Abstract
Patterning of developing tissues arises from a number of mechanisms, including cell shape change, cell proliferation, and cell sorting from differential cohesion or tension. Here, we reveal that differences in cell motility can also lead to cell sorting within tissues. Using mosaic engineered mammary epithelial tubules, we found that cells sorted depending on their expression level of the membrane-anchored collagenase matrix metalloproteinase (MMP)-14. These rearrangements were independent of the catalytic activity of MMP14 but absolutely required the hemopexin domain. We describe a signaling cascade downstream of MMP14 through Rho kinase that allows cells to sort within the model tissues. Cell speed and persistence time were enhanced by MMP14 expression, but only the latter motility parameter was required for sorting. These results indicate that differential directional persistence can give rise to patterns within model developing tissues.
Keywords: differential adhesion, morphogenesis, micropatterning, MT1-MMP, tissue patterning
Extensive cellular rearrangements take place during morphogenesis, both in vivo and in culture. In their landmark 1955 study, Townes and Holtfreter demonstrated that combinations of tissues reconstituted from amphibian embryos would spontaneously sort out according to their germ layers of origin, and in some cases the final configuration resembled that of their native structures in vivo (1). Similar spontaneous sorting events re-established histological patterns from species as divergent as chickens and sponges (2). At that time, the underlying mechanism was hypothesized to be a combination of differential tissue cohesion and differential motility. Subsequent investigations revealed that differential intercellular adhesion mediated by quantitative differences in cell-surface cadherins induced sorting of embryonic cells as well as mammalian cell lines (3, 4). The differential adhesion hypothesis was originally inspired by the similarity of the sorting process to the immiscibility of liquid droplets with different surface tensions (5), a phenomenon that was also consistent with differential contraction rather than adhesion (6). Recently, numerical simulations resurrected the idea that sorting can be mediated also by differences in contractility (7, 8), and experimental analyses have suggested that differential cortical tension may contribute to sorting of the germ layers in zebrafish embryos (9, 10). Differential motility as a mechanism for sorting and self-organization of tissues has been largely ignored, except as a possible explanation for slug formation by Dictyostelium amoebae (11).
Tracking individual cells within whole organ cultures has revealed that vertebrate cells move dynamically against each other and the surrounding extracellular matrix (12–14). In the context of a 3D developing tissue, motility requires the generation of a propulsive force and, in some cases, an active proteolytic mechanism to remove steric barriers. Membrane type-1 matrix metalloproteinase (MT1-MMP; also known as MMP14) binds to or cleaves multiple targets, including the zymogen form of matrix metalloproteinase (MMP)-2; extracellular matrix proteins such as collagen, laminin, and fibronectin; and cell surface receptors including CD44 (reviewed in refs. 15 and 16). MMP14 is up-regulated also in many epithelial tumors, including those from breast, lung, and colon (17–19), and confers cancer cells with the pernicious ability to degrade and penetrate the basement membrane and metastasize to distant sites (20–23). Intriguingly, cells at the invasive front of metastatic cohorts express the highest levels of MMP14 (24, 25). Understanding how the expression pattern of this protease is determined will likely yield insights into possible mechanisms of cancer progression and invasion.
Here we present evidence to suggest that cellular rearrangements generated by differential cellular motility determine the pattern of MMP14-expressing cells within a model mammary epithelial tissue. We use lithography-based culture models that mimic the architecture of mammary epithelial ductal trees to generate mammary tubules mosaic for MMP14 expression. We find that cells rearrange with respect to each other such that the subpopulation highest for MMP14 expression segregates to the ends of tubules. MMP14 levels correlate with directional persistence, which is sufficient to induce sorting in silico. Surprisingly, we find that MMP14-driven sorting is independent of its catalytic activity and requires signaling through Rho kinase (ROCK). Cells within model tissues thus appear to organize depending on differences in their relative motilities.
Results
MMP14-Expressing Cells Sort to the Leading Edge of Engineered Mammary Ducts.
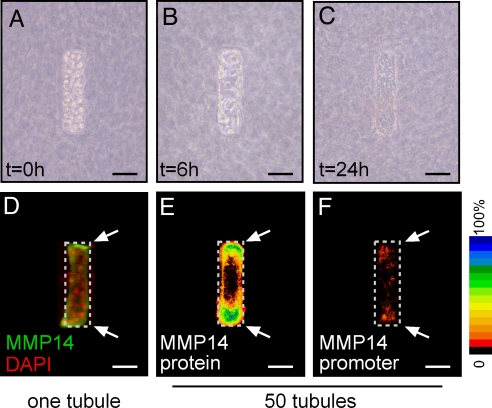
We previously developed an engineered tissue model of the mammary epithelial duct comprised of murine mammary epithelial tubules of arbitrary geometry embedded within a 3D type I collagen gel (26). To generate these tissues, a concentrated suspension of single mammary epithelial cells is placed within micro-scale collagen cavities prepared by replica micro-molding. Initially, individual cells are randomly dispersed within the cavities (Fig. 1A). Over a period of 24 h, the cells form contacts with their neighbors, synthesize and assemble a basement membrane, and rearrange into a polarized epithelial tubule (Fig. 1 B and C) (27). Despite their simplicity, these model tissues recapitulate several aspects of normal mammary histology and morphogenesis (27). Here, we found that, after the 24-h rearrangement period, the expression of MMP14 was highest in the cells located at the ends of the tubules (Fig. 1 D, E). β-Galactosidase staining of tubules constructed of primary mammary epithelial cells isolated from mice heterozygous for LacZ inserted within the MMP14 gene (28) verified that MMP14 promoter activity was highest at the ends (Fig. 1F). MMP activity in general (27) and MMP14 expression in particular are necessary for later morphogenesis of these model tissues, as down-modulating MMP14 expression prevents branching (SI Materials and Methods and Fig. S1).
Fig. 1.
Cell rearrangement and differential expression of MMP14 in engineered mammary epithelial tubules. Phase-contrast images of model tissues at 0 (A), 6 (B), and 24 h (C) after construction. Immunofluorescence analysis of MMP14 in one tubule (D) and quantification of immunofluorescence intensity from 50 tubules (E) represented as a frequency map. (F) MMP14 promoter activity, as determined by β-galactosidase staining, in 50 micro-fabricated organoids, quantified and represented as a frequency map. (Scale bars, 50 μm.)
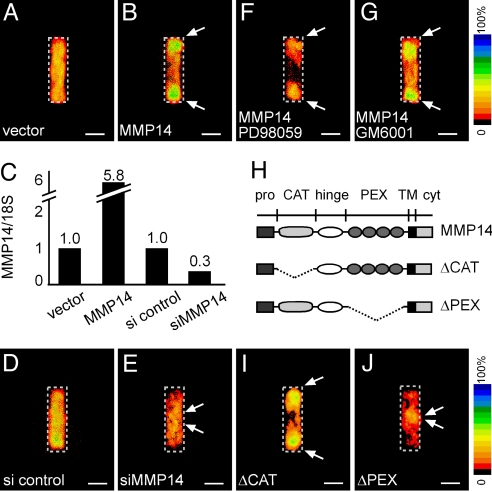
The observed expression pattern of MMP14 could result from either localized induction or cellular rearrangements as the tubule formed. To distinguish between these 2 possibilities, we constructed mosaic tissues in which a labeled subpopulation of cells exogenously expressed higher levels of MMP14 than the endogenous population. Cells were initially randomly distributed within mosaic tissues. However, after 24 h, the MMP14hi subpopulation was restricted to the ends of the tubules (Fig. 2 A–C). The number of cells per tubule did not change significantly over the time course of the experiment (24 h), indicating that the spatial segregation of the 2 populations of cells was caused by sorting rather than differential proliferation or cell death (Fig. S2). In tubules mosaic for siRNA-mediated depletion of MMP14 (siMMP14), the MMP14lo subpopulation was excluded from the ends (Fig. 2 C–E). These data suggest that cellular rearrangements within the tissues are sensitive to relative variations in endogenous levels of MMP14.
Fig. 2.
Tubules mosaic for MMP14 spontaneously self-organize. Frequency map quantifying location of YFP-expressing cells co-transfected with control vector (A) and mouse MMP14 (B). (C) Quantitative RT/PCR analysis for MMP14 expression in cells transfected with control vector, MMP14, siRNA control (si control), and siRNA against MMP14 (siMMP14), normalized to levels of 18S rRNA. Frequency maps quantifying location of YFP-expressing cells co-transfected with siRNA control (si control) (D) and siRNA against MMP14 (siMMP14) (E). Sorting does not require signaling through Erk or MMP proteolytic activity: frequency maps quantifying location of YFP-expressing cells co-transfected with MMP14 and treated with MEK inhibitor PD98059 (F) or broad-spectrum MMP inhibitor GM6001 (40 μM) (G). (H) Diagram of MMP14 constructs with the catalytic domain deleted (ΔCAT) or the hemopexin domain deleted (ΔPEX). Frequency maps quantifying location of YFP-expressing cells co-transfected with ΔCAT (I) or ΔPEX (J) demonstrate that sorting requires the hemopexin domain but is independent of the catalytic domain of MMP14. (Scale bars, 50 μm.)
Sorting of MMP14-Expressing Cells Requires the Hemopexin Domain.
Branching morphogenesis of the model tubules requires coordination between exogenous agonists and endogenous antagonists (27). These cues are surprisingly dispensable for the patterned rearrangements of MMP14-expressing cells. We found that MMP14-induced sorting occurred independently of addition of exogenous growth factors, did not require signaling through Erk (Fig. 2F and Fig. S3), and was unaffected by the previously identified TGF-β inhibitory morphogen gradient (Fig. S4). MMP14-induced sorting was also independent of its proteolytic activity, as sorting was unaffected by treatment with the broad-spectrum MMP inhibitor GM6001 (Fig. 2G). Furthermore, in mosaics comprised of a mutant MMP14 lacking the catalytic domain (ΔCAT), the MMP14hi cells still sorted to the ends of the tubules (Fig. 2 H and I). However, in tubules mosaic for MMP14 deleted of its hemopexin domain (ΔPEX), which mediates binding to extracellular substrates and cell surface receptors (29, 30), transfected cells were sequestered to the shafts of tubules and excluded from the ends (Fig. 2J). Importantly, these cellular rearrangements appeared to be specific to MMP14, as MMP3 was expressed evenly across the tubules, and over-expression or siRNA-mediated down-modulation of MMP3 did not induce sorting (Fig. S5).
MMP14-Mediated Sorting Requires Signaling Through ROCK.
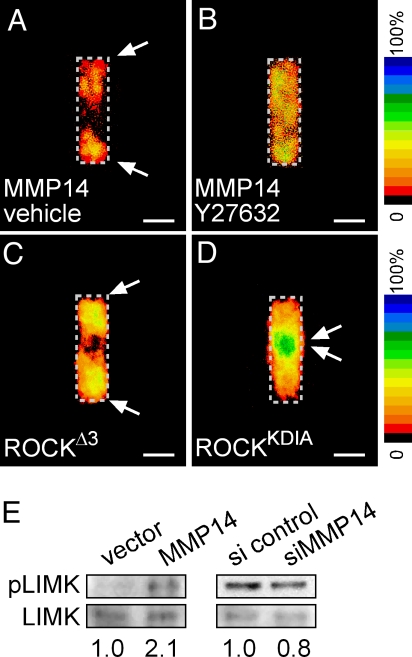
Proteolysis-independent cell motility has been shown to require signaling through the Rho-GTPase effector ROCK (31, 32). We found that MMP14-mediated sorting was blocked also by treatment of the tubules with the ROCK-specific inhibitor Y27632 (Fig. 3 A and B). Furthermore, tubules mosaic for constitutively active ROCKΔ3 exhibited sorting to the ends (Fig. 3C). Conversely, tubules mosaic for dominant negative ROCKKDIA exhibited sorting to the shafts (Fig. 3D). Therefore, ROCK mutant mosaics phenocopy MMP14 mosaics, suggesting that the MMP14 effect is mediated in part by signaling through ROCK. In support of this hypothesis, we found that over-expression of MMP14 resulted in a doubling of the activity of Lim kinase (LIMK), a downstream effector of ROCK, as measured by its relative phosphorylation (Fig. 3E). Conversely, down-modulating MMP14 with siRNA resulted in a modest decrease in LIMK activity (Fig. 3E). Mosaic tubules constructed by simultaneously over-expressing both MMP14 and ROCKKDIA, or siMMP14 and ROCKΔ3, revealed that ROCK was dominant over MMP14 (Fig. S6), confirming that MMP14-mediated sorting was a result of signaling through ROCK.
Fig. 3.
MMP14-mediated sorting requires signaling through ROCK. Frequency maps quantifying location of YFP-expressing cells co-transfected with MMP14 and treated with vehicle(A) or the ROCK inhibitor Y27632 (10 μM) (B). MMP14 sorting is phenocopied by ROCK, as shown in frequency maps of tubules mosaic for constitutively active ROCKΔ3 (C) or dominant negative ROCKKDIA (D). MMP14 activates ROCK signaling, as shown in E Western blots for phosphorylated LIMK (pLIMK) and total LIMK. (Scale bars, 50 μm.)
The PEX domain of MMP14 can induce cancer cell motility in 2D cultures by activating signaling through Rho GTPases, and the cell surface hyaluronan receptor, CD44, has been postulated to play a role in this process (33). MMP14 interacts with CD44 via its PEX domain (30). Furthermore, CD44 has been shown to associate with—and activate signaling through—Rho family GTPases in a number of different cell types (34–37). We found that CD44 was highly expressed at the ends of the tubules and that down-modulating CD44 by siRNA (siCD44) prevented sorting of MMP14 mosaics (Fig. S7). Furthermore, mosaic tubules constructed by over-expressing CD44 or siCD44 phenocopied tubules mosaic for MMP14 over-expression or siMMP14, respectively; CD44-mediated sorting required expression of the MMP14 PEX domain, was inhibited by simultaneous treatment with Y27632 (Fig. S7) and dominated by co-transfection with the ROCK mutants (Fig. S8). Consistent with these data, we found that modulating the level of CD44 altered signaling through ROCK-LIMK (Fig. S7). MMP14 therefore appears to elicit cell sorting in model tissues by signaling through ROCK via association with CD44.
MMP14-Mediated Sorting Involves Differential Cellular Motility.
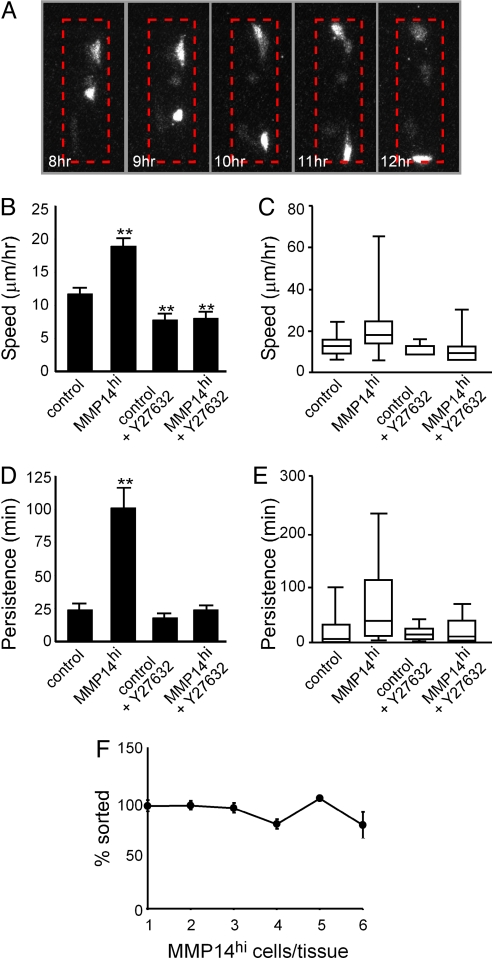
Time-lapse spinning disk confocal analysis confirmed that the MMP14hi subpopulation sorted to the ends of the tubules (Fig. 4A). To track individual cells within the engineered tissues, we developed a line of mammary epithelial cells that stably expressed nuclear localization sequence (NLS)-tagged YFP. NLS-YFP cells formed tubules and underwent branching morphogenesis identical to controls (data not shown). For mosaic tubules, MMP14hi cells were also tagged with CFP. Tracking individual cells within engineered tissues in both YFP and CFP channels demonstrated that MMP14hi cells moved significantly faster (50% increase) and with greater persistence time (>600% increase) than either WT cells or vector-transfected controls (Fig. 4 B–E). These differences disappeared upon treatment with Y27632 (Fig. 4 B–E), suggesting that differential cell motility was responsible for sorting of the tissues. Other mechanisms of sorting, such as differential adhesion, rely on mutual envelopment of cell types through cell-cell cohesion; reducing the number of MMP14hi cells would thus prevent sorting via differential adhesion (38). To distinguish between the various mechanisms, we engineered tissues with limiting numbers of randomly located MMP14hi cells and found that they still sorted, suggesting that differential adhesion was not involved (Fig. 4F).
Fig. 4.
MMP14 expression causes sorting by increasing cell motility. (A) Montage of 5 time points (among ≈90 frames over 20 h) of a single z-section in one mosaic tubule. Shown is the MMP14hi/CFP channel. Dashed red line indicates region of tubule. (B) Average speed of individual control cells, MMP14hi cells, and control and MMP14hi cells in tubules treated with the ROCK inhibitor Y27632. (C) Distribution of cell speed among populations in B. (D) Persistence time of individual control cells, MMP14hi cells, and control and MMP14hi cells in tubules treated with the ROCK inhibitor Y27632. (E) Distribution of persistence time among populations in D. (F) Graph of cell sorting as a function of MMP14hi cells within the tubule. For B, D, and F, error bars indicate SEM of 3 independent experiments. For C and E, edges represent 25th and 75th percentiles and error bars represent 10th and 90th percentiles. **P < 0.005 vs. controls, as determined by t test.
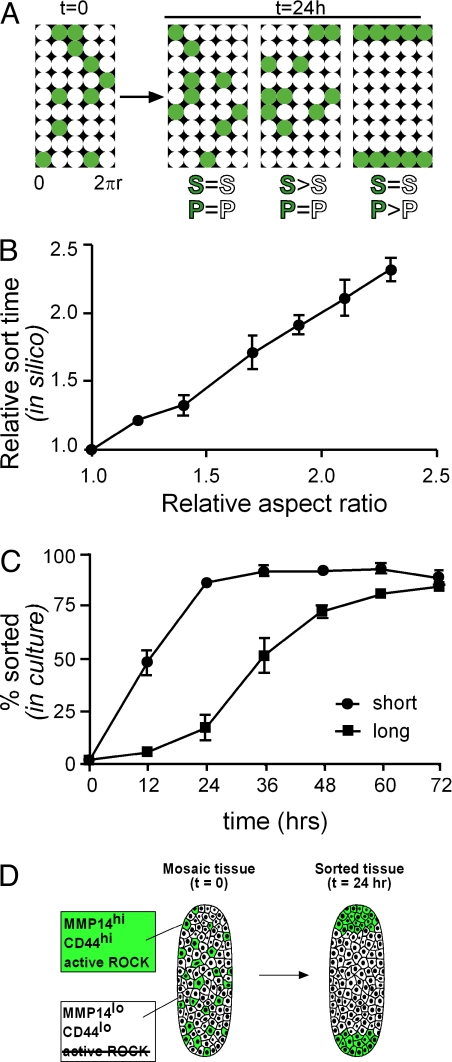
Do differences in cell motility alone lead to sorting? To address this question, we developed an agent-based model of the engineered tissues, comprised of 2 populations of cells each with characteristic speed and persistence time. Cells were initially randomly distributed within in silico tissues (Fig. 5A). We found that sorting occurred with differential persistence time; differential speed alone (within physiologically relevant limits) did not induce cell sorting, but did influence the time scale of the process. The agent-based model predicted that, given 2 populations of cells with differential motility parameters, rate of sorting would scale linearly with length of tissue; that is, short tissues should sort faster than long tissues (Fig. 5B). We tested this in silico-generated hypothesis experimentally by engineering mosaic tissues of short (200 μm) and long (500 μm) geometry, and found that long tissues indeed required approximately 2.5 times as long to sort (Fig. 5C). These results indicate that MMP14-mediated cell sorting depends primarily on differential persistence time.
Fig. 5.
Differential persistence leads to sorting of tissues in silico. (A) Simulations of cell sorting in mosaic tubules comprised of control cells (white) and MMP14hi cells (green). Speed (S) and persistence (P) were varied independently. (B) Average relative time for 20 simulated tissues to sort as a function of their length. (C) Experimental validation of simulation results, shown as percent of cell sorting as a function of time for short (200 μm) and long (500 μm) mosaic tubules. (D) Schematic of sorting within mammary epithelial tubules. Cells with highest levels of MMP14 and CD44 expression have highest levels of ROCK activity and move to the ends of tubules, regions competent for branching. Error bars indicate SEM of 3 independent experiments.
Discussion
This study examines the role of the collective dynamics of individual cells in generating patterns within model tissues. Cellular rearrangements are well accepted as being fundamental to embryonic development. During vertebrate gastrulation, distinct germ layers are formed by sorting of different types of progenitor cells. Cell sorting and tissue organization may result from a number of mechanisms, including differential intercellular adhesion (3, 4) and cortical tension (6, 7, 10). The data presented here demonstrate that differential cellular motility, specifically differential persistence time, can also give rise to distinct patterns of cellular arrangement. The sorting behavior of populations of cells with differences in persistence is akin to separations that result from differences in diffusion coefficients. The MMP14hi cells move in a directed manner, with greater directional persistence, and thus further over the same period; their increased persistence manifests as a reduced propensity to turn, so when they reach the limits of the tissue, they tend to stay there. The concept that differential motility could drive cell sorting within and between vertebrate tissues was proposed long ago by others (1, 39), but to our knowledge has never before been experimentally demonstrated. Sorting mediated by differential motility appears distinct from that mediated by differential adhesion: as predicted by Steinberg (38), tissues can sort via differential motility even if one population is limiting, although the number of MMP14hi cells influences the kinetics of the sorting process (Fig. S9), suggesting that the rate at which a tissue sorts depends on the product of the rates at which each MMP14hi cell moves toward the end. Furthermore, sorting by differential motility depends on tissue properties. The size, geometry, and boundary conditions of a tissue determine the final location of sorted cells and time scales of the sorting process.
Here, we uncovered cell sorting via differential motility using an engineered tissue model of the mammary epithelium. Mammary epithelial cells sort into end-regions of engineered tubules that are high for MMP14 expression, and trunk regions that are lower for MMP14 expression (Fig. 5D). MMP14 expression had no effect on E-cadherin transcript levels or protein distribution within the tissues (Fig. S10), again consistent with a sorting mechanism distinct from differential adhesion. MMP14 expression increases both cell speed and persistence, with a much greater increase (50% vs. >600%) in the latter. Agent-based modeling suggests that the increase in persistence time is sufficient for sorting to occur. In theory, an increase in speed alone could also to lead to sorting as long as the persistence length (i.e., the product of speed and persistence time) was comparable to the length of the tubule. However, the required median cell speeds (≥100 μm/h) are far greater than the median 3D migration speeds typically reported for normal or transformed mammary epithelial cells (≈10–20 μm/h), so it is very unlikely that differential cell speed could suffice for sorting. Recently, MMP14 expression was found to correlate with directional persistence in individual glioblastoma cells within 3D collagen gels (40) and with cell speed and polarized migration during zebrafish gastrulation (41). Persistence could not be separated from the proteolytic function of MMP14 in either of these experimental systems, although the latter showed a link between MMP14 and non-canonical Wnt signaling. Our results assign a novel proteolysis-independent role for MMP14 signaling to cellular persistence, although MMP-independent proteolytic mechanisms may be involved.
We show that MMP14-mediated cell sorting requires the hemopexin domain, which is also essential for MMP14-mediated cellular invasion through collagen (42) and for binding to molecules including CD44 (30). MMP14 and CD44 expression appear to be co-regulated in vivo, correlating with acquisition of a migratory mesenchymal phenotype and reduced time to metastasis in human breast cancers (43). Here, MMP14 and CD44 are both required for cell sorting, activating signaling, and increasing cell motility through ROCK. Cells at the ends of the tubules therefore express the highest levels of MMP14, CD44, and active ROCK. This mechanism may explain why MMP14-expressing cells segregate to the leading edge of metastatic cohorts, as a similar spatial requirement for ROCK activity has been uncovered recently in the collective invasion of cohorts of squamous carcinoma cells (44). It is tempting to speculate that directed migration and invasion of cancer cell collectives depends on sorting by differential motility. The mechanisms by which ROCK controls speed and persistence in mammary epithelial cells are unknown. In other systems, ROCK reorganizes the cytoskeleton, causing stress fiber formation in part through activation of actomyosin contractility (45) and front-rear polarization through activation of PTEN (46). Both could lead to increased motility (47). A complete understanding of patterning of the mammary gland and other organs—as well as engineered tissues and cancer collectives—will require determining how genetic programs (48, 49) and physical and geometric factors (27, 50) interact to regulate cellular rearrangements.
Are quantitative differences in cell motility actually used by developing tissues to control morphogenesis? Few experimental studies have been designed to answer this question, but recent results from a number of systems suggest a possible role for differential motility in tissue patterning. Time-lapse analyses of intact (13) and reconstituted (51) embryonic salivary epithelium and pubertal mammary epithelium (12) have revealed self-organizing dynamics amongst the cell populations. Salivary epithelial cells aggregate in culture and rearrange to form a branching tissue with a histology remarkably similar to that of the intact salivary gland (51); motility differences have been noted for the various epithelial cell types of this tissue (13). In vivo results consistent with the differential motility hypothesis are primarily limited to investigations of chemotaxis. Cells that express the highest levels of FGF receptor in the Drosophila trachea have a chemotactic advantage, allowing them to segregate to the tips of invading branches and to lead the growing branch to localized sources of FGF (52). Collective decisions based on individual differences in the strength of receptor signaling have also been observed in morphogenesis of Drosophila air sacs (53) and egg chambers (54). In the latter, uniform activation of EGF receptor in the border cells results in female infertility by impairing directed migration toward the oocyte (54, 55). Our data suggest that these cellular rearrangements may be driven in part by cell sorting via differential persistence and are not necessarily limited to chemotaxis per se. Recent technological advances in imaging in live animals (56, 57) should help to shed light on this possible mechanism of cell sorting during morphogenesis in vivo.
Materials and Methods
Cell Culture and Reagents.
Functionally normal EpH4 mouse mammary epithelial cells (58) were cultured in 1:1 DMEM/F12, 2% FBS, 5 μg/mL insulin, and 50 μg/mL gentamycin (Sigma). Primary epithelial organoids consisting mainly of luminal epithelial and myoepithelial cells were prepared from 10-week-old virgin MMP14+/lacZ C57BL/6 mice (28) as previously described (59). Micro-fabricated organoids were grown in DMEM/F12 supplemented with ITS and penicillin/streptomycin. For mosaic overexpression studies, EpH4 cells were transiently co-transfected with mouse MMP14, deletion mutants of mouse MMP14 created by PCR, mouse CD44, or ROCK mutants and YFP or YFP alone using Lipofectamine 2000 (Invitrogen) 1 d before micro-fabrication. For mosaic knockdown studies, predesigned siRNA sequences (Ambion) were verified for specific knockdown by at least 80% by quantitative RT-PCR, and co-transfected with YFP using Lipofectamine 2000 1 d before micro-fabrication. A clonal line of EpH4 cells which stably expressed NLS-YFP was selected and established using growth medium containing hygromycin. Tissues were treated with the following reagents diluted to the concentrations indicated in the text: GM6001, Y27632, and PD98059 (all from Calbiochem).
Micro-Fabricated Tubules.
Micro-fabricated cultures of epithelial cells embedded within collagen gels were formed by replica micro-molding as previously described (26, 27). Briefly, patterned elastomeric stamps of polydimethylsiloxane (i.e., Sylgard 184) rendered non-adhesive by coating with a 1% solution of BSA in PBS solution were placed on a drop of liquid neutralized collagen (4 mg/mL; ICN) at 37 °C until gelation. After removing stamps, a concentrated suspension of EpH4 cells or primary organoids was allowed to settle within the micro-molded collagen cavities. Excess cells were rinsed away with culture medium, leaving 65 ± 12 cells per cavity, and a second layer of collagen gel was gently placed on top of the pattern.
Reverse Transcription Followed by Real-Time PCR Analysis.
Total RNA was extracted from cells by using an RNeasy kit (Qiagen). cDNA was synthesized by using SuperScript III first strand synthesis kit (Invitrogen) from equal amounts of RNA. Quantitative real-time PCR analysis was performed with the Lightcycler System using the Lightcycler FastStart DNA Master SYBR Green I kit (Roche). Amplification was followed by melting curve analysis to verify the presence of a single PCR product.
Imaging and Statistical Analysis.
Samples were fixed, stained for nuclei with Hoechst 33258 (Invitrogen), and visualized using an Axiovert Mrm CCD camera attached to a Zeiss Axiovert 200 microscope. Total cumulative data were represented by stacking in registration binarized images of YFP signal from 50 samples, obtaining relative pixel frequency with Scion Image software, and color-coding images in Adobe Photoshop. All experiments were conducted at least 3 times.
For immunofluorescence analysis of MMP14, MMP3, and CD44, samples were fixed in 4% paraformaldehyde, permeabilized with 0.05% Triton X-100, and blocked in 5% goat serum. Antibodies against MMP14 (Chemicon), MMP3 (Chemicon), or CD44 (Santa Cruz Biotechnology) were diluted in 5% goat serum, applied to samples overnight, and removed by extensive washing in blocking buffer. Samples were incubated overnight with secondary antibodies diluted in blocking buffer, washed extensively, and visualized as described earlier.
β-Galactosidase Staining.
Transgenic mice carrying the LacZ gene under control of the MMP14 promoter were used (28). Tubules of primary cells from 12-week-old heterozygous mice (+/-) were collected 24 h after construction in ice-cold PBS solution and fixed for 15 min at room temperature in fix solution (2% formaldehyde, 0.2% glutaraldehyde, 0.02% Nonidet P-40, and 0.01% sodium deoxycholate in PBS solution). After fixation, samples were rinsed several times in PBS solution and then stained overnight at 37 °C in the dark with stain solution (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/ml X-gal, 2 mM MgCl2, 0.02% Nonidet P-40, and 0.01% sodium deoxycholate in PBS solution).
Real-Time Microscopy.
For real-time imaging, tubules were constructed of EpH4 cells that stably expressed NLS-YFP. Time-lapse movies were collected using a Stanford Photonics XR/Mega-10 ICCD camera attached to a Zeiss Axiovert S100 microscope customized with a Yokogawa spinning disk (Solamere Technology Group) and fitted with a humidified environmental chamber held at 37 °C and 5% CO2. Confocal stacks of 20 to 25 images (2 μm thick) were acquired using a Plan Apo 20 × 0.4 NA objective every 15 min beginning at 2 h after initial micro-fabrication for a total of 20 h. Movies were assembled and cells tracked in 3D using ImarisTrack (Bitplane). The average speed (S) and mean-squared displacements (〈d2(t)〉) of individual cells were used to calculate time of directional persistence (P) by fitting to the persistent random walk model (60):
Western Blotting.
Samples were lysed using modified RIPA buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 10 mM sodium pyrophosphate containing 1.5 mM MgCl2, 1 mM EGTA, 1% sodium deoxycholate, 0.25 mM Na3VO4, 100 mM NaF, and proteinase inhibitor mixture). Samples were mixed with Laemmli sample buffer, heated at 95 °C for 5 min, resolved by SDS/PAGE, and transferred to nitrocellulose. Membranes were blocked in milk and incubated overnight at 4 °C in 5% BSA, 0.1% Tween-20 in PBS solution containing antibodies specific to phosphorylated LIMK or total LIMK (Cell Signaling Technology). Primary antibodies were detected with the Pierce SuperSignal detection kit and signal was captured with the FluorChem 8900 analysis system (Alpha Innotech).
Agent-Based Modeling.
Cell dynamics simulations were performed by using NetLogo 4.0 (http://ccl.northwestern.edu/netlogo). The simulation environment consisted of a cylindrical space representing the collagen cavities. Two populations of cells, MMP14hi (green) and MMP14lo (white), were randomly placed in the simulated cavities to mimic the starting conditions of the tissue. The three parameters that could be measured in the culture experiments were duration of culture, cell speed, and directional persistence. These were matched to the 3 parameters that could be varied in silico, which were number of time steps, distance moved per time step, and random rotation at each time step.
Supplementary Material
Acknowledgments.
We thank Joe Tien, Mark Krasnow, Stas Shvartsman, and Cyrus Ghajar for illuminating discussions; and Motoharu Seiki for transgenic mice. This work was supported in part by Department of Energy Grants DE-AC03–76SF00098 and DE-AC02–05CH1123 and a Distinguished Fellow Award (to M.J.B.); by National Institutes of Health (NIH) Grants CA057621, CA064786, CA112970, and CA126552 (to M.J.B.); NIH Grants GM083997 and CA128660 (to C.M.N.); by Department of Defense Grants W81XWH0510338 and W81XWH0810736 (to M.J.B.); Department of Defense Grant W81XWH0410582 (to C.M.N.); by Susan G. Komen for the Cure Grants 02–1591 (to H.M.) and MODS0019923 (to C.M.N.); and by the David & Lucile Packard Foundation (C.M.N.). C.M.N. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.Y. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901269106/DCSupplemental.
References
- 1.Townes PL, Holtfreter J. Directed movements and selective adhesion of embryonic amphibian cells. J Exp Zool. 1955;128:53–120. doi: 10.1002/jez.a.114. [DOI] [PubMed] [Google Scholar]
- 2.Moscona A, Moscona H. The dissociation and aggregation of cells from organ rudiments of the early chick embryo. J Anat. 1952;86:287–301. [PMC free article] [PubMed] [Google Scholar]
- 3.Foty RA, Steinberg MS. The differential adhesion hypothesis: A direct evaluation. Dev Biol. 2005;278:255–263. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Steinberg MS. Differential adhesion in morphogenesis: A modern view. Curr Opin Gene Dev. 2007;17:281–286. doi: 10.1016/j.gde.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Steinberg MS. Reconstruction of tissues by dissociated cells. Some morphogenetic tissue movements and the sorting out of embryonic cells may have a common explanation. Science. 1963;141:401–408. doi: 10.1126/science.141.3579.401. [DOI] [PubMed] [Google Scholar]
- 6.Harris AK. Is cell sorting caused by differences in the work of intercellular adhesion? A critique of the Steinberg hypothesis. J Theor Biol. 1976;61:267–285. doi: 10.1016/0022-5193(76)90019-9. [DOI] [PubMed] [Google Scholar]
- 7.Brodland GW. The Differential Interfacial Tension Hypothesis (DITH): a comprehensive theory for the self-rearrangement of embryonic cells and tissues. J Biomech Eng. 2002;124:188–197. doi: 10.1115/1.1449491. [DOI] [PubMed] [Google Scholar]
- 8.Chen HH, Brodland GW. Cell-level finite element studies of viscous cells in planar aggregates. J Biomech Eng. 2000;122:394–401. doi: 10.1115/1.1286563. [DOI] [PubMed] [Google Scholar]
- 9.Schotz E-M, et al. Quantitative differences in tissue surface tension influence zebrafish germ layer positioning. HFSP J. 2008;2:1–56. doi: 10.2976/1.2834817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krieg M, et al. Tensile forces govern germ-layer organization in zebrafish. Nat Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 11.Umeda T, Inouye K. Cell sorting by differential cell motility: A model for pattern formation in Dictyostelium. J Theor Biol. 2004;226:215–224. doi: 10.1016/j.jtbi.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J Cell Sci. 2006;119:3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- 14.Shakya R, Watanabe T, Costantini F. The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev Cell. 2005;8:65–74. doi: 10.1016/j.devcel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Barbolina MV, Stack MS. Membrane type 1-matrix metalloproteinase: substrate diversity in pericellular proteolysis. Semin Cell Dev Biol. 2008;19:24–33. doi: 10.1016/j.semcdb.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh Y, Seiki M. MT1-MMP: A potent modifier of pericellular microenvironment. J Cell Physiol. 2006;206:1–8. doi: 10.1002/jcp.20431. [DOI] [PubMed] [Google Scholar]
- 17.Sato H, et al. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 18.Okada A, et al. Membrane-type matrix metalloproteinase (MT-MMP) gene is expressed in stromal cells of human colon, breast, and head and neck carcinomas. Proc Natl Acad Sci USA. 1995;92:2730–2734. doi: 10.1073/pnas.92.7.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein CA, et al. Combined transcriptome and genome analysis of single micrometastatic cells. Nat Biotech. 2002;20:387–392. doi: 10.1038/nbt0402-387. [DOI] [PubMed] [Google Scholar]
- 20.Sabeh F, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167:769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueda J, Kajita M, Suenaga N, Fujii K, Seiki M. Sequence-specific silencing of MT1-MMP expression suppresses tumor cell migration and invasion: Importance of MT1-MMP as a therapeutic target for invasive tumors. Oncogene. 2003;22:8716–8722. doi: 10.1038/sj.onc.1206962. [DOI] [PubMed] [Google Scholar]
- 22.Hotary K, Li XY, Allen E, Stevens SL, Weiss SJ. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 2006;20:2673–2686. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabova L, Chrysovergis K, Yamada SS, Holmbeck K. MT1-MMP is required for efficient tumor dissemination in experimental metastatic disease. Oncogene. 2008;27:3274–3281. doi: 10.1038/sj.onc.1210982. [DOI] [PubMed] [Google Scholar]
- 24.Nabeshima K, et al. Front-cell-specific expression of membrane-type 1 matrix metalloproteinase and gelatinase A during cohort migration of colon carcinoma cells induced by hepatocyte growth factor/scatter factor. Cancer Res. 2000;60:3364–3369. [PubMed] [Google Scholar]
- 25.Friedl P, Wolf K. Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 26.Nelson CM, Inman JL, Bissell MJ. Three-dimensional lithographically defined organotypic tissue arrays for quantitative analysis of morphogenesis and neoplastic progression. Nat Protoc. 2008;3:674–678. doi: 10.1038/nprot.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yana I, et al. Crosstalk between neovessels and mural cells directs the site-specific expression of MT1-MMP to endothelial tip cells. J Cell Sci. 2007;120:1607–1614. doi: 10.1242/jcs.000679. [DOI] [PubMed] [Google Scholar]
- 29.Tam EM, Wu YI, Butler GS, Stack MS, Overall CM. Collagen binding properties of the membrane type-1 matrix metalloproteinase (MT1-MMP) hemopexin C domain. The ectodomain of the 44-kDa autocatalytic product of MT1-MMP inhibits cell invasion by disrupting native type I collagen cleavage. J Biol Chem. 2002;277:39005–39014. doi: 10.1074/jbc.M206874200. [DOI] [PubMed] [Google Scholar]
- 30.Mori H, et al. CD44 directs membrane-type 1 matrix metalloproteinase to lamellipodia by associating with its hemopexin-like domain. EMBO J. 2002;21:3949–3959. doi: 10.1093/emboj/cdf411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 32.Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol. 2006;16:1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 33.Cao J, et al. Distinct roles for the catalytic and hemopexin domains of membrane type 1-matrix metalloproteinase in substrate degradation and cell migration. J Biol Chem. 2004;279:14129–14139. doi: 10.1074/jbc.M312120200. [DOI] [PubMed] [Google Scholar]
- 34.Bourguignon LY, Zhu H, Shao L, Zhu D, Chen YW. Rho-kinase (ROK) promotes CD44v(3,8–10)-ankyrin interaction and tumor cell migration in metastatic breast cancer cells. Cell Motil Cytoskel. 1999;43:269–287. doi: 10.1002/(SICI)1097-0169(1999)43:4<269::AID-CM1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 35.Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem. 2006;281:14026–14040. doi: 10.1074/jbc.M507734200. [DOI] [PubMed] [Google Scholar]
- 36.Bourguignon LY, Singleton PA, Zhu H, Diedrich F. Hyaluronan-mediated CD44 interaction with RhoGEF and Rho kinase promotes Grb2-associated binder-1 phosphorylation and phosphatidylinositol 3-kinase signaling leading to cytokine (macrophage-colony stimulating factor) production and breast tumor progression. J Biol Chem. 2003;278:29420–29434. doi: 10.1074/jbc.M301885200. [DOI] [PubMed] [Google Scholar]
- 37.Singleton PA, Bourguignon LY. CD44v10 interaction with Rho-kinase (ROK) activates inositol 1,4,5-triphosphate (IP3) receptor-mediated Ca2+ signaling during hyaluronan (HA)-induced endothelial cell migration. Cell Motil Cytoskel. 2002;53:293–316. doi: 10.1002/cm.10078. [DOI] [PubMed] [Google Scholar]
- 38.Steinberg MS. On the mechanism of tissue reconstruction by dissociated cells. I. Population kinetics, differential adhesiveness. and the absence of directed migration. Proc Natl Acad Sci USA. 1962;48:1577–1582. doi: 10.1073/pnas.48.9.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stefanelli A, Zacchei AM, Ceccherini V. Retinal reconstitution in vitro after disaggregation of embryonic chicken eyes (Translated from Italian) Acta Embryol Morphol Exper. 1961;4:47–55. [Google Scholar]
- 40.Kim HD, et al. Epidermal growth factor-induced enhancement of glioblastoma cell migration in 3D arises from an intrinsic increase in speed but an extrinsic matrix- and proteolysis-dependent increase in persistence. Mol Biol Cell. 2008;19:4249–4259. doi: 10.1091/mbc.E08-05-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coyle RC, Latimer A, Jessen JR. Membrane-type 1 matrix metalloproteinase regulates cell migration during zebrafish gastrulation: Evidence for an interaction with non-canonical Wnt signaling. Exp Cell Res. 2008;314:2150–2162. doi: 10.1016/j.yexcr.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 42.Wang P, Nie J, Pei D. The hemopexin domain of membrane-type matrix metalloproteinase-1 (MT1-MMP) Is not required for its activation of proMMP2 on cell surface but is essential for MT1-MMP-mediated invasion in three-dimensional type I collagen. J Biol Chem. 2004;279:51148–51155. doi: 10.1074/jbc.M409074200. [DOI] [PubMed] [Google Scholar]
- 43.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 45.Pellegrin S, Mellor H. Actin stress fibres. J Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, et al. Regulation of PTEN by Rho small GTPases. Nat Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 47.Worthylake RA, Burridge K. RhoA and ROCK promote migration by limiting membrane protrusions. J Biol Chem. 2003;278:13578–13584. doi: 10.1074/jbc.M211584200. [DOI] [PubMed] [Google Scholar]
- 48.Metzger RJ, Krasnow MA. Genetic control of branching morphogenesis. Science. 1999;284:1635–1639. doi: 10.1126/science.284.5420.1635. [DOI] [PubMed] [Google Scholar]
- 49.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson CM, et al. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci USA. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei C, Larsen M, Hoffman MP, Yamada KM. Self-organization and branching morphogenesis of primary salivary epithelial cells. Tissue Eng. 2007;13:721–735. doi: 10.1089/ten.2006.0123. [DOI] [PubMed] [Google Scholar]
- 52.Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441:746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- 53.Cabernard C, Affolter M. Distinct roles for two receptor tyrosine kinases in epithelial branching morphogenesis in Drosophila. Dev Cell. 2005;9:831–842. doi: 10.1016/j.devcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Bianco A, et al. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- 55.Duchek P, Rorth P. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science. 2001;291:131–133. doi: 10.1126/science.291.5501.131. [DOI] [PubMed] [Google Scholar]
- 56.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 57.Egeblad M, et al. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Model Mech. 2008;1:155–167. doi: 10.1242/dmm.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reichmann E, Ball R, Groner B, Friis RR. New mammary epithelial and fibroblastic cell clones in coculture form structures competent to differentiate functionally. J Cell Biol. 1989;108:1127–1138. doi: 10.1083/jcb.108.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simian M, et al. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128:3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stokes CL, Lauffenburger DA, Williams SK. Migration of individual microvessel endothelial cells: Stochastic model and parameter measurement. J Cell Sci. 1991;99(Pt 2):419–430. doi: 10.1242/jcs.99.2.419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.