Abstract
The textbook planar model of pentameric IgM, a potent activator of complement C1q, is based upon the crystallographic structure of IgG. Although widely accepted, key predictions of this model have not yet been directly confirmed, which is particularly important since IgG lacks a major Ig fold domain in its Fc region that is present in IgM. Here, we construct a homology-based structural model of the IgM pentamer using the recently obtained crystallographic structure of IgE Fc, which has this additional Ig domain, under the constraint that all of the cysteine residues known to form disulfide bridges both within each monomer and between monomers are bonded together. In contrast to the planar model, this model predicts a non-planar, mushroom-shaped complex, with the central portion formed by the C-terminal domains protruding out of the plane formed by the Fab domains. This unexpected conformation of IgM is, however, directly confirmed by cryo-atomic force microscopy of individual human IgM molecules. Further analysis of this model with free energy calculations of out-of-plane Fab domain rotations reveals a pronounced asymmetry favoring flexions toward the central protrusion. This bias, together with polyvalent attachment to cell surface antigen, would ensure that the IgM pentamer is oriented on the cell membrane with its C1q binding sites fully exposed to the solution, and thus provides a mechanistic explanation for the first steps of C1q activation by IgM.
Keywords: AFM, homology modeling, immunoglobulin, single molecule
Pentameric IgM is an important component of the first line of defense against foreign pathogens (1, 2) and possibly modified self-components (3), and is increasingly being developed in the diagnosis and therapy of malignancies (4–7). It is also implicated in the damage to organs and tissues following ischemia/reperfusion (8) and in multiple autoimmune diseases (9–11). Its best understood mechanism of action in the immune response is as the initiating component in the classical complement pathway mediated by C1q (12). In this, the binding of IgM to cell surface antigen enables C1q to bind to IgM, which thereby activates C1q for interactions with downstream components. The expected structural changes consequent to antigen-binding and associated with C1q activation, and how these might be modified in disease-conditions, have spurred a longstanding interest in the structure of this large multicomponent molecule (13–17).
Like other antibodies, IgM monomers consist of two light and two heavy chains. However, whereas the heavy chains of most antibodies (such as IgG) contain three constant Ig domains, the μ heavy chains of IgM have a fourth one, as do the ε heavy chains in IgE. These extra (Cμ2) domains are located in place of the proline-rich hinge region that is responsible for the rotational flexibility of the antigen-binding Fab domains (relative to the Fc domain).
Five IgM monomers complex with an additional small polypeptide (the J chain) to form the predominant oligomeric species in the human plasma (3). Obtaining crystals of such large flexible complexes is notoriously challenging, and to date, there have been no crystallizations reported for the pentamers (or the monomers). Our present understanding of the structure of pentameric IgM is largely derived from negative-stain electron microscopy (EM) images (13) which showed the pentamer, in the absence of antigen, to be a planar, star-shaped complex, with the Fab domains located at the extremity of each radial arm. When bound to bacterial flagella, the pentamer was found to adopt a table-like conformation, with each of the Fab domains bent away from the plane defined by the Fc domains. Similar structural features (of the antigen-free form) were also concluded from small angle x-ray scattering (SAXS) data (17), interpreted in terms of molecular models that took advantage of the high structural homology among antibodies. At the time of these scattering studies however, the only crystallographic structures available of antibody molecules were of IgG, and so the μ chains were modeled based on the principles observed in IgG, resulting in the commonly portrayed structure of the pentamer in which all of the Cμ domains lie within the same plane. However, subsequent fluorescence data of the homologous IgE molecule in solution seemed to indicate that the IgE Fc was actually sharply bent (18). These initial observations were then clearly detailed when the crystallographic structure of the IgE Fc was solved (19), showing that the Cε2 domains are bent back toward the Cε4 domains by ≈60°. These findings raise the possibility that the homologous IgM Fc region may also be bent, although it is not immediately obvious whether such a bent structure could possibly be assembled into a pentameric complex that is planar and in which all of the known disulfide bridges both within and between monomers are satisfied (20).
Here, we report the results of our homology modeling of the IgM pentamer based upon the structure of the IgE Fc domain and the known disulfide pairings. According to this analysis, we show that an exclusively planar structure is not tenable. Instead, for the Fc domain to remain bent and the pertinent cysteine residues to be in sufficiently close proximity to each other, the pentamer must form a non-planar, mushroom-shaped structure, with the C-terminal regions protruding out from the plane defined by the Fab and Cμ2 domains. We confirm this model with direct images of individual molecules using cryo-atomic force microscopy (cryo-AFM) (21). Furthermore, we show that free energy calculations of out-of-plane Fab domain rotations reveal a bias to flexions toward the central protrusion. This bias, together with the polyvalent attachment to the cell surface, would result in the table-like appearance, with the protrusion directed toward the cell surface. As this also leads to the presentation and exposure of the C1q-binding sites to the solution, the work presented here may provide a structural mechanism underlying the first steps of IgM-C1q association.
Results
Homology-Based Modeling of the IgM Pentamer.
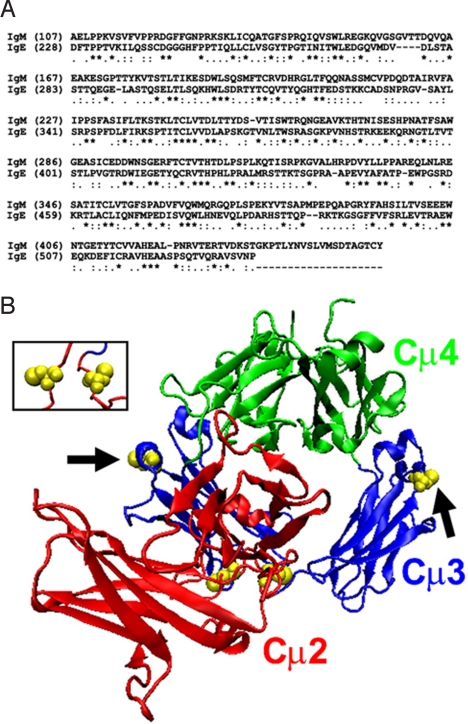
A BLAST search (22) for homologous proteins to the human IgM μ-chain identifies the human IgE ε-chain as an exceedingly similar match (E = 2 × 10−45). The alignment based on this BLAST result is shown in Fig. 1A. Excluding the unique C-terminal tail in IgM, the human IgM Fc domain exhibits an overall 29% identity and 86% similarity with the human IgE Fc domain, with a similar extent of homology across the entire protein. As a result, the IgE Fc crystal structure should serve as a useful model for the structure of the IgM Fc domain. Notably however, none of the cysteine residues in the IgM Fc that are known to be involved in inter- or intra-monomeric disulfide bridges (20) are conserved in the IgE Fc sequence. The disposition of these cysteine residues in the modeled IgM Fc structure is thus dependent upon the homology of the rest of the sequence, and so their locations serve as a useful test in evaluating the plausibility of any generated structure.
Fig. 1.
Homology model of the human IgM Fc based on the structure of the human IgE Fc. (A) The sequence alignment used to generate the model is based on a BLAST search (22) for sequences homologous to IgM that identified IgE as being a highly similar match (E = 2 × 10−45). An asterisk “*” below the alignment denotes identical amino acids while “.” and “:” denote similar and highly similar residues, respectively, as determined by DeepView. (B) Initial model of Fcμ before energy minimization. Shown as yellow van der Waals spheres are the cysteine residues that are known to be involved in intra-monomer (Cys-214) and inter-monomer (Cys-291, arrows) disulfide bridges. Even at this initial stage of modeling, the Cys-214 residues are close to each other (see Inset), while both Cys-291 residues are on the outermost surface of the complex and so could possibly interact with similar residues in neighboring molecules. None of these residues are conserved in IgE, and so their disposition in this structure at locations consistent with expectations lends strong support to the proposed model.
The model of the IgM Fc domain based only on this alignment, before any energy minimization, is shown in Fig. 1B. As shown in the Inset to Fig. 1B, the cysteine residues involved in intra-monomeric disulfide bridges (Cys-214) are in close proximity to each other even at this early stage of the modeling. In addition, the locations of the cysteines within the Cμ3 domains that are involved in inter-monomeric bridges (Cys-291) are on the exterior of the structure and are not buried, and are thus in a position to possibly interact with similar residues in the other monomers. This is particularly important since the C3 domains in this structure are not related by a two-fold dyad axis, and so the same residues in the two different chains are not in symmetrically equivalent positions.
Following energy minimization of the full-length IgM monomer (including the Fabs), the locations of these cysteine residues remain largely unchanged and each domain retains the expected β-sheet structure of typical Ig domains, providing validation for the model. Further inspection of this minimized structure reveals several somewhat unexpected similarities in the (non-covalent) inter-domain interactions with those of other Ig domain associations. In particular, the Cμ2-Cμ2 interaction is predominantly non-polar, and in fact the contact surface between this domain pair forms a pronounced hydrophobic patch on the surface of the Cμ2 domains (Fig. S1A). Such a predominantly non-polar interaction is common among many interacting Ig domains (19) (as shown for the Cε4-Cε4 domain pair in Fig. S1A). However, it is unexpected here since the Cε2-Cε2 interaction is actually predominantly polar (19), and so the non-polar character in the Cμ2-Cμ2 association is not a trivial consequence of the equivalence of the corresponding residues in the contact region. In addition, there were three salt bridges identified within the Cε2-Cε3,4 interface (19) and there are four in the Cμ2-Cμ3,4 interface [Asp121(B)-Lys315(A), Arg128(B)-Glu299(A), Lys150(B)-Glu385(A), Lys185(B)-Asp360(A)], none of which involve residues that are conserved with IgE. Still, two of the four salt bridges in Fcμ [Lys150(B)-Glu385(A) and Lys185(B)-Asp360(A)] involve residues that either coincide with or neighbor the residues at the corresponding locations of those involved in the salt bridges in Fcε (Fig. S1B). Further, a third salt bridge in Fcμ [Asp121(B)-Lys315(A)] may also have a correlate in the IgE Fc structure (Fig. S1B). These results suggest that there may actually be a higher degree of structural homology between IgM and IgE than sequence homology, particularly in those regions responsible for holding the Fc domain in a bent configuration, and further supports the plausibility of this structural model.
IgM monomers in the pentamer are known to interact via both their C-terminal tail region and residues within the Cμ3 domain, including Cys-291 (23, 24). The small J chain interacts with the C-terminal tail (23, 24) and could play a role in the assembly of the pentamer (24). This chain, however, is not expected to have a significant impact on the local structure of the monomers around the Cys-291 residues, and so it was not included in this modeling. As will be explained below, it is the disposition of the Cys-291 residues that most significantly determines the arrangement of the monomers in this pentameric structure.
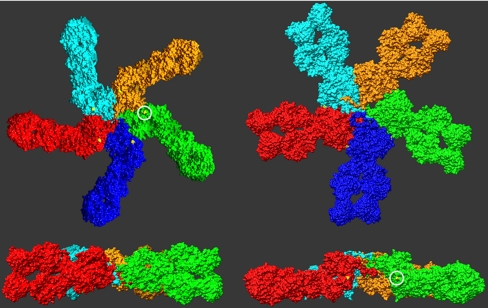
For the Cys-291 residues to form disulfide bonds between neighboring monomers in a circularized complex, they all must lie within a common plane. However, as demonstrated in the Left in Fig. 2, this is not possible for monomer orientations that produce an overall planar pentameric complex. In the monomer orientation depicted in this figure, these cysteines are located at the top and bottom of the complex. For these residues to interact with their corresponding residues in the neighboring monomers, each monomer must be rotated about its long axis by ≈90 °. Yet as a result of this rotation, the C-terminal Cμ4 and tail domains are rotated out of the plane defined by the Fab and Cμ2 domains, and the resulting pentamer complex adopts a non-planar, mushroom shaped structure as shown in the Right of Fig. 2 (see also Fig. S2).
Fig. 2.
Evaluation of the possible pentamer structures formed by the energy-minimized monomer, as judged by the relative disposition of the Cys-291 residues. The Left depicts a planar model, but the Cys-291 residues (circled) are not in a position to interact with a neighboring monomer. For this to occur, each must be rotated by 90 ° about its long axis. However, with this, as shown in the Right, the complex is not planar, but now exhibits a central protrusion, largely formed by the Cμ4 domains. In each of the Lower, the blue monomer was removed to facilitate visualization of the complex.
The location of the two Cys-291 residues in neighboring monomers in this non-planar structure is appropriate to permit the formation of disulfide bridges (Fig. S3A), yet there are other features of this interface that also appear advantageous for this interaction between monomers. In particular, the surfaces of the monomers in this region are highly complementary (Fig. S3A). There are also four salt bridges that span across the interface, above and below the disulfide bridge (Fig. S3B). Hence, while the disulfide bridge between Cys-291 residues undoubtedly plays a predominant role in linking together the monomers, these other features likely contribute to the relative angle of contact between the monomers, and thus to the observed pentameric stoichiometry.
Overall, then, there are a number of attributes of this modeled structure that favor a bent conformation for the IgM Fc domain and the assembly of the monomers into a complex that is mushroom-shaped, and not planar. Such a structure indeed differs significantly from the present planar model (17), however, it is not clear whether the previous methods used to establish this planar model could have readily discerned such a mushroom-shaped architecture. In the case of SAXS, it is well known that ab initio restoration of such large flat molecules by this technique is challenging (25, 26). Likewise, it would have been difficult for EM to clearly identify a central protrusion in the pentamer when absorbed flat on the grid (13). We therefore sought a different experimental technique to re-investigate the structure of the human IgM pentamer, and in particular to directly determine its surface topography.
Cryo-AFM of the Human IgM Pentamer.
Cryo-AFM is a particularly sensitive method to obtain surface contours of large, individual macromolecules (27, 28). Whereas the flexibility or softness of such complexes at room temperature could influence an accurate detection of height differences with conventional AFM (21, 29), the structural rigidity of proteins increases considerably at cryogenic temperatures, enabling the acquisition of perhaps otherwise unobtainable topographical profiles of macromolecular assemblies (30–33).
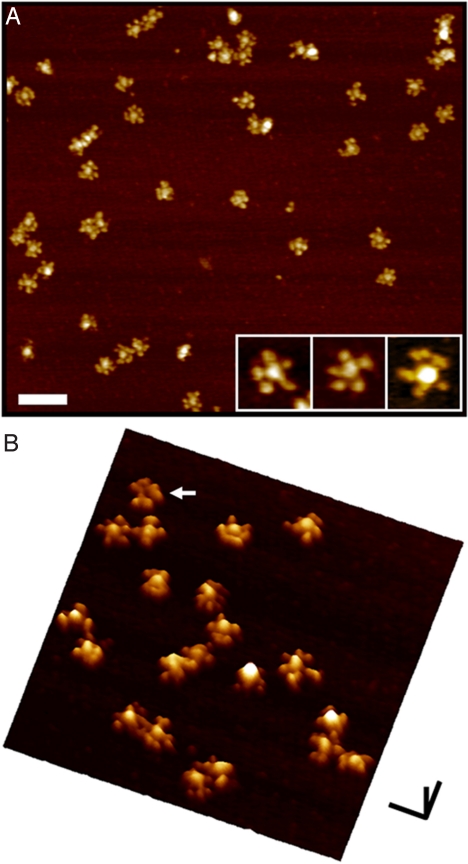
Typical cryo-AFM images of human IgM complexes deposited on a mica substrate are shown in Fig. 3. Each complex is star-shaped, with a central circular region (with a diameter of 19 ± 2 nm) from which a number of arms project out radially, each 11 ± 1 nm in length and 13 ± 3 nm wide. These dimensions are similar to those previously obtained by EM of similar complexes (13, 16), and the dimensions of the arms are consistent with those of Fab domains in previous cryo-AFM images of IgG (21). Hence, these images are in general agreement with the prevailing depiction of the IgM pentamer with the Fc domains located centrally and the Fab domains, all lying within a common plane, projecting out from the central region. The slightly variable number of arms visible between the different complexes is likely owing to variable distances and orientations between the pairs of Fab domains within each monomer.
Fig. 3.
Cryo-AFM images of individual IgM complexes deposited on mica. (A) Individual, well-separated pentamers are well resolved as star-shaped complexes, with a central ≈20 nm circular region, from which approximately 11 nm radial arms emerge. Most of the complexes have five radial arms that emanate from the central protrusion (see Inset), which are likely the closely apposed pairs of Fab domains of each monomer. Some of the oligomers exhibit more than five radial arms, which is likely a result of a slight separation of Fab domains within some of the monomers. These images are the so-called top-view representations, as if looking down onto the sample along the axis perpendicular to the substrate. (B) The surface view of the complexes clearly shows that the central region in most of the complexes protrudes out from the plane defined by the Fab domains. This is a demonstration of the non-planar topography of these IgM pentamers, and is in agreement with the predictions of the modeling presented here. A few (2%–3%) of the complexes appear to have a lower central region (arrow), which might be a result of mushroom-shaped complexes adsorbed in the opposite orientation on the mica surface. In this surface view representation, the data are presented as if viewed from the side, at an angle, thereby enabling an easier appreciation of the relative differences in height in the sample. [Scale bars (A) 100 nm; (B) x,y: 50 nm, z: 10 nm.]
However, rather than a completely planar pentamer, these images clearly show that, in most complexes, the height of the central region is larger than the height of the projecting arms (Fig. 3B). There are a few (2%–3%) complexes in which the central region appears to be lower than the radially projecting arms (arrowed in the figure), which could be complexes with the same shape as those with a central protrusion but that are oppositely oriented on the mica substrate. However, there are no complexes in which the central region is the same height as the arms. That is, none of the complexes are planar (in the manner presently believed). These direct images of the human IgM pentamer thus reveal that the complex is actually mushroom-shaped, in confirmation of the predictions from the modeling.
Comparison of the IgM Model with Previous Mutational Data.
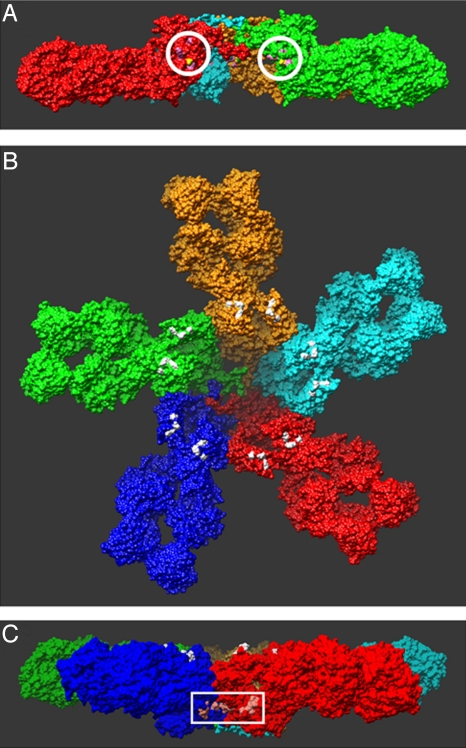
In addition to these direct images, there are at least three other independent lines of evidence in support of our model. First, previous studies of the effect of various single point mutations on the structure and stoichiometry of the IgM pentamer identified a number of residues (in addition to the aforementioned cysteines) that were likely to be directly involved in the inter-monomer interactions (34). As shown in Fig. 4A, in the mushroom-shaped complex described here, these residues indeed directly face the adjacent monomers. It should be emphasized that in none of the modeling was this information taken into consideration a priori. It should also be noted that two of these residues, Asp-293 and Lys-238, form a salt bridge between the monomers in this structure (Fig. S3B) as previously suggested (17).
Fig. 4.
Locations of critical residues previously identified in mutational analyses. (A) Encircled are the residues (pink) that were concluded to be involved in monomer-monomer interactions (34). Their location at the monomer-monomer interface in this structure lends further support to the proposed model. (B) Colored in white are the residues that are believed to directly interact with C1q (34–36). All of the residues are located on the surface of the complex, as they must be to interact with C1q, providing additional support for the proposed model. Also note that these residues are all located on the same side of the complex, which, as described in the text, places constraints on proposed mechanisms of C1q binding to cell-associated IgM. (c) The residues (light pink) in the white box were recently identified as likely directly interacting with a protein, PfEMP1 that is present in the membranes of erythrocytes infected with the bacteria that causes malaria, Plasmodium falcipaum (37). The location of these residues in this structure is consistent with the observed properties of the interaction with PfEMP1 (requiring oligomerization of IgM, not inhibited by binding by C1q) (37), further supporting the proposed structure.
Second, previous studies also identified a number of residues that were suggested to be directly involved in the binding to C1q (34–36). These residues thus must lie on the surface of the pentamer, at least when the pentamer is bound to antigen. In the model presented here, these residues form two localized regions on each monomer, on the same side of IgM complex, and indeed exposed to solution (Fig. 4B).
Finally, another just published study identified two additional sets of residues in IgM that were shown to directly interact with a protein (PfEMP1) present in erythrocyte membranes infected with the bacteria causing malaria, Plasmodium falcipaum (37). The study moreover showed that PfEMP1 only interacted with polymeric IgM, but not monomers, and that this interaction was not inhibited by the binding of C1q to IgM. As shown in Fig. 4C, these two sets of residues face each other at the junction between two monomers on the outer wall of the central protrusion in this model. It is clear that residues in adjacent monomers could form the binding pocket for PfEMP1, which would explain the requirement for pentameric IgM. Also, binding to this region should not be prohibited by the binding of C1q to IgM, as these residues are on the opposite side of the IgM complex from those involved in binding to C1q (Fig. 4B).
Energies Associated with Out-of-Plane Fab Rotations.
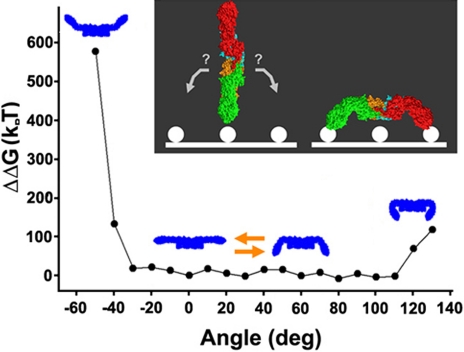
In the mutational data mentioned above, the C1q binding sites were not just located on the surface of the structure, but surprisingly, they were all localized to a single side (the flat side) of the complex (Fig. 4B). As mentioned in the introduction, when bound to bacterial flagella (and likely to cell surface antigens), the pentamer adopts a table-like structure (13), with each of the Fab domains bent out of the plane defined by the central portion of the complex, all in the same direction. Thus, the location of the C1q-binding sites in this model implies that when the IgM pentamer is bound to the cell surface antigens, it must be oriented in such a way that this flat side faces the solution (to interact with C1q). To address this issue, we sought to examine the energy associated with out-of-plane Fab domain rotations to determine whether there are any intrinsic properties that might favor or enhance such a bias. To do so, we rotated the two Fab domains within each monomer simultaneously, either toward or away from the central protrusion in increments of 10 °, and calculated the energy associated with the minimized structure at each angle of rotation (see Methods). While there is no formal hinge region in IgM, the previously observed branching of the antigen-free IgM monomers (13), which we have also observed, at the region that corresponds to the Fab-Fc junction in our model is consistent with some degree of flexibility at this location, as noted earlier (13). As shown in Fig. 5, there is, in fact, a strong steric hindrance preventing rotations of the Fab domains beyond 30 ° in the direction opposite to that of the central projection, but rotations up to 110° are possible (and roughly equivalent in energy) in the direction toward the central projection. In both cases, the steric hindrance is a clash between the Cμ1 and Cμ2 domains. The difference between the two directions is a result of the short, connecting loops between these two domains being closer to the side with the central projection than to the flat side. There is a slight minimum in the energy near 0 °, which could be responsible for the co-planar disposition of these arms in solution (13). Yet the most striking finding in these calculations is the restricted orientational space of the Fab domain flexions, biased in favor of rotations toward the central protrusion.
Fig. 5.
Energies associated with out-of-plane Fab domain rotations. The polyvalent attachment of the Fab domains to cell surface antigen can be accomplished if, after a first Fab-antigen interaction, the complex rotates in one of two directions, which results in one of two sides of the structure facing the solution (as portrayed in the Inset). The energies associated with the out-of-plane rotations of the Fab domains in the proposed model were calculated by manually rotating the Fab domains in increments of 10 °, followed by energy minimization. These results identify a pronounced steric hindrance to Fab flexions away from the central protrusion, and an absence of any similar prohibition to flexions toward the protrusion by up to 110 °. This result suggests that the complex would likely be oriented with the flat side facing the solution, when the Fab domains from more than a single monomer are associated with cell-surface antigen. This is also the side in which the C1q-binding sites are located (see Fig. 4B).
Discussion
The present work describes a model for the structure of the much-studied IgM pentamer. The most prominent feature of this model is the central region projecting out from the plane defined by the Fab domains, as shown directly in the cryo-AFM images (Fig. 3) and predicted by homology modeling using the structure of the highly similar IgE Fc domain. While the central protrusion in particular may prove to be a common target of Fc-like receptors for IgM (38) (as described above for the malarial protein), the structure as a whole, when taken with the previous studies and the free energy calculations preformed here, provides a structural explanation for the key first steps of C1q activation by pentameric IgM.
This structure exhibits a striking structural bias that results in a large orientational bias for the out-of-plane Fab domain rotations: rotations are limited to only 30 ° in the direction away from the central protrusion but those up to 110 ° are possible in the direction toward the protrusion. As the flexions within this range have roughly the same free energy, these angles thus delimit a broad (biased) orientational space that can be readily sampled through thermal fluctuations, without energetic penalty. When a single Fab domain binds to a cell surface antigen, these thermal fluctuations would tend to both bring the entire molecule closer to the cell with the central protrusion facing the cell surface and rotate each individual unbound Fab domain in the direction toward this protrusion. It is clear that the table-like conformation would then become “trapped” when the unbound Fab domains bind to the cell surface antigens. It should be noted that this large scale conformational change, derived from a transiently distorted antigen-free structure, differs from the conventional view in where the propagation of structural changes from the antigen-binding site to the Fc domain results in a significantly different conformation that is unique to the antigen-bound complex (39).
The orientation of the pentamer on the cell surface proposed here was found to agree with the location of the putative C1q binding sites in the complex (34–36). That these residues form such surface exposed, distinct regions in this structure is somewhat surprising, since it had been suggested that these binding sites might either be buried or might not yet be properly formed in the antigen-free complex, with antigen-binding leading to the formation of well-defined C1q binding sites on the surface of the complex (14, 15). Such conformational changes were suggested to underlie the requirement for antigen binding to IgM for subsequent C1q association with IgM (14, 15). While it is possible that much smaller scale structural changes occur at these sites with antigen binding, it may be that antigen binding to the Fab domain is more responsible for an increased accessibility of C1q for its binding sites. This could be achieved not just from the equilibrium position of the Fab domain, but also as a result of the larger region they effectively occupy through fluctuations. This reduced accessibility might play a role in any one of the many features of the highly complicated association between C1q and IgM (40, 41). Binding to surface antigen could then serve both to reduce the fluctuations of the Fab domains, and, as described above, sequester these domains in the direction away from the C1q binding sites. These possible effects of antigen-binding, that is, subtle changes in the conformation of C1q-binding sites, or accessibility because of the average location of the Fabs, are not mutually exclusive, and a structural understanding of which is dominant should now be possible with further interrogation of the model structure presented here.
Materials and Methods
Sample Preparation and Cryo-AFM Imaging.
Human IgM was obtained from Sigma. The purity of the protein was determined to be better than 95% using HPLC by the supplier. This quality was confirmed with SDS/PAGE, where degradation products were <10% (Fig. S4), and is consistent with the uniformity of the molecules in the cryo-AFM images. The stock solution (1 mg/mL) was diluted into a small droplet of low salt buffer (diluted PBS, 1:100) on a freshly cleaved mica surface to a final concentration of approximately 1 μg/mL. The low salt buffer is required to prevent the formation of salt crystals. To avoid aggregation, an incubation period of less than a minute was found sufficient, and no large aggregates were seen in the cryo-AFM images. After washing the specimen extensively, the excess solution was removed before it was transferred into the cryo-AFM, already adjusted to the imaging, cryogenic temperature. These procedures were all performed under dry nitrogen to avoid contamination. Our cryo-AFM is a homemade instrument, specifically designed for imaging biological molecules (21). The AFM operates in a liquid nitrogen vapor with an imaging temperature normally in the range of 80–85 °K. All AFM images were obtained with cantilevers purchased from Park Scientific. These were uncoated cantilevers with oxide-sharpened tips. The nominal spring constant was 0.03 N/m at room temperature, and measurements of the shift of the resonant frequencies indicated that the actual spring constant was only ≈15% greater at ≈100 K. The typical scanning speed was 1–2 Hz, and the adhesion force was typically between 1 and 2 nN. The instrument was calibrated from images of mica and gold ruling. It should be noted that the J-chain is expected to be located precisely in the center of the molecule, but as it is expected to be the size of a single Ig domain, it was not expected to change the measured height of the central protrusion significantly from expectations in cryo-AFM images. We did not observe any distinguishing topographic feature (an extra “bump”, for example) in the central protrusion in the AFM images, consistent with these expectations. We should also note that similar images as those described herein were obtained with several stocks of human IgM (all from Sigma).
Molecular Modeling.
The homology model was constructed with the automated homology modeling tools in DeepView v.3.7 (42), using the human IgM heavy chain (accession no. P01871.3) and the crystal structure of the IgE Fc (PDB accession codes: 1o0vA and 1o0vB), based on the alignment produced by the BLAST search (22). The IgM Fab domain (43) (PDB accession code 1DEE) and the C-terminal tail (generated as a random coil) were affixed to the Fc region using DeepView. The structure of each tail is different for the different monomers, and after an initial round of minimization, the closest pairs of cysteine residues on separate chains were linked together for subsequent minimization (20, 44). All energy calculations were performed using VMD/NAMD (45, 46). NAMD was developed by the Theoretical Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign. The structure was minimized in stages: first just the Fc domain, then the entire monomer, then a trimer, and finally the pentamer. With each case, the structure was minimized with all possible disulfide bridges formed. Each stage required between 15,000 and 30,000 rounds of minimization. An evaluation of the appropriateness of the amino acid dihedral angles was monitored with the Ramachandran plot during the course of energy minimization and (as is typical for such modeling procedures) found to be similar to the template structures used for the model. In initial trial calculations of the energies at different Fab domain rotations, it was observed that the presence of either the C-terminal tail regions or the VH-VL domains did not contribute significantly to the differences in energies at different angles, although the absence of the Cμ3 and Cμ4 domains did result in significantly different structures of the Cμ2 domains. Hence, for these calculations, we investigated a trimer consisting of the Fc domains of two monomers on either side of a nearly complete monomer (missing only the tail and VH-VL domains). The two Fc domains were included to ensure a proper structure of the Cμ3 and Cμ4 domain in the central monomer. An axis running between the two Leu-109 residues (see Fig. 1A) in the two different Cμ1-Cμ2 linker regions was identified as oriented perpendicular to the long axis of the monomer and within the middle of the plane of the pentamer, and so was chosen as the pivot. The region of both Fabs up to these residues was rotated about this axis in increments of 10 °. The energy of the structure was then minimized, generally requiring 1,000–2,000 rounds of calculations. Figs. 2, 4, and 5 were generated using Chimera (47), while the remaining structural models were generated with VMD. It should be noted that the early EM observations of IgM associated with antigen also observed some IgM complexes with the entire IgM monomer bent away from a central link (13). At present, we are not sure how this might be accommodated in this model but believe that further studies might be required to determine if some aspect of sample preparation in the previous studies might have contributed to this observation. Coordinates are available from the authors upon request.
Supplementary Material
Acknowledgments.
We thank Jie Liu for her help and useful discussions, and Yiyi Zhang and Sitong Sheng for their technical assistance. This work supported by National Institutes of Health Grants GM68729 and GM74495 (to Z.S.) and National 973 Project of China 2007CB936003.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903805106/DCSupplemental.
References
- 1.Boes M, Prodeus AP, Schmidt T, Carroll MC, Chen J. A critical role of natural immunoglobulin M in immediate defense against systemic bacterial infection. J Exp Med. 1998;188:2381–2386. doi: 10.1084/jem.188.12.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochsenbein AF, et al. Control of early viral and bacterial distribution and disease by natural antibodies. Science. 1999;286:2156–2159. doi: 10.1126/science.286.5447.2156. [DOI] [PubMed] [Google Scholar]
- 3.Vollmers HP, Brandlein S. Natural IgM antibodies: The orphaned molecules in immune surveillance. Adv Drug Deliv Rev. 2006;58:755–765. doi: 10.1016/j.addr.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Beutner U, et al. Neoadjuvant therapy of gastric cancer with the human monoclonal IgM antibody SC-1: Impact on the immune system. Oncol Rep. 2008;19:761–769. [PubMed] [Google Scholar]
- 5.Illert B, et al. Human antibody SC-1 reduces disseminated tumor cells in nude mice with human gastric cancer. Oncol Rep. 2005;13:765–770. [PubMed] [Google Scholar]
- 6.Vollmers HP, et al. Tumor-specific apoptosis induced by the human monoclonal antibody SC-1: A new therapeutical approach for stomach cancer. Oncol Rep. 1998;5:35–40. doi: 10.3892/or.5.1.35. [DOI] [PubMed] [Google Scholar]
- 7.Vollmers HP, et al. Adjuvant therapy for gastric adenocarcinoma with the apoptosis-inducing human monoclonal antibody SC-1: First clinical and histopathological results. Oncol Rep. 1998;5:549–552. doi: 10.3892/or.5.3.549. [DOI] [PubMed] [Google Scholar]
- 8.Fleming SD. Natural antibodies, autoantibodies and complement activation in tissue injury. Autoimmunity. 2006;39:379–386. doi: 10.1080/08916930600739381. [DOI] [PubMed] [Google Scholar]
- 9.Stahl D, et al. Altered antibody repertoires of plasma IgM and IgG toward nonself antigens in patients with warm autoimmune hemolytic anemia. Hum Immunol. 2001;62:348–361. doi: 10.1016/s0198-8859(01)00225-7. [DOI] [PubMed] [Google Scholar]
- 10.Baudino L, et al. IgM and IgA anti-erythrocyte autoantibodies induce anemia in a mouse model through multivalency-dependent hemagglutination but not through complement activation. Blood. 2007;109:5355–5362. doi: 10.1182/blood-2006-11-059899. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann M, Brennan FM, Maini RN. Rheumatoid arthritis. Cell. 1996;85:307–310. doi: 10.1016/s0092-8674(00)81109-5. [DOI] [PubMed] [Google Scholar]
- 12.Murphy KP, Travers P, Walport M. Janeway's immunobiology. 7th Ed. New York: Garland Science; 2008. [Google Scholar]
- 13.Feinstein A, Munn EA. Conformation of the free and antigen-bound IgM antibody molecules. Nature. 1969;224:1307–1309. doi: 10.1038/2241307a0. [DOI] [PubMed] [Google Scholar]
- 14.Feinstein A, Richardson N, Taussig MJ. Immunoglobulin flexibility in complement activation. Immunol Today. 1986;7:169–174. doi: 10.1016/0167-5699(86)90168-4. [DOI] [PubMed] [Google Scholar]
- 15.Feinstein A, Richardson NE. Tertiary structure of the constant regions of immunoglobins in relation to their function. Monogr Allergy. 1981;17:28–47. [Google Scholar]
- 16.Munn EA, Bachmann L, Feinstein A. Structure of hydrated immunoglobulins and antigen-antibody complexes. Electron microscopy of spray-freeze-etched specimens. Biochim Biophys Acta. 1980;625:1–9. doi: 10.1016/0005-2795(80)90102-6. [DOI] [PubMed] [Google Scholar]
- 17.Perkins SJ, Nealis AS, Sutton BJ, Feinstein A. Solution structure of human and mouse immunoglobulin M by synchrotron X-ray scattering and molecular graphics modelling. A possible mechanism for complement activation. J Mol Biol. 1991;221:1345–1366. doi: 10.1016/0022-2836(91)90937-2. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Y, Shopes B, Holowka D, Baird B. Conformations of IgE bound to its receptor Fc epsilon RI and in solution. Biochemistry. 1991;30:9125–9132. doi: 10.1021/bi00102a002. [DOI] [PubMed] [Google Scholar]
- 19.Wan T, et al. The crystal structure of IgE Fc reveals an asymmetrically bent conformation. Nat Immunol. 2002;3:681–686. doi: 10.1038/ni811. [DOI] [PubMed] [Google Scholar]
- 20.Wiersma EJ, Shulman MJ. Assembly of IgM. Role of disulfide bonding and noncovalent interactions. J Immunol. 1995;154:5265–5272. [PubMed] [Google Scholar]
- 21.Han W, Mou J, Sheng J, Yang J, Shao Z. Cryo atomic force microscopy: A new approach for biological imaging at high resolution. Biochemistry. 1995;34:8215–8220. doi: 10.1021/bi00026a001. [DOI] [PubMed] [Google Scholar]
- 22.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frutiger S, Hughes GJ, Paquet N, Luthy R, Jaton JC. Disulfide bond assignment in human J chain and its covalent pairing with immunoglobulin M. Biochemistry. 1992;31:12643–12647. doi: 10.1021/bi00165a014. [DOI] [PubMed] [Google Scholar]
- 24.Fazel S, Wiersma EJ, Shulman MJ. Interplay of J chain and disulfide bonding in assembly of polymeric IgM. Int Immunol. 1997;9:1149–1158. doi: 10.1093/intimm/9.8.1149. [DOI] [PubMed] [Google Scholar]
- 25.Volkov VV, et al. Low-resolution structure of immunoglobulins IgG, IgM and rheumatoid factor IgM-RF from solution X-ray scattering data. J Appl Cryst. 2003;36:503–508. [Google Scholar]
- 26.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng S, Shao Z. Cryo-atomic force microscopy. Methods Cell Biol. 2002;68:243–256. doi: 10.1016/s0091-679x(02)68013-6. [DOI] [PubMed] [Google Scholar]
- 28.Shao Z, Zhang Y. Biological cryo atomic force microscopy: A brief review. Ultramicroscopy. 1996;66:141–152. doi: 10.1016/s0304-3991(96)00087-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Sheng S, Shao Z. Imaging biological structures with the cryo atomic force microscope. Biophys J. 1996;71:2168–2176. doi: 10.1016/S0006-3495(96)79418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Shao Z, Somlyo AP, Somlyo AV. Cryo-atomic force microscopy of smooth muscle myosin. Biophys J. 1997;72:1308–1318. doi: 10.1016/S0006-3495(97)78777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheng S, Czajkowsky DM, Shao Z. Localization of linker histone in chromatosomes by cryo-atomic force microscopy. Biophys J. 2006;91:L35–37. doi: 10.1529/biophysj.106.090423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheng S, et al. Cryo-atomic force microscopy of unphosphorylated and thiophosphorylated single smooth muscle myosin molecules. J Biol Chem. 2003;278:39892–39896. doi: 10.1074/jbc.M306094200. [DOI] [PubMed] [Google Scholar]
- 33.Shao Z, Shi D, Somlyo AV. Cryoatomic force microscopy of filamentous actin. Biophys J. 2000;78:950–958. doi: 10.1016/S0006-3495(00)76652-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arya S, et al. Mapping of amino acid residues in the C mu 3 domain of mouse IgM important in macromolecular assembly and complement-dependent cytolysis. J Immunol. 1994;152:1206–1212. [PubMed] [Google Scholar]
- 35.Taylor B, et al. C1q binding properties of monomer and polymer forms of mouse IgM mu-chain variants. Pro544Gly and Pro434Ala. J Immunol. 1994;153:5303–5313. [PubMed] [Google Scholar]
- 36.Wright JF, Shulman MJ, Isenman DE, Painter RH. C1 binding by murine IgM. The effect of a Pro-to-Ser exchange at residue 436 of the mu-chain. J Biol Chem. 1988;263:11221–11226. [PubMed] [Google Scholar]
- 37.Ghumra A, et al. Identification of residues in the Cmu4 domain of polymeric IgM essential for interaction with Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) J Immunol. 2008;181:1988–2000. doi: 10.4049/jimmunol.181.3.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibuya A, et al. Fc alpha/mu receptor mediates endocytosis of IgM-coated microbes. Nat Immunol. 2000;1:441–446. doi: 10.1038/80886. [DOI] [PubMed] [Google Scholar]
- 39.Feinstein A, Munn EA, Richardson NE. The three-dimensional conformation of M and A globulin molecules. Ann N Y Acad Sci. 1971;190:104–121. doi: 10.1111/j.1749-6632.1971.tb13526.x. [DOI] [PubMed] [Google Scholar]
- 40.Zlatarova AS, et al. Existence of different but overlapping IgG- and IgM-binding sites on the globular domain of human C1q. Biochemistry. 2006;45:9979–9988. doi: 10.1021/bi060539v. [DOI] [PubMed] [Google Scholar]
- 41.Gaboriaud C, et al. Structure and activation of the C1 complex of complement: Unraveling the puzzle. Trends Immunol. 2004;25:368–373. doi: 10.1016/j.it.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 43.Graille M, et al. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: Structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci USA. 2000;97:5399–5404. doi: 10.1073/pnas.97.10.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorensen V, Sundvold V, Michaelsen TE, Sandlie I. Polymerization of IgA and IgM: Roles of Cys309/Cys414 and the secretory tailpiece. J Immunol. 1999;162:3448–3455. [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.