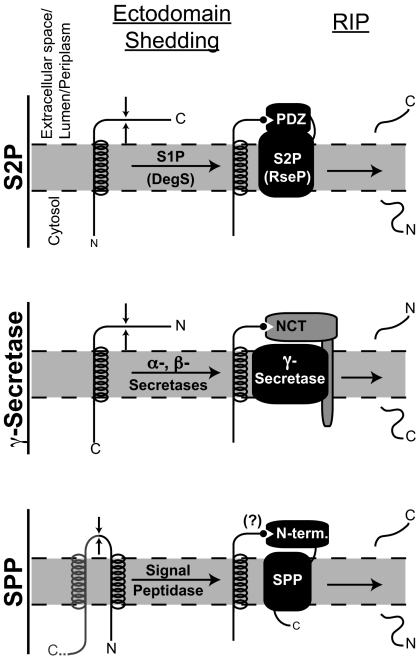
Regulated intramembrane proteolysis (Rip) controls a wide variety of cellular mechanisms such as cholesterol homeostasis, immune surveillance, cellular signaling, and β-amyloid formation in Alzheimer's disease (1). Rip of substrates is mediated by several families of intramembranously cleaving proteases (I-CLiPs), all of which perform the unique chemistry of hydrolysis within the hydrophobic lipid bilayer (2). There are four known families of I-CLiPs, each denoted by the protease that typifies each group: site-2 protease (S2P) metalloproteases, the γ-secretase and signal peptide peptidase (SPP) aspartyl proteases, and the rhomboid serine proteases. Rip cleavage of transmembrane substrates by S2P, SPP, and γ-secretase is preceded and regulated by an initial distinct cleavage in a process termed “ectodomain shedding” (Fig. 1). The reason ectodomain shedding is necessary in most Rip cases is not understood. An article in this issue of PNAS (3) has shed new light on this important question.
Fig. 1.
A common mechanism for the role of ectodomain shedding in Rip? Li et al. (3) report that upon ectodomain shedding of RseA by the site-1 protease DegS, the newly exposed C-terminal-most amino acid Val-148 binds one of the PDZ domains of RseP to facilitate the regulated intramembrane proteolytic (Rip) cleavage by the S2P RseP (Top). Such a mechanism is consistent with studies of γ-secretase-mediated Rip cleavage of the amyloid precursor protein and other substrates (Middle): upon ectodomain shedding by α- or β-secretase, the newly generated N terminus of the substrate binds to the NCT subunit of γ-secretase (11). A third family of I-CLiPs is typified by SPP, whose cleavage of substrates, like that of S2P and γ-secretase substrates, is also preceded by ectodomain shedding (Bottom). The N-terminal portion of SPP is not part of the catalytic core of SPP and may therefore function analogously to the PDZ domain of S2Ps and NCT of γ-secretase.
Li et al. (3) examine RseA, a bacterial transmembrane-anchored I-CLiP substrate whose proteolysis releases the transcription factor σE in times of envelope stress. Accumulation of unfolded outer membrane proteins in the bacterial envelope triggers a proteolytic cascade whereby DegS first cleaves RseA to shed its C-terminal ectodomain (4). The membrane-bound RseA fragment is then cleaved by the bacterial S2P family protease RseP (also called YaeL), thereby releasing the N-terminal RseA domain into the cytoplasm (Fig. 1). The cytoplasmic RseA fragment is further degraded by proteases, which in turn releases σE to enable the transcription of genes that respond to envelope stress (5).
So why must DegS cleavage precede RseP cleavage of RseA? Early studies implicated PSD95-Dlg-Zol (PDZ) domains in the regulation of RseP activity (6–9) but the underlying molecular logic remains elusive. Li et al. (3) combine reconstitution and biochemical analysis of the proteolytic cascade in vitro with X-ray crystallography to show that upon cleavage by DegS, the single terminal-most residue of the newly generated C terminus of RseA (Val-148) binds to one of the two PDZ domains of RseP, thereby facilitating its cleavage. Through mutation of Val-148 to all other amino acids, Li et al. demonstrate the remarkable power of the identity of this residue, as mutations fall into three groups: (i) those that permit RseA cleavage by RseP, (ii) those that permit cleavage by DegS but not RseP, and (iii) those that do not permit cleavage even by the upstream protease DegS. Moreover, X-ray crystallographic studies reveal that while the first PDZ domain of RseP is in a “closed” conformation, the second PDZ domain (PDZ2) is open to receive a ligand. Furthermore, the PDZ2 domain represents a circularly permutated PDZ domain in which the typical binding groove has been altered to form a binding pocket, accommodating only a single hydrophobic amino acid. Indeed, Li et al. provide structural evidence that RseA Val-148 interacts, via four specific hydrogen bonds and several van der Waals contacts, with the PDZ2 domain of RseP to facilitate its cleavage.
Interestingly, the findings of Li et al. (3) parallel early observations for the role of ectodomain shedding in γ-secretase (10, 11), a proteolytic complex of four membrane proteins [presenilin, nicastrin (NCT), Aph-1, and Pen-2] that is central to the production of β-amyloid peptides and Notch signaling. It has been suggested that NCT, via its catalytically crippled, peptidase-like DYIGS and peptidase (DAP) domain, interacts with the free α-amino group at the N terminus of γ-secretase substrates that have undergone ectodomain shedding (ref. 11 and Fig. 1). Thus, a common model for Rip emerges, whereby a single terminal residue of substrates newly freed by ectodomain cleavage interacts with an extracytoplasmic domain of I-CLiPs (i.e., the PDZ domain of S2P and NCT peptidase-like domain of γ-secretase) to facilitate or activate cleavage of substrates (Fig. 1). Analogous to the PDZ2 domain of RseP, the NCT DAP domain also appears to be noncanonical (12), probably containing a shallow substrate-binding pocket capable of accommodating a single substrate residue. In light of the findings of Li et al., it will be interesting to examine whether the identity of the N-terminal-most residue among the ever-increasing number of γ-secretase substrates similarly influences their binding to NCT to provide some selectivity.
Whereas Li et al. (3) address the reason intramembrane cleavage must be preceded and regulated by ectodomain cleavage in the bacterial envelope stress response, it is less clear precisely how this regulation is achieved. They propose two models whereby ectodomain cleavage of RseA by DegS facilitates its cleavage by RseP: the PDZ2 domain of RseP may simply be autoinhibitory, where binding by the RseA substrate activates the protease; alternatively, the PDZ2 domain may function as a receptor to recruit substrate. It is also conceivable that binding of RseA to the PDZ2 domain serves more than one purpose in activating RseP, for example by relieving autoinhibition, inducing a conformational change, and recruiting substrate. The interaction between the NCT-DAP domain and the free N terminus of substrates could be interpreted as NCT functioning to recruit substrates (11).
Interestingly, before becoming catalytically active, γ-secretase undergoes a complicated maturation process. The last step of this maturation is self-cleavage of presenilin within its seventh hydrophobic domain (HDVII). A mechanism whereby the NCT-DAP domain directly or indirectly recruits and facilitates cleavage of HDVII (the first γ-secretase substrate) would provide a potential explanation for the puzzling observation that a proposed substrate-binding residue of NCT also plays a role in γ-secretase maturation (12). The work of Li et al. and their parallels to γ-secretase biochemistry provide a platform from which the role of NCT in γ-secretase maturation and activation may also be evaluated.
Like RseP and γ-secretase, SPP and SPP-like (SPPL) aspartyl proteases require ectodomain shedding of their substrates. SPP shares weak sequence homology with the presenilin subunit of γ-secretase and, like γ-secretase, is an aspartyl protease. Unlike γ-secretase, however, SPP can perform catalysis without the need for other cofactors (13). It has been shown that the C-terminal half of SPP is responsible for catalysis (14). The N-terminal portion of SPP and SPPLs could in theory provide a putative binding pocket for the C-terminal-most residue of substrates exposed by ectodomain shedding by signal peptidase or its equivalents. It is thus tempting to speculate that a luminal N-terminal domain provides a PDZ/NCT-like functionality to SPP and SPPLs (Fig. 1). Among the known Rip cases, intramembrane cleavage by rhomboids is not preceded by ectodomain shedding. Did the rhomboids lose the need for ectodomain shedding during evolution, or did the other I-CLiPs gain this mode of regulation? From what common ancestral function did these classes of I-CLiPs arise, or did different families of I-CLiPs evolve separately? Is the molecular principle established for RseP cleavage of RseA applicable to other S2P proteases and even to SPP and SPPLs (Fig. 1)? These and other questions can now be asked and answered, thanks in large part to the work by Li et al. (3), who provide mechanistic insights not only relevant to bacterial envelope stress response but also to Rip cascades as a whole.
The findings of Li et al. (3) parallel early observations for the role of ectodomain shedding in γ-secretase.
Acknowledgments.
This work was supported by National Institutes of Health Grants F32 AG031625 (to D.R.D.), R01 AG023104, and R01 AG029547, Welch Foundation Grant I-1566, and the Ted Nash Long Life Foundation.
Footnotes
The authors declare no conflict of interest.
See companion article on page 14837.
References
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe MS. Intramembrane-cleaving proteases. J Biol Chem. 2009;284:13969–13973. doi: 10.1074/jbc.R800039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, et al. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proc Natl Acad Sci USA. 2009;106:14837–14842. doi: 10.1073/pnas.0903289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 5.Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 6.Kanehara K, Ito K, Akiyama Y. YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. EMBO J. 2003;22:6389–6398. doi: 10.1093/emboj/cdg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohn C, Collier J, Bouloc P. Dispensable PDZ domain of Escherichia coli YaeL essential protease. Mol Microbiol. 2004;52:427–435. doi: 10.1111/j.1365-2958.2004.03985.x. [DOI] [PubMed] [Google Scholar]
- 8.Inaba K, et al. A pair of circularly permutated PDZ domains control RseP, the S2P family intramembrane protease of Escherichia coli. J Biol Chem. 2008;283:35042–35052. doi: 10.1074/jbc.M806603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinch LN, Ginalski K, Grishin NV. Site-2 protease regulated intramembrane proteolysis: Sequence homologs suggest an ancient signaling cascade. Protein Sci. 2006;15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Struhl G, Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6:625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 11.Shah S, et al. Nicastrin functions as a γ-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Chavez-Gutierrez L, et al. Glu(332) in the Nicastrin ectodomain is essential for γ-secretase complex maturation but not for its activity. J Biol Chem. 2008;283:20096–20105. doi: 10.1074/jbc.M803040200. [DOI] [PubMed] [Google Scholar]
- 13.Fluhrer R, Steiner H, Haass C. Intramembrane proteolysis by signal peptide peptidases: A comparative discussion of GXGD-type aspartyl proteases. J Biol Chem. 2009;284:13975–13979. doi: 10.1074/jbc.R800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narayanan S, Sato T, Wolfe MS. A C-terminal region of signal peptide peptidase defines a functional domain for intramembrane aspartic protease catalysis. J Biol Chem. 2007;282:20172–20179. doi: 10.1074/jbc.M701536200. [DOI] [PubMed] [Google Scholar]