Abstract
Disruptions in local chromatin structure often indicate features of biological interest such as regulatory regions. We find that sonication of cross-linked chromatin, when combined with a size-selection step and massively parallel short-read sequencing, can be used as a method (Sono-Seq) to map locations of high chromatin accessibility in promoter regions. Sono-Seq sites frequently correspond to actively transcribed promoter regions, as evidenced by their co-association with RNA Polymerase II ChIP regions, transcription start sites, histone H3 lysine 4 trimethylation (H3K4me3) marks, and CpG islands; signals over other sites, such as those bound by the CTCF insulator, are also observed. The pattern of breakage by Sono-Seq overlaps with, but is distinct from, that observed for FAIRE and DNase I hypersensitive sites. Our results demonstrate that Sono-Seq can be a useful and simple method by which to map many local alterations in chromatin structure. Furthermore, our results provide insights into the mapping of binding sites by using ChIP–Seq experiments and the value of reference samples that should be used in such experiments.
Keywords: ChIP-Seq, ENCODE, formaldehyde cross–linking, sonication, DNA sequencing
The accessibility of regulatory elements in chromatin is critical for many aspects of gene regulation. Nucleosomes positioned over regulatory elements inhibit access of transcription factors to DNA; deprotection of the DNA arises from local changes in chromatin conformation. Previous methods for mapping chromatin accessibility include mapping DNase I hypersensitivity sites or formaldehyde-assisted isolation of regulatory elements (FAIRE) regions and analyzing the DNA using microarrays or DNA sequencing (1–3). These methods have mapped many open chromatin sites to promoters of actively transcribed genes as well as to enhancers.
The in vivo mapping of regulatory elements is often performed by chromatin immunoprecipitating of a factor of interest followed by analyzing the associated DNA (4–6). Chromatin complexes are preserved through cell fixation with formaldehyde, the chromatin is fragmented, and protein-bound DNA regions are isolated by using antibodies to a specific DNA-associated protein. DNA fragments are purified and used to probe DNA microarrays (ChIP–chip) or, more recently, identified by high-throughput DNA sequencing (ChIP–Seq), thereby locating transcription factor binding sites (TFBSs) on a genome-wide scale (4–7). In ChIP experiments, significant targets representing binding regions are found by analyzing signal levels produced by an experimental sample relative to a reference sample. Although several automated scoring algorithms exist for ChIP–Seq data (6, 8–11), an appreciation of the characteristics and biases inherent to different reference DNA samples and preparation methods is important for understanding the significance of the results obtained.
In the work presented here, we examine the signal distributions of commonly used reference samples including sonicated chromatin and investigate the aggregate signals relative to annotated regions (Table 1). We show that even without immunoprecipitation, cross-linked chromatin fragments can be size-selected for regions prone to physical breakage, many of which are proximal to promoters. We investigate the causes of these signals and develop this observation as a method for mapping these local alterations in chromatin structure at high resolution and on a genome-wide scale.
Table 1.
Data sources
| Library | Size selection | Cell conditions | Sonication conditions | Uniquely mapped reads | Biological replicates |
|---|---|---|---|---|---|
| RNA Pol II (HeLa S3) | 100–350 bp | formaldehyde-cross-linked | 7 × 30 sec | 29,060,928 | 3 |
| Sono-Seq (HeLa S3) | 100–350 bp | formaldehyde-cross-linked | 7 × 30 sec | 29,840,987 | 3 |
| Sono-Seq (HeLa S3) | 350–800 bp | formaldehyde-cross-linked | 7 × 30 sec | 19,729,371 | 3 |
| Naked DNA | 100–350 bp | not cross-linked | 1 × 30 sec | 34,550,812 | 3 |
| Normal IgG (mouse, HeLa S3) | 100–350 bp | formaldehyde-cross-linked | 7 × 30 sec | 28,960,961 | 2 |
| MNase (HeLa S3) | 100–200 bp | not cross-linked | not sonicated | 20,924,734 | 2 |
Results
Sonicated Chromatin Fragments Reveal Peaks over Promoter Regions.
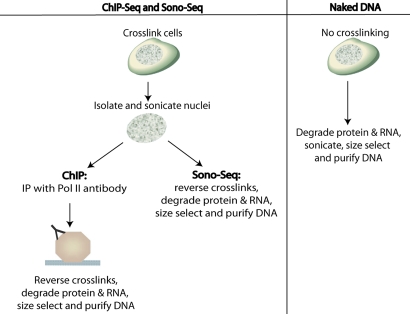
While examining the signal tracks of reference DNA samples for ChIP-Seq, we observed the presence of “peak” regions that appeared to have greater signal relative to the genome as a whole (Figs. S1 and S2). Sonicated chromatin was prepared from nuclear lysates of formaldehyde-cross-linked HeLa S3 cells. The sonicated chromatin was then either subjected to ChIP with an antibody specific to RNA polymerase II (ChIP DNA) or purified without immunoprecipitation (“Input” or “Sono-Seq” DNA) (Fig. 1). Both preparations of DNA were size-selected for 100–350-bp fragments and converted to libraries for sequencing on the Illumina Genome Analyzer II platform. A total of 29.0 M uniquely mapped reads were obtained for RNA polymerase II and 29.8 M reads for Sono-Seq DNA.
Fig. 1.
Steps used to prepare ChIP DNA, Sono-Seq DNA, and naked DNA.
The peaks in sonicated chromatin are often similar, albeit often of lower magnitude, than those obtained from the Pol II ChIP–Seq experiment (Fig. 2). For HeLa S3 cells, 106,958 Sono-Seq DNA peaks are observed as compared with 49,377 peaks for Pol II ChIP-Seq with a total coverage for the peaks of 27.7 and 36.7 Mb, respectively. Filtering for strong targets (see Materials and Methods) reduced the dataset to 27,773 peaks in Pol II and 21,762 peaks in Sono-Seq DNA. Using these strong targets, 65.0% of Sono-Seq regions are within 1 kb of a Pol II region and 49.4% of Sono-Seq regions are within 2.5 kb of a 5′ end of an Ensembl gene. To further investigate Sono-Seq characteristics, we examined its aggregated signal over the proximal promoter regions of expressed and nonexpressed Ensembl genes (12). In general, Sono-Seq DNA displays elevated signal at the 5′ ends of Ensembl genes compared with the background (see Materials and Methods; Fig. 3 A–C), although other peaks are also observed.
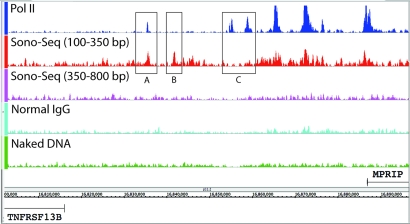
Fig. 2.
Signal map showing Pol II ChIP DNA, Sono-Seq DNA (small and large fragment sizes), normal IgG, and naked DNA. All signals are in HeLa S3 cells. Signal levels between positions 16,802,000 and 16,896,000 of chromosome 17 are shown. Tracks are scaled based on the number of uniquely mapped reads obtained for each sample type. Both TNFRSF13B and MPRIP are not expressed in HeLa S3 based on RNA-Seq data (14). Several regions of disagreement between Sono-Seq and Pol II signal are shown, such as a large Sono-Seq peak with a less pronounced Pol II peak (A), the absence of a Pol II peak and the presence of a Sono-Seq peak (B), and Pol II peaks without corresponding Sono-Seq peaks (C).
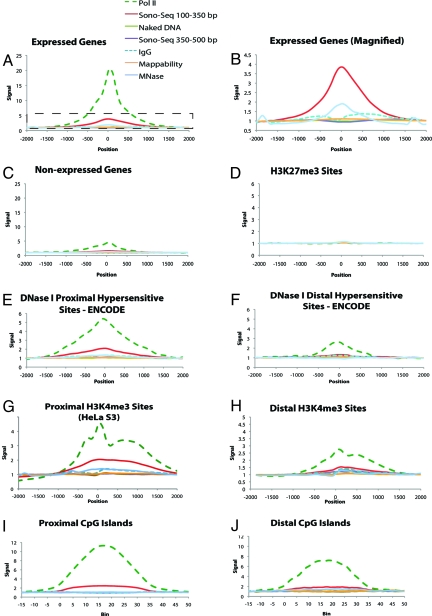
Fig. 3.
Aggregation plots depicting the average ChIP signal across a variety of genomic features. Vertical axis units are consistent between all plots. Horizontal axis units are given in nucleotides from the feature start site in plots A–H and in bins each representing 1/35th of the feature size in plots I and J. Frame B is a magnified view of the regions enclosed by the dotted box in frame A in which Pol II is removed and scales are altered to allow for better comparison between reference sample types. In all figures, position/bin 0 corresponds to the start of the target feature. For plots E–J, proximal is defined as lying within ±2.5 Kb of an Ensembl gene. Values are given in “fold enrichment” compared against a background signal (see Materials and Methods). A value of 1.0 indicates signal equal to background. The signal was calculated in Pol II, two different size selections of Sono-Seq DNA, naked DNA, normal IgG, and MNase-digested DNA. Mappability is a measure of how well reads are mapped to the features being compared (see Materials and Methods). A mappability of 1.0 indicates a mappability equal to that of background.
Sono-Seq DNA-enriched regions heavily overlap with those of Pol II (Fig. S3); however, approximately one-third of strong peaks do not co-occur with Pol II. Locations of Sono-Seq DNA peaks were intersected against Pol II peaks, and we found 6,892 peaks where no corresponding Pol II peaks were identified within ±1 kb. We found that some of these non-Pol II-associated Sono-Seq peaks (≈28%) correspond to either HeLa-derived CTCF sites, small RNAs, or enhancer regions predicted in HeLa (13), as shown in SI Text and Figs. S4 and S5. These results demonstrate that Sono-Seq peaks intersect a wide variety of biologically relevant regions.
Sono-Seq Requires Cross-Linked Chromatin.
The signals from Sono-Seq DNA could either be due to regions of preferential breakage intrinsic to the DNA sequence or breaks that occur in regions made accessible by biological activity. To further investigate the source of the Sono-Seq DNA signal, we prepared HeLa S3 genomic DNA from non-cross-linked, deproteinized cells and sonicated the DNA into fragments of 100–350 bp on average to produce “naked DNA” (Fig. 1). Naked DNA did not show visible peaks either in signal tracks or over promoter regions, and examination of its aggregated signal near transcription start sites did not reveal any enrichment at these regions (Fig. 2). These results indicate that Sono-Seq peaks require cross-linked chromatin, presumably because cross-linking preserves the in vivo state of DNA.
As an additional control, we prepared DNA according to the exact protocol as ChIP DNA, but substituted affinity-purified IgG from a nonimmunized animal for the antibody that recognizes Pol II. We call this dataset “normal IgG”. Interestingly, examination of the aggregated peak signals indicates that the normal IgG signal is near the baseline (0.9-fold) over transcriptional start sites (TSSs) (Fig. 3 A–C), lower than that of Sono-Seq DNA. We also prepared DNA in which chromatin from non-cross-linked cells was treated with micrococcal nuclease (MNase). MNase-treated DNA exhibits elevated signals over promoters analogous to Pol II and Sono-Seq DNA signals (Fig. 3 A and B).
Sono-Seq DNA Peaks Reside over Expressed Promoters.
By using HeLa S3 expression data determined from an RNA-Seq experiment (14), we examined Sono-Seq and Pol II ChIP aggregate signals over genes that are expressed in HeLa S3 cells as well as those that are not expressed. We define an expressed gene as having an average coverage of at least 1-fold across each nucleotide in a gene. The remaining genes were classified as nonexpressed. A total of 10,993 genes are expressed, and 19,273 genes are nonexpressed when using these criteria. Aggregated signals from Sono-Seq DNA are enriched 4-fold over expressed genes relative to nonexpressed genes (Fig. 3 A and B). MNase-treated DNA also showed increased signal levels over 5′ ends of expressed genes (2-fold vs. background), indicating that open chromatin is present in these regions. A total of 31.8% of all Ensembl genes and 67.9% of all expressed Ensembl genes in HeLa S3 possess a significant peak in Sono-Seq DNA proximal to the 5′ ends.
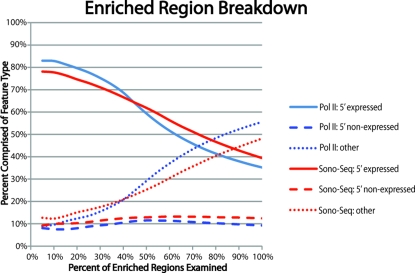
To ascertain relationships between peak significance and gene expression, we created rank-order lists for Pol II and Sono-Seq DNA peaks by sorting peaks, first by tag count and then by fold-enrichment over the corresponding signal in naked DNA. We then calculated the percentage of peaks occurring in promoters of expressed and nonexpressed genes as well as those occurring distal to promoter regions. The top of the list (i.e., the most significant peaks) contains a large percentage (90%) of peaks found in 5′ ends of expressed genes (Fig. 4). Toward the bottom of the rank-order list, the percentage of enriched regions found proximal to 5′ ends decreases whereas the percentage of enriched regions found distal to 5′ ends of genes increases. The percentage of enriched reads proximal to 5′ ends of nonexpressed genes, remains consistent throughout the data set (≈10%).
Fig. 4.
A rank-order plot depicting the percentage of Sono-Seq- and Pol II-enriched regions located proximal and distal to genes (see Materials and Methods). Regions most highly enriched by Sono-Seq typically lie proximal to the TSSs of expressed genes. Enrichment over promoter regions of nonexpressed genes remains constant, whereas enriched regions lying distal to known promoter regions are ranked lower (i.e., have lower scores).
Sono-Seq DNA Signals Are Enriched over Other Markers Associated with Gene Expression.
To further explore Sono-Seq signals, we compared the Sono-Seq peaks to several other chromosomal features, including DNase I hypersensitive sites, H3K4me3 sites, and CpG islands (Fig. 3 E–J). By using the ENCODE region data of Crawford et al. (2), we selected 2,060 DNase I hypersensitive sites from HeLa cells with a q-value <0.05. Of these, 958 were proximal (within 2.5 kb) and 1,103 were distal (>2.5 kb) to the TSSs of Ensembl genes. Aggregation of signals over proximal DNase I hypersensitive sites reveals Pol II signals increase more than 5-fold over these regions. Sono-Seq signal is also elevated (2-fold) over proximal DNase I hypersensitive sites. The signal is not enriched in naked DNA.
Mapping Sono-Seq DNA relative to distal DNase I hypersensitive sites reveals a different pattern. The Pol II signal is modestly elevated over these distal regions (2.5-fold). Small-fragment Sono-Seq DNA, normal IgG, and naked DNA all show minimal signal elevation over distal DNase I hypersensitive sites. Thus, higher Sono-Seq DNA signals are preferentially located over proximal DNase I hypersensitive sites as compared with distal ones.
To further investigate the Sono-Seq signal at promoters with proximal DNase I hypersensitive sites, we examined the association of Sono-Seq peaks with H3K4me3 sites, which are also correlated to gene expression level and promoter localization (15). For these analyses, we aggregated Sono-Seq signals over two different genome-wide H3K4me3 ChIP-Seq datasets, one containing a total of 54,467 H3K4me3 sites from HeLa cells (16) and another containing a random sample of 100,000 H3K4me3 sites from CD4+ cells (17). Aggregation of signals over either source of the H3K4me3 sites revealed that the Pol II and Sono-Seq DNA signals are significantly elevated at H3K4me3 sites (Fig. 3 G and H and Fig. S6). For the H3K4me3 sites identified in HeLa S3 cells, separate aggregations were performed for sites located distal and proximal to Ensembl genes. For proximal H3K4me3 sites, the Pol II signal is elevated 4.5-fold and the Sono-Seq DNA signal is elevated 2-fold. Normal IgG produced a lower enrichment signal (1.3-fold), whereas the signal was not elevated in naked DNA. Enrichment over distal H3K4me3 sites identified in HeLa S3 cells drops to 3-fold and 1.5-fold for Pol II and Sono-Seq DNA, respectively. Other reference samples exhibit signals comparable with those observed over proximal H3K4me3 sites.
Finally, we also analyzed Sono-Seq signals relative to CpG islands, which are associated with promoter regions (18). For this analysis, we used a coordinate list of unique CpG islands represented on the Illumina Infinium HumanMethylation27 BeadChip Assay (Illumina). The CpG islands on this array have a mean size of 1,388 bp and query 11,471 unique CpG islands. Of these regions, 7,101 sites lie within ±2.5 kb of TSSs of expressed Ensembl genes, whereas 4,029 lie within ±2.5 kb of TSSs of nonexpressed Ensembl genes. We observe signal enrichment over these CpG islands in Pol II, Sono-Seq (150–350 bp), and MNase-digested DNA. Other reference DNA types remain unenriched over these CpG islands (Fig. 3 I and J).
Sono-Seq DNA Signals Show Little Increase over H3K27me3 Sites.
H3K27me3 histone-modification sites represent a signature of closed-conformation, facultative heterochromatin and are established by Polycomb group proteins (17, 19). We compared Sono-Seq DNA signals with other signals over 100,000 H3K27me3 sites identified in CD4+ cells (17). The ChIP-Seq signals for all sample DNA types, including Pol II, remain flat over H3K27me3 sites (Fig. 3D). We also examined the sequences of these regions to determine if the observed lack of a ChIP–Seq signal was real or an artifact arising from an inability to map reads to these locations. As shown in Fig. 3D, the sequences in H3K27 trimethylation regions and other genome regions can be mapped equally well (i.e., the mappability line in the plots remains close to 1.0 at all times, representing complete mappability) and indicate that Sono-Seq signal is depleted over H3K27me3 sites. Thus, sonicated chromatin peaks preferentially lie near sites of active chromatin and are absent in closed chromatin regions.
Sono-Seq Signal Is Depressed over FAIRE Regions.
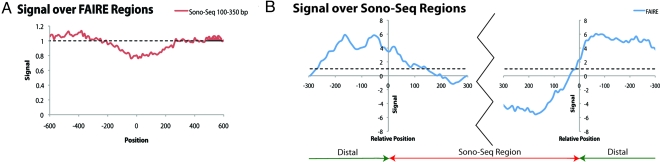
We next determined whether Sono-Seq regions coincide with FAIRE regions because both protocols rely upon sonication of cross-linked chromatin. We performed this comparison in S. cerevisiae, in which FAIRE was first described (1). Using data from Hogan et al. (20) that was generated from S. cerevisiae chromosome 3, we aggregated the Sono-Seq signal over FAIRE sites and found that Sono-Seq signal is depressed (Fig. 5A). When aggregating the FAIRE signal over Sono-Seq sites, we observe highly enriched FAIRE signal levels bordering Sono-Seq regions but depressed signal levels over the Sono-Seq regions themselves (Fig. 5B and Fig. S7). These findings indicate that Sono-Seq is different from FAIRE.
Fig. 5.
Sono-Seq differs from FAIRE. (A) Aggregation of signal from yeast Sono-Seq DNA (selected at 100–350 bp) over regions enriched in yeast FAIRE. The Sono-Seq signal is depressed over regions enriched by FAIRE. (B) Aggregation of FAIRE DNA signals over regions enriched in Sono-Seq. The FAIRE signal appears to be enriched in regions flanking Sono-Seq sites. All data shown in this figure originate from S. cerevisiae chromosome 3. In all figures, position 0 corresponds to the start of the target feature and the signal is given in fold-enrichment, as compared with background. Yeast FAIRE data are from Hogan et al. (20).
Hogan et al. (20) also found that FAIRE sites are anticorrelated with MNase-digested DNA signal. Our aggregation plots in HeLa S3 cells show that Sono-Seq and MNase-digested DNA signals exhibit trends similar to each other, further supporting that Sono-Seq and FAIRE experiments produce different results.
Sono-Seq DNA Peaks Are Affected by Fragment Size.
To further investigate the origin of the Sono-Seq DNA signals, we analyzed different fragment sizes. Instead of using only the small 100–350 bp size sample normally recommended for Illumina sequencing, we also analyzed a larger size fraction (350–800 bp) that was prepared from the same sonicated extracts as the 100–350 bp fragments. As shown in Fig. 3, the size of the fragments determines the presence and magnitude of the sonicated chromatin signals. The smallest fragments (100–350 bp) exhibit the largest signals, whereas the largest fragments (350–800 bp) give smaller signals. Greater signals were also observed when qPCR was performed by using electrophoretically separated small (100–500 bp), rather than large (1,000–6,000 bp), DNA fragments as a template (SI Text and Fig. S8). Thus, size selection is a critical step in the preparation of Sono-Seq DNA and the characterization of the signal obtained.
Discussion
We demonstrate that sonication of cross-linked chromatin causes breaks in localized and specific regions of the genome. By comparing aggregated Sono-Seq signals with TSSs, Pol II-bound regions, histone H3K4me3 sites, CpG islands and promoter-proximal DNase I hypersensitive sites, we show that many of these peaks are located within promoter regions. Further analysis reveals that higher Sono-Seq signals are observed over the promoters of expressed genes as compared to those with little or no expression. These results suggest that the breaks preferentially occur near regions of open chromatin of expressed genes; presumably these promoters have undergone chromatin remodeling, permitting access to transcriptional machinery but also allowing breakage by sonication of deprotected DNA. In addition, we show that Sono-Seq peaks are also found in intergenic regions where these signals are modestly enriched over features commonly associated with promoter activity of expressed genes such as CpG islands and H3K4me3 sites. Many distal regions enriched for Sono-Seq are enriched for Pol II (21.7%). We speculate that some of these distal peaks may lie proximal to genes that have not yet been annotated. Although many Sono-Seq peaks overlap with Pol II-bound loci, a number of Sono-Seq peaks do not and may represent binding regions of various other factors (SI Text and Fig. S9). Lastly we find that Sono-Seq peaks are preferentially depleted over regions of closed chromatin such as H3K27me3 sites.
The advent of ChIP–Seq permits mapping of DNA regulatory regions at high resolution and low cost and is rapidly replacing ChIP–chip for the mapping of transcription factor binding sites. However, there are important differences. For ChIP–chip, immunoprecipitated fragments are labeled along their entire length and hybridized to a microarray in a mixture containing a differentially labeled reference DNA, such that ratios of ChIP–DNA to reference DNA are typically recorded. In ChIP–Seq, breaks are generated in protein-bound DNA, short fragments are isolated and the ends sequenced. The combined effect of isolation of short fragments and high-resolution analysis of short fragment ends in ChIP–Seq reveals features of both the chromatin and the ChIP samples that have not been previously observed when ratios of ChIP to reference samples are analyzed. The implications of this are several-fold. First, reference DNA samples may exhibit increased signals over 5′ ends of genes and regions of open chromatin (Fig. 3 A–C). Second, we found that Sono-Seq DNA and normal IgG DNA are not equivalent; perhaps short fragments do not co-purify well with IgG beads. Regardless, we expect that normal IgG DNA may be more useful as a reference sample because its treatment closely parallels that of a ChIP DNA sample and signal levels will not be dampened as much over open regions, as would be the case when Sono-Seq DNA is used for scoring.
Fractionation of chromatin is nonrandom and may have an underlying biological basis depending on the method by which it is prepared. Studies similar to ours have also demonstrated that chromatin fragments may be associated with annotated regions. FAIRE has been shown to isolate regions correlated with nucleosome depletion, increased DNase I hypersensitivity, transcriptional start sites, and active promoters (1). Sono-Seq sites are different from FAIRE sites even though the Sono-Seq and FAIRE protocols share several common steps. Both protocols necessitate formaldehyde-cross-linking of proteins to DNA and sonication of the cross-linked DNA. The key difference between FAIRE and Sono-Seq is that in FAIRE phenol-chloroform extraction occurs before reverse-cross-linking, such that protein-protected DNA is trapped at the interface and the open regions of DNA are released into the aqueous phase. However, in Sono-Seq the protein-DNA cross-links are reversed before phenol-chloroform extraction, such that any protein-cross-linked DNA would be retained during purification and subsequently analyzed. Although both Sono-Seq and FAIRE are associated with active promoters, the aggregate Sono-Seq signal is depressed over FAIRE regions (Fig. 5A). Furthermore, the Sono-Seq signal parallels the MNase-digested DNA signal over promoter regions (Fig. 3 and figure 2C of ref. 20), the FAIRE signal is depressed over Sono-Seq regions, and Sono-Seq regions are bounded by the high FAIRE signal (Fig. 5B), all further differentiating Sono-Seq from FAIRE.
We speculate that Sono-Seq enriches regions that are both open and protein-bound, and that it detects breaks from neighboring open chromatin sites. For Sono-Seq regions to be recovered, they must have sensitivity to sonication, which may arise from regions that are undergoing chromatin remodeling or local denaturation (e.g., by Pol II), most likely in preparation for or during transcription. Interestingly, enrichment over distal DNase I hypersensitive sites was not observed; these regions may be smaller and not readily broken during sonication, or they may reside in areas where chromatin organization is relatively static. Regardless of the mechanism, our analyses illustrate the utility of Sono-Seq as an effective approach for detecting accessible chromatin regions by using less than 1/200th the DNA of a typical ChIP experiment, thus facilitating genome annotation and complementing existing experiments. In addition, we propose that Sono-Seq will be useful for detecting functionally important regions in new genomes that either are not well annotated, lack relevant reagents, or lack sufficient amounts of DNA such that ChIP experiments may not yet be practical. Another equally important implication of this work is that the choice of reference DNA type will directly influence the number of sites deemed significant when scoring ChIP–Seq data.
Materials and Methods
Preparation of DNA for ChIP–Seq and Sono-Seq.
Cell-growth protocols are available in the SI Text. For RNA Pol II ChIP–Seq, normal IgG ChIP–Seq and Sono-Seq, fixed HeLa S3 cells were washed in cold Dulbecco's PBS (Invitrogen) and swelled on ice in a 10-mL hypotonic lysis buffer [20 mM Hepes (pH 7.9), 10 mM KCl, 1 mM EDTA (pH 8.0), 10% glycerol, 1 mM DTT, 0.5 mM PMSF, and protease inhibitors]. Cell lysates were homogenized with 30 strokes in a Dounce homogenizer. Nuclear pellets were collected and lysed in 10 mL of RIPA buffer per 3 × 108 cells [RIPA buffer: 10 mM Tris-Cl (pH 8.0), 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholic acid, 0.5 mM PMSF, 1 mM DTT, and protease inhibitors]. Chromatin was sheared with an analog Branson 250 Sonifier (power setting 2, 100% duty cycle for 7 × 30-s intervals) to an average size of less than 500 bp, as verified on a 2% agarose gel. Lysates were then clarified by centrifugation at 20,000 × g for 15 min at 4 °C. For Sono-Seq, aliquots of clarified lysate were reserved for reversal of cross-linking, followed by RNase and proteinase K treatments. Sono-Seq DNA was further purified by phenol-chloroform extraction and ethanol precipitation. For RNA Pol II and normal mouse IgG ChIP samples, 12 μg of either the mouse monoclonal 8WG16 antibody (Covance MMS-126R) or normal mouse IgG (Santa-Cruz sc-2025) were added to 1 × 108 cells. ChIPs were conducted as previously described (21, 22). Libraries were constructed in a manner consistent with those from Rozowsky et al. (8). See SI Text.
Preparation of Naked DNA for Sequencing.
HeLa S3 cells were collected by centrifugation, resuspended in digestion buffer [100 mM NaCl, 10 mM Tris-HCl (pH 8.0), 25 mM EDTA (pH 8.0) and 0.5% SDS] and digested overnight at 50 °C with 0.1 mg/mL proteinase K (Ambion). The digest was extracted twice with phenol-chloroform, once with chloroform and ethanol precipitated. The DNA was recovered, treated with RNase (Qiagen) for 3 h at 37 °C, extracted once with phenol-chloroform, once with chloroform, ethanol precipitated and resuspended at 2.5 × 108 cell equivalents in 5 mL of 1X Tris-EDTA (TE) pH 7.5 (10 mM Tris, 1 mM EDTA). A 2.5-mL aliquot was sonicated once for 30 s with a Branson 250 Sonifier (power setting 2, 100% duty cycle) to an average size of less than 500 bp as verified on a 2% agarose gel.
Preparation of MNase-Treated DNA for Sequencing.
HeLa S3 cells were resuspended in MNase buffer [10 mM Tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 4% Nonidet P-40, and 1 mM DTT] and treated with 50 units of micrococcal nuclease (USB) at 37 °C for 1 h. The samples were treated with proteinase K for 2 h at 37 °C, extracted twice with phenol-chloroform, ethanol precipitated, treated with RNase A (Qiagen) and centrifuged through G50 sephadex spin columns. Each sample was treated with 30 units of calf intestinal alkaline phosphatase (NEB) for 2 h at 37 °C. After a second ethanol precipitation, the samples were treated with 30 units of T4 polynucleotide kinase (NEB).
Preparation of Yeast Sono-Seq DNA.
Saccharomyces cerevisiae strains CMY288–1B (BY background) and YJM339 (clinical isolate) were grown in 500 mL of YPAD to mid-log phase (OD600 = 1.0). Cells were fixed with 1% formaldehyde for 15 min, after which glycine was added to a final concentration of 125 mM. Cells were lysed with five 1-min bursts at 6.0m/s on a FastPrep-24 (MP Biomedicals). Chromatin was sonicated with a Branson 250 sonifier (Amplitude 50% for 5 × 30-s intervals) to an average size of 450–500 bp. For each biological replicate, 250 μL of clarified lysate were processed to reverse cross-links overnight, followed by a proteinase K treatment. The DNA was extracted three times in phenol:chloroform:isoamyl alcohol (25:24:1), and once in chloroform. After ethanol precipitation, DNA was resuspended in 1X TE (pH 8.0), RNase-treated and purified by using a Qiagen MinElute PCR purification column. Finally, 100–350 bp Sono-Seq DNA was size-selected by using a 2% agarose gel before Illumina library preparation. Sequencing libraries were generated as described above. Buffers are described in Aparicio et al. (23).
Computational Analysis of Illumina GA II Data.
Sequencing reads were analyzed by using Illumina's Genome Analysis Pipeline version 0.3. Reads were aligned to human genome build 18 by using the Eland aligner, and unique reads were used for ChIP–Seq scoring with PeakSeq (8). Signal maps and aggregation plots were generated as described in the SI Text and Table S1.
Data are available in NCBI's Gene Expression Omnibus (24) through accession numbers GSE12781 (Pol II and Sono-Seq) and GSE14022 (Naked DNA, DNA treated with MNase, large-fragment Sono-Seq, and normal IgG). Signal files and other data can be accessed at http://archive.gersteinlab.org/proj/Sono-Seq.
Creation of ChIP–Seq Mappability Aggregations.
A mappability profile for 30-nt reads was created and aggregations performed by using the same strategies presented in Rozowsky et al. (8). A mappability fraction of 1.0 for a given position means that a 30-nt read beginning at that position is fully mappable. Having all low ChIP signals from regions with high mappability indicates a true lack of reads from these regions.
Creation of FAIRE Signal Files and Enriched Regions.
Block normalized log2 normalized FAIRE data from yeast chromosome 3 tiling arrays from Hogan et al. were downloaded from the GEO (accession number GSE4721) (20, 24). Values for each probe were averaged across the four microarray experiments to produce a composite dataset and the probe IDs converted to genomic positions. These positions along with the corresponding average score were used to create a FAIRE signal file by averaging the values from overlapping tiles at each nucleotide position. Regions with a composite average score of at least 0.6 were deemed enriched and used to create a list of discrete FAIRE sites.
Scoring and Aggregating Sono-Seq DNA in Yeast.
A Sono-Seq DNA dataset for yeast was created by pooling data from two replicates and then scored against a randomized background by using PeakSeq (8). An aggregation of the FAIRE signal over Sono-Seq sites was then created by using the method described in Creation of FAIRE Signal Files and Enriched Regions. A bin size of 10 bp was used for all yeast aggregations. To create the FAIRE over the Sono-Seq plot, aggregation was performed over regions consisting of ±400 bp from the endpoints of each Sono-Seq region. The average of the furthest ten bins from each endpoint (corresponding to a region 300–400 bp distal of each Sono-Seq region endpoint) was used to normalize the remaining points.
Scoring Pol II and Reference DNA Samples Against Naked DNA and Intersecting Against Promoters of Ensembl Genes.
PeakSeq was used to score Pol II and each reference DNA type against naked DNA (8). Regions deemed to be enriched by PeakSeq were then intersected against promoter regions of Ensembl genes from Ensembl Release 50/NCBI36 by using a C program leveraging the BIOS library, and coverage statistics were generated by using the Active Region Comparer (25). For this analysis, we define promoter regions of Ensembl genes to be ±2.5 kb of the transcription start site and intersection as two sequences sharing at least one base position.
Calculating Percent Feature Composition and Creating a Rank-Order Plot for Sono-Seq DNA and Pol II DNA.
Enriched regions for Pol II and Sono-Seq DNA were ranked in descending order according to sequence tag count and fold enrichment versus naked DNA. These peaks were then classified by their proximity to known promoter regions and a rank-order plot produced (see SI Text).
Supplementary Material
Acknowledgments.
We thank Nick Carriero and Rob Bjornson for high-performance computing assistance, as well as Lukas Habegger for his consultation regarding the BIOS C library. This work was supported by grants from the National Institutes of Health (to M.S. and M.G.). The computational infrastructure is supported by the Yale University Biomedical High Performance Computing Center and National Institutes of Health Grant RR19895.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE14022 and GSE12781).
This article contains supporting information online at www.pnas.org/cgi/content/full/0905443106/DCSupplemental.
References
- 1.Nagy PL, Cleary ML, Brown PO, Lieb JD. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci USA. 2003;100:6364–6369. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford GE, et al. DNase-chip: A high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horak CE, Snyder M. ChIP-chip: A genomic approach for identifying transcription factor binding sites. Methods Enzymol. 2002;350:469–483. doi: 10.1016/s0076-6879(02)50979-4. [DOI] [PubMed] [Google Scholar]
- 5.Iyer VR, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 6.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 7.Lee TI, Johnstone SE, Young RA. ChIP and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotech. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flicek P, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Affymetrix and Cold Spring Harbor Laboratory ENCODE Transcriptome Projects. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morin R, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 15.Niwa H. Open conformation chromatin and pluripotency. Genes Dev. 2007;21:2671–2676. doi: 10.1101/gad.1615707. [DOI] [PubMed] [Google Scholar]
- 16.Robertson AG, et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18:1906–1917. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross P, et al. Polycomb genes expression and histone H3 lysine 27 tri-methylation changes during bovine preimplantation development. Reproduction. 2008;136:777–785. doi: 10.1530/REP-08-0045. [DOI] [PubMed] [Google Scholar]
- 20.Hogan GJ, Lee C, Lieb JD. Cell cycle–specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartman SE, et al. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Euskirchen GM, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aparicio O, Geisberg JV, Struhl K. ChIP for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Cell Biol. 2004 doi: 10.1002/0471143030.cb1707s23. Chapter 17:Unit 17.7. [DOI] [PubMed] [Google Scholar]
- 24.Barrett T, et al. NCBI GEO: Mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozowsky JS, et al. The DART classification of unannotated transcription within the ENCODE regions: associating transcription with known and novel loci. Genome Res. 2007;17:732–745. doi: 10.1101/gr.5696007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.