Malaria caused by Plasmodium falciparum (“malignant malaria”) is one of the most devastating pathogens of humans (1). Plasmodium reichenowi, which infects chimpanzees and gorillas, is the closest relative of P. falciparum (2). In early 20th-century experiments (never to be replicated), blood from P. reichenowi-infected chimpanzees was injected into humans, but failed to produce infections (3). Conversely, chimpanzees injected with P.falciparum-infected human blood suffered no in fection. Taken together, these data suggested that each parasite had coevolved with its host, but did not rule out chimpanzee to human transmission, or vice versa. In this issue of PNAS, Rich et al. (4) provide an answer to this malignant malaria “mystery” and confirm a prediction that we and our colleagues made earlier (5).
Several Anopheles mosquito species mediate the P. falciparum life cycle (6, 7). After an infected mosquito bite, the injected sporozoites infect host liver cells, generating merozoites that invade circulating erythrocytes (RBCs). Bouts of high fever follow when merozoites undergo cycles of asexual reproduction in RBCs. P. falciparum expresses multiple binding proteins that recognize specific targets on RBCs.* The major targets are terminal sialic acids (Sias) on O-linked glycan chains attached to glycophorins, the most abundant RBC surface glycoproteins. Although Sia-independent RBC invasion is known (8), most field isolates exhibit Sia dependency for RBC binding (9).
Sias are a family of sugars found at the outer termini of cell surface glycan chains of all cells of all vertebrates (10). Although Sias appear essential for development, they are also targets for many pathogens (10). Thus, the host “sialome” must constantly evolve to evade rapidly evolving pathogens. For example, an Alu-mediated exon deletion in the CMAH gene occurred ≈2 million to 3 million years ago in human ancestors, eventually eliminating biosynthesis of a common mammalian Sia called N-glycolylneuraminic acid (Neu5Gc) and causing accumulation of its precursor N-acetylneuraminic acid (Neu5Ac) (10). Thus, while chimpanzee and gorilla RBCs present a mixture of Neu5Gc and Neu5Ac, human RBCs are enriched in Neu5Ac. In keeping with this difference we found that the major merozoite RBC-binding protein EBA-175 of P. reichenowi and P. falciparum preferred to bind to Neu5Gc and Neu5Ac, respectively (5). Taken together with the apparently recent emergence of malignant P. falciparum (11), we (5) suggested that “… the falciparum/reichenowi common ancestor was more like P. reichenowi, preferring Neu5Gc over Neu5Ac… the loss of Neu5Gc in the human lineage might have provided our Homo ancestors with temporary relief from this form of malaria… we suggest that P. falciparum emerged later, through selective evolution of its EBA-175 toward preferentially recognizing the Neu5Ac-rich erythrocytes of humans.” While confirming this scenario, Rich et al. (4) point out that their results do not reveal the timing of the transfer. They suggest that it “must have happened much earlier… and it seems also likely that there would have been an intermediate stage, wherein EBA-175 of the P. reichenowi ancestor would have relaxed its specificity to accommodate binding of Neu5Ac. The final EBA-175 mutations potentially responsible for the malignancy and rapid expansion of P. falciparum may have occurred relatively recently.” And the fact that transfer apparently occurred only once “may reflect the difficulty in changing the Sia binding specificity of the plasmodial binding proteins” (4).
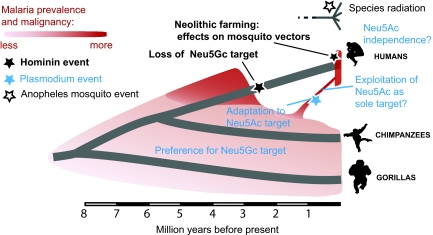
Lack of genetic diversity among P. falciparum strains (11) suggests an origin as recent as the Neolithic transition ≈10,000 years ago, when human farming practices augmented mosquito habitats near humans, with human settlement patterns favoring efficient transmission. In keeping with this notion, the main vector species complex Anopheles gambiae apparently evolved strong “anthropophily” and domesticity only since the Neolithic (7). We summarize all of these considerations in Fig. 1 and add some further speculations.
Fig. 1.
Proposed scenario for evolution of human and great ape malarias. The scenario accounts for most of the relevant facts and includes some speculation.
Until now, only one isolate of P. reichenowi had been sequenced. Recent sequences from two Plasmodium strains from pet chimpanzees in Gabon (12) indicated substantial divergence from the known P. reichenowi strain, suggesting a new species. Rich et al. (4) provide new data on chimpanzee malaria diversity, presenting DNA sequences from dried blood samples of dead wild chimpanzees or wild-born sanctuary ones. New sequence data from three distinct plasmodial genomes [mitochondrial (cytB), nuclear (18srRNA), and apicoplast (Csp)] from these eight new samples were analyzed together with corresponding sequences for 133 strains of P. falciparum, representing its worldwide populations. These data allowed the assessment of P. reichenowi diversity testing of the relative validity of different scenarios for the origin of hominid Plasmodia. Phylogenetic reconstructions were done for each of these three genomic fragments, along with data from the other three human malaria parasites (Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale), two rodent malaria strains, and one avian malaria strain. Although the amount of existing sequence data for each of these three genomic fragments differs and the three different Plasmodium genomes adhere to different models of molecular evolution, phylogenetic analyses strongly supported a monophyletic relationship of P. reichenowi and P. falciparum in each case. Overall, Rich et al. (4) conclude that all known P. falciparum strains arose from one of the many strains of P. reichenowi, i.e., a single chimpanzee to human transmission likely gave rise to P. falciparum.
Recent studies suggest the emergence of Sia-independent targets for P. falciparum (8), which is not surprising for a rapidly evolving pathogen spreading in epidemic proportions. Most such studies, however, are done in static ex vivo systems, involving long exposure times of merozoites to RBCs. This methodology can be misleading and does not rule out that Sias are still important for initial recognition phases in vivo. Regardless, this phenomenon may represent an ominous new phase in the ongoing “arms race” between this very successful pathogen and the human host.
Environmental factors may have been equally important. Chimpanzees sleep in tree nests in different locations daily, and adults sleep several meters apart. In contrast, humans have used permanent shelters and slept at higher densities since the Neolithic. Given the impact of P. falciparum on mosquito feeding, favoring blood meals from multiple hosts (6), these novel human behaviors would have amplified secondary infection rates among Neolithic humans and further favored greater virulence of the human-adapted P. falciparum. The young age of human genetic variants in genes such as glucose 6-phosphate dehydrogenase or hemoglobin, which convey partial resistance to malignant malaria, is a further indication of how recent the epidemic P. falciparum is in our species (13).
All known P. falciparum strains arose from one of the many strains of P. reichenowi.
More work is needed to dissect the scenario in Fig. 1. For example, the Sia-dependent binding proteins of P. falciparum and P. reichenowi other than EBA-175 are predicted to prefer Neu5Ac and Neu5Gc targets, respectively, and optimal recognition will likely require specific presentations on different glycophorins. Also, given the high frequency of positive PCR results (≈8%) in Rich et al. (4), further studies in extant African great ape populations are needed to assess the true distribution and virulence of P. reichenowi.
What are the practical implications? Considering how cowpox vaccination provides immunity against smallpox, is it possible that vaccination with P. reichenowi sporozoites or merozoites might give humans cross-immunity against P. falciparum? Indeed, might this already have occurred as a “natural experiment” in parts of Africa where both parasites and hosts coexist? Perhaps some humans living near ape habitats will have detectable antibodies specific for P. reichenowi, caused by abortive infections? If so, are such humans protected against P. falciparum?
Finally, Rich et al. (4) demonstrate the importance of noninvasive studies of wild and captive chimpanzee populations for understanding disease. There are currently >24 chimpanzee field research sites and 16 African sanctuaries housing >1,000 apes. These populations offer precious opportunities for studying uniquely human and chimpanzee diseases. Meanwhile, many of these sites are in dire need of financial and logistical support to which the biomedical research community could contribute. This is an example where all concerned stand to benefit from new knowledge. For example, surveillance of wild chimpanzee populations by veterinarians such as the Cote d'Ivoire site, where the World Health Organization is monitoring Ebola virus has resulted in precious information about the health of wild chimpanzees and the threats they face from anthroponotic pathogens such as measles and respiratory diseases (14).
Even today, ethically acceptable studies in human volunteers are commonplace, including recent studies of malaria vaccines (1). In contrast, the conflict between “ape advocates” and researchers used to treating chimpanzees as laboratory animals (15) has gone to one extreme, where all research on great apes has been banned in most parts of the world and possibly soon in the United States as well. Considering the crucial need for optimal surveillance and care of both wild and captive chimpanzees, would it not be better to establish ethically acceptable standards for biomedical care and research in these precious populations? Indeed, would it be ethical or rational to ban all future research on humans?
Footnotes
The authors declare no conflict of interest.
See companion article on page 14902.
We avoid the terms “receptor” and “ligand,” because of confusion about their meanings. We instead use “target” for the entity on the host cell that is recognized and “binding protein” for the pathogen protein that mediates target recognition.
References
- 1.Roestenberg M, et al. Protection against a malaria challenge by sporozoite inoculation. N Engl J Med. 2009;361:468–477. doi: 10.1056/NEJMoa0805832. [DOI] [PubMed] [Google Scholar]
- 2.Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodhain J. Les plasmodiums des anthropoides de I'Afrique centrale et leurs relations avec les plasmodiums humains. Ann Soc Belge Med Trop. 1939;19:563–572. [Google Scholar]
- 4.Rich S, et al. The origin of malignant malaria. Proc Natl Acad Sci USA. 2009;106:14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A. Evolution of human–chimpanzee differences in malaria susceptibility: Relationship to human genetic loss of N-glycolylneuraminic acid. Proc Natl Acad Sci USA. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koella JC, Sorensen FL, Anderson RA. The malaria parasite, Plasmodium falciparum, increases the frequency of multiple feeding of its mosquito vector, Anopheles gambiae. Proc Biol Sci. 1998;265:763–768. doi: 10.1098/rspb.1998.0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.della Torre A, et al. Speciation within Anopheles gambiae: The glass is half full. Science. 2002;298:115–117. doi: 10.1126/science.1078170. [DOI] [PubMed] [Google Scholar]
- 8.Gaur D, et al. Recombinant Plasmodium falciparum reticulocyte homology protein 4 binds to erythrocytes and blocks invasion. Proc Natl Acad Sci USA. 2007;104:17789–17794. doi: 10.1073/pnas.0708772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baum J, Pinder M, Conway DJ. Erythrocyte invasion phenotypes of Plasmodium falciparum in the Gambia. Infect Immun. 2003;71:1856–1863. doi: 10.1128/IAI.71.4.1856-1863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 11.Rich SM, Ayala FJ. Population structure and recent evolution of Plasmodium falciparum. Proc Natl Acad Sci USA. 2000;97:6994–7001. doi: 10.1073/pnas.97.13.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ollomo B, et al. A new malaria agent in African hominids. PLoS Pathog. 2009;5:e1000446. doi: 10.1371/journal.ppat.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tishkoff SA, et al. Haplotype diversity and linkage disequilibrium at human G6PD: Recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 14.Kondgen S, et al. Pandemic human viruses cause decline of endangered great apes. Curr Biol. 2008;18:260–264. doi: 10.1016/j.cub.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Gagneux P, Moore JJ, Varki A. The ethics of research on great apes. Nature. 2005;437:27–29. doi: 10.1038/437027a. [DOI] [PubMed] [Google Scholar]