Abstract
Amyloid β-protein (Aβ) oligomers may be the proximate neurotoxins in Alzheimer's disease (AD). “Oligomer” is an ill-defined term because many kinds have been reported and they often exist in rapid equilibrium with monomers and higher-order assemblies. We report here results of studies in which specific oligomers have been stabilized structurally, fractionated in pure form, and then studied by using a combination of CD spectroscopy, Thioflavin T fluorescence, EM, atomic force microscopy (AFM), and neurotoxicity assays. Aβ monomers were largely unstructured, but oligomers exhibited order-dependent increases in β-sheet content. EM and AFM data suggest that dimerization and subsequent monomer addition are processes in which significant and asymmetric monomer conformational changes occur. Oligomer secondary structure and order correlated directly with fibril nucleation activity. Neurotoxic activity increased disproportionately (order dependence >1) with oligomer order. The structure–activity correlations reported here significantly extend our understanding of the conformational dynamics, structure, and relative toxicity of pure Aβ oligomers of specific order.
Keywords: Alzheimer's disease, toxicity
Alzheimer's disease (AD) is the most common form of late-life dementia. Current estimates of AD incidence are >24 million worldwide, a number that is expected to double every 20 years, reaching 81 million in 2040 (1). AD is a slowly progressive disorder with insidious onset and progressive impairment of episodic memory and executive function coupled with aphasia, apraxia, and agnosia (1).
The amyloid β-protein (Aβ) appears to play an essential role in the pathogenesis of AD. Aβ is produced throughout life through posttranslational processing of the Aβ precursor (APP). Familial forms of AD increase Aβ production or the propensity of Aβ to aggregate (2). The “amyloid cascade hypothesis” proposes that assemblies of Aβ initiate a process leading to neuronal dysfunction and cell death (2). The most potent neurotoxic assemblies appear to be oligomeric, rather than fibrillar, in nature (3, 4). For example, oligomers extracted from AD brain potently impair synapse structure and function (5). The smallest of these oligomers appears to be dimeric (5). However, a systematic correlation of oligomer structure and neurotoxic potency has not been reported. This correlation is critical for the targeting and design of disease-modifying therapeutic agents.
Efforts to establish rigorous structure–toxicity correlations have been hindered by the complex, dynamic equilibria displayed by Aβ (for recent reviews, see refs. 6 and 7). To enable determination of the oligomer frequency distribution, we have used in situ chemical cross-linking to prevent oligomer dissociation or growth (8–10). Oligomers thus stabilized can be visualized and quantified by SDS/PAGE. However, in theory, the method also could be used to produce pure populations of oligomers of defined order†, enabling the biophysical and biological studies necessary to establish structure–toxicity correlations. We report here that this goal is attainable in practice and discuss insights into the structural biology of Aβ thus obtained.
Results
Preparation and Characterization of Aβ40 Oligomers of Defined Order.
To study specific Aβ oligomers, we used the technique of photo-induced cross-linking of unmodified proteins (PICUP) (10) to chemically stabilize oligomers. PICUP has been shown to rapidly (<1 s) and efficiently (80–90%) cross-link native Aβ oligomers, allowing accurate determination of the oligomer size distribution using SDS/PAGE (10). We found, as reported (9), that cross-linked Aβ40 comprises a mixture of monomers and oligomers of order 2–5 (Fig. 1, lane 1).
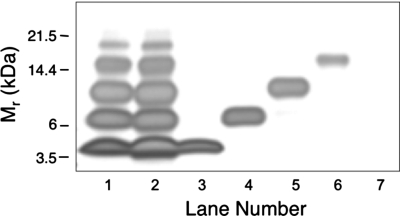
Fig. 1.
Stability of purified oligomers. Aβ samples were subjected to PICUP and SDS/PAGE. Individual gel bands were stained with Coomassie blue, excised, and then extracted under alkaline conditions. The extracts then were reconstituted in F12K medium and analyzed by SDS/PAGE and silver staining. Lane 1, cross-linked Aβ immediately after PICUP (cross-linking control). Lane 2, cross-linked Aβ subjected to the entire protocol, but with all bands pooled together (control for unfractionated Aβ subjected to alkaline extraction). Lane 3, monomer band. Lane 4, dimer band. Lane 5, trimer band. Lane 6, tetramer band. Lane 7, a “band-equivalent”-sized piece of gel (“no protein” control). The data are representative of results from each of three independent experiments.
To obtain pure oligomer populations, we developed a method to extract the protein component from a gel band and remove the accompanying SDS (see Materials and Methods). We performed this procedure first on an entire gel lane in which cross-linked Aβ had been electrophoresed. When the material thus isolated was electrophoresed on a second SDS gel, the oligomer distribution was identical, within experimental error, to that of the original cross-linked preparation (Fig. 1, lane 2). Proteins extracted in this manner thus “ran true,” as determined by SDS/PAGE behavior‡. We next purified monomers through tetramers by using this method (Fig. 1, lanes 3–6). Highly pure preparations were obtained. Percentage purities determined by densitometric analysis of the monomer, dimer, trimer, and tetramer preparations were 99.9%, 97.0%, 94.3%, and 99.6%. The low abundance (<5%) of pentamers and higher-order oligomers precluded their isolation and study.
To determine whether the purified, cross-linked species self-associated to produce Thioflavin T (ThT)-positive assemblies, we incubated them in F12K medium at 37 °C and monitored β-sheet content periodically. In addition to the pure oligomers, we studied unfractionated, cross-linked Aβ. All of the samples displayed low initial ThT fluorescence intensities that did not change significantly over a 2-day period (data not shown). These data show that cross-linking prevents the continued evolution of the β-sheet structures that are characteristic of Aβ assembly.
To determine whether dissociation of the purified oligomers occurred during incubation in culture medium, an important question for interpretation of cytotoxicity assays, aliquots of pure, cross-linked monomers, dimers, and trimers were removed from the F12K medium at the end of the incubation (2 days) and were analyzed by SDS/PAGE (Fig. S1). Densitometry then was done to quantify the relative amounts of each band in each lane. In this experiment, before incubation, monomers, dimers, and trimers displayed purities of 91.2%, 93.5%, and 100%, respectively. After incubation, the purities were 80.0%, 89.7%, and 94.5%. The decreases in purity reflected small increases (4–12%) in the intensity of oligomers of order 2n, where n was the initial order of the purified oligomer. The similarity in oligomer states for each of the “before and after” samples means that experimental differences observed among the pure oligomer populations in subsequent experiments must be caused primarily by oligomer order and not postincubation alterations thereof.
Oligomer Conformation.
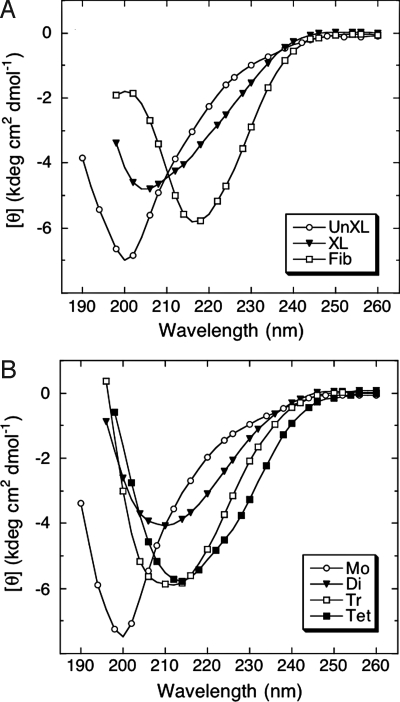
Studies have revealed that Aβ exists predominately as a statistical coil (SC) if prepared under conditions designed to prevent its folding and self-assembly (7, 11). To determine whether initial Aβ oligomerization involves changes in peptide secondary structure, each purified oligomer population was studied by using CD (Fig. 2). Uncross-linked Aβ (Fig. 2A) and purified monomer (Fig. 2B) produced initial spectra characteristic of SCs. The major feature of these spectra was a large magnitude minimum centered at ≈200 nm. In contrast, the major spectral feature of fibrils was a large magnitude minimum centered at ≈216 nm, indicative of β-sheet structure (Fig. 2A). The major feature of the spectrum produced by cross-linked, unfractionated Aβ was a minimum at ≈205 nm, a wavelength between those of the minima of Aβ monomers and fibrils. This finding indicates that oligomerization is accompanied by an increase in structural order§. Spectra from dimers, trimers, and tetramers all displayed single minima, the wavelengths of which (≈210, ≈212, and ≈214 nm, respectively) correlated directly with oligomer order (Fig. 2B). The tetramer wavelength minimum approached that of fibrils (≈216 nm). Deconvolution of the spectra (Table 1) revealed that the sum of the percentages of β-sheet and SC remained almost constant (≈90%) among monomers and oligomers, but that β-sheet content increased from ≈25% in the monomer to ≈45% in the tetramer (Fig. S2). The largest relative change in secondary structure content was between monomer and dimer. Cross-correlation of the SC and β-sheet structural elements yielded r2 = 0.995, suggesting that the increased β-sheet could have formed directly from disordered elements. However, it also is possible that an SC → α-helix → β-sheet transition could occur.
Fig. 2.
Secondary structure dynamics of Aβ assemblies. Uncross-linked (UnXL), cross-linked (XL), or fibrillar (Fib) Aβ (A) or isolated oligomers [monomer (Mo), dimer (Di), trimer (Tr), and tetramer (Te)] (B) were prepared in 10 mM PBS, pH 7.4, and then monitored immediately by CD. Data are representative of three independent experiments.
Table 1.
Characteristics of Aβ assemblies
| Sample | α-Helix* | β-Sheet* | SC* | Diameter† | Height‡ | EC50§ |
|---|---|---|---|---|---|---|
| Uncross-linked | 9.4 | 25.2 | 65.4 | 1.15 ± 0.17 (79) | 0.14 ± 0.01 (135) | 102.5 ± 5.6 |
| Cross-linked | 11.1 | 33.6 | 55.3 | 12.39 ± 2.13 (55) | 2.93 ± 0.40 (56) | 43.0 ± 2.7 |
| Pure monomer | 8.7 | 24.0 | 67.3 | 1.30 ± 0.13 (127) | 0.24 ± 0.01 (178) | 67.3 ± 8.7 |
| Pure dimer | 10.5 | 38.6 | 50.9 | 1.78 ± 0.23 (116) | 0.53 ± 0.03 (165) | 41.6 ± 3.9 |
| Pure trimer | 10.3 | 40.8 | 48.9 | 7.22 ± 1.12 (64) | 0.94 ± 0.13 (67) | 24.5 ± 1.9 |
| Pure tetramer | 12.7 | 44.9 | 42.4 | 11.00 ± 2.08 (39) | 1.51 ± 0.30 (38) | 20.5 ± 0.4 |
| Fibrils | ND | 57.0 | 43.0 | ND | ND | 57.6 ± 2.2 |
ND, not determined.
*CD data are expressed as percent of each secondary structure element.
†Mean diameter ± SE, in nm, is listed for (n) Aβ assemblies visualized by EM.
‡Mean height ± SE, in nm, is listed for (n) Aβ assemblies visualized by AFM.
§Effective concentration (EC50) is the concentration of a particular Aβ assembly that produced a level of toxicity in MTT assays that was half maximal. EC50 [mean concentration (μM) ± SE] values were calculated after sigmoidal curve fitting of the data shown in Fig. S5, using GraphPad Prism software (version 4.0a).
Aβ Assembly Morphology.
We next determined the morphologies of the oligomers by using EM (Fig. S3 and Table 1). Uncross-linked Aβ and purified monomers displayed irregular thread-like and globular shapes that had average diameters of 1.15–1.30 nm. As oligomer order increased, the relative amounts of thread-like structures decreased and the amounts of quasi-spherical structures increased (compare Fig. S3 C and F). Diameter increased progressively with oligomer order, from 1.78 nm for dimers to 11.00 nm for tetramers. Unfractionated, cross-linked species had the largest average diameter, 12.39 nm, but this value was not significantly different from that of tetramers.
In prior studies of Aβ assembly, we have found that EM and atomic force microscopy (AFM) sometimes reveal different features in a single population, which is not surprising considering the differences in substrates (carbon-coated formvar versus mica) to which assemblies must adhere and in visualization physics (electron transmission versus physical displacement). Therefore, in addition to EM, we studied oligomer morphology by using AFM (Fig. S4 and Table 1). The rank order of heights was identical to that for diameter. The smallest assemblies were in the uncross-linked population, and the largest were in the unfractionated, cross-linked population. Analysis of the diameter and height data revealed that these parameters were correlated (r2 > 0.95). EM and AFM thus revealed the same order dependency of assembly geometry, although the absolute values of the respective diameters and heights were unequal.
Nucleation of Fibril Formation.
Aβ fibril assembly is a nucleation-dependent elongation process (12–14). To determine the effects of specific oligomers on this process, we used ThT to monitor temporal changes in β-sheet content in samples of Aβ to which were added preformed fibrils, unfractionated oligomer mixtures, or pure monomers, dimers, trimers, or tetramers. ThT fluorescence is not a measure of fibril content per se, but because β-sheet formation correlates with amyloid fibril formation, ThT fluorescence is a useful surrogate marker (15–17). The assay also provides the means to monitor assembly kinetics.
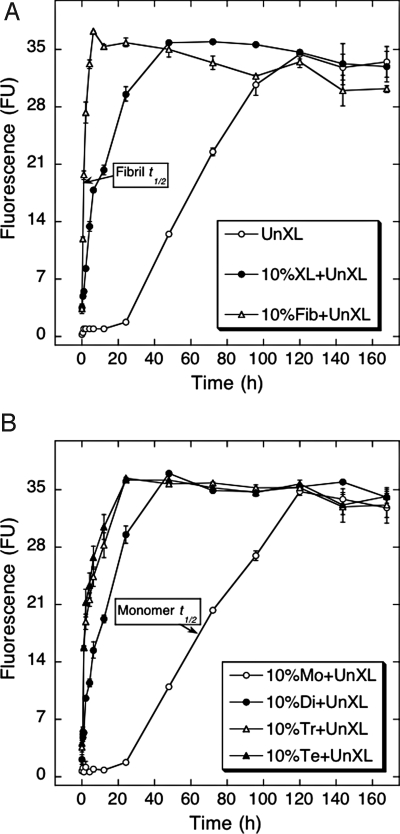
Aβ displayed a sigmoidal process curve characterized by a ≈1-day lag time, a ≈4-day period of increasing ThT binding, and a binding plateau occurring after ≈5 d (Fig. 3A), results consistent with a nucleation-dependent polymerization process (18). The addition of preformed fibrils (nuclei) eliminated the lag period and produced a rapid increase in ThT binding that reached maximal levels after ≈4 h. Similar effects were observed after addition of unfractionated, cross-linked Aβ, but the rate of increase of ThT fluorescence was lower (maximal signal occurred after ≈2 days). These data showed that stable oligomers function as fibril nuclei.
Fig. 3.
Nucleation of Aβ assembly. The nucleation activity of different Aβ preparations was assessed by addition of each preparation to uncross-linked Aβ, which then was incubated for 7 d at 37 °C in 10 mM PBS, pH 7.4. Aliquots were assayed periodically by using ThT. The preparations were uncross-linked Aβ (○, UnXL), 10% (vol/vol) cross-linked Aβ (●, XL), or 10% (vol/vol) sonicated Aβ fibrils (△, Fib) (A) or 10% (vol/vol) Aβ monomer (○, Mo), dimer (●, Di), trimer (△, Tr), or tetramer (▲, Te) (B). Binding is expressed as mean fluorescence [in arbitrary fluorescence units (FU)] ± SE. Data were obtained in three independent experiments. Arrows indicate times at which half maximal ThT binding was observed.
To differentiate effects among the oligomers, the experiments were repeated with the pure cross-linked monomer and oligomer preparations (Fig. 3B). Monomers had no effect, as would be expected because monomers cannot nucleate fibril formation (12, 13). However, each stabilized oligomer nucleated assembly growth. Dimers displayed a process curve similar to that of unfractionated, cross-linked oligomers (compare Fig. 3 A and B), demonstrating that they nucleate assembly significantly. However, substantially greater effects were observed with trimers and tetramers. Initial rates of ThT signal increase for these latter oligomers were ≈75% that of fibrils (Fig. 3A), showing that these oligomeric assemblies were almost as efficient as preformed fibrils in nucleating assembly. If one defines t1/2 as the time at which half maximal ThT binding is observed, t1/2 values ranged from 1 h for fibril seeds to 65 h for monomers (Fig. 3, arrows). In contrast, dimers displayed t1/2 = 10 h and trimers and tetramers had identical (within experimental error) t1/2 = 2 h. Dimers thus can act as seeds, and trimers and tetramers are almost as active as preformed fibrils themselves.
It is important to recognize that the nominal total Aβ monomer concentration in each experiment was identical. This means that the molar concentration of each assembly differed. If one calculates effectiveness relative to that of the monomer (relative activity; Table S1), the data suggest that dimers are at least an order of magnitude more effective in nucleating fibril formation than are monomers (which must assemble de novo to produce nuclei). Trimers and tetramers are at least 100-fold more effective. Fibrils are more effective than oligomers, but fibril-specific activity cannot be determined accurately because fibril structure is not static, but is represented by a quaternary structure frequency distribution that itself comprises dynamic lower-order structural equilibria.
Biological Activities of Oligomers.
To establish structure–activity relationships, we determined the effects of specific oligomers on cellular metabolism and survival by using 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) (Table 1 and Fig. S5) and lactate dehydrogenase (LDH) (Fig. 4) assays (19, 20), respectively. For the MTT experiments, differentiated PC12 cells were treated with oligomers of varying concentration and then the concentration at which a 50% effect was observed (EC50) was determined for each (see Materials and Methods). Uncross-linked Aβ, cross-linked unfractionated Aβ, and Aβ fibrils yielded EC50 values of 102.5 ± 5.6, 43.0 ± 2.7, and 57.6 ± 2.2 μM (mean ± SE), respectively (Fig. S5A). Both cross-linked oligomeric Aβ and fibrillar Aβ thus were significantly more toxic than unassembled Aβ.
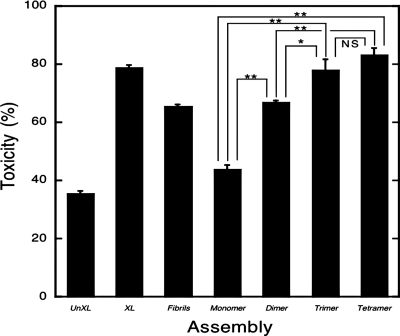
Fig. 4.
LDH activity. Uncross-linked (UnXL), cross-linked (XL), fibrillar, monomeric, and purified oligomeric (dimers, trimers, tetramers) Aβ samples were added at final nominal concentrations of 25 μM to differentiated PC12 cells. LDH activity in the supernatant fluid then was measured after 48 h. Data are representative of that obtained in three independent experiments. Each column represents means ± SE. The statistical significance of the toxicity differences among samples was determined by one-way fractional ANOVA and multiple comparison tests. *, P < 0.01; **, P < 0.001. NS, not significant.
To determine the contributions of each oligomer species to that of the cross-linked mixture, EC50 values were determined for each pure population. As observed in the biophysical studies discussed above, EC50 was proportional (in this case, inversely) to oligomer order (monomer = 67.3 ± 8.7 μM, dimer = 41.6 ± 3.9 μM, trimer = 24.5 ± 1.9 μM, and tetramer = 20.5 ± 0.4 μM) (Fig. S5B).
We next determined the cytotoxic ability of each assembly by measuring LDH release from differentiated PC12 cells (Fig. 4). Uncross-linked Aβ produced modest toxicity, averaging ≈35%. Unfractionated, cross-linked Aβ was significantly (P < 0.001) more toxic, producing almost 80% LDH release, and this level of release was significantly (P < 0.05) greater than that of fibrils (≈66%). Purified monomers were slightly more toxic than was Aβ alone (≈44% versus ≈35%), although this toxicity difference was not significant statistically. However, dimers were substantially and significantly (P < 0.001) more toxic and their toxicity was equal, within experimental error, to that of fibrils. These data show that toxicity was directly related to oligomer order. This was a consistent trend that was statistically significant between each assembly pair, except trimers and tetramers. In summary, the MTT and LDH assays produced identical rank orders of toxicity for uncross-linked Aβ and pure monomer and oligomer populations, namely tetramer > trimer > dimer > monomer = uncross-linked monomer. In addition, unfractionated, cross-linked Aβ consistently was more toxic than were fibrils.
We postulated that if the neurotoxic activity (t) of each Aβ assembly i (ti) was constant, regardless of whether it was determined in pure form or in mixtures, that total neurotoxic activity T would be a function of ti and fi, the occurrence frequency of oligomer of order i, according to the following equation:
 |
To determine whether this were true, we quantified the oligomer frequency distribution by densitometry, obtaining f1 = 0.39, f2 = 0.27, f3 = 0.17, and f4 = 0.13, and determined that T = 44.3 μM. Cross-linked, unfractionated Aβ, which includes additional (<4% more) higher-order oligomers, had an EC50 = 43.0 ± 2.7 μM, a value identical, within experimental error, to the calculated total neurotoxicity T. This result shows that there exists “conservation of toxicity” between unfractionated and fractionated populations of oligomers and supports the supposition that each oligomer maintains its unique specific activity within heterogeneous Aβ oligomer populations.
Maintenance of activity suggests maintenance of structure. To explore this question, we applied the same analytical reasoning used in our analysis of toxicity (Eq. 1) to the CD data (Table 1). We found that the weighted sums for α-helix for the monomer through tetramer states equaled the α-helix content in an unfractionated, cross-linked sample. The same relationship was observed for β-sheet and SC secondary structure elements. These structural data thus were consistent with our examination of conservation of toxicity.
To address the question of oligomer-specific toxic activity, we calculated the toxicity of each oligomer relative to monomer (relative toxicity; Table S2). Quantitative relationships were found that were similar to those for nucleation activity. Trimer relative toxicity was ≈3 (3-fold higher than monomer), whereas trimer and tetramer exhibited values of ≈8 and ≈13, respectively. The unfractionated cross-linked oligomer population was ≈3-fold more toxic than monomer. This result is consistent with the relative nucleation activity of this population and is consistent with the fact that the number average oligomer order in the mixed population is ≈2 (Table S2) and the toxicity of this population was identical, within experimental error, to that of the pure dimer.
Discussion
The supplanting of the amyloid cascade hypothesis by what now may be called the “oligomer cascade hypothesis” has led to enormous efforts to identify and characterize intermediates in the pathways of Aβ assembly and determine their biological activities (5, 21–27). A pervasive and fundamental problem with these efforts is the relatively imprecise and sometimes nebulous determination of oligomer structure and dynamics (2, 28). The use of pure, well-defined, structurally stable, oligomer populations is a prerequisite for establishing rigorous structure–activity correlations leading to knowledge-based therapeutic drug design. Here, we have reported results of studies using pure (94.3–99.6%), low-order, oligomer populations of constant quaternary structure and restricted conformational complexity. Structural studies revealed that a consistent feature of Aβ assembly space was a correlation between oligomer structure and order. This correlation extended not only from an intuitively obvious relationship between size (determined by EM and AFM) and oligomer order, but to the level of secondary structure, fibril nucleation activity, and neurotoxicity. Interestingly, and potentially importantly, nonlinear correlations were observed (see below).
Nascent Aβ in vivo and in vitro is significantly unstructured (6). In vivo, this is a consequence of the necessity for peptide α-helix denaturation during β-secretase and γ-secretase function (29). In vitro, strongly denaturing conditions are used before experiments to eliminate preexistent aggregates and other amyloid-like assemblies and conformers (7). In contrast, protofibrils and fibrils are structures exhibiting high β-sheet content (≥50%) (30). Given this continuum in secondary structure from SC to extended β-sheet, we began the studies reported here by asking the question, where do specific oligomers lie within the conformational continuum?
We observed a direct relationship between oligomer and structural (secondary) order (Table 1 and Fig. S2). The relative magnitudes of conformational change between each oligomer are informative. The data suggest that the monomer → dimer transition involves substantial secondary structure change, significantly more than in the higher-order transitions. Our data do not reveal whether this change occurs in the monomer before its dimerization, within a nascent dimer, or both. It is interesting that fibril elongation also has been found to involve structural rearrangement in monomers and these rearrangements were suggested to occur in the incoming monomer or the monomers comprising the fibril ends themselves (31).
Examination of the quaternary structure data in Table 1 suggests, as did the results from CD analyses, that the monomer → dimer transition is qualitatively different from higher-order transitions. In both the EM and AFM experiments, the dimer → trimer → tetramer transitions involved increases in assembly size that significantly exceeded that in the monomer → dimer transition. This finding suggests that monomer addition to dimeric and higher-order assemblies is associated with tertiary conformational changes producing relatively extended conformers. For example, in the AFM experiments, each monomer added to a dimer increases assembly height by ≈0.5 nm. This relationship [h = 0.5(o −1)], where h is height and o is order) predicts h = 0 for monomer, which is impossible. Alternatively, if monomer height were constant (≈0.25 nm), then the maximum height increase per monomer addition would be 0.25 nm. This relationship (h = 0.25 o) holds for monomer and dimer, but not for higher-order oligomers, again suggesting oligomerization-linked conformational change. Taken together, the data show a nonlinear dependence of oligomer geometry on oligomer order.
A fundamental question in any nucleated assembly process is whether particular assemblies can act as nuclei. Many oligomeric assemblies are intermediates in fibril assembly (see Table S1 in ref. 6). What has not been clear is whether the smallest of these oligomers, in pure form, function as fibril “seeds,” i.e., can eliminate the lag phase in fibril elongation process. We found here that purified oligomers as small as dimers seed fibril formation. However, they are not as effective as trimers or tetramers, which have an effectiveness closer to that of fibrils than that of monomers.
An important aspect of the analysis of the nucleation data was the consideration of number average nucleation activity, i.e., what is the quotient of activity/number of oligomers? This is an important question in the Aβ system, because experiments usually are done, and analyzed, at a constant total Aβ concentration, without consideration for the number of moles of each assembly present. When the nucleation data were analyzed in this manner, the relative activity of dimers was an order of magnitude higher than that of monomers, and trimer and tetramer activity was two orders of magnitude higher.
Once static and dynamic features of the purified oligomers were determined, we next sought to correlate these features with biological activities. This structure–activity correlation is central to investigation of the pathobiology of oligomer-mediated neurotoxicity in vivo. We observed an order dependence of toxicity (Table S2) that correlated with the dependencies observed in the prior structural studies, namely tetramers ≥ trimers > dimers ≥ fibrils > monomers > uncross-linked Aβ. Analysis of the molar toxicity of each oligomer, determined similarly to the molar nucleation activity, showed that dimers are 3-fold more toxic than monomers and that the toxicity of trimers and tetramers are up to ≈13-fold more toxic.
Recently, Chimon et al. (32) reported results of structure–toxicity studies of nonfibrillar Aβ assemblies. These studies focused on a high-order (n = 150) spherical assembly that had substantial β-sheet content and a fibril-like β-strand organization, Iβ. They suggested that Iβ was an immediate precursor to fibrils and showed that its toxicity was significantly greater than monomers but significantly less than fibrils. Our results show that the lowest-order (n = 2–4) Aβ oligomers, which evidence suggests are the most clinically relevant, are less organized structurally than Iβ and likely are the link between the SC state of the monomer and that in Iβ and fibrils.
Insights and Implications
Our results provide significant insights into the biophysical and pathobiological behavior of Aβ, and importantly, into strategies for developing therapeutics for AD. The “specific activity” of Aβ assemblies depends nonlinearly on oligomer order. In fact, dimers were ≈3-fold more toxic than monomers, and tetramers were ≈13-fold more toxic. Are tetramers the “most toxic” assembly? We do not know. Determination of a complete toxicity/order correlation is not possible because of the progressive decrease in occurrence frequency of higher-order oligomers, which precludes their isolation and study.
A broader consideration of the pathobiology of AD suggests that determination of a toxicity/oligomer-order frequency distribution may not be necessary. Fibrils, the most toxic assemblies on a molar basis, accumulate in plaques, a process now thought to be beneficial by sequestering these toxic entities (2). Monomers have very low toxic activity. Only when Aβ self-associates does toxicity rise substantially. How likely to occur are these associations? Because the occurrence frequency has a negative exponential dependence on oligomer order, and because initial assembly can nucleate rapid Aβ polymerization, the steady-state levels of any oligomer must be relatively low. This logic suggests that a strategy targeting low-order oligomers, e.g., dimers through hexamers, is reasonable because these targets likely contribute most substantially to AD neurotoxicity (according to Eq. 1). The larger of these assemblies is significantly more toxic than the smaller, but the concentration of these highly toxic larger oligomers will be lower (assuming they do not exist in covalently associated form).
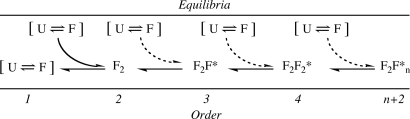
How should one target low-order oligomers? Two points in the oligomerization process may be particularly attractive (Fig. 5). The first point is the conformational organization of the monomer during its dimerization. Therapeutic agents could stabilize a disordered state of the monomer or destabilize the dimer state, resulting in complete blockage of oligomerization and fibril formation. The second point would be the stabilization of the less extended form of the Aβ monomer within the dimer or in incoming monomers (preventing trimerization and further assembly, both of which would produce significantly more toxic assemblies than dimers). A requirement in this and related strategies is that the strategy must not stabilize toxic oligomers, unless inhibitor binding blocks toxicity.
Fig. 5.
Aβ assembly. The data are consistent with an initial oligomerization process in which dimerization involves the self-association of two monomers, each of which exists in a folded (F) state in the dimer. It is not clear when folding from the unfolded (U) state occurs, e.g., before dimerization, contemporaneous with dimerization, or through conformational rearrangement within the dimer. Once the dimer forms, subsequent addition of a monomer to form the trimer involves accommodation (dotted arrow) of the incoming monomer into the dimer structure. The structure of this third monomer within trimers (F*) is different from that of free monomer or of monomers comprising a dimer because the size of the trimer is not thrice that of the monomer or 150% that of the dimer. Each monomer addition past the dimer stage produces the same size increase, and this increase is larger than that observed in dimerization; thus each of these stages would involve monomer accommodation, i.e., F*. It should be noted that every step of peptide oligomerization or fibril formation may not involve simple monomer addition. Other pathways are possible and likely occur (e.g., tetramer formation by dimer association). The scheme presented here illustrates one pathway that is both reasonable and consistent with the experimental data.
Materials and Methods
Complete discussions of chemicals and reagents, peptide preparation, Aβ aggregation, preparation of fibrils, PICUP, extraction of Aβ from SDS gels, determination of oligomer frequency distributions, ThT fluorescence, CD spectroscopy, EM, AFM, toxicity assays, and statistical analysis are in SI Text.
Supplementary Material
Acknowledgments.
We thank Drs. Erica Fradinger, Mohammed Inayathullah, Kazuma Murakami, Robin Roychaudhuri, and Mingfeng Yang, and Ghiam Yamin and Eric Pang for valuable advice and critical comments; Clark Rosensweig, Jason Reed, Adam Stieg, Tuan Le, Masami Saito, and Tsuyoshi Terayama for technical assistance; and Drs. Kazumasa Sakurai and Yuji Goto for assistance in the deconvolution and quantitative secondary structure determination of CD data. This work was supported by the Japan Human Science Foundation, a Pergolide Fellowship from Eli Lilly Japan, and the Mochida Memorial Foundation for Medical and Pharmaceutical Research (K.O.) and National Institutes of Health Grant AG027818, the Alzheimer's Association, and the Jim Easton Consortium for Alzheimer's Drug Discovery and Biomarkers at the University of California, Los Angeles (D.B.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
In this context, “order” refers to the number of monomers comprising a specific assembly.
Similar experiments done using Aβ42 revealed that some, but not all, of the oligomers formed were stable upon re-electrophoresis. For the purpose of structure–activity studies, the partial instability of the repurified Aβ42 oligomers precludes structure–activity studies. This is an important difference between Aβ40 and Aβ42. This difference requires further investigation to develop the means to eliminate the instability of the Aβ42 oligomers and to understand this fundamental difference in biochemical behavior, which likely correlates with known differences in pathophysiologic behavior.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905127106/DCSupplemental.
“Order,” in this context, refers to secondary and tertiary structure, not to “oligomer order,” the number of monomers in an assembly.
References
- 1.Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 2.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid β-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 3.Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders: The emerging role of oligomeric assemblies. J Neurosci Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 4.Walsh DM, Selkoe DJ. Aβ oligomers: A decade of discovery. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 5.Shankar GM, et al. Amyloid-β protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. Amyloid β-protein assembly and Alzheimer's disease. J Biol Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teplow DB. Preparation of amyloid β-protein for structural and functional studies. Methods Enzymol. 2006;413:20–33. doi: 10.1016/S0076-6879(06)13002-5. [DOI] [PubMed] [Google Scholar]
- 8.Bitan G, Lomakin A, Teplow DB. Amyloid β-protein oligomerization: Prenucleation interactions revealed by photo-induced cross-linking of unmodified proteins. J Biol Chem. 2001;276:35176–35184. doi: 10.1074/jbc.M102223200. [DOI] [PubMed] [Google Scholar]
- 9.Bitan G, et al. Amyloid β-protein (Aβ) assembly: Aβ40 and Aβ42 oligomerize through distinct pathways. Proc Natl Acad Sci USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitan G, Teplow DB. Rapid photochemical cross-linking: A new tool for studies of metastable, amyloidogenic protein assemblies. Acc Chem Res. 2004;37:357–364. doi: 10.1021/ar000214l. [DOI] [PubMed] [Google Scholar]
- 11.Yang M, Teplow DB. Amyloid β-protein monomer folding: Free-energy surfaces reveal alloform-specific differences. J Mol Biol. 2008;384:450–464. doi: 10.1016/j.jmb.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. On the nucleation and growth of amyloid amyloid β-protein fibrils: Detection of nuclei and quantitation of rate constants. Proc Natl Acad Sci USA. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lomakin A, Teplow DB, Kirschner DA, Benedek GB. Kinetic theory of fibrillogenesis of amyloid β-protein. Proc Natl Acad Sci USA. 1997;94:7942–7947. doi: 10.1073/pnas.94.15.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarrett JT, Lansbury PT., Jr Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer's disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 15.LeVine H., 3rd Thioflavine T interaction with synthetic Alzheimer's disease β-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeVine H., 3rd Quantification of β-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 17.Naiki H, Nakakuki K. First-order kinetic model of Alzheimer's β-amyloid fibril extension in vitro. Lab Invest. 1996;74:374–383. [PubMed] [Google Scholar]
- 18.Harper JD, Lansbury PT., Jr Models of amyloid seeding in Alzheimer's disease and scrapie: Mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 19.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 20.Abe K, Saito H. Amyloid β protein inhibits cellular MTT reduction not by suppression of mitochondrial succinate dehydrogenase but by acceleration of MTT formazan exocytosis in cultured rat cortical astrocytes. Neurosci Res. 1998;31:295–305. doi: 10.1016/s0168-0102(98)00055-8. [DOI] [PubMed] [Google Scholar]
- 21.Vigo-Pelfrey C, Lee D, Keim P, Lieberburg I, Schenk DB. Characterization of β-amyloid peptide from human cerebrospinal fluid. J Neurochem. 1993;61:1965–1968. doi: 10.1111/j.1471-4159.1993.tb09841.x. [DOI] [PubMed] [Google Scholar]
- 22.Roher AE, et al. Morphology and toxicity of Aβ-(1-42) dimer derived from neuritic and vascular amyloid deposits of Alzheimer's disease. J Biol Chem. 1996;271:20631–20635. doi: 10.1074/jbc.271.34.20631. [DOI] [PubMed] [Google Scholar]
- 23.Walsh DM, et al. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 24.Cleary JP, et al. Natural oligomers of the amyloid-β protein specifically disrupt cognitive function. Nat Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 25.Lesné S, et al. A specific amyloid-β protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 26.Townsend M, Shankar GM, Mehta T, Walsh DM, Selkoe DJ. Effects of secreted oligomers of amyloid β-protein on hippocampal synaptic plasticity: A potent role for trimers. J Physiol (London) 2006;572:477–492. doi: 10.1113/jphysiol.2005.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung LW, et al. Amyloid-β peptide (Aβ) neurotoxicity is modulated by the rate of peptide aggregation: Aβ dimers and trimers correlate with neurotoxicity. J Neurosci. 2008;28:11950–11958. doi: 10.1523/JNEUROSCI.3916-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glabe CG. Structural classification of toxic amyloid oligomers. J Biol Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato T, et al. A helix-to-coil transition at the ϵ-cut site in the transmembrane dimer of the amyloid precursor protein is required for proteolysis. Proc Natl Acad Sci USA. 2009;106:1421–1426. doi: 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh DM, et al. Amyloid β-protein fibrillogenesis: Structure and biological activity of protofibrillar intermediates. J Biol Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 31.Kusumoto Y, Lomakin A, Teplow DB, Benedek GB. Temperature dependence of amyloid β-protein fibrillization. Proc Natl Acad Sci USA. 1998;95:12277–12282. doi: 10.1073/pnas.95.21.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chimon S, et al. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer's β-amyloid. Nat Struct Mol Biol. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.