Abstract
The retinoblastoma tumor-suppressor protein, pRb, is a member of the pocket protein family that includes p107 and p130. These proteins have well-defined roles in regulating entry into and exit from the cell cycle and also have cell cycle–independent roles in facilitating differentiation. Here we investigate the overlap between pocket protein's function during embryonic development by using conditional mutant alleles to generate Rb;p107 double-mutant embryos (DKOs) that develop in the absence of placental defects. These DKOs die between e13.5 and e14.5, much earlier than either the conditional Rb or the germline p107 single mutants, which survive to birth or are largely viable, respectively. Analyses of the e13.5 DKOs shows that p107 mutation exacerbates the phenotypes resulting from pRb loss in the central nervous system and lens, but not in the peripheral nervous system. In addition, these embryos exhibit novel phenotypes, including increased proliferation of blood vessel endothelial cells, and heart defects, including double-outlet right ventricle (DORV). The DORV is caused, at least in part, by a defect in blood vessel endothelial cells and/or heart mesenchymal cells. These findings demonstrate novel, overlapping functions for pRb and p107 in numerous murine tissues.
Keywords: cell cycle, heart, p107, retinoblastoma
The human retinoblastoma gene, RB-1, which encodes pRB, was the first identified tumor suppressor. pRB belongs to the pocket protein family that includes p107 and p130 (1). All 3 pocket proteins have some ability to promote G0/G1 arrest, in part through repression of E2F-responsive genes that encode key regulators of cell proliferation (1). But the mechanisms of action of these proteins have significant differences, with p107 and p130 being more similar to each other than to pRB (1). First, the pocket proteins target different subsets of the E2F family. p107 and p130 bind specifically to the repressive E2Fs, E2F4 and E2F5, and the resulting complexes bind and repress E2F-responsive promoters in G0/G1 cells via recruitment of histone deacetylases. pRB regulates E2F4 in a similar manner to p107 and p130, but it also binds the activating E2Fs, E2F1, 2, 3A and 3B during G1 and inhibits their transcriptional activity. Mitogenic signaling induces phosphorylation of pRB, p107, and p130 by the cyclin–dependent kinases. This induces dissociation of the pocket protein–E2F complexes, thereby relieving repression and allowing the activating E2Fs to promote transcription. The second distinct feature of the pocket proteins is that pRB associates with various transcription factors that drive tissue-specific differentiation, but p107 and p130 do not seem to share this function. Finally, there is a fundamental difference in the tumor-suppressive properties of pRB versus p107 and p130: RB-1 is inactivated in approximately one-third of all human tumors, whereas p107 and p130 are rarely disrupted. This has led to intense interest in establishing the relative roles of the pocket proteins in vivo.
Mouse models have been a key tool in probing the pocket proteins' tumor-suppressive properties. Rb+/− mice develop tumors with near-complete penetrance, but inactivation of p107 or p130 does not yield tumors (2–4). However, chimeric mouse studies show that a loss of both Rb and p107 or Rb and p130 causes a broader spectrum of tumors than that resulting from mutation of Rb alone (5). Thus, p107 and p130 can substitute for pRB in suppressing tumor formation in some tissues. Germline mutant mice also have yielded key insights into the pocket proteins' roles in normal development (6). In mixed C57BL/6 × 129/Sv and pure 129/Sv backgrounds, p107−/− and p130−/− mutant mice are largely viable and have few or no defects (3, 4). Even the combined mutation of p107 and p130 yields live-born mice in which most tissues appear normal (4); the few exceptions include defects in long-bone development, abnormal epidermal differentiation, and early neonatal lethality of p107−/−;p130−/− mice (4, 6). In stark contrast, in the same genetic backgrounds, the Rb−/− mice die in mid-gestation (between e13.5 and e15.5), with ectopic proliferation and apoptosis in the central nervous system (CNS), peripheral nervous system (PNS), and lens (2, 7). These Rb−/− mice also have an erythroid defect that was initially thought to account for their death (8). Early studies showed that Rb−/−;p107−/− embryos died only slightly earlier (1–2 days) than the Rb germline mutants (3). This suggested that little functional overlap exists between pRb and p107 and supported the view of Rb as the most important pocket protein in vivo. But later studies showed that the mid-gestational lethality of the germline Rb−/− embryos is due to poor placental function (9). Indeed, Rb−/− embryos survive to birth when provided with a wild-type placenta. Although ectopic proliferation still occurs in the lens, CNS, and PNS, the apoptosis is largely suppressed, suggesting that this is due in large part to placental insufficiency (9). The erythroid defect is also reduced but not fully rescued, indicating that this has both cell and noncell autonomous origins (8). Finally, this and other mouse models revealed a key role for Rb in the development of skeletal muscle, bone, and skin and intestinal epithelia (6, 10, 11). Many tissues develop completely normally in the absence of pRB, however. Given this revised appreciation of the developmental role of Rb, we used conditional mice to examine the relationship between Rb and p107 when embryos develop in the context of a wild-type Rb placenta.
Results
Rbc−/c−;p107−/−;Mox2+/Cre Double-Mutant Embryos Die During Mid-Gestation.
To assess the potential overlap between Rb and p107 in embryos developing without placental deficiency, we crossed germline p107 mutants (12) with lines carrying conditional alleles of Rb (Rbc/c; ref. 12) or the Mox2-Cre transgene (Mox2+/Cre; ref. 13) to generate 4 test genotypes: Rb+/c;p107+/− (control), Rb+/c;p107−/− (p107 mutant), Rbc−/c−;p107+/+;Mox2+/Cre (Rb mutant), and Rbc−/c−;p107−/−;Mox2+/Cre (DKO). The Mox2-Cre strain expresses the Cre recombinase specifically in the embryo proper, beginning around e6.5 (13), and thus the extraembryonic tissues are Rb wild-type irrespective of the embryo's genotype. Accordingly, the placental tissues were found to be completely normal in the Rb mutants, as well as in all other genotypes studied [supporting information (SI) Fig. S1]. Having verified placental integrity, we next examined the embryos' lifespan. As reported previously (9), the presence of a wild-type placenta allowed Rb mutant embryos to survive to birth. In contrast, viable DKOs were not observed at this time point (data not shown). Timed pregnancies showed that DKOs were present at the expected Mendelian ratio at e13.5 (Table 1) and were alive, as judged by the presence of a heartbeat. DKO embryos were present but mostly dead at e14.5 (Table 1) and were absent at later time points. These results indicate that loss of p107 shortens the lifespan of Rb mutant embryos from birth to between e13.5 and 14.5.
Table 1.
DKO's (Rbc−/c−;p107−/−;Cre+) die between e13.5 and e14.5
| Genotype* | p107+/− Rb+/cCre− | p107+/−Rb+/c−Cre+ | p107+/−Rbc/c−Cre− | p107+/− Rbc−/c−Cre+ | p107−/− Rb+/cCre− | p107−/− Rb+/c−Cre+ | p107−/− Rbc/c−Cre− | p107−/− Rbc−/c−Cre+ | Total |
|---|---|---|---|---|---|---|---|---|---|
| Obs e13.5 | 30 | 35 | 19 | 13 | 25 | 26 | 18 | 25 | 191 |
| Exp e13.5 | 23.875 | 23.875 | 23.875 | 23.875 | 23.875 | 23.875 | 23.875 | 23.875 | 191 |
| Obs e14.5 | 12 | 11 | 13 | 12 | 14 | 12 | 10 | 9 (7)† | 93 |
| Exp e14.5 | 11.625 | 11.625 | 11.625 | 11.625 | 11.625 | 11.625 | 11.625 | 11.625 | 93 |
*Mice were generated from crosses of Rb+/c−;p107+/−;Mox2+/Cre males with Rbc/c;p107−/− females.
†Number in parentheses indicates dead embryos.
p107 Loss Exacerbates Proliferation and Apoptosis Defects in the CNS and Lens of Rb Mutants.
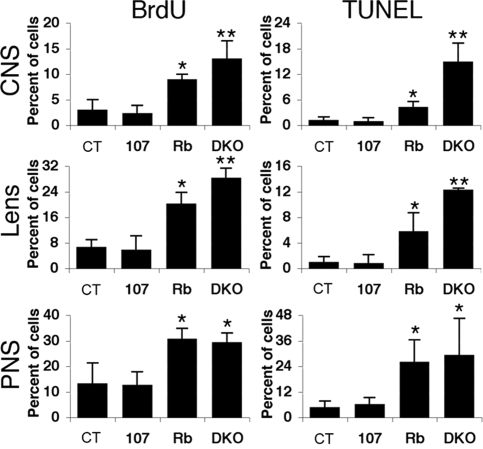
We next explored whether p107 loss alters the known phenotypes of Rb mutants. First, we screened the CNS, PNS, and ocular lens of viable e13.5 embryos for proliferation and apoptosis (Fig. 1A and Fig. S2). As expected (9), the Rb mutants displayed a significantly higher level of proliferating cells in the CNS, PNS, and lens than either the control or p107 mutants (which were indistinguishable from one another). Loss of Rb also caused a low but significant level of apoptosis in all 3 tissues. The additional loss of p107 had no detectable effect on either the proliferation or apoptosis defects in the dorsal root ganglia of the PNS of Rb mutants. In contrast, proliferation and apoptosis levels were significantly higher in the hindbrain (CNS) and the ocular lens of DKOs versus Rb mutants. In addition, sporadic apoptosis was detected in the liver and skeletal muscle of DKOs but not of other genotypes (data not shown). Together, our data indicate that p107 is able to substitute, either partially or fully, for critical pRB functions in the CNS, ocular lens, liver, and skeletal muscle.
Fig. 1.
Mutation of p107 exacerbates the proliferative and apoptotic defects observed in the CNS and lens, but not the PNS, of Rbc−/c− embryos. Average percentage of BrdU- and TUNEL-positive nuclei in indicated tissues from e13.5 embryos (n ≥ 3 for each genotype). Error bars represent 1 standard deviation. The asterisks denote statistical significance (P < .05) compared with controls and p107−/− (*) or Rbc−/c− (**).
Because Rb inactivation is known to disrupt erythroid development (8), we also examined the fetal erythroid cells present in the placental sections. We could not assess enucleation (which is known to be defective in Rb mutants), because the erythroid cells were mostly nucleated at e13.5; however, we could screen for erythroid abnormalities, such as irregularly shaped, multiple, or fragmented nuclei. This analysis revealed the expected spectrum of erythroid defects in the Rb mutant and showed that the percentage of affected cells was not altered in the DKOs (Fig. S3). Thus, at least at this developmental stage, p107 does not synergize with Rb in fetal erythroid development.
DKO Embryos Exhibit Edema and Severe Heart Defects.
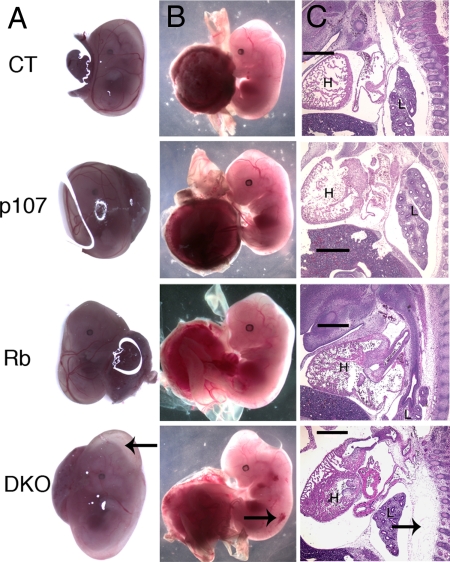
The analysis of e13.5 and e14.5 embryos also revealed macroscopic phenotypes specific to the DKOs. First, the yolk sacs of DKOs were bloated and abnormally large, and some (although not all) were abnormally pale (Fig. 2; data not shown). Compared with all other genotypes, the e13.5 and e14.5 DKOs exhibited a more severely hunched appearance and a bloated midsection, and a subset had hemorrhaging. These phenotypes led us to investigate the embryos in more detail. Sagittal sections revealed severe edema in the vast majority of DKOs examined (10/14 at e13.5; 5/6 at e14.5) that was most prevalent along the backbone but was also seen in the midsection and around the skull (Fig. 2 and data not shown). In contrast, edema was completely absent in the control (n = 5 at e13.5; n = 2 at e14.5) and p107 mutant (n = 2 at e13.5; n = 4 at e14.5) embryos and was present only in a much milder form in a subset of the Rb mutants (1/4 at e13.5; 1/2 at e14.5).
Fig. 2.
DKOs display several abnormal macroscopic phenotypes. (A) At e13.5, the yolk sacs of DKOs are enlarged (arrow) compared with other genotypes. (B) DKO embryos have a bloated midsection, a more severely hunched neck, and occasional hemorrhaging (arrow). (C) Edema (arrow) in sagittal sections of the DKO embryos. (Magnification: X4.) (Scale bar: 500 μm.) H, heart; L, liver.
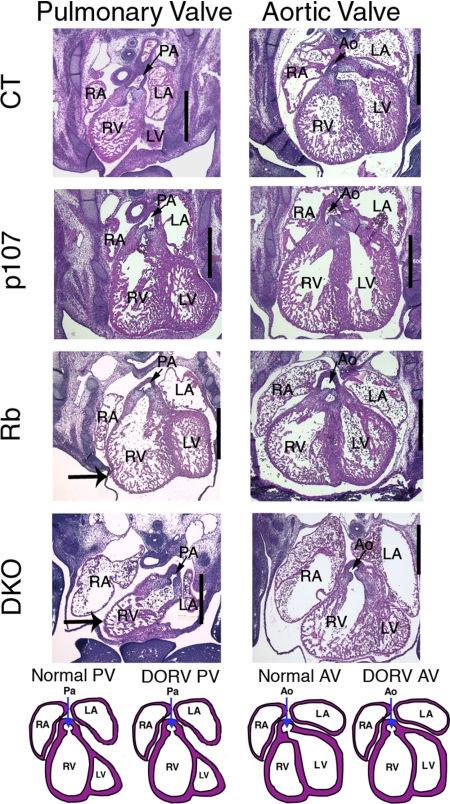
There are several known causes of edema, including kidney, lung, and heart dysfunction, as well as anemia. We ruled out kidney and lung dysfunction, because these organs were histologically normal in the DKOs (data not shown) and also are known to be dispensable for fetal development. We next considered erythroid defects and anemia. Although the DKOs had abnormal erythroid cells, the frequency was the same as that seen in nonedematous Rb mutant embryos (Fig. S3). Some DKO yolk sacs were pale, indicating mild anemia, but the DKO embryos showed no evidence of severe anemia (data not shown). Thus, these defects seemed insufficient to explain the edema. Finally, we screened for heart defects, a common cause of edema in humans (14), by comparing serial transverse sections of e13.5 hearts from DKOs with those of other test genotypes (Fig. 3 and data not shown). Control (n = 6), p107 (n = 3), and Rb (n = 3) mutant embryos displayed relatively normal architecture and development of the heart, great arteries (pulmonary artery and aorta), and valves, with the exception of a marginally thinner myocardial wall in the Rb mutants. In contrast, all 7 of the DKOs serially examined exhibited double-outlet right ventricle (DORV), in which both the pulmonary artery and the aorta exit from the right ventricle. Some DKOs also displayed dilated atria and thin myocardial walls, while others had atrial and ventricular septum formation defects (data not shown). Because ventricular septum formation is only just completed by e13.5 in wild-type embryos and its timing can vary somewhat, this finding could reflect either a specific septal defect or a subtle developmental delay in the DKOs. Overall, our data indicate that pRb and p107 play overlapping roles in embryonic heart development.
Fig. 3.
DKO hearts display DORV. Transverse section of e13.5 embryonic hearts showing the pulmonary and aortic valves. (Scale bar: 500 μm.) Schematic diagrams show normal and DORV hearts from both views. The DKO hearts exhibit dilated atria, a thin myocardial wall (large arrow) and DORV. The Rbc−/c− hearts show only slightly thinner myocardial walls compared with control hearts (large arrow). Ao, aorta and aortic valve; LA, left atrium; LV, left ventricle; RA, right atrium; RV, right ventricle; PA, pulmonary artery and pulmonary valve.
It has been reported that Rb−/− embryonic stem (ES) cells have impaired capacity to induce cardiomyocyte genes and differentiate in vitro (15). Thus, we isolated RNA from the hearts of e13.5 embryos and analyzed the mRNA levels of key cardiac transcription factors and differentiation markers by quantitative RT-PCR (qRT-PCR) (Fig. S4). In contrast to the in vitro ES cell analysis, we found no down-regulation of cardiomyocyte markers in the Rb mutants or DKOs. In fact, several cardiomyocyte markers of the Mesp1/2 pathway (i.e., Mesp2, Nkx2.5, Gata4, and ANF) had higher mRNA levels in the DKOs than in the other genotypes. These findings suggest that the heart defects are not due to any general defect or delay in cardiomyocyte differentiation.
p107 is a known E2f target gene, and in some settings pRb loss has been shown to increase p107 mRNA levels, presumably as a direct result of E2f activation (16). This is thought to contribute to p107's ability to compensate for pRb loss. Thus, we investigated whether there was any up-regulation of p107 mRNA in Rb mutant hearts that might contribute to the very modest heart phenotype in Rb mutants versus the DKOs. Using qRT-PCR to compare p107 and Rb mRNA levels in the e13.5 hearts of wild-type, p107, and Rb mutant embryos, we found no detectable increase in p107 mRNA in the Rb mutants (Fig. S4). Together, these data show that pRb plays a key role in heart development in vivo, but that normal levels of p107 can largely substitute for this function, and thus major heart defects occur only after combined loss of pRb and p107.
DORV in the DKOs Is Likely Due To Endothelial or Mesenchymal Cell Defects.
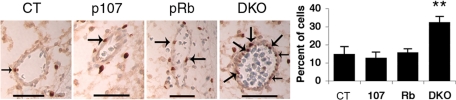
Given the relatively normal expression of cardiomyocyte markers in the DKO hearts, we next examined the development of the great arteries and/or other blood vessels. Immunohistochemical analyses of several blood vessel markers, including smooth muscle actin, laminin, and fibronectin, revealed no major disruption of blood vessel differentiation in the DKOs (Fig. S5 and data not shown). Keeping in mind the consequences of pocket protein loss in other tissues, we next examined cellular proliferation and apoptosis. We detected no apoptosis in any blood vessel cells (data not shown) irrespective of genotype; however, we did detect a significant increase in the number of proliferating endothelial cells lining the blood vessels in the DKOs relative to all of the other genotypes (n = 4 for each) (Fig. 4). Thus, while the DKO blood vessels display the appropriate differentiation markers, the endothelial cells are undergoing ectopic proliferation. It seems possible that this defect could disrupt development or function of the blood vessels, perhaps by interfering with vessel integrity or microvascular stability, thereby contributing to the development of edema and DORV.
Fig. 4.
DKO blood vessels exhibit increased proliferation. BrdU incorporation in blood vessels of e13.5 embryo heads. Arrows indicate positively stained cells. (Scale bar: 50 μm.) Statistical significance is as noted in Fig. 1.
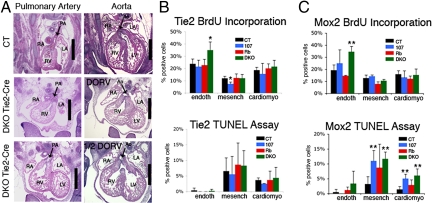
To further examine the basis of the heart defect, we crossed Rbc/+;p107+/− mice with the Tie2-Cre mouse strain (17, 18). We selected this model because it expresses Cre recombinase in 2 different locations that could be relevant to the DKO heart defect: all endothelial cells within the embryo, as well as the mesenchymal cells of the developing pulmonary valves, aortic valves, and atrioventricular canal (17, 18). This latter expression seems to result from endothelial–mesenchymal transdifferentiation in the development of both the atrioventricular canal and outflow tract regions (17). We used timed pregnancies to isolate Tie2-Cre+ mutants at e13.5. There were no detectable cardiac defects in Rbc−/c−;Tie2-Cre+ embryos or any other control genotypes (n ≥ 3 for each class; data not shown). In contrast, of the 7 Rbc−/c−;p107−/−;Tie2-Cre+ embryos (Tie2-Cre DKOs) analyzed, 3 exhibited a partial DORV phenotype and 3 displayed full DORV (Fig. 5A). Notably, like the Mox2-Cre DKOs, the Tie2-Cre DKOs died in utero (data not shown). Thus, we conclude that both the heart defect and embryonic lethality of the DKOs results from the loss of pRb and p107 in cells that express Tie2-Cre.
Fig. 5.
Tie2-Cre DKO hearts display DORV phenotypes and show increased proliferation of the heart endocardium. (A and B) Transverse sections of e13.5 Tie2-Cre DKOs through the pulmonary valves (A) and aortic valves (B). Examples of full DORV (Middle) and partial DORV (Bottom) are shown. (Scale bar: 500 μm.) Abbreviations are as in Fig. 3. (C and D) Quantification of BrdU-positive (C) and TUNEL-positive (D) endothelial cells, mesenchymal cells, and cardiomyocytes in the aortic valves from e13.5 Tie2-Cre and Mox2-Cre models. * and ** denote significant increases (P < .02 and < .01, respectively) relative to controls.
We used BrdU incorporation and TUNEL assays to determine the levels of proliferation and apoptosis in the endothelial, mesenchymal, and cardiomyocyte populations in the hearts of e13.5 embryos taken from both the Tie2-Cre and Mox2-Cre models (n ≥ 3 for each genotype) (Fig. 5 B and C and Fig. S6). This analysis revealed a statistically significant increase in the level of proliferating cells in both models that was specific to the DKO endothelial cells near the aortic valve. We also detected significantly higher levels of apoptosis in the mesenchyme and myocardium of both the DKO and p107 mutants in the Mox2-Cre model, but not in the Tie2-Cre model (Fig. 5 B and C and Fig. S6). Thus, we conclude that elevated apoptosis is not required for DORV to occur in DKOs, although it could contribute to the greater penetrance of the DORV phenotype in the Mox2-Cre versus the Tie2-Cre DKOs. Taken together, these data suggest that the incorrect positioning of the aorta, and hence the resulting DORV, is correlated with and likely due primarily to ectopic proliferation of the DKO endothelial cells.
Discussion
Our analysis reveals considerable overlap between Rb and p107 function during embryogenesis. First, the DKOs die between e13.5 and e14.5, much earlier than either Rb or p107 single mutants, which survive to birth or are largely viable, respectively. Second, p107 loss exacerbates some, but not all, of the known defects in Rb-deficient embryos. Specifically, we found a statistically significant increase in ectopic proliferation and apoptosis in both the CNS and ocular lens of DKOs versus Rb mutants, but both genotypes had similar PNS defects. We speculate that the lack of functional overlap in the PNS is due to the poor expression of p107 in this tissue (19) and/or that proliferation and apoptosis are already maximally induced by the loss of pRb function. Consistent with both of these hypotheses, we found that Rb loss yielded a much stronger response in the PNS than in either the CNS or lens with regard to both proliferation (30.8% vs. 9.0% and 21.7%) and apoptosis (26.1% vs. 4.3% and 6.1%) defects. Notably, p107 inactivation does not alter the erythroid phenotype of e13.5 Rb mutants. However, e13.5 tissues that are completely unaffected by Rb loss display ectopic proliferation (blood vessels) or apoptosis (liver and skeletal muscle) defects in the DKOs. Thus, we conclude that p107 can either partially or fully suppress ectopic proliferation and apoptosis in some, but not all, Rb-deficient tissues.
Our analyses also revealed gross abnormalities in DKOs that were absent in Rb mutants. External examination showed that the yolk sacs of DKOs were enlarged and bloated and that some appeared abnormally pale. Moreover, the DKO embryos exhibited a bloated midsection, and a subset showed hemorrhaging. We believe that all of these abnormalities are caused by edema. We also detected major heart defects in the DKOs, specifically severely dilated atria and thin myocardial walls. Finally, the DKOs exhibited DORV, in which both great arteries (the pulmonary artery and the aorta) originate from the right ventricle. DORV is a clinically important congenital heart defect resulting from various cellular events, including proliferation and migration defects of neural crest cells, delayed or increased apoptosis in the myocardium of the conotruncal area, and faulty left/right signaling (20). We have been able to narrow down the defective cell types that likely underlie the DKO DORV phenotype to the endothelial cells and/or the mesenchymal cells of the pulmonary and aortic valves/atrioventricular canal, where the Tie2-Cre is known to be expressed (17, 18). Interestingly, these mesenchymal cells are thought to arise via endothelial to mesenchymal transdifferentiation of a subset of endothelial cells in the vicinity of the cardiac cushion, the future atrioventricular canal and outflow tract (21). Interestingly, we found no impairment in the expression of either blood vessel or cardiac differentiation markers in the DKOs; however, our analysis revealed a significant increase in the percentage of proliferating endothelial cells in both the Tie2-Cre and Mox2-Cre DKOs. This raises the possibility that loss of pRb and p107 allows differentiated cells to remain in cycle, as has been reported previously for Rb-deficient hair cells (22).
Apoptosis can play a crucial role in the alignment of the arteries (20). Of note, although increased apoptosis was noted in the myocardium or the mesenchyme in the Mox2-Cre DKOs, this did not occur in the Tie2-Cre model at e13.5. Thus, abnormal proliferation of the blood vessel and/or heart endothelial cells seems to be sufficient to account for the development of DORV in the DKOs, presumably by disrupting the appropriate positioning of the outflow tract. But elevated apoptosis may be contributing to the greater penetrance of the DORV phenotype in the Mox2-Cre versus the Tie2-Cre DKOs. It seems unlikely that DORV is the sole cause of the observed embryonic lethality of the DKOs; however, we note that the Tie2-Cre DKOs also die in utero. Thus, we conclude that the embryonic lethality results from a defect in one or more of the cell types in which Tie2-Cre is expressed. We speculate that the proliferation defect of the endothelial cells may somehow compromise blood vessel function, and that this acts together with the heart defect to cause edema and embryonic lethality.
In conclusion, our analyses demonstrate that pRb and p107 act together to control the proliferation of many embryonic tissues. In particular, we have identified novel, overlapping functions for pRb and p107 in blood vessel endothelium and endocardial cell proliferation, where loss of these proteins results in DORV, a clinically important congenital heart defect. Interestingly, the combined loss of pRB and p130 pocket proteins is known to disrupt cardiomyocyte differentiation and lead to increased heart size (23). The differential ability of p107 versus p130 to synergize with Rb in heart endothelial cells versus cardiomyocytes likely reflects differences in the expression patterns of p107 and p130 during heart development (23). Importantly, ectopic proliferation was observed in all of the tissues affected by the loss of pRb, pRb and p107, or pRb and p130. This strongly suggests that the overlapping role of the pocket proteins largely reflects their shared role in promoting cell cycle exit. We speculate that the ectopic proliferation results, at least in part, from the deregulation of E2F, but this remains to be established. Similarly, we have yet to understand how pocket protein–deficient cells can differentiate and cycle at the same time.
Materials and Methods
Animal Maintenance and Tissue Analysis.
The Rbc/c, p107−/−, Mox2-Cre, and Tie2-Cre strains have been described previously (12, 13, 18). All mice were crossed in the mixed C57BL/6 × 129/Sv background for 2 or more generations, and the Cre transgenes were transmitted through the males to avoid germline deletion (18). Pregnant mice were injected with 5 mg/mL of BrdU in PBS (10 μL/g of body weight) 2 hours before tissue harvest. Embryos were fixed in 4% paraformaldehyde and embedded in paraffin. Serial 6-μm transverse sections were used to analyze the embryonic heart. All other sections were cut sagittally. Immunohistochemical analysis was performed as described previously (11) using antibodies for BrdU (1:50 347580; BD Biosciences), Ki67 (1:50 550609; BD Biosciences), smooth muscle actin (undiluted U7033; DakoCytomation), and laminin and fibronectin (1:100 rabbit Ab, provided by R. Hynes, MIT). TUNEL analysis was performed as directed by the equipment manufacturer (Roche). The percentage of BrdU- or TUNEL-positive cells was assessed by counting all of the large (>8 cell) blood vessels in the field of view (typically multiple vessels) for blood vessel endothelial cells, 50–100 endothelial cells around the aortic valve, and 250–1,000 nuclei for all other tissues. Significance was determined by the 2-sample t test. Error bars in the graphs represent SDs except for blood vessels, where the SEM was used. For qRT-PCR, e13.5 hearts were homogenized in TRIzol (Invitrogen). First-strand cDNA was transcribed from 1 μg of RNA using SuperScript III Reverse Transcriptase (Invitrogen), and qRT-PCR was performed with 20 ng cDNA using SYBR Green (Applied Biosystems). Primer sequences are listed in Table S1.
Supplementary Material
Acknowledgments.
We thank T. Jacks for the Rb+/c and p107+/− mice, R. Hynes for antibodies, R. Bronson for invaluable histopathologic analysis, and Lees laboratory members for helpful discussions. This work was supported by a National Cancer Institute/National Institutes of Health Grant CA121921 (to J.A.L.). S.D.B. was a David H. Koch Graduate Fellow and J.A.L. is a Ludwig Scholar at MIT.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902408106/DCSupplemental.
References
- 1.Dimova DK, Dyson NJ. The E2F transcriptional network: Old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 2.Jacks T, et al. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 3.Lee MH, et al. Targeted disruption of p107: Functional overlap between p107 and Rb. Genes Dev. 1996;10:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- 4.Cobrinik D, et al. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 5.Dannenberg JH, Schuijff L, Dekker M, van der Valk M, te Riele H. Tissue-specific tumor-suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 2004;18:2952–2962. doi: 10.1101/gad.322004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wikenheiser-Brokamp KA. Retinoblastoma family proteins: Insights gained through genetic manipulation of mice. Cell Mol Life Sci. 2006;63:767–780. doi: 10.1007/s00018-005-5487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macleod KF, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 8.Macleod KF. The role of the RB tumour-suppressor pathway in oxidative stress responses in the haematopoietic system. Nat Rev Cancer. 2008;8:769–781. doi: 10.1038/nrc2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bruin A, et al. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci USA. 2003;100:6546–6551. doi: 10.1073/pnas.1031853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang HS, Hinds PW. pRb-mediated control of epithelial cell proliferation and Indian hedgehog expression in mouse intestinal development. BMC Dev Biol. 2007;7:6. doi: 10.1186/1471-213X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman SD, et al. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res. 2008;6:1440–1451. doi: 10.1158/1541-7786.MCR-08-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sage J, et al. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: A tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Cho S, Atwood JE. Peripheral edema. Am J Med. 2002;113:580–586. doi: 10.1016/s0002-9343(02)01322-0. [DOI] [PubMed] [Google Scholar]
- 15.Papadimou E, Menard C, Grey C, Puceat M. Interplay between the retinoblastoma protein and LEK1 specifies stem cells toward the cardiac lineage. EMBO J. 2005;24:1750–1761. doi: 10.1038/sj.emboj.7600652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 17.Kisanuki YY, et al. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 18.Koni PA, et al. Conditional vascular cell adhesion molecule 1 deletion in mice: Impaired lymphocyte migration to bone marrow. J Exp Med. 2001;193:741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Z, Zacksenhaus E, Gallie BL, Phillips RA. The retinoblastoma gene family is differentially expressed during embryogenesis. Oncogene. 1997;14:1789–1797. doi: 10.1038/sj.onc.1201014. [DOI] [PubMed] [Google Scholar]
- 20.Obler D, Juraszek AL, Smoot LB, Natowicz MR. Double-outlet right ventricle: Aetiologies and associations. J Med Genet. 2008;45:481–497. doi: 10.1136/jmg.2008.057984. [DOI] [PubMed] [Google Scholar]
- 21.Armstrong EJ, Bischoff J. Heart valve development: Endothelial cell signaling and differentiation. Circ Res. 2004;95:459–470. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sage C, et al. Essential role of retinoblastoma protein in mammalian hair cell development and hearing. Proc Natl Acad Sci USA. 2006;103:7345–7350. doi: 10.1073/pnas.0510631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLellan WR, et al. Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol Cell Biol. 2005;25:2486–2497. doi: 10.1128/MCB.25.6.2486-2497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.