Abstract
Radical S-adenosine-L-methionine (SAM or AdoMet) proteins are involved in chemically difficult reactions including the synthesis of cofactors, the generation of protein radicals, and the maturation of complex organometallic catalytic sites. In the first and common step of the reaction, a conserved [Fe4S4] cluster donates an electron to perform the reductive cleavage of AdoMet into methionine and a reactive radical 5′-dA· species. The latter extracts a hydrogen atom from substrate eliciting one of the about 40 reactions so far characterized for this family of proteins. It has been suggested that the radical-generating mechanism differs depending on whether AdoMet is a cofactor or a substrate. It has also been speculated that electron transfer from the [Fe4S4] cluster to AdoMet is sulfur-based. Here we have used protein crystallography and theoretical calculations to show that regardless whether AdoMet serves as a cofactor or a substrate, the 5′-dA· generating mechanism should be common to the radical SAM proteins studied so far, and that electron transfer is mediated by a unique Fe from the conserved [Fe4S4] cluster. This unusual electron transfer is determined by the sulfonium ion in AdoMet.
Keywords: density functional theory, hybrid potentials, iron sulfur cluster, redox chemistry, S-adenosyl-L-methionine
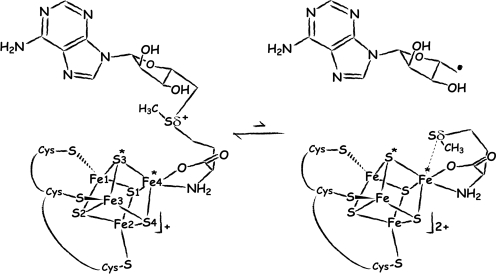
The catalytic cycle of all of the radical S-adenosyl-L-methionine (SAM or AdoMet) enzymes (1–3) involves the formation of a 5′-deoxyadenosyl radical species (5′-dA·), resulting from the reductive cleavage of AdoMet. Diverse reactions are catalyzed by this family of proteins, such as sulfur insertion, isomerization and protein radical formation; all initiated by the abstraction of a hydrogen atom from the substrate by the highly reactive 5′-dA·. The electron donor to form the radical species is a reduced [Fe4S4]+ cluster, coordinated by a generally conserved Cys-X3-Cys-X2-Cys motif (4). Model chemistry has shown that reductive cleavage of sulfonium-bearing compounds by reduced [Fe4S4] clusters is a facile reaction, although it is generally a two-electron process (5). In addition, electrophilic attack of sulfonium to thiolates has been also noted in these model systems but not in the enzymes. These observations underscore the role of the protein in modulating the one-electron reaction that leads to 5′-dA· formation. A 2H and 13C ENDOR study of pyruvate formate-lyase activating enzyme (PFL-AE) predicted the distances between the closest iron atom of the [Fe4S4] cluster and the methyl protons and carbon atom of AdoMet to be approximately 3.0 Å to 3.8 Å and approximately 4–5 Å, respectively (6). Using these data, a model for the interaction between AdoMet and the cubane was postulated where the carboxylate moiety of AdoMet established a bidentate interaction with an iron ion from the cluster. The ENDOR-based model was subsequently refined using additional isotopic labeling in the same enzyme to include an interaction of the carboxy and amino groups of AdoMet (7) with a unique iron site, identified by Mössbauer spectroscopy (8). An additional interaction between one of the sulfides of the cluster and the sulfonium ion (Sδ+) of AdoMet was also postulated and it was argued that this interaction could be involved in sulfur-based inner sphere electron transfer from the cluster to AdoMet (6, 7). The X-ray structures of the AdoMet-bound Radical SAM enzymes HemN (9), BioB (10), MoaA (11), and LAM (12) have confirmed the prediction that the [Fe4S4] cluster interacts with the carboxy and amino groups of AdoMet. However, as it will be discussed below, no close interaction between S (S3* in Fig. 1) and Sδ+ (6, 7) has been observed in these structures.
Fig. 1.
Depiction of AdoMet radical cleavage. Fe4* indicates the unique iron site; S3* and Sδ+/Sδ are discussed in the text ([Fe4S4] cluster numbering as in PDB code 3CIW).
Selenium K-edge X-ray absorption spectroscopy (XAS) experiments carried out with lysine 2,3-aminomutase (LAM) revealed a Se-Fe interaction at 2.7 Å after cleavage of S-adenosyl-L-selenomethionine (13). The absence of this interaction in PFL-AE and BioB (14), was thought to reflect the role of AdoMet during the reaction. AdoMet is a substrate for both PFL-AE that generates a glycyl radical in PFL (15) and for BioB that inserts a sulfur atom in dethiobiotin (16). Conversely, AdoMet is a cofactor for LAM that catalyzes the interconversion of α-L-lysine and β-L-lysine (17) and uses it as a reversible source of 5′-dA·. A structural basis for the XAS-based difference in selenomethionine binding after AdoMet cleavage was proposed using the ENDOR model described above: after cleavage, the methionine Sδ would interact with one iron ion from the cluster in LAM (13) and it would be either more distant or dissociated from the [Fe4S4] cluster in PFL-AE and BioB (14). In all these structures, and in the recently published structures of the [FeFe]-hydrogenase active site maturase HydE from Thermotoga maritima (18) and PFL-AE complexed with a peptide (19), AdoMet binds the conserved [Fe4S4] cluster in the same way. This observation favors methionine dissociation because it is not obvious why, upon AdoMet cleavage, methionine should bind differently in any of these proteins.
The 1.35-Å resolution HydE structure (18) provided a high resolution image of a radical AdoMet enzyme with bound S-adenosyl-L-homocysteine (SAH) (in the remainder the AdoMet cleavage reaction site will be called the active site). Following our report on that crystal structure, and to better define the binding of cleavage products in a Radical SAM protein, we have solved the structures of HydE bound to AdoMet and [5′-dA + Met] up to 1.62 Å and 1.25-Å resolution, respectively (Fig. 2). These high resolution structures have subsequently been used to investigate in detail the AdoMet cleavage mechanism by computational methods.
Fig. 2.
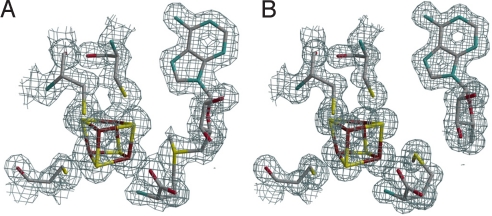
Active site X-ray structures. 2Fo-Fc electron density maps on the x-ray model of (A) AdoMet-bound HydE and (B) [5′-dA + Met]-bound HydE. Map contours were drawn at the 1 σ level with a cover radius of 1.8 Å. The three cysteine residues, the iron sulfur cluster and AdoMet or [5′-dA + Met] are represented as sticks; C gray, Fe brown, N blue, O red, S yellow.
Results
Crystal Structures of the AdoMet-Bound and [5′-dA + Met]-Bound HydE.
Table 1 summarizes data collection and refinement statistics (see Materials and Methods). Fig. 2 A and B depict the active sites of AdoMet-bound and [5′-dA + Met]-bound HydE, respectively. We observe that, as in all other available AdoMet-bound X-ray structures (Table 2), the distance between the AdoMet sulfonium ion and the unique iron of the cluster is shorter than the S3*–Sδ+ distance. As shown in Fig. 2, there are some differences between the [5′-dA + Met]-HydE complex and its AdoMet counterpart. 5′-dA binds to HydE approximately as the adenosine part of AdoMet does (SI Text). However, free methionine binding differs significantly from its covalently bound counterpart in AdoMet. Whereas the Sδ from methionine is still far from the closest [Fe4S4] sulfur atom S3* at 3.50 Å, its distance to the unique iron, Fe4*, is only 2.67 Å (Fig. 1). When AdoMet-bound and [5′-dA + Met]-bound HydE structures are compared, the distortion of the cluster is more pronounced in the latter, resulting from pseudooctahedral coordination at Fe4* (SI Text). We have already discussed a similar situation in the SAH-bound HydE structure (18). A structure of the active site after AdoMet cleavage has been postulated based on a crystallographic study of MoaA in complex with its substrate 5′GTP, obtained after reduction of the [Fe4S4] clusters (20). However, our examination of the corresponding electron density strongly suggests the presence of uncleaved AdoMet in one of the two MoaA molecules of the asymmetric unit and the absence of AdoMet-related species in the other (SI Text). A very recent report by Challand et al. (21) has shown that in the case of three AdoMet-dependent proteins [BioB, lipoyl synthase (LipA) and tyrosine lyase (ThiH)] that use AdoMet as a substrate, the products of its cleavage, methionine and 5′-dA, are inhibitory in vitro. The inhibition was removed by the enzyme 5′-methylthioadenosine S-adenosylhomocysteine nucleosidase that hydrolyses 5′-dA. BioB, LipA, and ThiH are expected to have lower binding constant for methionine and 5′-dA than enzymes that use AdoMet as a cofactor. Indeed, although it is not explicitly stated in their paper, at equimolar concentrations (30 μM) BioB was only about 10% inhibited by products [Fig. 2C in (21)]. This is to be compared with the XAS study of LAM (13) where, at equimolar concentrations, there was a strong Fe-Se peak at d = 2.7 Å resulting from the interaction of Se-Met and the [Fe4S4] cluster. Experiments equivalent to those of Challand et al. (21) have not been reported for LAM or other reversible radical AdoMet enzymes. However, we have observed extremely tight binding of methionine and 5′-dA to HydE in our crystals. Even after a prolonged back-soak of a crystal in its crystallization solution devoid of these molecules, the electron density map was identical to the one shown in Fig. 2B. Although the role of this protein in [FeFe]-hydrogenase maturation is unknown, this observation suggests that HydE uses AdoMet as a cofactor.
Table 1.
Crystallographic data and refinement statistics
| Crystal |
||
|---|---|---|
| AdoMet | [5′-dA + Met] | |
| Space group | P212121 | |
| Data collection | ||
| Beamline | ID14-eh2 | ID29 |
| Cell parameters | ||
| a, Å | 50.63 | 51.05 |
| b, Å | 78.78 | 78.92 |
| c, Å | 85.94 | 86.19 |
| Wavelength, Å | 0.93 | 0.976 |
| Resolution, Å | 1.62 | 1.25 |
| Rsym | 0.066 (0.369) | 0.045 (0.407) |
| I/σ(Ι) | 13.39 (2.38) | 14.87 (2.10) |
| Completeness, % | 93.5 (72.9) | 95.1 (76.8) |
| Multiplicity | 2.5 (1.4) | 3.2 (1.45) |
| Refinement statistics | ||
| Rcryst | 0.137 | 0.139 |
| Rfree | 0.184 | 0.166 |
| No. of reflections | ||
| Work set | 38,939 | 83,859 |
| Test set | 2,204 | 4,675 |
| rmsd from ideal geometry | ||
| Bonds, Å | 0.014 | 0.012 |
| Angles, ° | 1.749 | 1.675 |
| No. of non-hydrogen atoms | 3,472 | 3,545 |
| Water molecules | 446 | 467 |
| Iron atoms | 6 | 4 |
| Others | 193 | 197 |
The two data sets are 97.5% and 98.8% complete at 1.72 Å and 1.33 Å with 93.9% and 96.7% completeness in their respective highest resolution data shells. However, data extended to 1.62 Å and 1.25 Å resolution, respectively, and were thus used up to these resolution limits for X-ray structure refinement. This allowed the inclusion of 101,20 and 24,234 additional reflections, respectively. Values in parentheses refer to the highest resolution shell (SI Text, Table S1).
Table 2.
AdoMet binding
| Protein Reference | HemN† (2.07) (ref. 9) | BioB (3.40) (ref. 10) | LAM (2.10) (ref. 12) | MoaA (2.20) (ref. 11) | PFL-AE (2.77) (ref. 19) | HydE (1.62) (this work) |
|---|---|---|---|---|---|---|
| Sδ-S3* | 3.64 | 4.77 | 3.79 | 3.84 | 3.97 | 3.66 |
| Sδ-Fe4* | 3.35 | 4.03 | 3.15 | 3.19 | 3.22 | 3.25 |
| OAdoMet-Fe4* | 2.13 | 2.51 | 1.98 | 1.97 | 2.17 | 2.25 |
| NAdoMet-Fe4* | 2.33 | 2.35 | 1.98 | 2.30 | 2.12 | 2.33 |
Resolution of the X-ray structures (in parentheses) and distances are given in Å.
†These distances correspond to the ones obtained after our re-setting of the HemN Fe-S distances (SI Text).
Calculations on Enzyme Models.
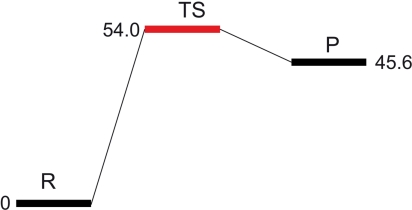
Hybrid quantum mechanical (QM)/molecular mechanical (MM) potentials have been used to investigate numerous enzymatic reaction mechanisms (22, 23). We have used the QM/MM approach as implemented in the Schrödinger Suite [(a) Maestro, version 8.5; (b) QSite, version 5.0; (c) Jaguar, version 7.5, Schrödinger, LLC, New York, NY, 2008] (see Materials and Methods), along with the crystal structures of AdoMet and [5′-dA + Met] HydE complexes, to model AdoMet cleavage (SI Text, Fig. S1). In Table 3, we report the key distances at the active site for the geometry-optimized structures of a model of the reactant (R) and the radical product (P), as depicted in Fig. 1. Table 3 also shows key distances for the transition state structure (TS). Its energy, relative to the R state, corresponds to the electron transfer activation barrier between R and P. We observed only minor deviations between the QM/MM optimized models of R and P and the starting X-ray models; thus, the N/O anchoring is maintained, the AdoMet binding does not vary and 5′-dA· and 5′-dA adopt similar conformations. The distortion of the [Fe4S4] cluster due to methionine binding to Fe4* is slightly more pronounced in the calculated models than in the X-ray structures (Table 3) while the other S-Fe iron distances in the cluster are standard (d = 2.3 Å). The calculated barrier height for AdoMet cleavage is about 54.0 kJ/mol (Fig. 3), similar to the estimated 37.7 and 54.4 kJ/mol barriers for AdoMet cleavage in LAM, in the presence and absence of substrate, respectively (24). The long Sδ-C5′ distance in the TS state (3.3 Å) indicates that the bond is already broken and the spin density at C5′ confirms that the electron has already been transferred (Fig. 4A). In addition, the TS and P structures are very similar, the only difference residing in the position of Sδ that is further away from the radical C5′ in the latter. This similarity explains the small 8.4 kJ/mol difference between the two states (Fig. 3).
Table 3.
QM/MM models
| X-ray† | X-ray‡ | R§ | TS¶ | P‖ | |
|---|---|---|---|---|---|
| NAdoMet-Fe4* | 2.33 | 2.26 | 2.30 | 2.24 | 2.25 |
| OAdoMet-Fe4* | 2.25 | 2.27 | 2.15 | 2.13 | 2.14 |
| Sδ-Fe4* | 3.25 | 2.67 | 3.20 | 2.62 | 2.63 |
| Sδ-C5′ | 1.82 | 3.78 | 1.84 | 3.33 | 4.00 |
| Sδ-S3* | 3.66 | 3.50 | 3.70 | 3.61 | 3.58 |
| S3*-Fe4* | 2.28 | 2.35 | 2.42 | 2.48 | 2.47 |
| S1-Fe4* | 2.25 | 2.39 | 2.58 | 2.61 | 2.62 |
| S4-Fe4* | 2.33 | 2.44 | 2.43 | 2.60 | 2.62 |
Key distances (ångstoms) in the active site for the reactant (R = AdoMet-bound HydE), the transition state (TS) and the product (P = [5′-dA·+ Met]-bound HydE) compared to X-ray (see Fig. 1 for atom labels).
†X-ray model of AdoMet-bound to HydE.
‡X-ray model of [5′-dA· + Met]-bound to HydE.
§QM/MM optimize structure of R.
¶QM/MM optimized structure of TS.
‖QM/MM optimized structure of P.
Fig. 3.
Barrier height for the formation of [5′-dA· + Met] from AdoMet. Energies relative to that of R are in kJ/mol.
Fig. 4.
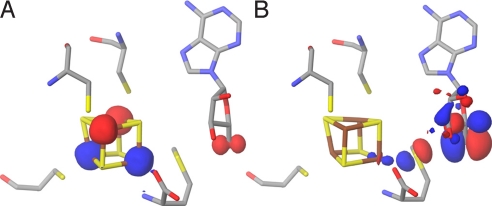
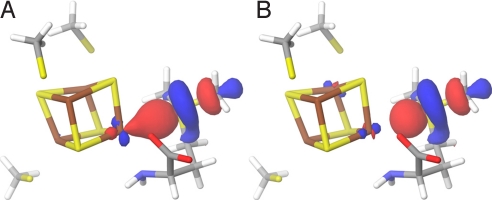
Transition state. (A) The spin density at the active site of the QM/MM-optimized structure of the transition state. (B) HOMO at the active site of the QM/MM-optimized structure of the transition state (surfaces in red [+] and blue [-]; atom color codes are as in Fig. 1).
The highest occupied molecular orbital (HOMO) of the TS gives an indication of how the reducing electron is delocalized when going from R to P. As can be seen in the TS HOMO of Fig. 4B, the main contributions to the orbital come from the radical-bearing C5′ (see Table S2), the intercalating Sδ from methionine and the unique Fe4*. To better understand how the reducing electron would preferentially be transferred during cleavage, we looked at the HOMO and the lowest unoccupied molecular orbital (LUMO) of R. As a first approximation, these delocalized orbitals can be conceptualized as resulting from the interaction between the putative donor (centered on Fe4*) and acceptor (centered on Sδ+ and C5′) localized orbitals (SI Text). However, technically, the perturbation induced by the protein matrix treated with MM prevents us from analyzing the LUMOs. Because we wanted to further investigate the role of Fe4* in the electron transfer upon AdoMet cleavage (Fig. 4B), we constructed a small model of the reactant that could be treated quantum mechanically (Fig. S2A). We extracted from the QM/MM optimized reactant structure the three cysteines, the iron sulfur cluster and we simplified AdoMet to S-methylmethionine (SI Text). Consistent with the composition of the TS HOMO (Fig. 4B), the R HOMO is mainly centered on Fe4*. As expected, both α (spin up) and β ((spin down) LUMOs are centered on the S-methylmethionine, with a small Fe4* contribution (Fig. 5). However, Sδ+ does not contribute significantly to the HOMO of this reactant model system. Moreover, we found little spin density on Sδ+ (Table S3), although for electron transfer to occur, non-zero orbital overlap between donor and acceptor is required.
Fig. 5.
Active site model. LUMO (A) α (spin up) and (B) β (spin down) for the system composed of [Fe4S4]+ coordinated by three methyl thiolates to model the cysteine residues and S-methylmethionine to model AdoMet.
This result can be rationalized by the well known fact that the B3LYP potential used for these QM calculations tends to underestimate the extent of covalency, and, as a consequence, it minimizes orbital overlap, notably in iron sulfur clusters (25). Consequently, we performed additional calculations on the same model with the more delocalized VBP potential implemented in the ADF QM code (26) (see Materials and Methods). This potential is expected to give a more accurate description of the orbitals (SI Text). Using this approach, 17% of the total spin population was found to reside on Sδ+ (Table S4). In addition, the Sδ+ pz character appears in both the HOMO and LUMO, indicating that the electron transfer to C5′ involves a direct path from Fe4* via the Sδ+ orbitals. Finally, given that the X-ray structures of SAH (18) and AdoMet superimpose well (SI Text), we built a second minimal model to evaluate the effect of changing the charge on Sδ from + 1 in the sulfonium of the R model to 0 in the thioether of SAH (Fig. S2B). In this model, we found no spin population on Sδ (even when using the VBP potential), indicating that when this atom is neutral it does not contribute to the HOMO/LUMO (Table S3 and Table S4). In both minimal models, the orbitals corresponding to the N/O atoms of methionine lie at a lower energy and, consequently, do not contribute to the HOMO/LUMO involving Fe4* and Sδ.
Discussion
The calculated barrier for AdoMet cleavage of 54.0 kJ/mol agrees well with an estimate of the corresponding barrier in LAM (24). This value should be compared to the reported 133.9 kJ/mol for AdoMet cleavage in solution. The reaction energy profile obtained in this study, with a TS close in energy to the P state, clearly indicates that the reaction is concerted and can easily proceed backwards, regenerating AdoMet. This is in agreement with the fact that, in all radical SAM proteins that have been studied so far, AdoMet is stable and difficult to cleave in the absence of substrate (27).
The model built from distances found between the methyl group of AdoMet and the [Fe4S4] cluster in the 2H and 13C ENDOR study of PFL-AE (6) needs to be revised. Firstly, this model postulated that only one of the iron ions (corresponding to Fe1 in Fig. 1) interacted with the C atom from the methyl group. However, in all available AdoMet-bound radical SAM enzyme crystal structures, both Fe4* and Fe1 are at about 4 Å from the methyl C atom of AdoMet and therefore, both are expected to contribute to the distant dipolar hyperfine tensor of the 13C atom. Secondly, the ENDOR-based model predicted that the redox chemistry of electron transfer from the cluster to the AdoMet was sulfur-centered (6, 28). Our calculations show that it is not the case in HydE and, given the common AdoMet binding, it should not apply to any of the radical SAM proteins of known three-dimensional structure. Indeed, there is no evidence for anything more than a van der Waals contact between S3* and Sδ (Fig. 1) in any of these proteins. The sulfur-centered mechanism (29) has been justified using data published by Noodleman and Case (30). In their study of a [Fe4S4]+ cluster coordinated by four cysteines, the reducing electron charge was found to be mainly located on the sulfide ions, suggesting they play a role in electron transfer from the cluster. Our system differs from Noodleman and Case's in that AdoMet bidentate binding to one of the Fe ions replaces cysteine ligation and places a sulfonium ion close to the cluster (Fig. S3). We found that in the bound AdoMet the Sδ+ orbital is lower in energy than the S2− orbital (SI Text) and the Sδ+ and Fe4* orbitals have matching energies, a necessary condition for electron transfer between two atoms to occur. In our [5′-dA + Met]-bound HydE structure the distance between Sδ and Fe4* is 2.67 Å. This compares well with the 2.7 Å distance between Se and the unique iron found in the LAM XAS study mentioned above (13). In addition, the reported models for AdoMet and cleavage product complexes with LAM (12, 24) are stereochemically correct, as ascertained by our HydE structure. In fact, the two HydE structures reported here satisfy all of the spectroscopic data reported so far for radical SAM proteins.
Taken together, our results indicate that the AdoMet cleavage mechanism should be common to, at least, those radical SAM proteins with known three-dimensional structures (Table 2). We also think that the most parsimonious conclusion would be that the freed methionine binds to the [Fe4S4] cluster, even if only transiently, always as observed in our [5′-dA + Met]-bound HydE complex. Consequently, we postulate that, besides the nature of the reaction, the role of AdoMet as either a cofactor or a substrate is determined by the relative affinity of the enzyme for [5′-dA + Met] and AdoMet and not by the cleavage mechanism itself. We interpret the fact that a Se-Fe interaction was not detected in the XAS study of PFL-AE and BioB using equimolar concentrations of protein and products (14), as an indication of the low affinity of methionine for the AdoMet binding site in these enzymes. The recent report by Challand et al. (21) on product inhibition of irreversible AdoMet-dependent radical proteins gives an idea of this affinity. Total inhibition of BioB with products was observed when about a 10-fold excess of the latter was used. It would be of interest to repeat the XAS studies on BioB (14) using an equivalent excess of products. Knowing the relative affinities of reversible and irreversible AdoMet enzymes for their products would help in the classification of new members of this family.
Our calculations highlight the crucial role of the unique iron site Fe4* from which the electron is transferred to AdoMet. They also underscore the influence of its Sδ+ that specifically changes the electronic distribution and favors iron-centered over the standard sulfur-centered (30) redox chemistry for electron transfer. Structural and functional data from other radical SAM proteins will be necessary to establish whether, as may be expected from our results, the cleavage mechanism postulated here is common to all of the members of this family.
Materials and Methods
For details see SI Text.
Crystal Sample Preparation.
The HydE protein from Thermotoga maritima (TmHydE) protein was purified as previously described (18, 31). Before crystallization either S-adenosyl-L-methionine (AdoMet) (2 mM) or 5′-deoxyadenosine (5′-dA) (10 mM) was added to a 10 mg/mL TmHydE protein solution. To obtain the [5′-dA + Methionine]-HydE complex, methionine was added to the pretreated 5′-dA-HydE protein solution in the crystallization drops (final concentration 30 mM). Crystals were grown and flash-cooled as previously described (18).
Data Collection and Model Refinement.
The two X-ray data sets used here were collected at the European Synchrotron Radiation Facility. Data were processed with XDS (32) (see Table 1 for statistics). Both the AdoMet and the [5′-dA + Met] complex structures were solved using our previously published 1.35-Å resolution structure (18) (PDB code 3CIW). Because all of the crystals are isomorphous, there was no need to apply an initial rigid-body refinement procedure. The S-adenosyl-L-homocysteine (SAH) and the water molecules filling the barrel cavity were removed from structure factor and phase calculations, before refinement. Subsequent manual building, including fitting of the AdoMet or [5′-dA + Met] into the observed electron density map, followed by refinement with REFMAC (33), led to the final models with statistics presented in Table 1.
Theoretical Calculations.
Calculations on the enzyme models were performed with the QSite code in the Schrödinger Suite [(a) Maestro, version 8.5; (b) QSite, version 5.0; (c) Jaguar, version 7.5, Schrödinger, LLC, New York, NY, 2008]. A hybrid potential combining (i) density functional theory (DFT) with the B3LYP functional to describe the atoms at the active site and (ii) molecular mechanics to include the effect of the protein matrix were used to model the reactant R and the product P of the AdoMet cleavage reaction. We used the LACVP** basis set (34) on metals and 6–31G** on all other atoms for geometry optimization. For single-point energy calculations at geometry-optimized structures, we chose the larger basis set of triple-ζ quality, LACV3P** (Schrödinger Suite) for metals and cc-pVTZ(-f) (35) for all other QM atoms. The rest of the system was treated with the OPLS2005 force field (36).
To comfort the interpretation of our results with the enzyme, we also performed the above computations on small QM models (Fig. S2A and B). Along with using the Gaussian-based Jaguar code in the Schrödinger suite, we ran the Slater-based ADF code (26) (triple-ζ basis sets for all atoms) with the Vosko, Wilk, and Nusair (VWN) LDA functional (37, 38) and Becke-Perdew exchange-correlation potential (39, 40) (combination referred to as VBP). While the mixed Hartree-Fock/VWN B3LYP potential gives good energetics, the standard ‘pure’ DFT VBP potential is better in terms of orbital characterization (25, 41). Reality (so to speak) lies in between.
Supplementary Material
Acknowledgments.
We thank Ms. Carole Mathevon and Pr. Marc Fontecave for providing purified HydE protein and for fruitful discussions. This work was financed by contract ANR-08-BLAN-0224-01 from the French Agence Nationale pour la Recherche and institutional funds from the Commissariat à l'Energie Atomique et le Centre National de la Recherche Scientifique.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3IIZ and 3IIX).
This article contains supporting information online at www.pnas.org/cgi/content/full/0904385106/DCSupplemental.
References
- 1.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicolet Y, Drennan CL. AdoMet radical proteins—from structure to evolution—alignment of divergent protein sequences reveals strong secondary structure element conservation. Nucleic Acids Res. 2004;32:4015–4025. doi: 10.1093/nar/gkh728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frey PA, Hegeman AD, Ruzicka FJ. The radical SAM superfamily. Crit Rev Biochem Mol Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 4.Ollagnier-de Choudens S, et al. Reductive cleavage of S-adenosylmethionine by biotin synthase from Escherichia coli. J Biol Chem. 2002;277:13449–13454. doi: 10.1074/jbc.M111324200. [DOI] [PubMed] [Google Scholar]
- 5.Daley CJA, Holm RH. Reactions of site-differentiated [Fe4S4](2+,1+) clusters with sulfonium cations: Reactivity analogues of biotin synthase and other members of the S-adenosylmethionine enzyme family. J Inorg Biochem. 2003;97:287–298. doi: 10.1016/s0162-0134(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 6.Walsby CJ, et al. Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe-4S](+) cluster of pyruvate formate-lyase activating enzyme. J Am Chem Soc. 2002;124:3143–3151. doi: 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- 7.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. An anchoring role for FeS clusters: Chelation of the amino acid moiety of S-adenosylmethionine to the unique iron site of the [4Fe-4S] cluster of pyruvate formate-lyase activating enzyme. J Am Chem Soc. 2002;124:11270–11271. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 8.Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH. Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase activating enzyme: A Mossbauer spectroscopic study. J Am Chem Soc. 2002;124:912–913. doi: 10.1021/ja017562i. [DOI] [PubMed] [Google Scholar]
- 9.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes. EMBO J. 2003;22:6214–6224. doi: 10.1093/emboj/cdg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanzelmann P, Schindelin H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci USA. 2004;101:12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lepore BW, Ruzicka FJ, Frey PA, Ringe D. The X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale. Proc Natl Acad Sci USA. 2005;102:13819–13824. doi: 10.1073/pnas.0505726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosper NJ, Booker SJ, Ruzicka F, Frey PA, Scott RA. Direct FeS cluster involvement in generation of a radical in lysine 2,3-aminomutase. Biochemistry. 2000;39:15668–15673. doi: 10.1021/bi0022184. [DOI] [PubMed] [Google Scholar]
- 14.Cosper MM, et al. Structural studies of the interaction of S-adenosylmethionine with the [4Fe-4S] clusters in biotin synthase and pyruvate formate-lyase activating enzyme. Protein Sci. 2003;12:1573–1577. doi: 10.1110/ps.0302203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey M, Rothe M, Wagner AFV, Knappe J. Adenosylmethionine-dependent synthesis of the glycyl radical in pyruvate formate-lyase by abstraction of the glycine C-2 pro-S hydrogen atom—Studies of [H-2]glycine-substituted enzyme and peptides homologous to the glycine-734 site. J Biol Chem. 1994;269:12432–12437. [PubMed] [Google Scholar]
- 16.Guianvarch D, Florentin D, Bui BTS, Nunzi F, Marquet A. Biotin synthase, a new member of the family of enzymes which uses S-adenosylmethionine as a source of deoxyadenosyl radical. Biochem Biophys Res Com. 1997;236:402–406. doi: 10.1006/bbrc.1997.6952. [DOI] [PubMed] [Google Scholar]
- 17.Frey PA, Magnusson OT. S-Adenosylmethionine: A wolf in sheep's clothing, or a rich man's adenosylcobalamin? Chem Rev. 2003;103:2129–2148. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- 18.Nicolet Y, et al. X-ray structure of the [FeFe]-hydrogenase maturase hydE from Thermotoga maritima. J Biol Chem. 2008;283:18861–18872. doi: 10.1074/jbc.M801161200. [DOI] [PubMed] [Google Scholar]
- 19.Vey JL, et al. Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc Natl Acad Sci USA. 2008;105:16137–16141. doi: 10.1073/pnas.0806640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanzelmann P, Schindelin H. Binding of 5 '-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc Natl Acad Sci USA. 2006;103:6829–6834. doi: 10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Challand ML, et al. Product inhibition in the radical S-adenosylmethionine family. FEBS Lett. 2009;583:1358–1362. doi: 10.1016/j.febslet.2009.03.044. [DOI] [PubMed] [Google Scholar]
- 22.Friesner RA, Guallar V. Ab initio quantum chemical and mixed quantum mechanics/molecular mechanics (QM/MM) methods for studying enzymatic catalysis. Annu Rev Phys Chem. 2005;56:389–427. doi: 10.1146/annurev.physchem.55.091602.094410. [DOI] [PubMed] [Google Scholar]
- 23.Senn HM, Thiel W. Atomistic Approaches in Modern Biology: From Quantum Chemistry to Mol Simuls. Berlin: Springer-Verlag Berlin; 2007. pp. 173–290. [Google Scholar]
- 24.Wang SC, Frey PA. Binding energy in the one-electron reductive cleavage of S-adenosylmethionine in lysine 2,3-aminomutase, a radical SAM enzyme. Biochemistry. 2007;46:12889–12895. doi: 10.1021/bi701745h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szilagyi RK, Winslow MA. On the accuracy of density functional theory for Iron-Sulfur clusters. J Comp Chem. 2006;27:1385–1397. doi: 10.1002/jcc.20449. [DOI] [PubMed] [Google Scholar]
- 26.te Velde GT, Baerends EJ. Numerical-integration for polyatomic systems. J Comp Phys. 1992;99:84–98. [Google Scholar]
- 27.Jarrett JT. The generation of 5 '-deoxyadenosyl radicals by adenosylmethionine-dependent radical enzymes. Curr Opin Chem Biol. 2003;7:174–182. doi: 10.1016/s1367-5931(03)00022-x. [DOI] [PubMed] [Google Scholar]
- 28.Buis JM, Broderick JB. Pyruvate formate-lyase activating enzyme: Elucidation of a novel mechanism for glycyl radical formation. Arch Biochem Biophys. 2005;433:288–296. doi: 10.1016/j.abb.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 29.Walsby CJ, et al. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “Radical-SAM” protein superfamily. Inorg Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 30.Noodleman L, Case DA. Density-functional theory of spin polarization and spin coupling in iron-sulfur clusters. Adv Inorg Chem. 1992;38:423–470. [Google Scholar]
- 31.Rubach JK, Brazzolotto X, Gaillard J, Fontecave M. Biochemical characterization of the HydE and HydG iron-only hydrogenase maturation enzymes from Thermotoga maritima. FEBS Letts. 2005;579:5055–5060. doi: 10.1016/j.febslet.2005.07.092. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Hay PJ, Wadt WR. Ab initio effective core potentials for molecular calculations—Potentials for K to Au including the outermost core orbitals. J Chem Phys. 1985;82:299–310. [Google Scholar]
- 35.Dunning TH. Gaussian-basis sets for use in correlated molecular calculations. 1. The atoms boron through neon and hydrogen. J Chem Phys. 1989;90:1007–1023. [Google Scholar]
- 36.Jorgensen WL, Maxwell DS, Tirade-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 37.Painter GS. Density functional description of molecular bonding within the local spin-density approximation. J Phys Chem. 1986;90:5530–5535. [Google Scholar]
- 38.Vosko SH, Wilk L, Nusair M. Accurate spin-dependent electron liquid correlation energies for local spin-density calculations—A critical analysis. Can J Phys. 1980;58:1200–1211. [Google Scholar]
- 39.Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 40.Perdew JP, Yue W. Accurate and simple density functional for the electronic exchange energy—Generalized gradient approximation. Phys Rev B. 1986;33:8800–8802. doi: 10.1103/physrevb.33.8800. [DOI] [PubMed] [Google Scholar]
- 41.Orio M, Mouesca JM. Variation of average g values and effective exchange coupling constants among [2Fe-2S] clusters: A density functional theory study of the impact of localization (trapping forces) versus delocalization (double-exchange) as competing factors. Inorg Chem. 2008;47:5394–5416. doi: 10.1021/ic701730h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.