Ghanaian clade from the Japanese strain occurred 394 to 529 thousand years ago. The Ghanaian subtypes diverged about 1000 to 3000 years ago, or even much more recently, because we found evidence that they evolved significantly faster than average. Our results offer significant insight into the evolution of M. ulcerans and provide a comprehensive report on genetic diversity within a highly clonal M. ulcerans population from a Buruli ulcer endemic region, which can facilitate further epidemiological studies of this pathogen through the development of high-resolution tools.
Author Summary
Mycobacterium ulcerans is the causative agent of Buruli ulcer (BU), a necrotizing skin disease and the third most common mycobacterial disease after tuberculosis and leprosy. It is an emerging infectious disease that afflicts mainly children and youths in West Africa. The disease is also found in tropical and subtropical regions of Asia, the Western Pacific, and Latin America. Limited knowledge of this neglected tropical disease is partially due to the lack of known genetic polymorphisms among isolates, which hinder the study of transmission, epidemiology, and evolution of M. ulcerans. Our aim is to systematically profile genetic diversity among M. ulcerans isolates by sequencing and comparing the genomes of selected strains. We identified single nucleotide polymorphisms (SNPs) within a highly clonal M. ulcerans population from a Buruli ulcer endemic region. Based on the SNPs discovered, we developed SNP typing assays and were able to differentiate a collection of M. ulcerans isolates from this Buruli ulcer endemic region into 13 SNP haplotypes. Our results lay the ground for developing a highly discriminatory and cost-effective tool to study M. ulcerans evolution and epidemiology at a population level.
Introduction
Mycobacterium ulcerans causes Buruli ulcer (BU), a necrotizing skin disease and the third most common mycobacterial disease, after tuberculosis and leprosy [1]. In the past decade the incidence of BU has increased dramatically in West African countries, but the disease is also found in tropical and subtropical regions of Asia, the Western Pacific, and Latin America [2]. Due to the focal distribution of the disease and the fact that it affects mainly poor rural communities, BU belongs to the neglected tropical diseases. Limited knowledge about the disease is partially caused by the lack of molecular fine-typing methods, which hinder the study of transmission, epidemiology, and evolution of the clonal pathogen.
Genetic analyses suggested that M. ulcerans diverged from the fish pathogen M. marinum between 470,000 and 1,200,000 years ago by acquiring the virulence plasmid pMUM001 [3],[4]. Whole genome comparison of M. marinum strain M and M. ulcerans strain Agy99 revealed that the two strains share >98% nucleotide sequence identity, although extensive DNA insertions and deletions have been observed [4]. Our recent comparative genomic hybridization study found extensive large sequence polymorphisms (LSPs) among M. ulcerans clinical isolates of diverse geographic origins. Two distinct M. ulcerans lineages have been defined: the ancestral lineage of strains from Asia, South America and Mexico, which are genetically closer to the progenitor M. marinum, and the classical lineage of strains from Africa, Australia and South East Asia [5],[6].
Although continental types of M. ulcerans strains have been well established, differentiation between isolates within a geographic region, such as strains from African countries, has remained a challenge [7],[8]. Different genotyping methods have been applied to M. ulcerans, including IS2426 polymerase chain reaction (PCR) [9],[10], amplified fragment length polymorphism (AFLP) [11], IS2404 restriction fragment length polymorphism (RFLP) [12],[13], multi-locus sequence typing (MLST) [3],[14],[15], variable-number tandem repeat (VNTR) typing [7],[8],[16],[17] and IS2404-Mtb2 PCR [18]. Among these, AFLP [11] and recently established VNTR typing methods [7],[8] were the only techniques that have provided some resolution among clinical strains of M. ulcerans from Africa, confirming that genotypic diversity attributable to insertions, deletions, and duplications of variable DNA fragments exists among African strains. However, protein coding genes in M. ulcerans African populations harbor extremely low levels of polymorphisms. No single nucleotide polymorphisms (SNPs) were detected in a multi locus sequence typing of a few chromosomal and plasmid genes [3],[14],[15].
To systematically and comprehensively study the genetic diversity and the evolution of M. ulcerans strains, a genome wide profiling is needed. The complete genome sequence of M. ulcerans Agy99 consists of a circular chromosome of 5632 Kb and a plasmid pMUM001 of 174 Kb [4], which can be used as the reference for comparative genome analysis. The ongoing revolution in massively parallel sequencing technology [19],[20], such as the availability of Roche 454 Life Sciences Genome Sequencer FLX [21], Applied Biosystems SOLiD System, and Illumina Solexa Genome Analyzer [22], has made it possible to sequence large numbers of bacterial strains within days [23]. Next generation sequencing platforms have been used for genome wide profiling of novel genetic variations in many different organisms, including viruses [24], bacteria [25],[26],[27], plants [28],[29], worms [30] and humans [31],[32]. Here we report the sequencing of the genomes of three selected M. ulcerans strains using pyrosequencing (Roche 454 Life Science) and Solexa (Illumina) sequencing by synthesis technologies. Based on these sequences we identified SNPs, which we used to estimate evolutionary times for the emergence of M. ulcerans. We also developed SNP typing assays as high-resolution genotyping methods for M. ulcerans. Genetic fingerprinting of bacterial isolates will be a valuable tool for distinguishing relapses from new infections, tracing infection chains, and identifying environmental reservoirs. Molecular epidemiological analyses based on SNP typing may finally contribute to better disease control by identifying preventable risks for infection.
Results
Selection of M. ulcerans strains for genome sequencing
With the aim to comprehensively investigate genome diversity of M. ulcerans strains from an individual geographical region, we selected two Ghanaian patient isolates from different residential districts and sequenced their genomes with 454 and Solexa technologies, respectively (Table 1). These two strains were isolated from the same African country as the fully sequenced reference strain Agy99 [4]. Whereas Agy99 was isolated in 1999 from a BU patient, the two selected Ghanaian patient isolates NM20/02 and NM31/04 were isolated after an apparent process of replacement of VNTR types in Ghana [7] in the years 2002 and 2004, respectively. Agy99, NM31/04 and NM20/02 represent the three VNTR types (Table 1) previously identified in Ghana [7]. While the Ghanaian strains belong to the classical lineage, we also included a Japanese patient isolate, ITM Japan8756 (denoted as Jp8756 from here on in the paper), as a representative of the ancestral lineage in our analysis. Its genome was sequenced with the Solexa Genome Analyzer. Selected genomic regions of this strain were also sequenced with a NimbleGen comparative genome sequencing (CGS) microarray and the results were compared.
Table 1. Mycobacterium ulcerans strains sequenced in this study.
Single nucleotide polymorphisms in M. ulcerans strains
We sequenced three M. ulcerans strains with single end reads generated by two different next–generation sequencing platforms. For NM20/02, we obtained 424,494 GS FLX reads (Roche 454) with an average length of 213 bases. For NM31/04 and Jp8756, we obtained 2.5 and 2.7 million 35-bp Solexa reads, respectively (Table 2). To identify SNPs, we mapped the reads to the reference genome, including both the Agy99 chromosome and the plasmid pMUM001.
Table 2. Summary of next generation sequencing results.
| Strain | NM20/02 | NM31/04 | Jp8756 | |
| Sequencing method | Roche 454 GS FLX | Illumina Solexa GA | Illumina Solexa GA | |
| Total no. Reads | 424,494 | 2,538,429 | 2,651,276 | |
| Averaged read length (nt) | 213 | 35 | 35 | |
| Total sequences (nt) | 90,299,836 | 88,845,015 | 92,794,660 | |
| Map to Agy99 chromosome | Total no. reads mapped (%) | 382,116 (90.01) | 2,343,269 (92.31) | 2,279,741 (85.99) |
| % genome mapped | 93.72 | 99.99 | 94.47 | |
| Average depth of mapped regions | 14.5 | 14.1 | 13.6 | |
| Map to Agy99 plasmid pMUM001 | Total no. reads mapped (%) | 5,269 (1.24) | 326,541 (12.86) | 89,325 (3.37) |
| % genome mapped | 32.56 | 100 | 20.35 | |
| Average depth of mapped regions | 19.8 | 63.4 | 17.2 | |
We used the 454 software gsMapper for GS FLX reads and MAQ [33] for Solexa reads. The MAQ places Solexa reads mapped to multiple locations randomly, while gsMapper excludes reads mapped to repeated regions, such as insertion sequences (IS)2404 and IS2606, which are present in high copy numbers in the M. ulcerans genome [4]. Therefore, the Agy99 chromosome was better covered by Solexa reads than by GS FLX reads. 94% of the Agy99 chromosome was mapped with NM20/02 GS FLX reads, 99.99% with NM31/04 Solexa reads, 94.47% with Jp8756 Solexa reads. The average depth was 14 to 15 fold. We identified 135 chromosomal SNPs in NM20/02, 83 SNPs in NM31/04, and 26,564 SNPs in Jp8756 (Table S1).
The coverage for pMUM001 varied a lot from strain to strain. 33% of the pMUM001 was mapped with NM20/02 reads, 100% with NM31/04 reads, and 20% with Jp8756 reads. The average depth ranged from 63 for NM31/04 reads to 17 for Jp8756 reads (Table 2). Because the low coverage of the plasmid in NM20/02 could be an artifact from the gsMapper, which excluded reads mapped to non-unique regions, we mapped NM20/02 reads to pMUM001 using another software, MOSAIK (http://bioinformatics.bc.edu/marthlab/Mosaik), which allowed us to compare mapping results with non-uniquely mapped reads included or excluded. When all mapped reads were assembled regardless of their uniqueness, the full length of pMUM001 was well covered (Figure S1 A), suggesting the presence of a pMUM001-like plasmid in NM20/02. When only uniquely mapped reads were recorded, pMUM001 was partially covered (Figure S1 B). Reads mapped to regions such as those encoding Type I modular polyketide synthase genes, transposase genes, IS elements (IS2606 and IS2404) were excluded due to their non-uniqueness. Although mapping analysis using MOSAIK confirmed that the lack of pMUM001 coverage in NM20/02 was a data analysis artifact, the lack of pMUM001 in Jp8756 was confirmed. MAQ didn't exclude non-uniquely mapped reads and was able to map the full length of pMUM001 with NM31/04 reads, which were analyzed exactly the same way as Jp8756 reads. Mapping using MOSAIK not only confirmed the lack of pMUM001 coverage by Jp8756 reads (Figure S1 E), but also revealed that the depth of pMUM001 regions covered by uniquely mapped Jp8756 reads were very low, which ranged from one read to four reads (Figure S1 F), while the depth of pMUM001 regions covered by uniquely mapped reads ranged from 10 to 50 in NM20/02 and NM31/04 (Figure S1 B and D). Most likely the under-representation of plasmid DNA in the total DNA sample is due to either complete or partial loss of plasmid sequences, which is frequently found in M. ulcerans strains that have been cultured over extended periods of time. Previous plasmid sequences analyses have found that plasmids in Japanese and African M. ulcerans strains are highly conserved in size and sequence [15],[34]. However, a recent study found different strains of M. ulcerans were capable of producing structurally distinct mycolactones, which could be due to presence of sequence variations in pMUM001[35]. Our study suggests that the plasmids from all three African strains are highly similar. We only found one intergenic SNP and one synonymous SNP shared by NM31/04 and NM20/02 plasmids. The two non- synonymous SNPs found in the NM20/02 plasmid were within IS2606 genes. In Jp8756 we identified one intergenic SNPs in the plasmid regions with sufficient coverage (Table S1). To elucidate how the sequence variations affect mycolacton production, future experiments in the lab will be needed to enrich and analyze the plasmids in each test strain, together with characterization of mycolacton production.
Before next generation sequencing technologies became widely available, we have sequenced Jp8756 using comparative genome sequencing microarrays covering selected regions of the Agy99 chromosome and pMUM001. A total of 1,618 SNPs were identified in the selected 1.2 Mb chromosomal protein coding regions. 1,389 (86%) of these SNPs were confirmed by Solexa sequencing (Table S2). On both mutation mapping arrays and re-sequencing arrays, probes targeting pMUM001 showed very low signals (Figure S2), as compared to probes targeting the Agy99 chromosome, which also confirmed the lack of plasmid DNA in the Jp8756 DNA sample.
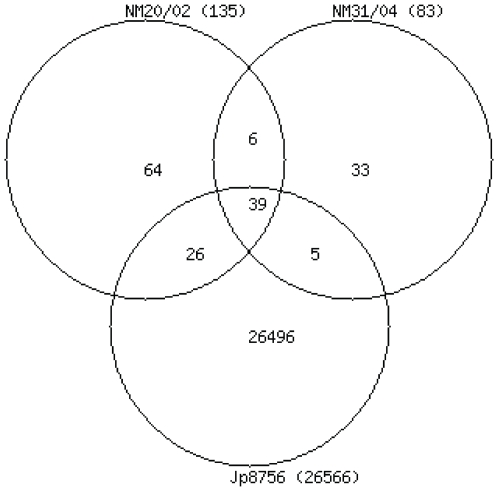
In total 26,669 SNPs were identified by comparing the Jp8756 to the Agy99 chromosome; 18,510 in 3,597 protein coding genes, and 8,159 in 1,768 different intergenic regions (Table S1). 99.35% of the SNPs were found only in the Japanese strain (Figure 1). In comparison to Agy99, the average number of SNPs ranged from 1 per 210 bp in Jp8756 to 1 per 68 Kb in NM31/04. While the Japanese strain Jp8756 and the Ghanaian strain Agy99 share 99.53% nucleotide sequence identity, the average percentage of polymorphic nucleotide sites between Ghanaian strains is only 0.0015% (NM31/04 vs. Agy99) and 0.0024 (NM20/02 vs. Agy99), respectively. These results are consistent with previous findings that strains of the ancestral lineage are genetically distant to the classical lineage strains [6]. There are 103 SNPs specific to the Ghanaian strains with an average of 0.0018% of polymorphic nucleotide site differences from the Agy99 genome (Figure 1).
Figure 1. Venn diagram of single nucleotide polymorphisms in M. ulcerans strains.
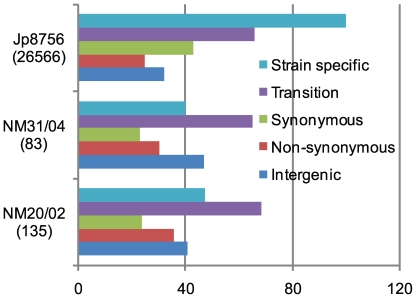
We classified identified SNPs into five different categories: strain specific SNPs, transversions, synonymous SNPs (sSNPs), non-synonymous SNPs (non-sSNPs), and intergenic SNPs. Figure 2 shows the total numbers of SNPs and the percentage of different categories of SNPs found in each strain. Overall, only 34 SNPs were found parsimoniously informative (PI), common to at least two strains or “non-strain-specific.” The numbers of “strain-specific” SNPs varied from 33 (40% of 83 SNPs) in NM31/04 to 26,496 (99.7% of 26564 SNPs) in Jp8756. In all three strains the percentage of intergenic SNPs was around 40%, indicating a similar distribution of point mutations in coding and non-coding regions. Interestingly, the majority of coding region SNPs (63%) found in Jp8756 was synonymous, while in the two Ghanaian strains the majority was non-synonymous. About 100 non-synonymous SNPs found in strain Jp8756, but none of the non-synonymous SNPs found in the two Ghanaian strains caused premature stop codons. While the accumulation of pseudogenes seems to play an important role in both the divergence of M. ulcerans from M. marinum and the emergence of M. ulcerans continental types [4], our observation suggests that there is no further formation of pseudogenes within the studied Ghanaian strains. The percentage of SNP transition ranged between 65% and 68%, suggesting a substitution bias in favor of nucleotide substitution within the purine or pyrimidine group.
Figure 2. Summary of single nucleotide polymorphisms in M. ulcerans strains.
The number in parentheses represents the total number of SNPs in each sequenced strain. The color bars show the distributions of different SNP categories in each strain.
We did not try to identify insertions or deletions (indels) of any size. First of all, single-base/small indels were difficult to identify based on 454 data due to higher indel error rate observed in pyrosequencing [36]. Secondly, identification of single-base/small indels using short reads, such as solexa or SOLiD data, requires paired-end data [33], which were not provided in our study. Without paired-end data, discovery of single-base/small indels using short reads requires gapped alignments of millions of short reads, which is computational challenged. Most currently available short read mapping tools supports only non-gapped alignments. Gapped alignment tools such as BLAST is not feasible for mapping short reads in a reasonable time scale and not guaranteed to give the right answer [37]. Last but not least, identification of large indels with high confidence also requires paired-end data. This is because missing regions can be explained by other factors, such as insufficient sequencing coverage, differences in mapping algorithms when dealing with reads mapped to repeated regions, and so on.
Genes potentially under selections
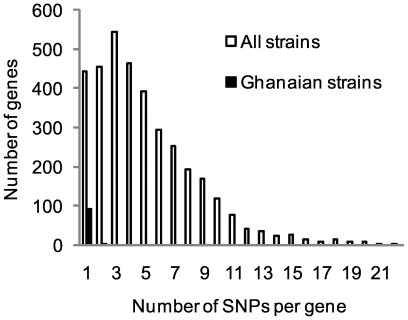
Among the 94 protein coding genes containing SNPs found in the Ghanaian strains, most genes harbored only one SNP (Figure 3). Only three genes contained two SNPs: MUL_2118 (hypothetical protein), MUL_3524 (diphosphomevalonate decarboxylase) and MUL_3716 (nucleoside diphosphate kinase). When considering all strains, 3,597 genes harbored SNPs, most of which contained one to five SNPs (Figure 3), but much higher numbers of SNPs per gene were also observed. For example, the peptide synthetase Nrp gene MUL_2638 contained 59 SNPs, the Pks12 gene MUL_2266 harbored 55 SNPs, and the fatty acid synthase Fas gene MUL_3818 had 30 SNPs. However, all three genes were over 9 Kb in length, which may account for the high number of SNPs. We did find 70 genes with a SNP density higher than 1 per 80 bp, which was one standard deviation higher than the average SNP density (Table 3). More than 50% of these consisted of genes encoding hypothetical proteins. Others included genes associated with antigenic proteins (i.e. esxE, esxF, mpt64), lipoproteins (i.e. dsbF, lppN, lpqV), PE/PE-PGRS family proteins (MUL_4359, MUL_0355), membrane proteins (mmr) and transcription regulators (MUL_2645, MUL_0993). The high number of SNPs may represent evidence for selection pressure on these genes. We thus calculated values of synonymous differences per synonymous site (pS) and nonsynonymous differences per nonsynonymous site (pN) across all SNP harboring loci. The average pN−pS value was −0.00276±0.0073 (P<0.001, pS = 0.0073±0.0063, pN = 0.0046±0.0033), suggesting that on average the frequency of synonymous mutation was significantly higher than the non-synonymous mutation frequency; i.e. there was no evidence for diversifying selection. Among the 70 genes found earlier with high SNP density, five genes (esxE, mmr, MUL_0355, as well as the hypothetical protein genes MUL_0161 and MUL_2106) showed significantly high pN−pS values (above the mean + 3× standard deviations) and might be under diversifying selection, while six genes (glbO, mcmA2b, MUL_1435, MUL_2645, MUL_4312 and MUL_4846) showed significantly low pN−pS values (less than the mean−3× standard deviations), which might be under negative selection (Table 3). Meanwhile, we found 32 genes with average SNP density but significant low pN−pS values, including genes related to transcription and translation (whiB4, trxC, trpG, MUL_0058, MUL_2937, MUL_4776) and another ESAT-6 family protein gene, esxR (Table 3).
Figure 3. Distribution of number of SNPs per gene.
Table 3. Genes potentially under selection ordered by SNP density.
| Locus_tag | Locus | Product | COG | SNP density (bp per SNP) | Selection1 |
| MUL_3769 | - | hypothetical protein | - | 39 | |
| MUL_4312 | - | hypothetical protein | - | 41 | − |
| MUL_4235 | - | hypothetical protein | - | 45 | |
| MUL_5054 | esxE | ESAT-6 like protein EsxE | COG1314U | 46 | + |
| MUL_3425 | mmr | multidrug-transport integral membrane protein Mmr | - | 46 | + |
| MUL_1135 | - | hypothetical protein | COG1902C | 46 | |
| MUL_5072 | gid | glucose-inhibited division protein B Gid | - | 47 | |
| MUL_4359 | - | PE family protein | COG0357M | 48 | |
| MUL_3746 | glbO | globin (oxygen-binding protein) GlbO | - | 48 | − |
| MUL_4906 | - | hypothetical protein | - | 50 | |
| MUL_0630 | - | hypothetical protein | COG0500QR | 51 | |
| MUL_5017 | - | hypothetical protein | - | 52 | |
| MUL_4764 | - | hypothetical protein | COG0500QR | 54 | |
| MUL_2201 | - | hypothetical protein | - | 55 | |
| MUL_0760 | - | hypothetical protein | - | 56 | |
| MUL_3596 | - | hypothetical protein | - | 56 | |
| MUL_1662 | gloA | lactoylglutathione lyase, GloA | - | 56 | |
| MUL_2106 | - | hypothetical protein | COG0315H | 57 | + |
| MUL_4509 | - | hypothetical protein | - | 57 | |
| MUL_4133 | dsbF | lipoprotein DsbF | - | 58 | |
| MUL_0355 | - | PE-PGRS family protein family protein | COG2346R | 60 | + |
| MUL_3885 | echA4_2 | enoyl-CoA hydratase, EchA4_2 | COG1773C | 60 | |
| MUL_5108 | - | transposase | COG1119P | 60 | |
| MUL_0384 | - | hypothetical protein | - | 61 | |
| MUL_5010 | - | phosphoglycerate mutase | COG0406G | 62 | |
| MUL_0161 | - | hypothetical protein | COG2076P | 63 | + |
| MUL_4655 | cysE_1 | serine acetyltransferase CysE_1 | COG1278K | 63 | |
| MUL_2839 | - | hypothetical protein | - | 64 | |
| MUL_2263 | - | hypothetical protein | COG0793M | 64 | |
| MUL_1479 | trxB1 | thioredoxin TrxB1 | COG0526OC | 66 | |
| MUL_4386 | - | hypothetical protein | COG0620E,COG1309K | 67 | |
| MUL_5055 | esxF | ESAT-6 like protein EsxF | COG4842S | 67 | |
| MUL_0327 | - | oxidoreductase | COG1028IQR | 67 | |
| MUL_3206 | - | hypothetical protein | - | 67 | |
| MUL_3717 | - | hypothetical protein | - | 68 | |
| MUL_0010 | - | hypothetical protein | - | 70 | |
| MUL_3264 | fdxA_1 | ferredoxin FdxA_1 | COG1146C | 70 | |
| MUL_1435 | - | exported protein | COG0704P | 70 | − |
| MUL_3277 | - | hypothetical protein | - | 71 | |
| MUL_3581 | - | phage-related integrase | COG0582L | 71 | |
| MUL_1216 | tam | trans-aconitate methyltransferase Tam | COG1522K | 72 | |
| MUL_2917 | - | hypothetical protein | - | 72 | |
| MUL_1771 | - | hypothetical protein | COG1670J | 72 | |
| MUL_0951 | - | hypothetical protein | - | 73 | |
| MUL_0889 | - | hypothetical protein | - | 73 | |
| MUL_0993 | - | transcriptional regulatory protein | COG0236IQ | 73 | |
| MUL_4846 | - | hypothetical protein | - | 73 | − |
| MUL_0366 | mcmA2b | methylmalonyl-CoA mutase alpha subunit, McmA2b | - | 73 | − |
| MUL_1003 | - | hypothetical protein | - | 74 | |
| MUL_4682 | - | hypothetical protein | COG3391S | 74 | |
| MUL_4870 | - | short chain dehydrogenase | COG1028IQR | 74 | |
| MUL_0457 | - | hypothetical protein | COG2351R | 74 | |
| MUL_5109 | - | hypothetical protein | - | 75 | |
| MUL_0761 | - | hypothetical protein | - | 76 | |
| MUL_1274 | lppN | lipoprotein LppN | - | 76 | |
| MUL_2490 | - | hypothetical protein | - | 76 | |
| MUL_0820 | - | methyltransferase | COG0500QR,COG2226H | 76 | |
| MUL_2645 | - | AsnC family transcriptional regulator | COG0526OC | 76 | − |
| MUL_3440 | - | hypothetical protein | COG2185I | 77 | |
| MUL_3194 | - | hypothetical protein | COG2261S | 77 | |
| MUL_0424 | - | hypothetical protein | - | 77 | |
| MUL_5032 | mpt64 | immunogenic protein Mpt64 | COG0425O | 77 | |
| MUL_4365 | - | hypothetical protein | COG0393S | 77 | |
| MUL_4394 | - | hypothetical protein | COG0526OC | 78 | |
| MUL_3305 | ribD | hypothetical protein | COG1985H | 78 | |
| MUL_3524 | - | diphosphomevalonate decarboxylase | COG3407I | 78 | |
| MUL_0217 | lpqV | lipoprotein LpqV | - | 78 | |
| MUL_0241 | bioF2_1 | 8-amino-7-oxononanoate synthase BioF2_1 | COG0156H | 79 | |
| MUL_5058 | - | hypothetical protein | - | 79 | |
| MUL_4336 | - | PE family protein | - | 79 | |
| MUL_4899 | - | hypothetical protein | - | 80 | − |
| MUL_4670 | - | hypothetical protein | - | 82 | − |
| MUL_2937 | - | ArsR-type repressor | COG1846K | 82 | − |
| MUL_0017 | trpG | para-aminobenzoate synthase component II | COG1695K | 86 | − |
| MUL_5067 | trxC | thioredoxin TrxC | COG1522K | 89 | − |
| MUL_2060 | - | hypothetical protein | - | 92 | − |
| MUL_0670 | rimL | acetyltransferase, RimL | COG0664T | 92 | − |
| MUL_4330 | - | hypothetical protein | - | 96 | − |
| MUL_0430 | - | hypothetical protein | COG2608P | 99 | − |
| MUL_1434 | - | hypothetical protein | - | 99 | − |
| MUL_0058 | - | transcriptional regulatory protein | COG1670J | 101 | − |
| MUL_1897 | - | ABC transporter ATP-binding protein | - | 105 | − |
| MUL_5123 | - | hypothetical protein | COG3576R | 105 | − |
| MUL_4776 | - | hypothetical protein | COG1309K | 107 | − |
| MUL_2966 | - | hypothetical protein | - | 110 | − |
| MUL_5035 | - | hypothetical protein | COG0792L | 111 | − |
| MUL_0441 | phoY2 | phosphate-transport system regulatory protein, PhoY2 | - | 112 | − |
| MUL_1835 | secG | preprotein translocase subunit SecG | - | 117 | − |
| MUL_0825 | - | hypothetical protein | COG1359S | 122 | − |
| MUL_4918 | mce6B | MCE-family protein Mce6B | COG1463Q | 129 | − |
| MUL_2369 | - | hypothetical protein | - | 132 | − |
| MUL_0065 | - | hypothetical protein | COG2353S | 136 | − |
| MUL_0035 | - | DNA-binding protein | COG1045E | 136 | − |
| MUL_3556 | - | integral membrane protein | COG0454KR | 140 | − |
| MUL_0015 | - | putative septation inhibitor protein | COG4842S | 141 | − |
| MUL_2243 | esxR | ESAT-6 family protein | - | 146 | − |
| MUL_4627 | - | hypothetical protein | - | 147 | − |
| MUL_2232 | - | molecular chaperone (small heat shock protein) | COG3585H | 150 | − |
| MUL_3337 | - | hypothetical protein | - | 152 | − |
| MUL_3714 | rplU | 50S ribosomal protein L21 | COG0261J | 156 | − |
| MUL_3030 | ureB | urease beta subunit UreB | COG0832E | 156 | − |
| MUL_4051 | - | hypothetical protein | - | 165 | − |
| MUL_4256 | whiB4 | transcriptional regulatory protein Whib-like WhiB4 | - | 183 | − |
“+” represents potential diversifying selection indicated by pN−pS values higher than mean + 3× standard deviation; “−” represents potential negative selection indicated by pN−pS values lower than mean−3× standard deviation.
There has been growing evidence suggesting that some of the ESAT-6 family proteins are involved in the interplay between host and pathogen via either antigenic variation or antigenic drift [38]. ESAT-6 protein encoding genes (esxA esxB) are deleted in M. ulcerans strains of the classical lineage [4],[39], which could contribute to antigen variation and enable the pathogen to escape the immune defense of the host. The Agy99 genome contains 12 genes encoding ESAT-6 like proteins [4]. This may represent a genomic basis for antigenic variation. The duplicated genes might encode antigenically different proteins with the same function. The differential expression of individual genes could enable them to substitute for each other functionally but escape from host immune recognition. The high number of SNPs and significantly high/low pN−pS values we observed here on ESAT-6 family protein genes such as exsE, esxF, esxR, and another secreted antigenic protein gene mpt64, may be the result of selective pressure imposed by the immune system of the host. Mutations that lead to replacement of amino acids within the immunodominant epitopes have been proposed as a mechanism producing antigenic drift [40]. However, it remains to be investigated whether the esx genes are expressed, if their expression is controlled and coordinated, and if the mutations identified here lead to antigenic drift.
We further examined whether certain functional classes of genes were under positive selection by comparing pN−pS values according to the Clusters of Orthologous Group (COG) classification. We found no significant overrepresentation of any functional class in gene group with positive pN−pS values (under diversifying selection), except the “Function unknown” class (p value <0.05). Five functional classes showed overrepresentation of genes with negative pN−pS values, indicating possible selections against amino acid changes, including functional classes J (translation, ribosomal structure and biogenesis), O (posttranslational modification, protein turnover, chaperones), F (nucleotide transport and metabolism), H (coenzyme transport and metabolism) and V (defense mechanisms).
Phylogeny and estimation of the divergence time of M. ulcerans strains
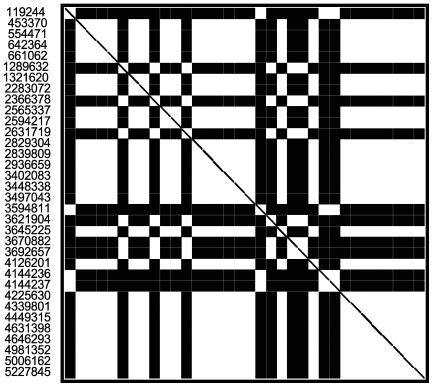
To evaluate the phylogenetic relatedness of the M. ulcerans strains sequenced in this study, we first analyzed the 34 PI SNP sites using the compatibility matrix program [41] to detect the effects of recombination on sequence divergence among the genes harboring these SNPs (Figure 4). In the square matrix, each white square corresponds to two compatible nucleotide sites, at which all nucleotide changes can be inferred to have occurred only once in a phylogeny. Black squares represent incompatible sites, where nucleotide changes are inferred to have occurred multiple times either due to recombination or repeated mutation. We found the 34 PI SNPs formed three groups: SNPs shared by the two Ghanaian test strains (at genomic locations of 1289632, 2366378, 2631719, 3621904, 3670882, 3692657), SNPs shared by NM31/04 and Jp8756 (at genomic locations of 119244, 3594811, 4144236, 4144237) and the rest of the SNPs shared by NM20/02 and Jp8756. The SNPs were compatible within each group but not across groups. The overall compatibility score of all 34 PI SNP sites was 0.5294, which measures the extent of the sites consistent with one phylogeny. The neighbor similarity score of the matrix was 0.6506, which was not significantly higher than scores of 1000 random matrices produced by shuffling the order of sites (mean score 0.6168, P = 0.30), suggesting that recombination among these regions has been rare.
Figure 4. Compatibility matrix of parsimony informative SNPs.
The genome positions are numbered to the left of the matrix. Black squares indicate incompatible sites, where nucleotide changes are inferred to have occurred multiple times either due to recombination or repeated mutation. White squares represent compatible sites, at which all nucleotide changes can be inferred to have occurred only once in a phylogeny.
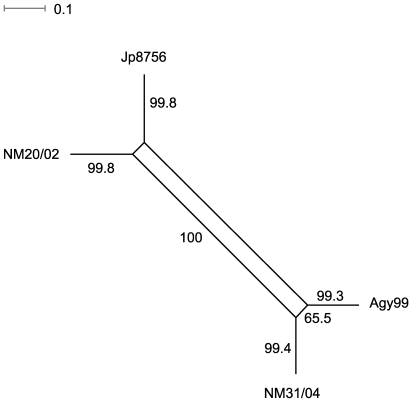
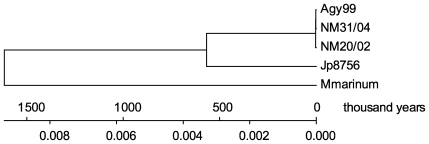
We then used the split decomposition method to detect possible conflicting phylogenetic signals (Bandelt and Dress 1992). NeighborNet Network analysis of the 34 PI SNP sites revealed two parallel paths indicative of the presence of phylogenetic incompatibilities in the divergence of M. ulcerans strains, which could arise from recurrent mutation or recombination in the genomes. However, one parallel path showed much lower bootstrap support value (65.5%) than the other parallel path (100%). The paths with high bootstrap support values (over 99.5%) showed a tree-like network (Figure 5), suggesting that a bifurcating tree is an appropriate model for constructing strain phylogeny. Therefore, we constructed a minimum evolution tree of M. ulcerans strains rooted with M. marinum M to estimate the divergence time (Figure 6). We first compared the 3,597 SNP harboring M. ulcerans genes with the M. marinum M genome and identified 3,059 genes that have homologous genes in M. marinum M [42]. We then generated concatenated allelic sequences of the 3,059 genes for the three test strains and two reference strains (a combined total of 1,032,790 codons in 3,113 kb of coding sequences per strain), based on which a minimum evolution tree of M. ulcerans strains was constructed. Estimated divergence time based on comparison of numbers of synonymous substitutions per nucleotide site (dS) in the 1,032,790 allelic codons are shown in Table 4. The calculations were based on the estimated rate of synonymous substitution in bacteria of 5.8×10−9 to 7.8×10−9 substitution per site per year [43]. Our results suggest that Agy99 and M. marinum M diverged from a common ancestor about 1.13 to 1.52 million years ago, which confirms the recent divergence of M. ulcerans from its M. marinum progenitor, and seems more precise than the earlier estimation of between 1.2 and 4.7 million years [3]. The African classical lineage strain Agy99 and the Japanese ancestral lineage strain Jp8756 diverged about 394 to 529 thousand years ago (Table 4). Although the ancestral lineage was found genetically closer to the progenitor M. marinum in regions of difference (RD) composition [6], the SNP data suggested the Japanese strain was closer to the Ghanaian strains than to M. marinum. The discrepancy between the SNP phylogeny and the RD phylogeny could indicate that regions of M. ulcerans genomes harboring these genetic variations have diversified through different mechanisms at variable rates. The discrepancy could also be due to the small number of strains analyzed in our study. Among Ghanaian M. ulcerans strains the divergence times is less than 3,000 years. However, there are uncertainties in our phylogenetic estimates because of rate heterogeneity. The two-cluster test in LINTREE showed that all the interior nodes within the Ghanaian clade evolved at a uniform rate, while the Japanese strain evolved at a significantly different rate (CP = 99.96%). The branch-length test further indicated that the Japanese strain evolved significantly slower than average, and that all three Ghanaian strains evolved significantly faster than average. Thus, the divergence time of three Ghanaian strains is likely to be shorter than estimated here.
Figure 5. NeighborNet network of the M. ulcerans strains based on the parsimony informative SNPs.
Bootstrap values shown close to branches are based on 1000 bootstrap replicates.
Figure 6. Minimum evolution tree based on 1,032,790 allelic codons of the M. ulcerans and M. marinum strains.
The scale shows the divergence time frame and the number of synonymous substitutions per nucleotide site. The rate of synonymous substitution used for time calibration was 5.8×10−9 substitution per site per year.
Table 4. Estimation of time of divergence between M. ulcerans Agy99 and other M. ulcerans and M. marinum strains.
| Strain | ds 1 | Est. divergence time2 (yr) | Est. divergence time3 (yr) |
| NM31/04 | 0.000017±0.000004 | 1,466±345 | 1,090±256 |
| NM20/02 | 0.00003±0.000006 | 2,586±517 | 1,923±385 |
| Jp8756 | 0.006141±0.000081 | 529,397±6,983 | 393,654±5,192 |
| M. marinum M | 0.017642±0.000138 | 1,520,862±11,897 | 1,130,897±8,846 |
mean±standard error
Based on the rate of synonymous substitution of 5.8×10−9 per site per year
Based on the rate of synonymous substitution of 7.8×10−9 per site per year
Genetic diversity among Ghanaian clinical isolates
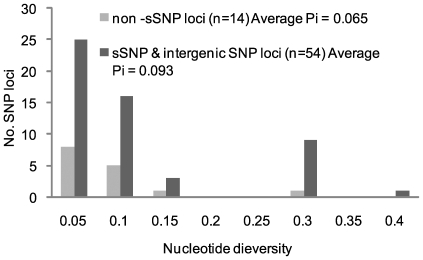
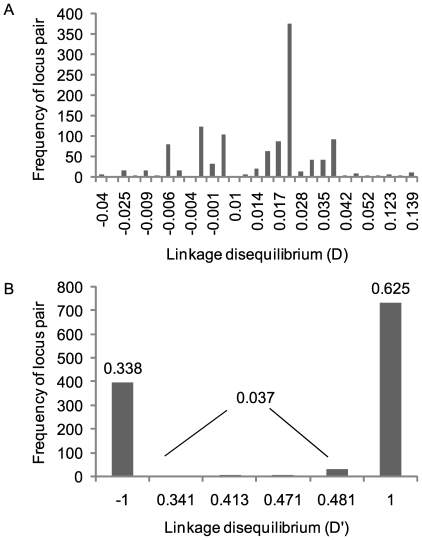
We are developing hairpin primer SNP assays [44] based on the 173 SNP loci discovered through pairwise comparisons of the three M. ulcerans Ghanaian strains and are analyzing our collection of Ghanaian M. ulcerans isolates at these loci. At the current stage it is possible to resolve the nucleotides at 68 SNP loci in 54 Ghanaian strains with 13 distinct SNP haplotypes identified (including the three reference strains). Previously only 3 haplotypes were found in this strain collection by VNTR typing, which was the best resolution achieved [7]. The nucleotide diversity ranged from 0.05 to 0.40 across the 23 sSNP and 31 intergenic SNP loci with an average diversity of 0.09 (Figure 7). Across the 14 non-sSNP, the average nucleotide diversity was 0.06. This level of nucleotide diversity means that two isolates selected at random from this collection will differ at a SNP locus in 6–9% of the cases. The alleles at the sSNP loci were highly nonrandom in their haplotype distribution. This statistical association can be seen in the distribution of the linkage disequilibrium coefficient (D) for 1,176 pairwise comparisons of alleles at 54 sSNP and intergenic SNP loci (Figure 8). A total of 751 (64%) of these comparisons were significant by a chi-squared test, and 600 (51%) were significant using a highly conservative Bonferroni correction for multiple tests [45]. The standardized coefficient of linkage disequilibrium (D′) was strongly U-shaped with 96% of the locus pairs in complete linkage disequilibrium. This observation indicates that there are at most three out of the four possible haplotypes for most locus pairs. It also suggests that recurrent mutation and recombination events have played only a minor role in generating haplotype diversity. Detailed epidemiological analysis of the typing results is out of the scope of this paper and will be summarized elsewhere (manuscript in preparation).
Figure 7. Nucleotide diversity among SNPs identified through genome comparison of three Ghanaian strains, for which complete SNP data have been collected in 54 Ghanaian M. ulcerans strains.
Figure 8. Linkage disequilibrium among study loci.
A). The distribution of the linkage disequilibrium coefficient (D) for 1,176 pairwise comparisons of alleles at 54 sSNP and intergenic SNP loci. A total of 751 (64%) of these comparisons are significant by a chi-squared test, and600 (51%) remained significant using a Bonferroni correction for multiple tests. B). The distribution of the standardized coefficient of linkage disequilibrium (D'). Ninety-six percent of the locus pairs are in complete linkage disequilibrium.
Discussion
In this study we have sequenced genomes of two Ghanaian and one Japanese M. ulcerans disease isolates by using two different massively parallel sequencing platforms. By comparison of genome sequences of these strains with the genome of the Ghanaian reference strain Agy99, we have identified over a hundred SNPs within Ghanaian strains and about 26,000 SNPs in the Japanese strain. The SNPs identified among Ghanaian strains for the first time allow to resolve the population structure and evolutionary relationship of an intra-continental population of M. ulcerans. The SNP data confirmed the recent divergence of M. ulcerans and M. marinum from a common ancestor and refined the estimated time of emergence to about 1.3 million years. We also estimated that the classical M. ulcerans lineage (represented by the Ghanaian strains) diverged from the ancestral lineage (represented by the Japanese strain) about 400,000 years ago, which is about the time when the modern human species, Homo sapiens, evolved (∼250,000 to 400,000 years). However, we need to point out two limiting factors of our current estimates. First is the limited number of strains analyzed. Future studies should include many more strains from different countries and continents representing branches of the two lineages to improve the phylogenetic resolution and accuracy of the dating. Second is that our dating was based on the estimated rate of synonymous substitution in bacteria of 5.8×10−9 to 7.8×10−9 substitution per site per year, which was not the most appropriate clock rate for calculating the age of genetically monomorphic pathogens [46]. However, till now studies trying to establish the ideal molecular clock calibrated against a fossil record have not yielded much usable new information [46]. The clock rate we used has been used to date other monomorphic pathogens such as M. ulcerans [3], M. tuberculosis [43],[47] and Escherichia coli [48], and our estimated age of M. ulcerans is thus still informative and comparable to these previous estimations.
The SNP analysis suggested that the Japanese strain was closer to the Ghanaian strains than to M. marinum, which is contradictory to the previous finding based on RD composition, where the ancestral lineage was found genetically closer to the progenitor M. marinum [6]. The discrepancy might suggest that regions of M. ulcerans genomes harboring these genetic variations have diversified through different mechanisms at variable rates. However, more strains within the ancestral lineage should be analyzed before firm conclusions can be drawn. Among Ghanaian strains the divergence time was less than 3000 years. Two-cluster and branch-length tests in this study revealed that the Ghanaian strains evolved significantly faster than average, thus the actual divergence time within Ghana could be much shorter than our current estimate. M. ulcerans was first isolated in 1948 [49], but large ulcers almost certainly caused by M. ulcerans have been already described by Sir Albert Cook in 1897. Since the late 1980s the number of reported cases has increased dramatically in West Africa [50],[51],[52],[53] and in Australia [54]. Increased disease incidence has been primarily attributed to factors such as environmental changes, increased exposure of the affected populations, and improved surveillance. VNTR typing results gave indications for the emergence and spreading of new genetic variants of M. ulcerans within Ghana [7]. More in-depth phylogenetic and functional analyses are needed to test if mechanisms such as virulence evolution and host adaptation of M. ulcerans play a role in the increasing incidence of BU.
Genome comparison of M. ulcerans Agy99 and M. marinum M revealed that M. ulcerans underwent reductive evolution with genomic signatures such as proliferation of ISE, accumulation of pseudogenes, chromosomal rearrangements, genome downsizing, and acquisition of foreign genes by acquisition of plasmids or bacteriophages [4]. Microarray based comparative genomic hybridization with a worldwide set of M. ulcerans isolates identified genomic regions of difference and demonstrated that the two major M. ulcerans lineages can be distinguished based on the location and size of genomic deletions [5],[6]. Due to the lack of paired-end sequencing data, we did not carry out systematic analysis on chromosomal deletions and rearrangements. But when we tried to close sequencing gaps using the Agy99 chromosome as the reference, there were more successful gap closure reactions in Ghanaian strains than in the Japanese strain. Failed gap closure attempts in Ghanaian strains were mostly around IS elements, while this was not the case for the Japanese strain (data not shown). These results suggest a high frequency of large chromosomal rearrangement events in the Japanese strain compared to the African classical lineage strains. In the two Ghanaian strain, NM31/04 and NM20/02, the plasmid was fully covered by sequence reads and almost identical to pMUM001 in Agy99, while the plasmid in the Japanese strain was only partially covered with low depth. Altogether these observations suggest a more stabilized genome and a less important role of reductive evolution within M. ulcerans Ghanaian strains.
SNP analysis revealed no further pseudogene formation within the Ghanaian strains, and the Ghanaian strains were found to evolve significantly faster than average. While the majority of coding region SNPs found in Jp8756 was synonymous, the majority found in the two Ghanaian strains was non-synonymous. Further functional analysis of genes containing these non-sSNPs would help to elucidate if they could lead to “pathoadaptive” niche expansion, or provide a selective advantage in both sporadic infection and epidemic spread, which have been found in other bacterial pathogens [55],[56] and suggested for M. ulcerans [39],[57].
The low genetic diversity and high linkage disequilibrium within Ghanaian isolates supports the hypothesis that the M. ulcerans population spread over the African continent has gone through a severe bottleneck during adaptation to a possibly host-specific environment and has not yet accumulated much sequence diversity [15]. SNP typing of Ghanaian isolates was consistent with VNTR typing [7], but allowed to further differentiate between isolates coming from the same BU endemic focus. Closely related, but distinct clonal complexes including strains with minor variation seem to dominate in different BU endemic areas. Diversity of these local clonal complexes is indicative for ongoing microevolution.
Although there has been impressive recent progress in studying the transmission of BU, the precise environmental reservoirs and mode(s) of transmission are not fully understood [58],[59]. High-throughput genotyping platforms, such as Hairpin primer real time PCR assays [44], BeadArray [60] and OpenArray [61] will make genome wide SNP typing a highly discriminatory and cost-effective tool to study M. ulcerans evolution and epidemiology at a population scale. More in-depth phylogenetic and phenotypic analyses of a large number of disease isolates and environmental strains (once becoming available) is expected to shed more light into transmission and virulence evolution of M. ulcerans after its divergence from M. marinum. The study will also help to identify SNPs associated with host specificity and geographical origins. However, the current set of SNP markers were obtained by comparing genomes of three very closely related Ghanaian strains and one very distant Japanese strain. Future genome sequencing of more representative strains from diverse locations around the world will be necessary to identify additional SNP markers to delineate the origin and spread of M. ulcerans at both local and global level.
Materials and Methods
Bacterial strains and genomic DNA isolation
The three M. ulcerans strains sequenced with various sequencing platforms are listed in Table 1. Strain Jp8756 (ATCC 33728) from Japan was provided by Francoise Portaels (Institute of Tropical Medicine, Antwerp, Belgium). Isolation and characterization of strains NM31/04 and NM20/02 from Ghana has been described elsewhere [7],[8],[62],[63]. We used the complete Agy99 genome sequence (chromosome, NC_008611, and the plasmid pMUM001, NC_005916) as the reference sequences. Genomic DNA was isolated as described [64].
Pyrosequencing of M. ulcerans NM20/02
We sequenced the genome of M. ulcerans NM20/02 using the Roche 454 Life Sciences Genome Sequencer FLX following the manufacturer's instructions (Roche 454 Life Science, Branford, CT, USA). The shotgun library was prepared with 5 µg genomic DNA using the “Standard DNA Library Preparation Kit” (04852265001, Roche). Nebulized, purified, and adaptors attached single strand DNA fragments were clonally amplified using the “Emulsion PCR Kit I” (04852290001, Roche). Sequencing on the GS FLX was performed using the “Standard LR70 Sequencing Kit” (04932315001, Roche). Images were processed using the “Genome Sequencer FLX Data Processing Pipeline 1.1.02.15”. A total of 424,494 reads with average length of 213 bases were obtained.
We mapped 454 reads to the reference sequence using gsMapper (v1.1.02.15, Roche) to make SNP calls. To identify high confidence SNPs, we filtered substitutions reported by gsMapper using two rules: (1) supported by at least three non-duplicated reads, with at least one aligned in the forward direction and at least one aligned in the reverse direction. (2) Requirement for at least five conserved bases on both sides of the SNPs. We randomly selected 37 SNPs distributed across the genome for PCR and capillary sequencing. 29 SNPs were validated, three were miscalled due to sequencing errors in the Agy99 genome, and five failed due to unspecific PCR reactions (Table S3). After manual inspection, we found that each of the five SNPs was supported by all the reads (7 to 16) uniquely mapped to the region, thus was likely to be a true SNP. Unspecific PCR reactions might be due to the presence of genes paralogous to genes harboring or flanking the SNPs.
Solexa sequencing of M. ulcerans NM31/04 and Jp8756
We sequenced the genomes of M. ulcerans NM31/04 and Jp8756 with the Illumina Genome Analyzer according to the manufacturer's instructions (Illumina, San Diego, CA, USA). The DNA Colony/Cluster Template Library was prepared using the Illumina “Genomic DNA Sample Prep Kit” (Illumina). Briefly, 5 µg of genomic DNA was broken into fragments of approximately 100 bp by nebulization. After end repairing and adaptor ligation, the samples were gel-purified to recover fragments of 150–250 bp, which were PCR amplified for 15 cycles. For quality control, an aliquot of the library was cloned into a TOPO plasmid. Six clones from each bacterial strain were picked and subjected to capillary sequencing. The DNA Colony/Cluster Template Library was then used for flow-cell preparation using the “Standard Cluster Generation Kit” (Illumina). Sequencing on the Illumina Genome Analyzer was performed using “Genomic DNA sequencing primer V2” for 36 cycles. At the end of the run, images were processed using the “Solexa Data Analysis Pipeline 0.2.2.6”. A total of 2.538 and 2.651 million reads of 35 bases in length were obtained for NM31/04 and Jp8756, respectively.
We mapped Solexa reads to the reference sequences using MAQ v0.6.3, which is particularly developed for building mapping assemblies from Illumina Solexa reads. For each read, un-gapped alignment against the reference was performed; all hits with up to 2 mismatches in the first 24 bp were found. Each read was placed to the position where the sum of quality values of the mismatched nucleotides is minimum [33]. For detection of high confidence SNPs, we first filtered the SNP calls reported by MAQ based on two rules: (1) supported by more than three non-duplicated reads; (2) covered by at least one read with a mapping quality higher than 40. We then randomly selected 84 SNPs called in NM31/04 or Jp8756 for PCR and capillary sequencing to determine the cutoff value of the consensus quality. We found that the consensus quality equal or higher than 40 excluded most false positive SNP calls and that the false positive rate was about 1% (Figure S3).
NimbleGen comparative genome sequencing (CGS) of selective regions in M. ulcerans Jp8756
Before next-generation sequencing became widely available, NimbleGen CGS was the cost effective tool for the comparative analysis of microbial genomes to identify SNPs, insertions, or deletions with high speed and accuracy [65]. We first used a mutation mapping array tiling the reference genome to locate potential mutation harboring sites. Then a high density re-sequencing array tiling the putative sites was produced to identify the mutations [66]. The method has been applied successfully to survey the entire or partial genomes of several bacteria [48],[67],[68], including M. tuberculosis [69].
For CGS analysis of M. ulcerans Jp8756, we selected 1,265 of the 4,160 protein coding genes across the chromosome of Agy99 (1,210,734 bp out of 5,805,761 bp, 20%) and 51 of the 81 protein coding genes on the plasmid pMUM001 (135,612 bp out of 174,155 bp, 78%) (Table S2). These genes include drug resistance genes, known antigens, genes with housekeeping roles, and genes of hypothetical proteins. To avoid cross hybridization and ambiguous SNP calls, we excluded protein coding genes with paralogs in the M. ulcerans genome. Highly conserved genes between M. ulcerans and its ancestor M. marinum were also excluded. Mutation mapping and re-sequencing probes were designed for selected gene using the ArrayScribe software, synthesized using the Maskless Array Synthesis (MAS) technology [70],[71], and printed in a random layout. Genomic DNA samples from Jp8756 and the reference Agy99 were hybridized to the arrays separately following the NimbleGen protocol. Data were analyzed using NimbleScan software (NimbleGen). A list of 1,619 identified SNPs is provided in Table S2.
SNP validation
We used Sanger sequencing of PCR products to validate a selected subset of SNPs. Primer sequences used for PCR and sequencing are provided in Table S3. PCR products were purified using the NucleoSpin Extract II Kit (Clontech Laboratories, Mountain View, CA).
SNP typing
We are developing hairpin primer (HP) assays [44] for SNPs discovered through pairwise comparisons of the three M. ulcerans Ghanaian strains (Agy99, NM20/02, and NM31/04) and analyzing our collection of clinical isolates from Ghana [7]. All HP assays were tested on the three reference strains to confirm the presence of each allele and to verify the performance of the SNP assays. Assays on the clinical DNA samples were considered reliable only if the cycle thresholds generated in the paired wells differed by three or more cycles. At the reported stage, it was possible to assign alleles to 68 SNP loci in 54 Ghanaian isolates (including the reference strains) using this approach.
Phylogenetic analysis
To identify putative regions of recombination or gene conversion, we used the Reticulate program and constructed a compatibility matrix [41]. We also used the SplitsTree program to detect conflicting phylogenetic information and determine if a bifurcating tree is an appropriate model to construct strain phylogeny [72]. A minimum evolution tree rooted with M. marinum was constructed by using the MEGA [73] software based on the numbers of synonymous substitutions per nucleotide site in concatenated SNP harboring protein coding genes in all strains. The numbers of synonymous substitutions per synonymous site were calculated from the concatenated nucleotide sequences using the modified Nei-Gojobori Jukes Cantor method. The Complete deletion method was used for handling alignment gaps. Because rate heterogeneity may have contributed to uncertainty in phylogeny estimates, we applied the two-cluster and branch-length tests in LINTREE [74] to identify significant rate heterogeneity in the phylogeny. The two cluster test was used to test the molecular clock hypothesis for the two lineages above each interior node of a tree and the branch-length test was used to examine the deviation of each root-to-tip branch length relative to the average length [74]. To generate the concatenated sequences, we first determined homologous genes in the M. marinum M genome [42] for each of the 3,597 protein coding gene harboring SNPs by standalone BLAST search [75]. Using the threshold of 90% nucleotide sequence identity over a minimum alignment length of 90% of both query and hit genes, 3,059 homologous genes were identified. Protein sequences of the homologous genes were aligned using the CLUSTALW program [76]. Homologous genes were then aligned using the EMBOSS [77] Tranalign program so that the corresponding amino acid sequence alignment was imposed on the DNA sequence alignment. Allelic genes in M. ulcerans strains and homologous genes in M. marinum M with alignment gaps were then concatenated.
Accession numbers
The sequencing reads of the NM20/02, NM31/04 and Jp8756 genomes have been deposited in the NCBI Short Read Archive database under the accession number SRA008258.
Supporting Information
Coverage of the plasmid pMUM001 by NM20/02 GS FLX reads (A, B), NM31/04 Solexa reads (C, D) and Jp8756 Solexa reads (E, F). X axes represent genomic region of pMUM001 (1 to 174,155 ). The Y axes show the coverage depth. In A, C, and E, all mapped reads were recorded and reads mapped to multiple locations were counted multiple times, while only uniquely mapped reads were counted in B, D, and F.
(0.30 MB PDF)
Absence of pMUM001 plasmid genes in Jp8756 revealed by NimbleGen microarrays. Most hybridization signal intensities from Jp8756 (in Green) were close to zero in both mutation mapping and re-sequencing of selected pMUM001 plasmid genes (A and B), while hybridization signal intensities from Agy99 (in Blue) were properly detected. Hybridization signal intensities from both strains were properly detected in mutation mapping and re-sequencing of selected chromosomal genes (C and D). X axes represent genomic location of oligo probes (1 to 174,155 in A and B, 1 to 5,631,606 in C and D). Y axes show h hybridization signal intensities.
(2.45 MB EPS)
Empirically determined MAQ consensus quality scores improved the accuracy of SNP calls in M. ulcerans Solexa data. Sanger sequencing of potential SNP harboring regions in Jp8756 and NM31/04 revealed that MAQ consensus quality scores equal or higher than 40 excluded most false positive SNP calls.
(0.71 MB EPS)
SNPs detected among four M. ulcerans genomes. Coordinates correspond to Agy99 finished genome sequence (NCBI:NC_008611 for Agy99 chromosome and NCBI:NC_005916 for Agy99 pMUM001).
(7.21 MB XLS)
SNPs identified by NimbleGen re-sequencing array in selected chromosomal and plasmid genes. SNPs confirmed by Solexa Genome Analyzer are indicated in the last column.
(0.52 MB XLS)
Primers used for PCR and sequencing of SNP harboring regions.
(0.05 MB XLS)
Acknowledgments
We thank the Michigan State University Research Technology Support Facility, Fasteris, and Roche NimbleGen for sequencing services, and the [BC]2 Basel Computational Biology Center at the Biozentrum of the University of Basel for hardware and software support. We thank David Alland, Manzour Hernando Hazbón, and Alifiya S. Motiwala at the University of Medicine and Dentistry of New Jersey, as well as David W. Lacher at the US Food and Drug Administration for original protocols on hair-pin primer design and real time PCR experiments. We also thank Francoise Portaels at the Institute of Tropical Medicine, Antwerp, for providing us with strain ITM Japan8756, and Dieter Ebert of the Zoological Institute at the University of Basel for helpful discussions and a critical reviewing of the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the Swiss Tropical Institute Seed Money Fund (http://www.sti.ch/) and the Stop Buruli initiative funded by UBS Optimus Foundation (http://www.ubs.com/1/e/wealthmanagement/optimusfoundation/commitment/global_research/buruli.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. Buruli ulcer. Relevé épidémiologique hebdomadaire / Section d'hygiène du Secrétariat de la Société des Nations (Weekly Epidemiological Record / Health Section of the Secretariat of the League of Nations) 2000;75:106–108. [Google Scholar]
- 2.Johnson PDR, Stinear T, Small PLC, Pluschke G, Merritt RW, et al. Buruli ulcer (M. ulcerans infection): new insights, new hope for disease control. PLoS Med. 2005;2:e108. doi: 10.1371/journal.pmed.0020108. doi: 10.1371/journal.pmed.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stinear TP, Jenkin GA, Johnson PD, Davies JK. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. Journal of Bacteriology. 2000;182:6322–6330. doi: 10.1128/jb.182.22.6322-6330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Research. 2007;17:192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rondini S, Käser M, Stinear T, Tessier M, Mangold C, et al. Ongoing genome reduction in Mycobacterium ulcerans. Emerging Infectious Diseases. 2007;13:1008–1015. doi: 10.3201/eid1307.060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Käser M, Rondini S, Naegeli M, Stinear T, Portaels F, et al. Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans. BMC Evolutionary Biology. 2007;7:177–177. doi: 10.1186/1471-2148-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilty M, Yeboah-Manu D, Boakye D, Mensah-Quainoo E, Rondini S, et al. Genetic diversity in Mycobacterium ulcerans isolates from Ghana revealed by a newly identified locus containing a variable number of tandem repeats. Journal of Bacteriology. 2006;188:1462–1465. doi: 10.1128/JB.188.4.1462-1465.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stragier P, Ablordey A, Bayonne LM, Lugor YL, Sindani IS, et al. Heterogeneity among Mycobacterium ulcerans isolates from Africa. Emerging Infectious Diseases. 2006;12:844–847. doi: 10.3201/eid1205.051191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stinear T, Ross BC, Davies JK, Marino L, Robins-Browne RM, et al. Identification and Characterization of IS2404 and IS2606: Two Distinct Repeated Sequences for Detection of Mycobacterium ulcerans by PCR. J Clin Microbiol. 1999;37:1018–1023. doi: 10.1128/jcm.37.4.1018-1023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stinear T, Davies JK, Jenkin GA, Portaels F, Ross BC, et al. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. Journal of Clinical Microbiology. 2000;38:1482–1487. doi: 10.1128/jcm.38.4.1482-1487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huys G, Rigouts L, Chemlal K, Portaels F, Swings J. Evaluation of amplified fragment length polymorphism analysis for inter- and intraspecific differentiation of Mycobacterium bovis, M. tuberculosis, and M. ulcerans. Journal of Clinical Microbiology. 2000;38:3675–3680. doi: 10.1128/jcm.38.10.3675-3680.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chemlal K, Huys G, Fonteyne PA, Vincent V, Lopez AG, et al. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. Journal of Clinical Microbiology. 2001;39:3272–3278. doi: 10.1128/JCM.39.9.3272-3278.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chemlal K, De Ridder K, Fonteyne PA, Meyers WM, Swings J, et al. The use of IS2404 restriction fragment length polymorphisms suggests the diversity of Mycobacterium ulcerans from different geographical areas. The American Journal of Tropical Medicine and Hygiene. 2001;64:270–273. doi: 10.4269/ajtmh.2001.64.270. [DOI] [PubMed] [Google Scholar]
- 14.Stinear TP, Hong H, Frigui W, Pryor MJ, Brosch R, et al. Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. Journal of Bacteriology. 2005;187:1668–1676. doi: 10.1128/JB.187.5.1668-1676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yip MJ, Porter JL, Fyfe JAM, Lavender CJ, Portaels Fo, et al. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. Journal of Bacteriology. 2007;189:2021–2029. doi: 10.1128/JB.01442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ablordey A, Hilty M, Stragier P, Swings J, Portaels Fo. Comparative nucleotide sequence analysis of polymorphic variable-number tandem-repeat Loci in Mycobacterium ulcerans. Journal of Clinical Microbiology. 2005;43:5281–5284. doi: 10.1128/JCM.43.10.5281-5284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stragier P, Ablordey A, Meyers WM, Portaels Fo. Genotyping Mycobacterium ulcerans and Mycobacterium marinum by using mycobacterial interspersed repetitive units. Journal of Bacteriology. 2005;187:1639–1647. doi: 10.1128/JB.187.5.1639-1647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ablordey A, Kotlowski R, Swings J, Portaels Fo. PCR amplification with primers based on IS2404 and GC-rich repeated sequence reveals polymorphism in Mycobacterium ulcerans. Journal of Clinical Microbiology. 2005;43:448–451. doi: 10.1128/JCM.43.1.448-451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentley DR. Whole-genome re-sequencing. Current Opinion in Genetics & Development. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 20.von Bubnoff A. Next-Generation Sequencing: The Race Is On. Cell. 2008;132:721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 21.Droege M, Hill B. The Genome Sequencer FLX System–longer reads, more applications, straight forward bioinformatics and more complete data sets. Journal of Biotechnology. 2008;136:3–10. doi: 10.1016/j.jbiotec.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 22.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 23.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends in Genetics: TIG. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Spatz SJ, Rue CA. Sequence determination of a mildly virulent strain (CU-2) of Gallid herpesvirus type 2 using 454 pyrosequencing. Virus Genes. 2008;36:479–489. doi: 10.1007/s11262-008-0213-5. [DOI] [PubMed] [Google Scholar]
- 25.Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill Fo-X, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nature Genetics. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotewicz ML, Mammel MK, LeClerc JE, Cebula TA. Optical mapping and 454 sequencing of Escherichia coli O157:H7 isolates linked to the US 2006 spinach-associated outbreak. Microbiology (Reading, England) 2008;154:3518–3528. doi: 10.1099/mic.0.2008/019026-0. [DOI] [PubMed] [Google Scholar]
- 27.Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4868–4873. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bekal S, Craig JP, Hudson ME, Niblack TL, Domier LL, et al. Genomic DNA sequence comparison between two inbred soybean cyst nematode biotypes facilitated by massively parallel 454 micro-bead sequencing. Molecular Genetics and Genomics: MGG. 2008;279:535–543. doi: 10.1007/s00438-008-0331-8. [DOI] [PubMed] [Google Scholar]
- 29.Novaes E, Drost DR, Farmerie WG, Pappas GJ, Grattapaglia D, et al. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics. 2008;9:312–312. doi: 10.1186/1471-2164-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, et al. Whole-genome sequencing and variant discovery in C. elegans. Nature Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 31.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Wang W, Li R, Li Y, Tian G, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Research. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stinear TP, Mve-Obiang A, Small PLC, Frigui W, Pryor MJ, et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong H, Demangel C, Pidot SJ, Leadlay PF, Stinear T. Mycolactones: immunosuppressive and cytotoxic polyketides produced by aquatic mycobacteria. Natural Product Reports. 2008;25:447–454. doi: 10.1039/b803101k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan AR, Stewart DA, Strömberg MP, Marth GbT. Pyrobayes: an improved base caller for SNP discovery in pyrosequences. Nature Methods. 2008;5:179–181. doi: 10.1038/nmeth.1172. [DOI] [PubMed] [Google Scholar]
- 37.Bloksberg LN. Next-Generation Sequence Data Analysis. In: Janize M, editor. Next-Generation Genome Sequencing. Weinheim: WILEY-VCH Verlag GmbH & Co KGaA; 2008. pp. 79–88. [Google Scholar]
- 38.Brodin P, Rosenkrands I, Andersen P, Cole ST, Brosch R. ESAT-6 proteins: protective antigens and virulence factors? Trends in Microbiology. 2004;12:500–508. doi: 10.1016/j.tim.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Huber CA, Ruf M-T, Pluschke G, Käser M. Independent loss of immunogenic proteins in Mycobacterium ulcerans suggests immune evasion. Clinical and Vaccine Immunology: CVI. 2008;15:598–606. doi: 10.1128/CVI.00472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skjot RLV, Brock I, Arend SM, Munk ME, Theisen M, et al. Epitope Mapping of the Immunodominant Antigen TB10.4 and the Two Homologous Proteins TB10.3 and TB12.9, Which Constitute a Subfamily of the esat-6 Gene Family. Infect Immun. 2002;70:5446–5453. doi: 10.1128/IAI.70.10.5446-5453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jakobsen IB, Easteal S. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput Appl Biosci. 1996;12:291–295. doi: 10.1093/bioinformatics/12.4.291. [DOI] [PubMed] [Google Scholar]
- 42.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Research. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kapur V, Whittam TS, Musser JM. Is Mycobacterium tuberculosis 15,000 years old? The Journal of Infectious Diseases. 1994;170:1348–1349. doi: 10.1093/infdis/170.5.1348. [DOI] [PubMed] [Google Scholar]
- 44.Hazbon MH, Alland D. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. Journal of Clinical Microbiology. 2004;42:1236–1242. doi: 10.1128/JCM.42.3.1236-1242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 46.Achtman M. Evolution, Population Structure, and Phylogeography of Genetically Monomorphic Bacterial Pathogens. Annual Review of Microbiology. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 47.Hughes AL, Friedman R, Murray M. Genomewide pattern of synonymous nucleotide substitution in two complete genomes of Mycobacterium tuberculosis. Emerging Infectious Diseases. 2002;8:1342–1346. doi: 10.3201/eid0811.020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, et al. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Research. 2006;16:757–767. doi: 10.1101/gr.4759706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacCullum P. A new mycobacterial infection in man; clinical aspects. The Journal of Pathology and Bacteriology. 1948;60:93–102. [PubMed] [Google Scholar]
- 50.Debacker M, Aguiar J, Steunou C, Zinsou C, Meyers WM, et al. Buruli ulcer recurrence, Benin. Emerging Infectious Diseases. 2005;11:584–589. doi: 10.3201/eid1104.041000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marston BJ, Diallo MO, Horsburgh CR, Diomande I, Saki MZ, et al. Emergence of Buruli ulcer disease in the Daloa region of Cote d'Ivoire. The American Journal of Tropical Medicine and Hygiene. 1995;52:219–224. doi: 10.4269/ajtmh.1995.52.219. [DOI] [PubMed] [Google Scholar]
- 52.Debacker M, Aguiar J, Steunou C, Zinsou C, Meyers WM, et al. Mycobacterium ulcerans disease (Buruli ulcer) in rural hospital, Southern Benin, 1997–2001. Emerging Infectious Diseases. 2004;10:1391–1398. doi: 10.3201/eid1008.030886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amofah G, Bonsu F, Tetteh C, Okrah J, Asamoa K, et al. Buruli ulcer in Ghana: results of a national case search. Emerging Infectious Diseases. 2002;8:167–170. doi: 10.3201/eid0802.010119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson PD, Veitch MG, Leslie DE, Flood PE, Hayman JA. The emergence of Mycobacterium ulcerans infection near Melbourne. The Medical Journal of Australia. 1996;164:76–78. doi: 10.5694/j.1326-5377.1996.tb101352.x. [DOI] [PubMed] [Google Scholar]
- 55.Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu X-R, et al. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weissman SJ, Moseley SL, Dykhuizen DE, Sokurenko EV. Enterobacterial adhesins and the case for studying SNPs in bacteria. Trends in Microbiology. 2003;11:115–117. doi: 10.1016/s0966-842x(03)00010-6. [DOI] [PubMed] [Google Scholar]
- 57.Käser M, Pluschke G. Differential Gene Repertoire in Mycobacterium ulcerans Identifies Candidate Genes for Patho-Adaptation. PLoS Negl Trop Dis. 2008;2:e353. doi: 10.1371/journal.pntd.0000353. doi: 10.1371/journal.pntd.0000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benbow ME, Williamson H, Kimbirauskas R, McIntosh MD, Kolar R, et al. Aquatic invertebrates as unlikely vectors of Buruli ulcer disease. Emerging Infectious Diseases. 2008;14:1247–1254. doi: 10.3201/eid1408.071503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stragier P, Hermans K, Stinear T, Portaels Fo. First report of a mycolactone-producing Mycobacterium infection in fish agriculture in Belgium. FEMS Microbiology Letters. 2008;286:93–95. doi: 10.1111/j.1574-6968.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- 60.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. BioTechniques. 2002;(Suppl):56–58, 60-51-56-58, 60-51. [PubMed] [Google Scholar]
- 61.Stedtfeld RD, Baushke SW, Tourlousse DM, Miller SM, Stedtfeld TM, et al. Development and experimental validation of a predictive threshold cycle equation for quantification of virulence and marker genes by high-throughput nanoliter-volume PCR on the OpenArray platform. Applied and Environmental Microbiology. 2008;74:3831–3838. doi: 10.1128/AEM.02743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeboah-Manu D, Bodmer T, Mensah-Quainoo E, Owusu S, Ofori-Adjei D, et al. Evaluation of decontamination methods and growth media for primary isolation of Mycobacterium ulcerans from surgical specimens. Journal of Clinical Microbiology. 2004;42:5875–5876. doi: 10.1128/JCM.42.12.5875-5876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mensah-Quainoo E, Yeboah-Manu D, Asebi C, Patafuor F, Ofori-Adjei D, et al. Diagnosis of Mycobacterium ulcerans infection (Buruli ulcer) at a treatment centre in Ghana: a retrospective analysis of laboratory results of clinically diagnosed cases. Tropical Medicine & International Health: TM & IH. 2008;13:191–198. doi: 10.1111/j.1365-3156.2007.01990.x. [DOI] [PubMed] [Google Scholar]
- 64.Käser M, Ruf M-T, Hauser J, Marsollier L, Pluschke G. Optimized Method for Preparation of DNA from Pathogenic and Environmental Mycobacteria. Appl Environ Microbiol. 2009;75:414–418. doi: 10.1128/AEM.01358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Albert TJ, Dailidiene D, Dailide G, Norton JE, Kalia A, et al. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nature Methods. 2005;2:951–953. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]
- 66.Albert TJ, Norton J, Ott M, Richmond T, Nuwaysir K, et al. Light-directed 5′→3′ synthesis of complex oligonucleotide microarrays. Nucleic Acids Research. 2003;31:e35–e35. doi: 10.1093/nar/gng035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 68.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-Wide Analysis of Group A Streptococci Reveals a Mutation That Modulates Global Phenotype and Disease Specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manjunatha UH, Boshoff H, Dowd CS, Zhang L, Albert TJ, et al. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:431–436. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nuwaysir EF, Huang W, Albert TJ, Singh J, Nuwaysir K, et al. Gene Expression Analysis Using Oligonucleotide Arrays Produced by Maskless Photolithography. Genome Research. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh-Gasson S, Green RD, Yue Y, Nelson C, Blattner F, et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nature Biotechnology. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- 72.Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 73.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 74.Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 75.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 76.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coverage of the plasmid pMUM001 by NM20/02 GS FLX reads (A, B), NM31/04 Solexa reads (C, D) and Jp8756 Solexa reads (E, F). X axes represent genomic region of pMUM001 (1 to 174,155 ). The Y axes show the coverage depth. In A, C, and E, all mapped reads were recorded and reads mapped to multiple locations were counted multiple times, while only uniquely mapped reads were counted in B, D, and F.
(0.30 MB PDF)
Absence of pMUM001 plasmid genes in Jp8756 revealed by NimbleGen microarrays. Most hybridization signal intensities from Jp8756 (in Green) were close to zero in both mutation mapping and re-sequencing of selected pMUM001 plasmid genes (A and B), while hybridization signal intensities from Agy99 (in Blue) were properly detected. Hybridization signal intensities from both strains were properly detected in mutation mapping and re-sequencing of selected chromosomal genes (C and D). X axes represent genomic location of oligo probes (1 to 174,155 in A and B, 1 to 5,631,606 in C and D). Y axes show h hybridization signal intensities.
(2.45 MB EPS)
Empirically determined MAQ consensus quality scores improved the accuracy of SNP calls in M. ulcerans Solexa data. Sanger sequencing of potential SNP harboring regions in Jp8756 and NM31/04 revealed that MAQ consensus quality scores equal or higher than 40 excluded most false positive SNP calls.
(0.71 MB EPS)
SNPs detected among four M. ulcerans genomes. Coordinates correspond to Agy99 finished genome sequence (NCBI:NC_008611 for Agy99 chromosome and NCBI:NC_005916 for Agy99 pMUM001).
(7.21 MB XLS)
SNPs identified by NimbleGen re-sequencing array in selected chromosomal and plasmid genes. SNPs confirmed by Solexa Genome Analyzer are indicated in the last column.
(0.52 MB XLS)
Primers used for PCR and sequencing of SNP harboring regions.
(0.05 MB XLS)