Abstract
The molecular chaperone Hsp104 is a crucial factor in the acquisition of thermotolerance in yeast. Under stress conditions, the disaggregase activity of Hsp104 facilitates the reactivation of misfolded proteins. Hsp104 is also involved in the propagation of fungal prions. For instance, the well-characterized [PSI+] prion of Saccharomyces cerevisiae does not propagate in Δhsp104 cells or in cells overexpressing Hsp104. In this study, we characterized the functional homolog of Hsp104 from Schizosaccharomyces pombe (Sp_Hsp104). As its S. cerevisiae counterpart, Sp_hsp104+ is heat-inducible and required for thermotolerance in S. pombe. Sp_Hsp104 displays low disaggregase activity and cannot propagate the [PSI+] prion in S. cerevisiae. When overexpressed in S. cerevisiae, Sp_Hsp104 confers thermotolerance to Δhsp104 cells and reactivates heat-aggregated proteins. However, overexpression of Sp_Hsp104 does not propagate nor eliminate [PSI+]. Strikingly, [PSI+] was cured by overexpression of a chimeric chaperone bearing the C-terminal domain (CTD) of the S. cerevisiae Hsp104 protein. Our study demonstrates that the ability to untangle aggregated proteins is conserved between the S. pombe and S. cerevisiae Hsp104 homologs, and points to a role of the CTD in the propagation of the S. cerevisiae [PSI+] prion.
Introduction
Yeasts have the ability to survive a broad spectrum of stress conditions. When exposed to mild stresses, cells trigger an adaptive response to enhance their protection. This adaptation endows the cells with the ability to survive more severe stresses. In the case of heat shock, this adaptive response is called thermotolerance. Hsp104 is an AAA+ protein (ATPase associated with various activities) involved in the acquisition of thermotolerance in yeasts [1]. Unlike classical molecular chaperones that assist protein folding and prevent their aggregation, Hsp104 is a disaggregase that dissolves preformed protein aggregates [2]. The Saccharomyces cerevisiae Hsp104 protein contains two ATPase domains that are both involved in disaggregation. The first nucleotide-binding domain (NBD1) is crucial for substrate binding, while the second NBD2 domain is involved in the processing of protein aggregates and in the oligomerization of Hsp104 [3], [4], [5], [6]. The activities of the two NBDs are modulated by the middle (M) domain [7]. In vitro experiments demonstrated that Hsp104 binds and untangles aggregated proteins, releasing them in a folding-competent state [5], [8]. These observations are consistent with data obtained for ClpB, the bacterial homolog of Hsp104, suggesting that the mechanisms of protein disaggregation are well conserved across the ClpB/Hsp100 family [9], [10]. Although Hsp104 and ClpB are able to untangle protein aggregates by themselves, collaboration with the Hsp40 and Hsp70 families of molecular chaperones greatly enhances the disaggregating activities of both Hsp104 and ClpB [11], [12], [13], [14]. Interestingly, this chaperone interaction is species-specific since bacterial chaperones cannot collaborate efficiently with Hsp104 [11], [13].
Apart from its role as a disaggregase during heat stress, Hsp104 is also essential for the propagation of several prions in S. cerevisiae. Prions are proteins that can adopt at least two conformations, one of which is self-replicative. The best characterized yeast prions are [PSI+], [URE3] and [PIN+] from the budding yeast. In S. cerevisiae, these prions form amyloid fibers, and cannot be propagated in cells devoid of Hsp104 [15], [16], [17], [18]. [PSI+] is the most studied yeast prion. [PSI+] results from the amyloidogenic conformational switch of the Sup35 protein, a factor involved in the fidelity of translation termination [19].
The role of Hsp104 in prion propagation is not completely understood. The current model suggest that prion fibers need to be shortened or cleaved by Hsp104 in order to be correctly transmitted to the mitotic progeny during cell division [20], [21]. This model is supported by in vivo experiments showing that a reduction of the quantity of Hsp104 results in an increase of the length of the [PSI+] fibrils as well as a reduction in their ability to be transmitted from to daughter cells [22], [23]. On the other hand, overexpression of Hsp104 eliminates (or “cures”) the [PSI+] prion [15]. Moreover, Hsp104 is able to fragment Sup35p amyloid fibers [24]. Interestingly, a number of studies showed that mutants of Hsp104 can differentially affect thermotolerance, prion propagation and prion curing [25], [26], [27]. The Hsp104 homolog of the yeast Candida albicans also propagates [PSI+] in S. cerevisiae, suggesting that this ability is conserved among yeast species [28].
Whereas protein folding has been extensively studied in budding yeast, less is known about the function of molecular chaperones in Schizosaccharomyces pombe. Some studies reported that the fission yeast can acquire thermotolerance [29], [30], but the involvement of a putative Hsp104 homolog in this process has not been confirmed so far. In this study, we characterized for the first time an S. pombe protein (Sp_Hsp104) showing a high level of sequence identity with the S. cerevisiae Hsp104 protein. Our results demonstrate that Sp_Hsp104 is a heat-inducible disaggregase and a crucial factor in the acquisition of thermotolerance in fission yeast. Heterologous expression of Sp_Hsp104 in S. cerevisiae confirmed that this protein is a functional homolog of Hsp104 for thermotolerance. However, unlike the budding yeast Hsp104, Sp_Hsp104 did not support the propagation of the [PSI+] prion under basal expression levels. Furthermore, overexpression of Sp_Hsp104 did not cure [PSI+]. Remarkably, a chimeric Sp_Hsp104 bearing the CTD of S. cerevisiae Hsp104 gained the ability to cure the [PSI+] prion. These observations suggest that the CTD region of Hsp104 plays an important role in the propagation of prions.
Results
S. pombe encodes a single Hsp104 homolog
To identify a putative S. pombe homolog of Hsp104, we performed a protein-protein BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/) against the S. pombe proteome. A single S. pombe ORF, SPBC16D10.08c [31], encoded a predicted polypeptide with more than 52% identity and 83% similarity with the S. cerevisiae Hsp104 protein (Figure 1). This predicted S. pombe protein was not functionally characterized before. However, genome-wide expression analyses demonstrated that the gene is expressed and that the protein is translated and localizes to both the cytoplasm and the nucleus [32], [33], [34], [35], [36]. We named the gene hsp104+ according to the S. pombe terminology. However, to avoid any confusion with the HSP104 gene from S. cerevisiae (Sc_HSP104), in this article the hsp104 + gene will be referred to as Sp_hsp104+, and the synthesized protein Sp_Hsp104. Accordingly, proteins from S. pombe and S. cerevisiae will be referred to as Sp_Hsp104 and Sc_Hsp104, respectively.
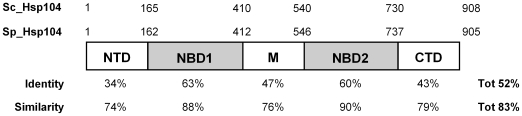
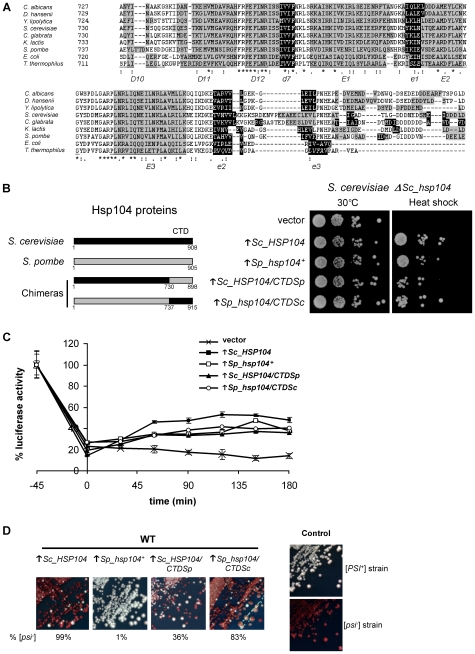
Figure 1. Sequences comparison between Hsp104 homologs.
Schematic representation of the five domains of Hsp104 as described by Hung et al. (2006) [25]. Hsp104 has two nucleotide-binding domains (NBD1 and NBD2), which are well conserved among AAA+ proteins. These domains are separated by a middle (M) domain that is specific to the ClpB/Hsp100 subfamily. The N- and C-terminal domains (NTD and CTD) are the least conserved domains. The position of the first amino acid of each region is indicated above the representation of the S. cerevisiae and S. pombe Hsp104 homologs (Sc_Hsp104 and Sp_Hsp104, respectively). The percentage of identity and similarity between both Hsp104 homologs is indicated below each domain.
Like all members of the ClpB/Hsp100 family, Sp_Hsp104 contains two putative nucleotide-binding domains (NBD1 and NBD2) that are well conserved between S. pombe and S. cerevisiae (63% and 60% amino acid identity, respectively). All the critical residues required for the ATPase activity of those domains are perfectly conserved between these two proteins [28]. The M domain, which is important for the disaggregase activity, shares 47% amino acid identity between Sp_Hsp104 and Sc_Hsp104 (Figure 1). The M domain of Sp_Hsp104 also possesses the typical leucine-zipper motifs, suggesting that this domain adopts a functional structure (not shown). Hence, the fission yeast homolog encoded by SPBC16D10.08c exhibits all the ClpB/Hsp100 characteristics required for the disaggregase activity.
The Sp_hsp104+ gene is required for thermotolerance in S. pombe
Sc_Hsp104 is a major factor involved in the acquisition of thermotolerance in the budding yeast [1]. Interestingly, it was shown in an S. pombe genome-wide microarray study that the SPBC16D10.08c gene (Sp_hsp104+) is overexpressed 54 fold by a mild heat shock of 15 minutes at 39°C [33]. This demonstrates that the expression of Sp_hsp104+ is greatly enhanced by environmental stresses, similarly to Sc_HSP104 [37], [38].
To confirm that Sp_Hsp104 is heat-inducible, we carried out immunoblots in different conditions with polyclonal antibodies raised against the full-length Sc_Hsp104 protein [6]. These antibodies detected a ∼100 kDa protein in the wild-type fission yeast strain (SP3220), but no protein in the ΔSp_hsp104 strain (SP12422); thus confirming the specificity of the immunodetection (Figure 2A). Quantification of each band revealed that a 1-hour pre-treatment at 37°C increased the level of Sp_Hsp104 protein by ∼100-fold, as compared to unstressed cells (Figure 2A). Thus, like its S. cerevisiae counterpart, Sp_Hsp104 expression is induced by heat.
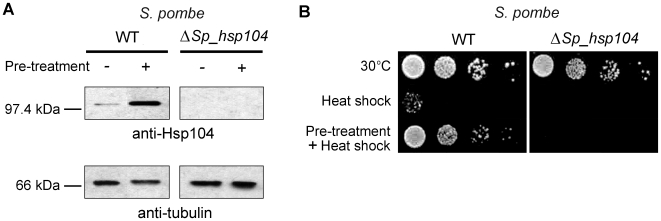
Figure 2. Sp_Hsp104 is required for thermotolerance in S. pombe.
(A) The Sp_hsp104+ gene is heat-inducible. WT (SP3220) and Δhsp104 (SP12422) S. pombe cells were treated or not for 1 hour at 37°C. Cell extracts were separated by SDS-PAGE, transferred onto a nitrocellulose membrane and immunoblotted with rabbit polyclonal antibodies against Sc_Hsp104 [6]. Immunoblot against tubulin is shown as a loading control. (B) The Sp_hsp104+ gene is required for thermotolerance. WT (SP3220) and Δhsp104 (SP12422) S. pombe cells were submitted to a thermotolerance assay. Pre-treatment of 1 hour at 37°C was applied or not before heat shock at 50°C for 20 minutes. Cells were then briefly cooled on ice, serially diluted (10−1, 10−2, 10−3, 10−4), spotted on minimal media and incubated for 5 days at 30°C. Results are representative of three independent experiments.
To investigate whether Sp_Hsp104 is able to confer protection against heat in fission yeast, we performed thermotolerance assays. As shown in Figure 2B, ΔSp_hsp104 cells did not show any significant growth defect when maintained at 30°C. Likewise, neither wild-type nor ΔSp_hp104 cells showed growth defects at 37°C (not shown). In contrast, most WT and ΔSp_hsp104 cells did not survive a severe heat shock of 20 minutes at 50°C. When WT cells were pre-treated at 37°C for 1 hour before being submitted to a severe heat shock, they acquired thermotolerance and survived (Figure 2B). However, ΔSp_hsp104 cells did not form colonies following heat shock at 50°C, even after pre-treatment at 37°C. This demonstrates that Sp_Hsp104 is required for thermotolerance in S. pombe.
Sp_Hsp104 is a functional disaggregase in S. cerevisiae
To verify that the S. pombe Hsp104 homolog exhibits disaggregase activity, the Sp_hsp104+ gene was cloned into a centromeric plasmid in which expression is driven by the native promoter of Sc_HSP104. The resulting construct was transformed into Δhsp104 S. cerevisiae cells. Expression of Sp_hsp104 + was confirmed by Western blotting using polyclonal anti-Hsp104 antibodies (Supplemental Figure S1A). To assess the ability of Sp_hsp104+ to complement thermotolerance in S. cerevisiae, we performed serial-dilution assays. As shown in Figure 3A (left panel), heterologous expression of Sc_Hsp104 had no effect on growth at 30°C as compared to the endogenous Hsp104. As expected, exposing the cells to a severe heat shock at 50°C resulted in the death of most Δhsp104 S. cerevisiae cells (not shown). However, after a pre-treatment of 1 hour at 37°C, cells expressing either the fission yeast or the budding yeast Hsp104 survived as well as the untreated cells (Figure 3A, left panel). To verify if thermotolerance could also be achieved by constitutive overproduction of Sp_Hsp104, we cloned the Sp_hsp104+ gene in a centromeric plasmid under the control of the GAL1 promoter. The GAL1 promoter increases gene expression 1000 fold in the presence of galactose in media without glucose, thus providing a major overproduction of the corresponding protein [39]. Overexpression of each Hsp104 homolog was confirmed by Western blotting (Supplemental Figure S1B). Overproduction of either Sp_Hsp104 or Sc_Hsp104 allowed survival of S. cerevisiae cells after a severe heat shock at 50°C, without any pre-treatment (Figure 3A, right panel). Hence, the S. pombe Hsp104 homolog complements thermotolerance defects of Δhsp104 S. cerevisiae cells as well as Sc_Hsp104.
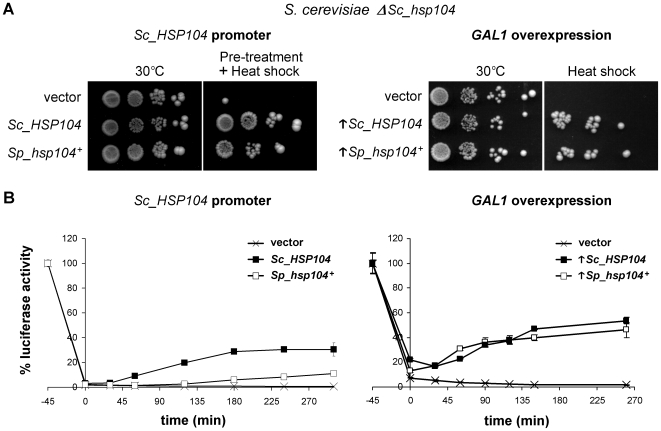
Figure 3. Sp_Hsp104 complements the knockout of Sc_HSP104 in S. cerevisiae.
(A) Heterologous complementation of thermotolerance. Δhsp104 S. cerevisiae cells (SL303a) bearing an empty vector or a plasmid expressing either Sc_HSP104 or Sp_hsp104+ under the control of the S. cerevisiae HSP104 promoter (left panel), or under the control of the GAL1 overexpression promoter (right panel) were pre-treated or not for 1 h at 37°C before a heat shock of 20 minutes at 50°C. Cells were then briefly cooled on ice, serially diluted (10−1, 10−2, 10−3, 10−4), spotted on minimal media and incubated for 5 days at 30°C. Results are representative of three independent experiments. (B) Luciferase reactivation assay. Δhsp104 S. cerevisiae cells (SL303a) expressing one of the Hsp104 homologs and bacterial luciferase were submitted to a heat shock of 46°C for 45 minutes to inactivate the protein. Reactivation was assessed by measuring luciferase activity at the indicated time points and reporting the value on the initial activity (100%). Reactivation curves of cells bearing an empty vector (X), expressing either Sc_HSP104 (▪) or Sp_hsp104+ (□) (left panel), or overexpressing (↑) these genes (right panel) are indicated. Each point is the mean±standard deviation of three independent measurements.
To confirm that the thermotolerance supported by Sp_hsp104+ is due to disaggregase activity of the protein, we performed luciferase reactivation assays [2]. As shown in Figure 3B, both Hsp104 homologs showed disaggregating activity when expressed under the control of the Sc_HSP104 promoter. Under basal expression conditions, Sp_Hsp104 homolog is significantly less efficient than Sc_Hsp104 to reactivate luciferase. When overexpressed however, both Sp_Hsp104 and Sc_Hsp104 displayed a similar disaggregating activity. Together, these results indicate that the fission yeast Hsp104 homolog is a functional disaggregase in the budding yeast. Since Sp_Hsp104 provides thermotolerance to both S. pombe and S. cerevisiae, and is also able to reactivate bacterial luciferase, a protein without any close homolog in yeasts, these experiments indicate that Sp_Hsp104 is a disaggregase of broad specificity, as was shown for Sc_Hsp104 [2].
The S. pombe Hsp104 homolog does not propagate nor cure the [PSI+] prion
Another well-known function of Sc_Hsp104 is the propagation of S. cerevisiae prions [40]. Since propagation of [PSI+] requires the disaggregase activity of Hsp104, we wondered if the low activity of Sp_Hsp104 in S. cerevisiae would be sufficient to propagate [PSI+]. Because [PSI+] cannot be maintained in the absence of Sc_Hsp104, we used a plasmid-shuffling strategy to evaluate the ability of Sp_Hsp104 to maintain this yeast prion. To this end, the Sp_hsp104+-encoding plasmid was transformed into a [PSI+] ΔSc_hsp104 strain (YJW532) complemented by an episomal copy of Sc_HSP104 [28]. Simultaneous expression of both Hsp104 homologs did not affect the propagation of [PSI+], suggesting that Sp_Hsp104 does not have a dominant negative effect on the endogenous protein (not shown). Next, we shuffled the Sc_HSP104-encoding plasmid with 5-FOA. Loss of Sc_Hsp104 was confirmed by auxotrophy on selective media and Western blotting using monoclonal antibodies specifically recognizing Sc_Hsp104 (not shown). After streaking on YPD¼ medium, the control strain expressing Sc_Hsp104 efficiently propagated the [PSI+] prion, as observed by the white color of the colonies. In contrast, all the cells bearing the Sp_hsp104+-encoding plasmid were undistinguishable from the [psi−] strain (Figure 4A). Thus, Sp_hsp104+ is unable to maintain the [PSI+] prion when expressed under the control of the endogenous Sc_HSP104 promoter. Overexpression of Sp_hsp104+ under the control of the GAL1 promoter was also tested, but failed to sustain [PSI+] propagation in absence of Sc_Hsp104 (Supplemental Figure S2). This demonstrates that Sp_Hsp104 is unable to propagate [PSI+] and/or that interaction with species-specific cofactors are required for this activity.
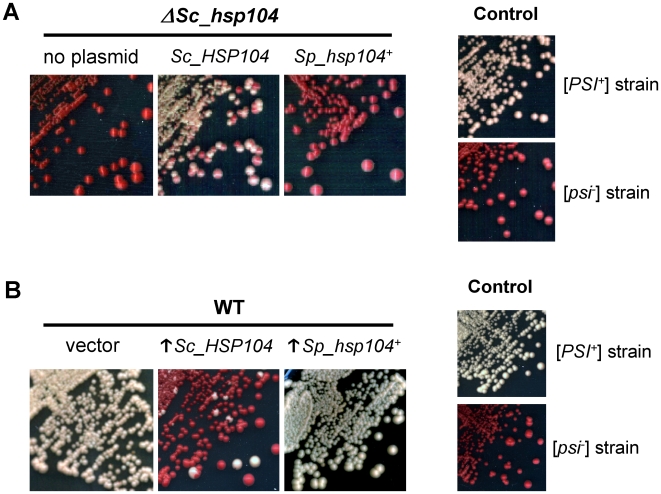
Figure 4. Sc_Hsp104 does not propagate or cure the S. cerevisiae [PSI+] prion.
(A) Sp_Hsp104 cannot sustain [PSI +] propagation. A [PSI +] ΔSc_hsp104 strain complemented by a plasmidic Sc_HSP104 gene (YJW532) was transformed with an empty vector or with a plasmid expressing Sp_hsp104+ under the control of the endogenous Sc_HSP104 promoter. After shuffling of the Sc_HSP104-encoding plasmid, cells were streaked on YPD¼ to test the maintenance of [PSI +]. Control strains show the expected white color of [PSI +] cells (Ψ-74-D694) and the red color of [psi −] cells (74-D694). (B) Overexpression of Sp_Hsp104 does not cure [PSI +]. A [PSI +] strain (Ψ-74-D694) was transformed with an empty vector or with plasmids overexpressing (↑) either Sc_HSP104 or Sp_hsp104+ under the control of the GAL1 promoter. Cells were streaked on YPD¼ to assess the appearance of red colonies, which indicates the curing of [PSI +]. [PSI +] (Ψ-74-D694) and [psi −] (74-D694) strains are shown as color controls.
Next, we assessed whether Sp_Hsp104 overexpression is able to cure [PSI+]. To this end, we transformed galactose-inducible plasmids encoding Sp_hsp104+ or Sc_HSP104 in [PSI+] cells. As expected, overexpression of Sc_Hsp104 cured the [PSI+] prion from most of the cells, resulting in the appearance of [psi-] colonies exhibiting the typical red color (Figure 4B). In contrast, heterologous overexpression of Sp_Hsp104 failed to cure [PSI+], as observed by the white color of colonies. Therefore, despite its disaggregating activity, Sp_Hsp104 is unable to block [PSI+] propagation when overexpressed.
The CTD of Sc_Hsp104 is crucial for the curing of [PSI+]
Sp_Hsp104 confers thermotolerance in both S. pombe and S. cerevisiae. However, unlike Sc_Hsp104, the S. pombe homolog is unable to propagate [PSI+] in Δhsp104 cells or to cure this prion by overexpression. Because the amino-acid sequences of Sp_Hsp104 and Sc_Hsp104 are not 100% identical (Figure 1A), some structural determinants must account for the inability of the S. pombe chaperone to propagate [PSI+]. To identify the less conserved structures between Sp_Hsp104 and Sc_Hsp104, we used the structural data obtained from ClpB [41] and Sc_Hsp104 [42] and a program for the prediction of secondary structures [43]. According to this analysis, Sc_Hsp104 and Sp_Hsp104 exhibit the least conserved secondary structure in the last segment of their CTD (Figure 5A). For instance, even though the NTD has a low level of amino acid identity (34% against 43% for the CTD), we were not able to identify any obvious secondary structure difference between the NTD of Sc_Hsp104 and that of Sp_Hsp104. On the other hand, the CTD has a very divergent region at the most C-terminal part of all Hsp104 homologs (Figure 5A). Further sequence alignments and secondary structure predictions confirmed that the CTD is the least conserved region between all ClpB/Hsp100 homologs that we analyzed.
Figure 5. Involvement of the CTD of Sc_Hsp104 in [PSI +] curing.
(A) Analysis of the CTD of Hsp104 homologs. The sequence of seven eukaryotic Hsp104 and two bacterial ClpB homologs were aligned using the Clustal W2 program [69]. Identical (*), conserved (:) and semi-conserved (.) residues are indicated. Predicted alpha helixes are shaded in gray and beta strands are shaded in black. Structural elements determined by crystallography of Thermus thermophilus ClpB are also indicated for comparison [41]. Alpha helixes are underlined and beta strands are in italic. (B) Heterologous complementation of thermotolerance by chimeric Hsp104 proteins. Hsp104 chimeras were created by interchanging the CTD of Sc_Hsp104 (amino acids 731–908) with the CTD of Sp_Hsp104 (amino acids 738–905) (left panel). Δhsp104 S. cerevisiae cells (SL303a) bearing an empty vector or overexpressing either Sc_HSP104, Sp_hsp104+, Sc_HSP104/CTDSp or Sp_hsp104/CTDSc under the control of the GAL1 promoter were submitted to a severe heat shock of 50°C for 20 minutes (right panel). Cells were then briefly cooled on ice, serially diluted (10−1, 10−2, 10−3, 10−4), spotted on minimal media and incubated for 5 days at 30°C. Results are representative of three independent experiments. (C) Luciferase reactivation assay. Δhsp104 S. cerevisiae cells (SL303a) expressing bacterial luciferase and an Hsp104 homolog or a chimera were submitted to a heat shock of 46°C for 45 minutes. Reactivation was assessed by measuring luciferase activity at the indicated time points and reporting the value on the initial activity (100%). Reactivation curves of cells bearing an empty vector (X) or overexpressing (↑) Sc_HSP104 (▪), Sp_hsp104+ (□), Sc_HSP104/CTDSp (▴) or Sp_hsp104/CTDSc (○) are indicated. Each point is the mean±standard deviation of three independent measures. (D) Curing of [PSI +] by Hsp104 chimeras. A [PSI +] strain (Ψ-74-D694) was transformed with an empty vector or with plasmids overexpressing (↑) Sc_HSP104, Sp_hsp104+, Sc_HSP104/CTDSp or Sp_hsp104/CTDSc. Cells were streaked on YPD¼ to assess the appearance of red colonies, which indicates the curing of [PSI +]. The rate of [PSI +] curing (% [psi −]) was estimated on ¼YPD on at least 500 colonies from two different assays. [PSI +] (Ψ-74-D694) and [psi −] (74-D694) strains are shown as color controls.
Because Sp_Hsp104 and Sc_Hsp104 exhibit significant differences in the primary and secondary structures of their CTD, we investigated the role of this domain in [PSI+] propagation. As described in Figure 1A, we defined the CTDs of Sc_Hsp104 and Sp_Hsp104 by the region encompassing the amino acids 731–908 and 738–905, respectively. Using homologous recombination, we modified the coding sequence of both Sc_Hsp104 and Sp_Hsp104 by interchanging their CTD. These chimeric genes were named Sc_HSP104/CTDSp and Sp_hsp104/CTDSc. The modified genes were cloned into an S. cerevisiae overexpression vector and transformed into [PSI +] cells. We confirmed that these plasmid-encoded chimeras (Sp_Hsp104/CTDSc and Sc_Hsp104/CTDSp) were expressed by Western blotting (Supplemental Figure S1C). Importantly, we confirmed that these overexpression chimeras were active in thermotolerance assays and retained disaggregase activity (Figure 5B and C).
Strikingly, the Sp_Hsp104/CTDSc chimera gained the ability to cure [PSI +] (Figure 5D) without increasing its disaggregase activity (Figure 5C). This could be explained if the CTD from S. cerevisiae confers the ability to interact with [PSI +] or if the chimeric chaperone has a dominant negative effect on the endogenous Sc_Hsp104. On the other hand, the Sc_Hsp104/CTDSp chimera was less efficient than WT Sc_Hsp104 to cure [PSI +] (Figure 5D). This could be due to the lower protein level of Sc_Hsp104/CTDSp as compared to the WT Sc_Hsp104 and the Sp_Hsp104/CTDSc chimera (Supplemental Figure S1C). Alternatively, the CTD of Sp_Hsp104 could interfere with the ability of Hsp104 to interact with [PSI +]. In any case, these results indicate that the CTD of Sc_Hsp104 plays an important role in the propagation of [PSI +].
Discussion
In this study we have characterized Sp_Hsp104, the fission yeast homolog of the Hsp104 chaperone. We demonstrated that the heat-inducible Sp_hsp104+ gene is required for the acquisition of thermotolerance in S. pombe and that the Sp_Hsp104 protein is a functional disaggregase in the budding yeast. Nevertheless, unlike its S. cerevisiae counterpart, Sp_Hsp104 is unable to maintain the [PSI+] prion in Δhsp104 cells and its overexpression does not cure [PSI+]. Interestingly, a chimera in which the CTD of Sp_Hsp104 was replaced with the corresponding domain of Sc_Hsp104 gained the ability to cure [PSI+].
Thermotolerance is a prime example of adaptation to environmental changes. In S. cerevisiae, specific heat-shock proteins and stress-activated response pathways are crucial for this process. In fission yeast, thermotolerance was originally linked to the metabolism of trehalose, a reserve sugar acting as a chemical chaperone [29]. Synergic effects between trehalose metabolism and Hsp104 were discovered in the budding yeast, suggesting a conserved thermotolerance pathway [44], [45]. Nevertheless, some differences were also observed between these two yeast species. For instance, the small heat shock protein Hsp16 was linked to thermotolerance in nuclear mRNA export in S. pombe, whereas the S. cerevisiae homolog of this protein, Hsp26, was shown to be dispensable for thermotolerance [46], [47]. Hence, it remained possible that each yeast species possesses specific chaperone machineries devoted to thermotolerance. Here we show that like in S. cerevisiae and C. albicans [28], the fission yeast Hsp104 exhibits disaggregating activity and is essential for thermotolerance. This suggests that Hsp104 has a conserved thermotolerance function in fungi.
Sc_Hsp104 is involved in the propagation of S. cerevisiae prions. Indeed, among the 24 yeast proteins containing a prion-forming domain recently identified in S. cerevisiae, only one was independent of Sc_Hsp104 for its propagation [48]. Because Hsp104 homologs from S. pombe and S. cerevisiae share a high level of sequence identity and are both functional disaggregases, we were surprised that Sp_Hsp104 failed to propagate or cure [PSI+]. Sp_Hsp104 confers thermotolerance in budding yeast when expressed under the control of the heat-inducible Sc_HSP104 promoter or under GAL1 overexpression. However, whereas the level of luciferase reactivation by Sp_Hsp104 was close to that of Sc_Hsp104 when overexpressed, Sp_Hsp104 was less efficient when expressed under the control of the Sc_HSP104 promoter on a centromeric plasmid. Therefore, the differences in disaggregating activities of the Hsp104 homologs could be, at least in part, responsible for the inability of Sp_Hsp104 to propagate and cure [PSI+].
Our analysis of the protein sequence of Hsp104 homologs prompted us to investigate the possible involvement of the CTD in [PSI+] propagation. Intriguingly, we found that a chimeric Sp_Hsp104 protein bearing the CTD from S. cerevisiae gained the ability to cure [PSI+]. This suggests that the CTD of Sc_Hsp104 modulates its prion-curing activity. Four non-exclusive possibilities may explain this gain of activity. First, the CTD of Hsp104 could be involved in [PSI+] recognition or binding. Indeed, previous studies proposed that the acidic C-terminal region of Hsp104 acts as a substrate-binding determinant for prion recognition [7]. Secondly, the CTD of Hsp104 could be involved in the recruitment of co-factors required for prion propagation. For instance, the Hsp90 co-factors Sti1p and Sgt2p interact with Hsp104 through the C-terminal tetratricopeptide-repeat (TPR)-like motif [49], [50]. A third possibility could be that the CTD of Sc_Hsp104 specifically increases the prion-propagation activity of the disaggregase without affecting its ability to untangle heat-aggregated proteins. Supporting this idea, the unstructured polypeptide poly-L-lysine was shown to stimulate the ATPase activity of both Sc_Hsp104 and ClpB by binding to the CTD region [51]. Finally, it is also possible that the curing of [PSI+] by overexpression of Sp_Hsp104/CTDSc is due to a dominant-negative effect of this chimera by interfering with the endogenous WT Sc_Hsp104 in the replication of [PSI+]. For instance, the CTD of the Sp_Hsp104/CTDSc chimera might interact with [PSI+] without promoting further replication of the prion.
Recently, Tipton et al. addressed the function of each Hsp104 domain by constructing chimeras with ClpB [13]. In contrast with our study, they concluded that the CTD of Hsp104 was dispensable for thermotolerance, disaggregation and prion propagation. Interestingly, a chimeric Hsp104 protein bearing the C-terminal end of ClpB was as efficient as the WT protein to propagate [PSI+]. ClpB lacks most of the amino acids corresponding to the CTD of Hsp104 (Figure 5A). Thus, it is unlikely that the Hsp104-ClpB chimera interacts with any Hsp104 CTD-specific binding factor. However, the chimera created by Tipton et al. also contained the NBD2 of ClpB. Unlike the NBD2 from S. cerevisiae, which has poor ATPase activity [27], the NBD2 of ClpB exhibits ATPase activity on its own [52]. Hence, it remains possible that the NBD2 of ClpB synergistically increases the prion-propagation activity of Hsp104. As such, the CTD of Hsp104 could be dispensable for prion propagation by Hsp104-ClpB because this chimera is a more efficient disaggregase than the WT Hsp104. On the other hand, Sp_Hsp104, which has a lower disaggregase activity in S. cerevisiae (Figure 3B), could require the CTD of Sc_Hsp104 to stimulate its ATPase activity in order to cure [PSI+]. This being said, although our CTD chimeras revealed the importance of this domain in prion curing, we cannot exclude that other structural differences between the fission and budding yeast homologs also account for the inability of Sp_Hsp104 to propagate [PSI+]. For instance, the NTD domain of Sc_Hsp104 was shown to be required for the curing of [PSI+] [25], and our sequence analysis revealed some disparities between Sp_Hsp104 and Sc_Hsp104 in this region. According to cryo-EM structural studies, both N- and C-terminal domains cap the cavity of the hexameric Hsp104 ring [42]. Therefore, N-terminal contributions to prion disaggregation activity might explain why the Sc_Hsp104/CTDSp chimera cures [PSI+] despite bearing the CTD from Sp_Hsp104.
Whereas Sp_Hsp104 is unable to propagate the [PSI+] in S. cerevisiae, does it assist in the propagation of prions in S. pombe? To date no prions have been identified in S. pombe. One prion-like element, [cif], was reported [53], [54], but its molecular nature remains to be elucidated. The knockout of Sp_hsp104+ had no effect on the maintenance of [cif], indicating that Sp_Hsp104 is not involved in the propagation of this epigenetic element (not shown). Since one of the 24 prions reported in S. cerevisiae is independent of Sc_Hsp104 for replication [48], it remains possible that some yet to be identified fission yeast prions require Sp_Hsp104 for their inheritance. Interestingly, genome-wide analyses showed that proteins bearing Q/N-rich regions characteristic of most yeast prions are almost inexistent in S. pombe, in contrast to S. cerevisiae [55], [56]. One can thus envision that this proteome unusually low in aggregation-prone proteins has favoured a divergent evolution for Sp_Hsp104, thus making it less efficient to untangle aggregates of Q/N-rich proteins.
In conclusion, our research demonstrates that Sp_Hsp104 is the first wild-type yeast AAA+ protein able to complement thermotolerance in S. cerevisiae but unable to propagate [PSI+]. Hence, the fission yeast Hsp104 could be used in further studies to discriminate the molecular requirements of prion propagation from those responsible of disaggregation of heat-aggregated proteins.
Materials and Methods
Strains and media
Yeast strains used in this study are listed in Table 1. Unless otherwise indicated, S. pombe strains were grown at 30°C in Edinburgh minimal medium (MM) supplemented with the required nutrients [57]. For growth of the various S. cerevisiae strains, standard growth media were used, and cells were routinely cultured at 30°C as previously described [58]. To ensure plasmid retention, transformed cells were grown on a selective synthetic media (SD, also called YNBD) containing all necessary supplements (Sigma). The [PSI+]-mediated suppression of the ade1-14 marker was routinely assessed by the color of colonies formed on YPD¼ medium (YEPD with 2.5 g/L of yeast extract rather than 10 g/L) and confirmed on SD-adenine defined medium supplemented with 2.5% (vol/vol) YEPD. The GAL1 promoter was induced by incubating cells on SDGal (SD containing 20 g of galactose/L instead of glucose as the carbon source) after preliminary growth in SDLG (YEPD containing 3% of lactate and 3% of glycerol instead of glucose) to eliminate all glucose from the liquid medium [59].
Table 1. Yeast strains used in this study.
| Species | Strain | Genotype | Reference |
| S. pombe | SP3220 (WT) | h − his3-D1 ade6-M216 ura4-D18 leu1-32 Δcnx1::his3+pREP41cnx1 + | Elagoz et al. (1999) [70] |
| S. pombe | SP12422 (ΔSp_hsp104) | SP3220 Δhsp104::neoR | This study |
| S. cerevisiae | W303a (WT) | MATa leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 lys2Δ can1-100 | Thomas et al. (1989) [71] |
| S. cerevisiae | SL303a (ΔSc_hsp104) | W303a Δhsp104::LEU2 | Sanchez et al. (1990) [1] |
| S. cerevisiae | YJW532 [PSI+] | MATa ade1-14 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 hsp104::HIS3+pRS316-Sc_HSP104 | Zenthon et al. (2006) [28] |
| S. cerevisiae | Ψ-74-D694 [PSI+] | MATa ade1-14 his3Δ-200 leu2-3 trp1-289 ura3-52 | Chernoff et al. (1995) [15] |
| S. cerevisiae | 74-D694 [psi-] | MATa ade1-14 his3Δ-200 leu2-3 trp1-289 ura3-52 | Chernoff et al. (1995) [15] |
Identification and deletion of the S. pombe hsp104+ gene
Using the S. cerevisiae Hsp104 (YLL026W) amino acid sequence, the S. pombe genome database was searched with the BLASTP alignment program [60]. A single sequence at locus NP_596503 encoding the hypothetical protein SPBC16D10.08c showed significant sequence identity (52%) to the Sc_ Hsp104 protein sequence. The deletion of Sp_hsp104+ was carried out by the method described in Krawchuk and Wahls [61]. Using the pFA6a-KanMX6 plasmid [62] as a template, the neomycin resistance gene was amplified with the Neo_FW and Neo_REV primers (Table 2). Sp_hsp104+ was amplified with its flanking regions from fission yeast genomic DNA using the primers Sp_Hsp104_FL_FW and Sp_Hsp104_FL_REV and cloned into the pCR-XL-TOPO vector. The coding region of Sp_hsp104 + was extracted by a AgeI/PacI digestion and replaced with the neomycin resistance gene. The Sp_hsp104+ knockout cassette was extracted from pCR-XL-TOPO by XhoI digestion and transformed into strain SP3220. Southern blot analyses were performed to confirm the correct and unique insertion of the cassette in the S. pombe genome using standard methods [57], [63] (not shown).
Table 2. Oligonucleotides used in this study.
| Name | Sequence* | Restriction site |
| Neo_FW | 5′-CACCGGTCCGGGTTAATTAA-3′ | AgeI |
| Neo_REV | 5′-CCTTAATTAACGAGCTCGTTTAAACTGG-3′ | PacI |
| Sp_Hsp104_FL_FW | 5′-CTCGAGCAACCTCTTCATCCTCAG-3′ | XhoI |
| Sp_Hsp104_FL_REV | 5′-CTCGAGCCATATTAGCTGCTACCG-3′ | XhoI |
| Sp_Hsp104_REC_FW | 5′-CAAAGAAAAAAGAAATCAACTACACGTACCATAAAATATACAGAATATATGGCTGATTATCCTTTTACTGAC-3′ | |
| Sp_Hsp104_REC_REV | 5′-AGAGTTCCAATTCTTCTTGCAATGAAGCTTCCTTCTGCCTAGCTAACTTTATTCCAATTCTTCATCATTAAC-3′ | |
| Sc_Hsp104_UTR_FW | 5′-ATACATATCCATATCTAATCTTACTTATATGTTGTGGAAATGTAAAGAGCGCGGCCGCATCGATTCAAAGGCGTTATTCAGC-3′ | NotI |
| Sc_Hsp104_UTR_REV | 5′-CATATATTCTGTATATTTTATGGTACGTG-3′ | |
| Sp_Hsp104_CTD_FW | 5′-AAGACGATCGACTGTTCCAATTGTATTGTCATCATGACTTCCAATCTAGGTGCTGAATACTTGACAACAGACAATGAGTCT-3′ | |
| Sp_Hsp104_CTD_REV | 5′-TTATATTACTGATTCTTGTTCGAAAGTTTTTAAAAATCACACTATATTAAATTATTCCAATTCTTCATCATTAACATCGTC-3′ | |
| Sc_Hsp104_CTD_FW | 5′-CAGGTTGTTGATGCCAAGAATGCTGTTATCATTATGACTTCTAACTTGGGCGCTGAATTTATCAATTCTCAACAAGGATCA-3′ | |
| Sc_Hsp104_CTD_REV | 5′-ATATTACTGATTCTTGTTCGAAAGTTTTTAAAAATCACACTATATTAAACTTTAATCTAGGTCATCATCAATTTCCATACT-3′ |
Restriction sites are in italic.
Plasmid constructions and transformation
Plasmids used in this study are described in Table 3. All plasmids expressing the S. cerevisiae HSP104 gene are from the Susan Lindquist lab. Plasmids expressing the S. pombe hsp104 + gene were created by in vivo recombination in S. cerevisiae. First, the Sp_hsp104+ gene was amplified using the Sp_Hsp104_REC_FW and Sp_Hsp104_REC_REV primers to add 50 bp corresponding to the flanking regions of the pYSGAL-Sc_HSP104 plasmid on either side of the coding sequence. The BglII-linearized pYSGAL-Sc_HSP104 vector and the PCR amplification of Sp_hsp104+ were then transformed into W303a competent S. cerevisiae cells with the specifications described in Knop et al. (1999) [64] for recombination. After selection for URA3, the pYSGAL-Sp_hsp104+ plasmid was extracted using the lyticase extraction protocol of Ling et al. (1995) [65]. For creation of the pRS315 and pRS316 plasmids, the GAL1 promoter of the pYSGAL-Sp_hsp104+ plasmid was replaced by 640 pb of the genomic 5′UTR of S. cerevisiae HSP104. The 5′UTR was amplified using the Sc_HSP104_UTR_FW and Sc_HSP104_UTR_REV primers. The Sp_hsp104+ gene under the control of the endogenous S. cerevisiae promoter was then extracted by NotI digestion and cloned into pRS315 and pRS316. The chimeric genes were constructed using the same recombination approach using PCR amplification of the CTDs of each gene. The CTD from S. pombe Hsp104 was amplified using the Sp_Hsp104_CTD_FW and Sp_Hsp104_CTD_REV primers, while the CTD from S. cerevisiae Hsp104 was amplified using the Sc_Hsp104_CTD_FW and Sc_Hsp104_CTD_REV primers. All PCR amplifications were performed with the Phusion™ High-Fidelity DNA Polymerase (NEB, Ipswich, MA, USA). All plasmid constructions were verified by standard sequencing methods (IRIC genomic platform, Montréal, Canada). In addition, all plasmids were tested for protein expression by immunoblotting with appropriate antibodies. DNA transformations into S. pombe and S. cerevisiae cells were performed by the polyethylenglycol (PEG)-lithium acetate procedure [66].
Table 3. Plasmids used in this study.
| Plasmid | Features | Source |
| pRS316 | pBluescript-based centromeric yeast expression vector with URA3 gene for selection | ATCC |
| pRS316-Sc_HSP104 | pRS316 expressing the HSP104 gene from S. cerevisiae under the control of the endogenous promoter (588 bp) | S. Lindquist Lab |
| pRS316-Sp_hsp104+ | pRS316 expressing the hsp104+ gene from S. pombe under the control of the endogenous S. cerevisiae HSP104 promoter (588 bp) | This study |
| pRS315 | pBluescript-based centromeric yeast expression vector with LEU2 gene for selection | ATCC |
| pRS315-Sp_hsp104+ | pRS315 expressing the hsp104+ gene from S. pombe under the control of the endogenous S. cerevisiae HSP104 promoter (588 bp) | This study |
| pGPD-luxAB(HIS) | p426GPD vector expressing a bacterial temperature-sensitive Vibrio harveyi luciferase | S. Lindquist Lab |
| pYSGAL | pRS316 with the galactose-inducible GAL1 overexpression promoter | S. Lindquist Lab |
| pYSGAL-Sc_HSP104 | pYSGAL overexpressing the HSP104 gene from S. cerevisiae | S. Lindquist Lab |
| pYSGAL-Sp_hsp104+ | pYSGAL overexpressing the hsp104+ gene from S. pombe | This study |
| pYSGAL-Sc_HSP104/CTDSp | pYSGAL overexpressing a chimeric HSP104 gene from S. cerevisiae (first 2190 bp) with the CTD of the hsp104 + gene from S. pombe (last 507 bp) | This study |
| pYSGAL-Sp_hsp104/CTDSc | pYSGAL overexpressing a chimeric hsp104 + gene from S. pombe (first 2211 bp) with the CTD of the HSP104 gene from S. cerevisiae (last 537 bp) | This study |
Antibodies and immunoblotting
For the specific detection of Hsp104 from S. cerevisiae, we used the commercially available polyclonal rabbit antibodies directed against the last residues of Sc_Hsp104 (Stressgen) or the monoclonal antibodies described in Cashikar et al. (2002) [7], which specifically recognize the M domain of Sc_Hsp104. In contrast to a previous report [67], we were not able to detect Sp_Hsp104 with the commercial anti-Hsp104 antibodies. For the detection of Sp_Hsp104, we used antibodies raised against the whole recombinant His-tagged Sc_Hsp104 protein described in Tkach et al. (2004) [6]. These polyclonal antibodies were able to detect Sp_Hsp104 in a specific manner when used at a dilution of 1∶5000. To eliminate background, we pre-blotted these antibodies with an empty nitrocellulose membrane and we treated them with acetone powder of the ΔSp_hsp104 strain, as described in Sambrook et al. (1989) [63]. For Western blotting, standard immunoblotting procedures were used [68]. Bands were quantified using the Quantity One software (BioRad).
Thermotolerance assay
Exponentially growing cells were adjusted to an OD595 of 0.5, serially diluted (10−1 to 10−4), spotted on solid media and grown for 5 days at 30°C. Heat shock was performed on exponentially growing cells adjusted to an OD595 of 0.5. Cells were pre-treated or not at 37°C for 1 hour, and then incubated with slight agitation at 50°C for 20 minutes and cooled on ice for 5 minutes. Cells were mixed by vortexing, serially diluted and subsequently spotted on the corresponding solid media.
Luciferase reactivation assay
The luciferase reactivation assay was essentially performed as described in Zenthon et al. (2006) [28]. Briefly, the relevant strains were transformed with the plasmid pGPD-luxAB(HIS) (AddGene #1106), which expresses a temperature-sensitive Vibrio harveyi luciferase [2]. The transformed cells were grown to an OD595 of approximately 0.5. The luciferase activity was determined before treatment as a control. The culture was then transferred to 46°C, and after 30 minutes of incubation at this temperature, cycloheximide was added to a final concentration of 10 µg/mL. The culture was then incubated for further 15 minutes, after which the cell culture was transferred back to 25°C to allow the cells to recover. Cell samples were taken immediately to determine the level of luciferase activity and then collected every 30 to 45 minutes for up to 4 hours. The luciferase activity was determined by using 200 µL of cells plus 5 µL decylaldehyde (Sigma), and the resulting luminescence was immediately quantified using a Lumat LB 9507 luminometer (EG&G Berthold). Three independent samples were taken per time point.
Supporting Information
Expression and overexpression of Hsp104 homologs and chimeras (A) Expression of Sc_Hsp104 and Sp_Hsp104 was verified by immunoblotting. Protein extracts from S. cerevisiae Δhsp104 strains bearing an empty vector or expressing Sc_HSP104 or Sp_hsp104+ under the control of the endogenous Sc_HSP104 promoter were separated by SDS-PAGE and immunoblotted using monoclonal anti-Hsp104 antibodies (left panel) or polyclonal antibodies raised against the full-length protein (right panel). The monoclonal antibodies specifically recognized the Sc_Hsp104 protein, while the polyclonal antibodies from Tkach and Gover (2004) were the only ones able to detect Sp_Hsp104, when concentrated at a dilution of 1∶5000. Immunoblotting of Pgk1p (phosphoglycerate kinase) is shown as a loading control. (B) Overexpression of Sc_Hsp104 and Sp_Hsp104 was verified by immunoblotting. Protein extracts from S. cerevisiae Δhsp104 strains bearing an empty vector or overexpressing Sc_HSP104 or Sp_hsp104+ under the control of the GAL1 promoter were separated by SDS-PAGE and immunoblotted using monoclonal anti-Hsp104 antibodies (left panel) or polyclonal antibodies raised against the full-length protein (right panel). Immunoblotting of Pgk1p is shown as a loading control. (C) Overexpression of Hsp104 chimeras was verified by immunoblotting. Protein extracts from S. cerevisiae Δhsp104 strains bearing an empty vector or overexpressing either Sc_HSP104, Sp_hsp104+, Sc_HSP104/CTDSp or Sp_hsp104/CTDSc under the control of the GAL1 promoter were separated by SDS-PAGE and immunoblotted using polyclonal antibodies directed against the CTD of Sc_Hsp104 (Stressgen, upper panel) or monoclonal anti-Hsp104 antibodies (middle panel). Immunoblotting of Pgk1p is shown as a loading control (lower panel).
(0.59 MB TIF)
Overexpression of Sp_Hsp104 cannot sustain [PSI+] propagation A [PSI+] ΔSc_hsp104 strain complemented by a plasmidic Sc_HSP104 gene (YJW532) was transformed with an empty vector or with a plasmid overexpressing Sp_hsp104+ under the control of the GAL1 promoter. After shuffling of the Sc_HSP104-encoding plasmid, cells were streaked on YPG1/4 to test the maintenance of [PSI+]. Control strains show the expected white color of [PSI+] cells (Ψ-74-D694) and the red color of [psi−] cells (74-D694)
(3.84 MB TIF)
Acknowledgments
We thank Mick Tuite for the YJW532 strain and John Glover for the polyclonal anti-Hsp104 antibodies. We also thank Pascal Chartrand for the strain W303a and Pierre Belhumeur for the pRS315 plasmid. We thank Nicolas Paquin from the Pascal Chartrand lab for technical advice on in vivo recombination cloning and for fruitful discussions. We also thank all the Rokeach lab members for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Canadian Institutes of Health Research (Grant MOP-89702) and National Science and Engineering Council of Canada (171325) to L.A.R. P.S. received a FRSQ MSc studentship scholarship and A.L. received a NSERC CGS-M scholarship. P.S. and A.L. both received a scholarship from the Faculte des Etudes Superieures, Departement de biochimie, Universite de Montreal. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 2.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 3.Bosl B, Grimminger V, Walter S. Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J Biol Chem. 2005;280:38170–38176. doi: 10.1074/jbc.M506149200. [DOI] [PubMed] [Google Scholar]
- 4.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 5.Schaupp A, Marcinowski M, Grimminger V, Bosl B, Walter S. Processing of proteins by the molecular chaperone Hsp104. J Mol Biol. 2007;370:674–686. doi: 10.1016/j.jmb.2007.04.070. [DOI] [PubMed] [Google Scholar]
- 6.Tkach JM, Glover JR. Amino acid substitutions in the C-terminal AAA+ module of Hsp104 prevent substrate recognition by disrupting oligomerization and cause high temperature inactivation. J Biol Chem. 2004;279:35692–35701. doi: 10.1074/jbc.M400782200. [DOI] [PubMed] [Google Scholar]
- 7.Cashikar AG, Schirmer EC, Hattendorf DA, Glover JR, Ramakrishnan MS, et al. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol Cell. 2002;9:751–760. doi: 10.1016/s1097-2765(02)00499-9. [DOI] [PubMed] [Google Scholar]
- 8.Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, et al. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosl B, Grimminger V, Walter S. The molecular chaperone Hsp104–a molecular machine for protein disaggregation. J Struct Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Zolkiewski M. A camel passes through the eye of a needle: protein unfolding activity of Clp ATPases. Mol Microbiol. 2006;61:1094–1100. doi: 10.1111/j.1365-2958.2006.05309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 12.Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci U S A. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 16.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 18.Taneja V, Maddelein ML, Talarek N, Saupe SJ, Liebman SW. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol Cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 20.Byrne LJ, Cox BS, Cole DJ, Ridout MS, Morgan BJ, et al. Cell division is essential for elimination of the yeast [PSI+] prion by guanidine hydrochloride. Proc Natl Acad Sci U S A. 2007;104:11688–11693. doi: 10.1073/pnas.0701392104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones GW, Tuite MF. Chaperoning prions: the cellular machinery for propagating an infectious protein? Bioessays. 2005;27:823–832. doi: 10.1002/bies.20267. [DOI] [PubMed] [Google Scholar]
- 22.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 23.Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5:e24. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurahashi H, Nakamura Y. Channel mutations in Hsp104 hexamer distinctively affect thermotolerance and prion-specific propagation. Mol Microbiol. 2007;63:1669–1683. doi: 10.1111/j.1365-2958.2007.05629.x. [DOI] [PubMed] [Google Scholar]
- 27.Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. Embo J. 2002;21:12–21. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zenthon JF, Ness F, Cox B, Tuite MF. The [PSI+] prion of Saccharomyces cerevisiae can be propagated by an Hsp104 orthologue from Candida albicans. Eukaryot Cell. 2006;5:217–225. doi: 10.1128/EC.5.2.217-225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribeiro MJ, Reinders A, Boller T, Wiemken A, De Virgilio C. Trehalose synthesis is important for the acquisition of thermotolerance in Schizosaccharomyces pombe. Mol Microbiol. 1997;25:571–581. doi: 10.1046/j.1365-2958.1997.4961856.x. [DOI] [PubMed] [Google Scholar]
- 30.De Virgilio C, Simmen U, Hottiger T, Boller T, Wiemken A. Heat shock induces enzymes of trehalose metabolism, trehalose accumulation, and thermotolerance in Schizosaccharomyces pombe, even in the presence of cycloheximide. FEBS Lett. 1990;273:107–110. doi: 10.1016/0014-5793(90)81062-s. [DOI] [PubMed] [Google Scholar]
- 31.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 32.Mata J, Lyne R, Burns G, Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 33.Chen D, Toone WM, Mata J, Lyne R, Burns G, et al. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, et al. Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 35.Mata J, Bahler J. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc Natl Acad Sci U S A. 2006;103:15517–15522. doi: 10.1073/pnas.0603403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 37.Grably MR, Stanhill A, Tell O, Engelberg D. HSF and Msn2/4p can exclusively or cooperatively activate the yeast HSP104 gene. Mol Microbiol. 2002;44:21–35. doi: 10.1046/j.1365-2958.2002.02860.x. [DOI] [PubMed] [Google Scholar]
- 38.Seppa L, Hanninen AL, Makarow M. Upregulation of the Hsp104 chaperone at physiological temperature during recovery from thermal insult. Mol Microbiol. 2004;52:217–225. doi: 10.1111/j.1365-2958.2003.03959.x. [DOI] [PubMed] [Google Scholar]
- 39.Ronicke V, Graulich W, Mumberg D, Muller R, Funk M. Use of conditional promoters for expression of heterologous proteins in Saccharomyces cerevisiae. Methods Enzymol. 1997;283:313–322. doi: 10.1016/s0076-6879(97)83025-x. [DOI] [PubMed] [Google Scholar]
- 40.True HL. The battle of the fold: chaperones take on prions. Trends Genet. 2006;22:110–117. doi: 10.1016/j.tig.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 42.Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, et al. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 44.Elliott B, Haltiwanger RS, Futcher B. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics. 1996;144:923–933. doi: 10.1093/genetics/144.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwahashi H, Nwaka S, Obuchi K, Komatsu Y. Evidence for the interplay between trehalose metabolism and Hsp104 in yeast. Appl Environ Microbiol. 1998;64:4614–4617. doi: 10.1128/aem.64.11.4614-4617.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida J, Tani T. Hsp16p is required for thermotolerance in nuclear mRNA export in fission yeast Schizosaccharomyces pombe. Cell Struct Funct. 2005;29:125–138. doi: 10.1247/csf.29.125. [DOI] [PubMed] [Google Scholar]
- 47.Petko L, Lindquist S. Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell. 1986;45:885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- 48.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abbas-Terki T, Donze O, Briand PA, Picard D. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol Cell Biol. 2001;21:7569–7575. doi: 10.1128/MCB.21.22.7569-7575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liou ST, Cheng MY, Wang C. SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell Stress Chaperones. 2007;12:59–70. doi: 10.1379/CSC-220R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strub C, Schlieker C, Bukau B, Mogk A. Poly-L-lysine enhances the protein disaggregation activity of ClpB. FEBS Lett. 2003;553:125–130. doi: 10.1016/s0014-5793(03)00985-2. [DOI] [PubMed] [Google Scholar]
- 52.Kim KI, Woo KM, Seong IS, Lee ZW, Baek SH, et al. Mutational analysis of the two ATP-binding sites in ClpB, a heat shock protein with protein-activated ATPase activity in Escherichia coli. Biochem J. 1998;333(Pt 3):671–676. doi: 10.1042/bj3330671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collin P, Beauregard PB, Elagoz A, Rokeach LA. A non-chromosomal factor allows viability of Schizosaccharomyces pombe lacking the essential chaperone calnexin. J Cell Sci. 2004;117:907–918. doi: 10.1242/jcs.00943. [DOI] [PubMed] [Google Scholar]
- 54.Beauregard PB, Guerin R, Turcotte C, Lindquist S, Rokeach LA. A nucleolar protein allows viability in the absence of the essential ER-residing molecular chaperone calnexin. J Cell Sci. 2009 doi: 10.1242/jcs.040949. [DOI] [PubMed] [Google Scholar]
- 55.Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 58.Sherman F, Fink GR, Hicks JB Cold Spring Harbor Laboratory. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1981. Methods in yeast genetics.120 [Google Scholar]
- 59.Peng G, Hopper JE. Evidence for Gal3p's cytoplasmic location and Gal80p's dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5140–5148. doi: 10.1128/mcb.20.14.5140-5148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krawchuk MD, Wahls WP. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast. 1999;15:1419–1427. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1419::AID-YEA466>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 63.Sambrook J, Fritsch EF, Maniatis T. 3 v. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- 64.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 65.Ling M, Merante F, Robinson BH. A rapid and reliable DNA preparation method for screening a large number of yeast clones by polymerase chain reaction. Nucleic Acids Res. 1995;23:4924–4925. doi: 10.1093/nar/23.23.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 67.Parsell DA, Sanchez Y, Stitzel JD, Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- 68.Marechal A, Tanguay PL, Callejo M, Guerin R, Boileau G, et al. Cell viability and secretion of active proteins in Schizosaccharomyces pombe do not require the chaperone function of calnexin. Biochem J. 2004;380:441–448. doi: 10.1042/BJ20031546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elagoz A, Callejo M, Armstrong J, Rokeach LA. Although calnexin is essential in S. pombe, its highly conserved central domain is dispensable for viability. J Cell Sci. 1999;112 (Pt 23):4449–4460. doi: 10.1242/jcs.112.23.4449. [DOI] [PubMed] [Google Scholar]
- 71.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression and overexpression of Hsp104 homologs and chimeras (A) Expression of Sc_Hsp104 and Sp_Hsp104 was verified by immunoblotting. Protein extracts from S. cerevisiae Δhsp104 strains bearing an empty vector or expressing Sc_HSP104 or Sp_hsp104+ under the control of the endogenous Sc_HSP104 promoter were separated by SDS-PAGE and immunoblotted using monoclonal anti-Hsp104 antibodies (left panel) or polyclonal antibodies raised against the full-length protein (right panel). The monoclonal antibodies specifically recognized the Sc_Hsp104 protein, while the polyclonal antibodies from Tkach and Gover (2004) were the only ones able to detect Sp_Hsp104, when concentrated at a dilution of 1∶5000. Immunoblotting of Pgk1p (phosphoglycerate kinase) is shown as a loading control. (B) Overexpression of Sc_Hsp104 and Sp_Hsp104 was verified by immunoblotting. Protein extracts from S. cerevisiae Δhsp104 strains bearing an empty vector or overexpressing Sc_HSP104 or Sp_hsp104+ under the control of the GAL1 promoter were separated by SDS-PAGE and immunoblotted using monoclonal anti-Hsp104 antibodies (left panel) or polyclonal antibodies raised against the full-length protein (right panel). Immunoblotting of Pgk1p is shown as a loading control. (C) Overexpression of Hsp104 chimeras was verified by immunoblotting. Protein extracts from S. cerevisiae Δhsp104 strains bearing an empty vector or overexpressing either Sc_HSP104, Sp_hsp104+, Sc_HSP104/CTDSp or Sp_hsp104/CTDSc under the control of the GAL1 promoter were separated by SDS-PAGE and immunoblotted using polyclonal antibodies directed against the CTD of Sc_Hsp104 (Stressgen, upper panel) or monoclonal anti-Hsp104 antibodies (middle panel). Immunoblotting of Pgk1p is shown as a loading control (lower panel).
(0.59 MB TIF)
Overexpression of Sp_Hsp104 cannot sustain [PSI+] propagation A [PSI+] ΔSc_hsp104 strain complemented by a plasmidic Sc_HSP104 gene (YJW532) was transformed with an empty vector or with a plasmid overexpressing Sp_hsp104+ under the control of the GAL1 promoter. After shuffling of the Sc_HSP104-encoding plasmid, cells were streaked on YPG1/4 to test the maintenance of [PSI+]. Control strains show the expected white color of [PSI+] cells (Ψ-74-D694) and the red color of [psi−] cells (74-D694)
(3.84 MB TIF)