Abstract
The centromere of an eukaryotic chromosome can move to a new position during evolution, which may result in a major alteration of the chromosome morphology and karyotype. This centromere repositioning phenomenon has been extensively documented in mammalian species and was implicated to play an important role in mammalian genome evolution. Here we report a centromere repositioning event in plant species. Comparative fluorescence in situ hybridization mapping using common sets of fosmid clones between two pairs of cucumber (Cucumis sativus L.) and melon (Cucumis melo L.) chromosomes revealed changes in centromere positions during evolution. Pachytene chromosome analysis revealed that the current centromeres of all four cucumber and melon chromosomes are associated with distinct pericentromeric heterochromatin. Interestingly, inactivation of a centromere in the original centromeric region was associated with a loss or erosion of its affixed pericentromeric heterochromatin. Thus, both centromere activation and inactivation in cucurbit species were associated with a gain/loss of a large amount of pericentromeric heterochromatin.
Keywords: cucumber, melon, pericentromeric heterochromatin
The centromere governs chromosome segregation and transmission by serving as the chromosomal docking site for kinetochore assembly. Centromere function is not determined by its underlying DNA sequence, but by epigenetic mechanisms (1). Functional centromeres can emerge from non-centromeric regions due to poorly understood epigenetic mechanisms. Such new centromeres, also called neocentromeres, have been reported in both animal and plant species (2–6). Neocentromeres can also be induced by artificially deleting the original centromere on a chromosome (7, 8).
Neocentromere activation in humans is often associated with chromosomal rearrangements that are generally deleterious (5). In addition, human neocentromeres appear to be significantly smaller than native centromeres based on the size of the centromeric chromatin domain marked by the presence of centromere-specific histone CENP-A (CENH3 in plants) (9). Thus, neocentromeres do not seem to have any evolutionary advantage over existing native centromeres. However, comparative mapping among closely related mammalian species has revealed position changes of some centromeres during evolution, most likely via neocentromere formation (5, 10–15). Centromere repositioning (CR) events have occurred frequently in some mammalian lineages. For example, at least five CR events have occurred in donkey after its divergence from zebra approximately 1 million years ago (16).
Despite extensive documentation of CRs in mammalian species, CRs have been rarely reported in other eukaryotes. Thus far, only a single non-mammalian CR case was reported in birds (17). The lack of CR reports in non-mammal eukaryotes made us speculate if the CR phenomenon is associated with a structure unique to mammalian chromosomes. In addition, there has been little information on the genomic impact associated with CR. In this report, we describe a CR case in plants. Cucumber (Cucumis sativus L., 2n = 2x = 14) and melon (Cucumis melo L., 2n = 2x = 24) diverged from the same ancestor approximately 9 million years ago (18). Comparative fluorescence in situ hybridization (FISH) mapping using the same sets of genomic DNA clones revealed different centromeric positions between two pair of chromosomes from these two species. We also demonstrate that centromere activation or inactivation could have a major impact on the accumulation or loss of pericentromeric heterochromatin in the cucurbit species.
Results
Cucumber and Melon Centromeres Contain Different Satellite Repeats.
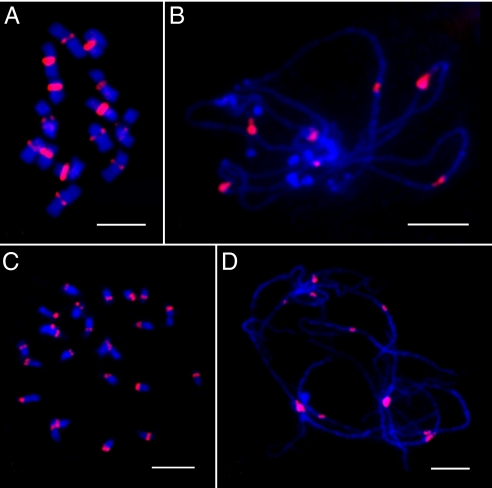
We previously demonstrated that cucumber centromeres contain a 177-bp Type III satellite repeat (19). The Type III repeat is highly abundant and accounts for approximately 4% of the cucumber genome. This satellite repeat is exclusively located in the centromeres of every cucumber chromosome, although the amount of repeat varies considerably among the different centromeres (Fig. 1 A and B).
Fig. 1.
Centromeric Satellite DNA repeats cucumber and melon. (A) FISH mapping of the Type III satellite repeat on the somatic metaphase chromosomes of cucumber. (B) FISH mapping of the Type III satellite repeat on the pachytene chromosomes of cucumber. (C) FISH mapping of the CentM satellite repeat on the somatic metaphase chromosomes of melon. (D) FISH mapping of the CentM satellite repeat on the pachytene chromosomes of melon. [Scale bars, 5 μm (A), 10 μm (B–D).]
We have identified a 352-bp satellite repeat in melon and named this repeat CentM. The CentM repeat is also highly abundant in the melon genome and is restricted within the centromeres of every melon chromosomes (Fig. 1 C and D). Sequence analysis revealed no similarity between the Type III and the CentM repeats. These two repeats did not generate any cross-hybridization signals when they were used reciprocally in the two species. These results indicate that the centromeric repeats of cucumber and melon chromosomes have completely diverged since these two species evolved from their common ancestor.
Comparative FISH Mapping of Cucumber Chromosome 6 and Melon Chromosome I.
We launched a project to develop pachytene chromosome-based molecular cytogenetic maps of individual cucumber and melon chromosomes. The maps were constructed by FISH mapping of cucumber fosmid clones on cucumber and melon pachytene chromosomes. The fosmid clones are anchored by Simple Sequence Repeat (SSR) markers that have been genetically mapped (20). A total of 8 and 12 SSR markers associated with cucumber linkage groups 3 and 7, respectively, were used to screen a fosmid library developed from the cucumber inbred line 9930. The SSR markers were dispersed at an average distance of ≈10 cM along the two linkage groups (Table S1). Individual cucumber chromosomes at mitotic metaphase can be identified by FISH mapping using the 45S rDNA probe and the Type III satellite repeat (19). FISH mapping of linkage group-specific fosmid clones together with the Type III repeat and 45S rDNA probes revealed that cucumber linkage groups 3 and 7 are associated with chromosome 7 and 6, respectively (Fig. S1).
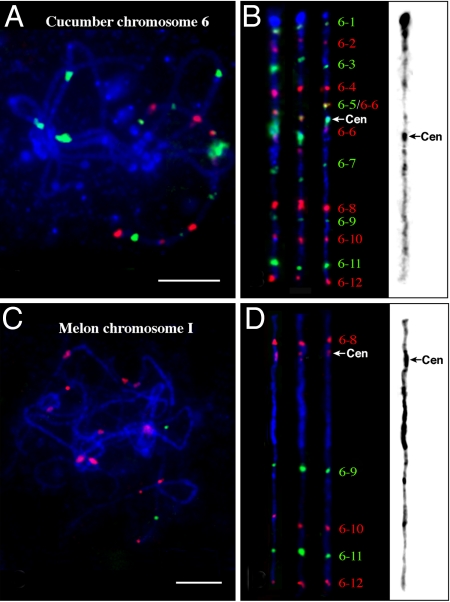
We first determined the physical order of adjacent fosmid clones based on the genetic positions of their corresponding SSR markers by dual-color FISH on somatic metaphase chromosomes. Multifosmid probe cocktails were then hybridized to the pachytene chromosomes along with the Type III repeat (Fig. 2A). Eleven of the 12 fosmid clones mapped to chromosome 6 generated a single FISH signal in the cucumber genome. Fosmid 6–6, however, generated two distinct signals flanking the centromere of cucumber chromosome 6 (Fig. 2B). The fosmid 6–6 signal on the short arm overlapped with the signal from fosmid 6–5. The centromere of chromosome 6 was located between fosmids 6–5 and 6–6 (Fig. 2B). The order of individual fosmids along chromosome 6 was concordant with the order of the SSR markers along the linkage map (Table S1).
Fig. 2.
Comparative FISH mapping of cucumber chromosome 6 and melon chromosome I. (A) Cucumber chromosomes at the pachytene stage were probed by a set of 12 fosmid clones together with the Type III satellite repeat. (B) Three straightened cucumber pachytene chromosome 6. One of the chromosomes was converted into a black-white image. Distinct heterochromatin is visible in the centromeric region and the distal end of the short arm. (C) Melon chromosomes at the pachytene stage were probed by a set of five fosmid clones together with the CentM satellite repeat. (D) Three straightened melon pachytene chromosome I. One of the chromosomes was converted into a black-white image. Distinct heterochromatin is visible in the centromeric region and the pericentromeric region on the long arm. (Scale bars, 10 μm.)
The 12 fosmids were then mapped on melon somatic chromosomes. Adjacent fosmid pairs were labeled in different colors and mapped on melon chromosomes, which revealed if a pair of adjacent fosmid clones were located on the same melon chromosome and whether their order is the same as that on cucumber chromosome (Fig. S2). Fosmids 6–1 to 6–7 mapped to several different melon chromosomes. However, five consecutive fosmids, 6–8 to 6–12, mapped to a single melon chromosome, which was named chromosome I (Fig. 2 C and D). These five fosmids were mapped to melon pachytene chromosomes together with the CentM repeat. The five fosmids covered almost the entire length of the melon chromosome I. The mapping order of these five fosmids was the same in cucumber and melon. Interestingly, the centromere of melon chromosome I is located between fosmid 6–8 and 6–9 (Fig. 2D). In addition, the physical distance between fosmid 8 and 9 was significantly expanded in melon compared to that in cucumber. Fosmids 6–8 and 6–9 were separated by a short euchromatic segment in cucumber chromosome 6. In contrast, these two fosmids were separated by a large heterochromatic segment that accounts for approximately 47% of the length of melon chromosome I (Fig. 2D).
Comparative FISH Mapping of Cucumber Chromosome 7 and Melon Chromosome II.
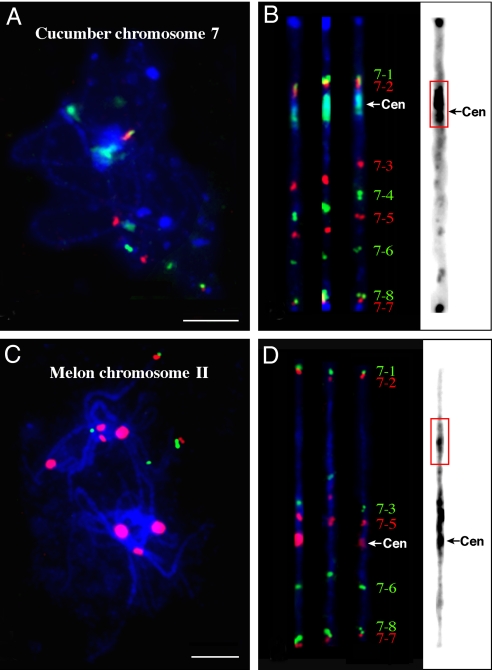
Eight fosmids associated with cucumber chromosome 7 were selected for FISH mapping (Table S1). Similarly, we first determined the physical order of the adjacent fosmids by dual-color FISH on cucumber somatic chromosomes. Multifosmid probe cocktails were then hybridized to the pachytene chromosomes together with the Type III repeat (Fig. 3A). The order of individual fosmids along chromosome 7 was concordant with the corresponding SSR markers on the linkage map, except that fosmids 7–7 and 7–8 showed an inverted positions compared to SSR20122 (61.0 cM, anchoring fosmid 7–8) and SSR17062 (66.5 cM, anchoring fosmid 7–7) (Table S1). The centromere was located between fosmids 7–2 and 7–3 (Fig. 3B).
Fig. 3.
Comparative FISH mapping of cucumber chromosome 7 and melon chromosome II. (A) Cucumber chromosomes at the pachytene stage were probed by a set of eight fosmid clones together with the Type III satellite repeat. (B) Three straightened cucumber pachytene chromosome 7. One of the chromosomes was converted into a black-white image. Distinct heterochromatin is visible in the centromeric region and the distal ends of both chromosome arms. (C) Melon chromosomes at the pachytene stage were probed by a set of seven fosmid clones together with the CentM satellite repeat. (D) Three straightened melon pachytene chromosome II. One of the chromosomes was converted into a black-white image. Distinct heterochromatin is visible in the centromeric region and in the region spanned by fosmids 7–2 and 7–3. The red boxes in B and D highlight the heterochromatin domains containing the centromere for the cucumber chromosome but not the melon chromosome. (Scale bars, 10 μm.)
Seven of the eight fosmids were mapped to a single melon chromosome, named chromosome II (Fig. 3 C and D; Fig. S2). Fosmid 7–4 generated dispersed FISH signals on all melon chromosomes without distinct signals associated with chromosome II (Fig. S3). The order of the other seven fosmids was identical between cucumber chromosome 7 and melon chromosome II. Interestingly, the centromere of melon chromosome II was located between fosmids 7–5 and 7–6 (Fig. 3D). Fosmids 7–5 and 7–6 span an euchromatic segment on cucumber chromosome 7. These two clones, however, span the centromere and a large section of the pericentromeric chromatin on melon chromosome II (Fig. 3D). The chromosomal domain spanned by fosmids 7–2 and 7–3, which includes the centromere of chromosome 7 in cucumber, showed a similar physical distance in melon chromosome II even though the centromere of chromosome II was not located within this domain. In addition, heterochromatin was associated with both chromosomes between these two fosmids (Fig. 3 B and D).
Discussion
Centromere Repositioning in Plant Species.
Comparative analysis of the melon genetic map with the cucumber sequence map revealed that cucumber chromosome 6 (Cu6) is an equivalent to a fusion of two melon chromosomes (21). Thus, a chromosomal fusion/fission event possibly occurred during the evolution of these cucurbit chromosomes. Such fusion/fission events have been well characterized in the Brassicaceae species by FISH using chromosome-specific painting probes (22, 23). Elimination of minichromosomes resulted from chromosomal translocations, rather than centromere inactivation, was used to explain the reduction of chromosome/centromere numbers during karyotype evolution of the Brassicaceae species (22, 23).
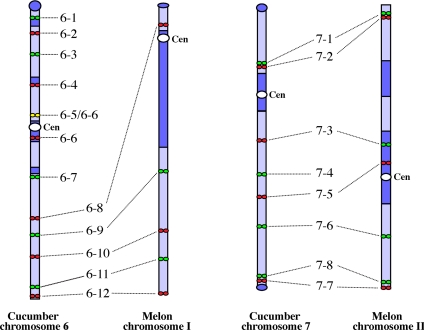
If the ancestor species for cucumber and melon contained 2n = 14 chromosomes (24), then melon chromosome I (MeI) would be derived from a chromosomal fission event. The centromere of MeI would have evolved from a neocentromere emerging between fosmids 6–8 and 6–9. Pericentromeric heterochromatin accumulation may have significantly expanded the distance between these two fosmids in MeI (Fig. 4). Alternatively, if the ancestor species contained 2n = 24 chromosomes (25, 26), Cu6 would be derived from a chromosomal fusion event. The current centromere of Cu6 would be derived from one of the two ancestor chromosomes, whereas the centromere between fosmids 6–8 and 6–9 from the second ancestor chromosome became inactivated. Interestingly, heterochromatin is not observed between fosmids 6–8 and 6–9 in Cu6, suggesting that the pericentromeric heterochromatin may have lost after the inactivation of the centromere. In either scenario, a dramatic loss/accumulation of a large block of heterochromatin was associated with the neocentromere emergence (after chromosome fission) or centromere inactivation (after chromosome fusion). In addition to the chromosomal fission/fusion-based hypothesis, the current centromere position on MeI may also be explained by an inversion, which spans the centromere and fosmid 6–8, followed by the loss or movement of the chromosomal region that spans from fosmids 6–1 to fosmid 6.7.
Fig. 4.
Diagrammatic illustration of the marker orders and centromere positions of two pairs of cucumber and melon chromosomes.
DNA markers and their order on Cu7 and melon chromosome II (MeII) are well conserved (21), suggesting that no major rearrangement events have occurred during the evolution of these two chromosomes. However, the centromeres in these two chromosomes are located in different positions, which is similar to the X chromosomes of primate species that also share DNA marker order except for the centromeric positions (11). Thus, the different centromeric position of Cu7 and MeII is best explained by a CR event that occurred during the evolution of these two chromosomes (Fig. 4). Nevertheless, we cannot exclude the possibility that the centromere position change between Cu7 and MeII was caused by two consecutive pericentromeric inversions with breakpoints located on different sides of the centromere.
Interestingly, MeII contains two interstitial heterochromatin domains. The first heterochromatin domain spans the current centromere and is located between fosmids 7–5 and 7–6 (Fig. 4). The second heterochromatin domain is spanned by fosmids 7–2 and 7–3 and is located within a similar position as the pericentromeric domain in Cu7 (Fig. 3 B and D). These results support the CR hypothesis that the original centromere of MeII was located in the second heterochromatin domain, but moved to the current position during evolution. Mapping the same centromere in another cucurbit species, such as watermelon, will provide additional evidence for this centromere repositioning event, because mapping results indicated that no fission or fusion event has occurred during the evolution of this chromosome in all three species (21). FISH mapping of additional fosmid clones close to the Cu7/MeII centromeres will also provide more evidence on whether pericentromeric inversions have occurred during the evolution of these two chromosomes.
Extensive comparative genome mapping and sequencing efforts in several model plant species have not revealed CR events. For example, the genetic colinearity between potato and tomato, which diverged ≈12 million years (27), was well documented by comparative genetic linkage mapping (28, 29). Potato and tomato chromosomes differ by several large chromosomal inversions (29, 30). However, the centromeric positions of potato and tomato chromosomes seem to be unchanged. Similarly, comparative mapping among several grass species, including rice, sorghum, and maize, has revealed extensive chromosomal rearrangements during the evolution of these species (31, 32). However, it is not clear if CRs have been associated with any chromosomes in these species.
The lack of CR reports in Solanaceae and grass species suggests that CRs may be less prevalent in certain eukaryotic lineages, including plant species, although there is no biological explanation for such lineage specificity. However, it is also possible that CR events in the Solanaceae and grass species may not be revealed by the previous DNA marker-based comparative mapping efforts, which often lacked precise locations of the centromeres. In mammalian species, comparative FISH mapping has proved to be the most effective methodology to reveal CR events. A number of cucurbit species are diploids and have not undergone whole genome duplications (21). These species also have relatively small genomes, thus, genomic DNA clones can be readily used in cross-species FISH mapping. These genomic characteristics of the cucurbit species make them ideal model plants for future CR research.
Potential Genomic Impact Associated with Centromere Repositioning in Plants.
Although CRs occurred frequently in the evolution of mammalian species and was implicated to play an important role in mammalian genome evolution (14), very little is known about the genomic consequences of re-seeding a centromere in an eukaryotic chromosome. Segmental duplication is a prominent feature associated with the pericentromeric regions of most human chromosomes (33). However, there has been limited evidence indicating that centromere re-seeding may trigger formation of such segmental duplications (14). Similar segmental duplications were not found in the pericentromeric regions of several well sequenced model plant species, including rice and Arabidopsis thaliana.
A major characteristic associated with plant chromosomes is the large amount of pericentromeric heterochromatin. The pericentromeric heterochromatin domains in many plant species are cytologically distinct compared to those of mammalian chromosomes and represent the majority of the heterochromatin in plant species with small genomes, such as A. thaliana (34). Pericentromeric heterochromatin domains account for more than 50% of the DNA of the chromosomes in some plants, such as tomato (35). Dominant pericentromeric heterochromatin was associated with all cucumber and melon chromosomes (36) (Figs. 2C and 3C). One general feature associated with the pericentromeric heterochromatin is the severe suppression of crossovers (37, 38). Pericentromeric heterochromatin probably also plays an important role in sister chromatid cohesion (39). Thus, one potential genomic impact of CRs in plants is inducing the accumulation of pericentromeric heterochromatin.
The CR events in the cucurbit species provide cytological evidence on the genomic impact of centromere re-seeding. Comparative mapping of Cu6 and MeI showed two possibilities after the chromosome fusion or fission event: either loss of the heterochromatin between fosmids 6–8 and 6–9 in Cu6 or gain of the pericentromeric heterochromatin in MeI. Thus, a centromere inactivation or centromere re-seeding event resulted in a dramatic loss in Cu6 or gain of the pericentromeric heterochromatin in MeI, which accounts almost 47% of the chromosome (Figs. 2D and 4). Similarly, a CR event in MeII may induce the accumulation of heterochromatin in the region spanned by fosmids 7–5 and 7–6. Interestingly, the DAPI staining of the chromatin domain spanned by fosmids 7–2 and 7–3 in MeII is clearly not as bright as the corresponding domain in Cu7 as well as the centromeric domain in MeII (Fig. 3 B and D). Thus, the inactivation of the original centromere in this region may cause the erosion of the heterochromatic characteristics of this chromosomal domain.
Methods
Plant Materials and Chromosome Preparation.
Cucumber inbred line 9930 and melon inbred line 3A832 were used for cytological studies. Root tips were harvested from germinated seeds, pretreated in 0.002 M 8-hydroxyquinoline at room temperature for 2 h, and fixed in methanol : glacial acetic acid (3:1). Root tips were macerated in the enzyme mixture of 2% cellulose and 1% pectolyase at 37 °C for 2 h, and squashes were made in the same fixative. Young panicles were harvested and fixed in 3:1 (100% ethanol:glacial acetic acid) Carnoy's solution. The procedure for meiotic chromosome preparation was the same as that used for preparing mitotic chromosomes from root tips with the following modification: Anthers were digested in the enzyme mixture for 4.5 h at 37 °C. The digested anthers were macerated on glass slides in 50% acetic acid solution with fine-pointed forceps and then “flame-dried” over an alcohol flame.
Fluorescence In Situ Hybridization.
All fosmid clones were from the cucumber fosmid library constructed from the cucumber inbred line 9930. SSR markers spaced ≈10 cM apart across linkage groups 3 and 7 (20) were used to select fosmids for FISH. Fosmid DNA was isolated using QIAGEN plasmid midi kit and further purified by Plant DNeasy spin columns (QIAGEN). The Type III repeat of cucumber (19) and the CentM repeat of melon (GenBank accession no. 3929695) were used for centromere identification. FISH was performed according to published protocols (40). DNA probes were labeled with digoxigenin-dUTP or biotin-dUTP via nick translation and detected with anti-digoxigenin antibody coupled with Rhodamine (Roche) or avidin-conjugated with FITC (Vector Laboratories), respectively. Chromosomes were counterstained by 4,6-diamidino-2-phenylindole (DAPI) in a VectaShield antifade solution (Vector Laboratories). Images were captured digitally using a CCD camera (QIMAGING, RETIGA-SRV, FAST 1394) attached to an Olympus BX61 epifluorescence microscope. Gray-scale images were captured for each color channel and then merged. Chromosome straightening was performed using the ‘straighten-curved-objects’ plug-in of Image J (41), and final image optimization was performed using Adobe Photoshop (Adobe Systems).
Supplementary Material
Acknowledgments.
We thank Dr. Paul Talbert for his valuable comments on the manuscript and Dr. H. S. Wang (Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences) for supplying young panicles of C. melon. This research was supported by the Natural Science Foundation of China (30771208), and Program for Changjiang Scholars and Innovative Research Team in University, Program of Introducing Talents of Discipline to Universities (111–2-03) to W.W.J.; by the Ministry of Agriculture (“948” program 2008-Z42) and the Ministry of Science and Technology (2006DFA32140) to S.W.H.; and U.S. National Science Foundation grant DBI-0603927 to J.M.J.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904833106/DCSupplemental.
References
- 1.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voullaire LE, Slater HR, Petrovic V, Choo KHA. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: Activation of a latent centromere? Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 3.Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 4.Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci USA. 2005;102:9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall OJ, Chueh AC, Wong LH, Choo KHA. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topp CN, et al. Identification of a maize neocentromere in an oat-maize addition line. Cytogenet Genome Res. 2009;124:228–238. doi: 10.1159/000218128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishii K, et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 8.Ketel C, et al. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alonso A, et al. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum Mol Genet. 2003;12:2711–2721. doi: 10.1093/hmg/ddg282. [DOI] [PubMed] [Google Scholar]
- 10.Montefalcone G, Tempesta S, Rocchi M, Archidiacono N. Centromere repositioning. Genome Res. 1999;9:1184–1188. doi: 10.1101/gr.9.12.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ventura M, Archidiacono N, Rocchi M. Centromere emergence in evolution. Genome Res. 2001;11:595–599. doi: 10.1101/gr.152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreri GC, Liscinsky DM, Mack JA, Eldridge MDB, O'Neill RJ. Retention of latent centromeres in the mammalian genome. J Hered. 2005;96:217–224. doi: 10.1093/jhered/esi029. [DOI] [PubMed] [Google Scholar]
- 13.Cardone MF, et al. Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol. 2006;7:R91. doi: 10.1186/gb-2006-7-10-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventura M, et al. Evolutionary formation of new centromeres in macaque. Science. 2007;316:243–246. doi: 10.1126/science.1140615. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, et al. Centromere repositioning in the X chromosome of XO/XO mammals, Ryukyu spiny rat. Chromosome Res. 2008;16:587–593. doi: 10.1007/s10577-008-1199-5. [DOI] [PubMed] [Google Scholar]
- 16.Carbone L, et al. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics. 2006;87:777–782. doi: 10.1016/j.ygeno.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Kasai F, Garcia C, Arruga MV, Ferguson-Smith MA. Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa): Evidence of the occurrence of a neocentromere during evolution. Cytogenet Genome Res. 2003;102:326–330. doi: 10.1159/000075770. [DOI] [PubMed] [Google Scholar]
- 18.Schaefer H, Heibl C, Renner SS. Gourds afloat: A dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc R Soc B. 2009;276:843–851. doi: 10.1098/rspb.2008.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han YH, et al. Distribution of the tandem repeat sequences and karyotyping in cucumber (Cucumis sativus L.) by fluorescence in situ hybridization. Cytogenet Genome Res. 2008;122:80–88. doi: 10.1159/000151320. [DOI] [PubMed] [Google Scholar]
- 20.Ren Y, et al. An integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE. 2009:e5795. doi: 10.1371/journal.pone.0005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang SW, Zhang ZH, Gu XF, Lucas WJ, Wang XW. The cucumber genome sequence reveals insights into biology and speciation of cucurbits and the evolution of the plant vascular system. Nat Genet. 2009 in press. [Google Scholar]
- 22.Lysak MA, et al. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA. 2006;103:5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandakova T, Lysak MA. Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae) Plant Cell. 2008;20:2559–2570. doi: 10.1105/tpc.108.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhaduri PN, Bose PC. Cyto-genetical investigations in some common cucurbits, with special reference to fragmentation of chromosomes as physical basis of speciation. J Genet. 1947;48:237–256. doi: 10.1007/BF02989384. [DOI] [PubMed] [Google Scholar]
- 25.Trivedi RN, Roy RP. Cytological studies in Cucumis and Citrullus. Cytologia. 1970;35:561–569. [Google Scholar]
- 26.Chung SM, Staub JE, Chen JF. Molecular phylogeny of Cucumis species as revealed by consensus chloroplast SSR marker length and sequence variation. Genome. 2006;49:219–229. doi: 10.1139/g05-101. [DOI] [PubMed] [Google Scholar]
- 27.deSa MM, Drouin G. Phylogeny and substitution rates of angiosperm actin genes. Mol Biol Evol. 1996;13:1198–1212. doi: 10.1093/oxfordjournals.molbev.a025685. [DOI] [PubMed] [Google Scholar]
- 28.Bonierbale MW, Plaisted RL, Tanksley SD. RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics. 1988;120:1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanksley SD, et al. High-density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iovene M, Wielgus SM, Simon PW, Buell CR, Jiang JM. Chromatin structure and physical mapping of chromosome 6 of potato and comparative analyses with tomato. Genetics. 2008;180:1307–1317. doi: 10.1534/genetics.108.093179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowers JE, et al. Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc Natl Acad Sci USA. 2005;102:13206–13211. doi: 10.1073/pnas.0502365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei F, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3:1254–1263. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.She XW, et al. The structure and evolution of centromeric transition regions within the human genome. Nature. 2004;430:857–864. doi: 10.1038/nature02806. [DOI] [PubMed] [Google Scholar]
- 34.Fransz P, et al. Cytogenetics for the model system Arabidopsis thaliana. Plant J. 1998;13:867–876. doi: 10.1046/j.1365-313x.1998.00086.x. [DOI] [PubMed] [Google Scholar]
- 35.Chang SB, et al. FISH mapping and molecular organization of the major repetitive sequences of tomato. Chromosome Res. 2008;16:919–933. doi: 10.1007/s10577-008-1249-z. [DOI] [PubMed] [Google Scholar]
- 36.Koo DH, Choi HW, Cho J, Hur Y, Bang JW. A high-resolution karyotype of cucumber (Cucumis sativus L. ‘Winter Long’) revealed by C-banding, pachytene analysis, and RAPD-aided fluorescence in situ hybridization. Genome. 2005;48:534–540. doi: 10.1139/g04-128. [DOI] [PubMed] [Google Scholar]
- 37.Sherman JD, Stack SM. Two-dimensional spreads of synaptonemal complexes from Solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum) Genetics. 1995;141:683–708. doi: 10.1093/genetics/141.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan HH, Jiang JM. Rice as a model for centromere and heterochromatin research. Chromosome Res. 2007;15:77–84. doi: 10.1007/s10577-006-1104-z. [DOI] [PubMed] [Google Scholar]
- 39.Topp CN, Dawe RK. Reinterpreting pericentromeric heterochromatin. Curr Opin Plant Biol. 2006;9:647–653. doi: 10.1016/j.pbi.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Jiang JM, Gill BS, Wang GL, Ronald PC, Ward DC. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA. 1995;92:4487–4491. doi: 10.1073/pnas.92.10.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kocsis E, Trus BL, Steer CJ, Bisher ME, Steven AC. Image averaging of flexible fibrous macromolecules: The clathrin triskelion has an elastic proximal segment. J Struct Biol. 1991;107:6–14. doi: 10.1016/1047-8477(91)90025-r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.