Abstract
Glutamate racemase (RacE) is responsible for converting L-glutamate to D-glutamate, which is an essential component of peptidoglycan biosynthesis, and the primary constituent of the poly-γ-D-glutamate capsule of the pathogen Bacillus anthracis. RacE enzymes are essential for bacterial growth and lack a human homolog, making them attractive targets for the design and development of antibacterial therapeutics. We have cloned, expressed and purified the two glutamate racemase isozymes, RacE1 and RacE2, from the B. anthracis genome.
Through a series of steady-state kinetic studies, and based upon the ability of both RacE1 and RacE2 to catalyze the rapid formation of D-glutamate, we have determined that RacE1 and RacE2 are bona fide isozymes. The x-ray structures of B. anthracis RacE1 and RacE2, in complex with D-glutamate, were determined to resolutions of 1.75 Å and 2.0 Å. Both enzymes are dimers with monomers arranged in a “tail-to-tail” orientation, similar to the B. subtilis RacE structure, but differing substantially from the A. pyrophilus RacE structure. The differences in quaternary structures produce differences in the active sites of racemases among the various species, which has important implications for structure-based, inhibitor design efforts within this class of enzymes. We found a Val to Ala variance at the entrance of the active site between RacE1 and RacE2 which results in the active site entrance being less sterically hindered for RacE1. Using a series of inhibitors, we show that this variance results in differences in the inhibitory activity against the two isozymes and suggest a strategy for structure-based inhibitor design to obtain broad-spectrum inhibitors for glutamate racemases.
Keywords: Bacillus anthracis, glutamate racemase isozymes, cofactor-independent racemase, antimicrobial drug design, broad spectrum inhibitors
Introduction
Bacillus anthracis is a gram-positive bacterium that is the causative agent of the disease anthrax. The Centers for Disease Control and Prevention categorize B. anthracis as a critical biothreat agent and have classified it as a high-priority, category A agent, based upon its potential for causing mass casualties in the event of a bioterrorist attack 1. The ability of microbes to acquire antibiotic resistance, as well as the feasibility of selecting multi-drug resistant strains of anthrax 2; 3; 4; 5, has created a need for novel antimicrobial drugs that target these lethal bacteria.
D-glutamate is an essential, biosynthetic building-block for all gram-positive and gram-negative bacteria. It is incorporated into the peptidoglycan monomeric unit by the MurD enzyme, and is necessary for the successful production of the peptidoglycan (murein) component of bacterial cell walls 6. In addition, D-glutamate is the primary constituent of the B. anthracis poly-γ-D-glutamyl capsule, which is one of the two virulence factors of the disease anthrax, and functions to protect the organism against the bactericidal components of serum and phagocytic engulfment 7. The enzyme glutamate racemase (RacE) appears to be the primary source of D-glutamate for cell wall biosynthesis, and is also unique to bacteria, making it a potentially attractive target for antimicrobial drug-design 8; 9; 10.
Many bacteria, including Escherichia coli, contain a single gene for glutamate racemase, which is the sole provider of D-glutamate for the cell and is thus vital for survival 11. A few bacteria, including B. cereus and B. anthracis, have two glutamate racemase genes that express different isoenzymes. Complete, independent inactivation of the two, putative glutamate racemase genes (racE1 and racE2) in the B. anthracis ΔANR strain 12, demonstrated that racE1 knock-outs lead to a moderate growth defect that can be fully restored by addition of D-glutamate 13. In contrast, the racE2 knock-out was found to severely inhibit growth that could only be partially restored by the addition of D-glutamate. These results suggest that the racE2 gene product is essential for survival of the pathogen, at least in standard laboratory rich medium.
Glutamate racemase is a member of a rare family of cofactor-independent racemases and epimerases 14. This family, which also includes aspartate racemase, proline racemase and diaminopimelate epimerase, undertakes the difficult task of abstracting the α-proton from substrates that have pKas as high as 21 15; 16. The well-studied, cofactor-dependent enzyme alanine racemase utilizes the cofactor pyridoxal 5′-phosphate (PLP) as an electron sink to acidify the α-proton and lower the free energy of activation 17. The ability of glutamate racemase and other racemases in this class to overcome such a hurdle without a cofactor is intriguing.
Extensive mechanistic studies on glutamate racemase from Lactobacillus fermenti demonstrated that racemization of glutamate proceeds via a deprotonation/reprotonation mechanism similar to that of alanine racemase 18. Additional experiments established that two cysteine residues function via a two-base mechanism in which one cysteine serves as a catalytic base, abstracting the C-2 proton from one face of glutamate, while the second catalytic cysteine delivers a proton to the opposite face, resulting in the inversion of configuration 19. The proposed catalytic cysteines, C73 and C184, were mutated to alanines, and the mutant enzymes were found to be devoid of any detectable activity with the natural substrates, indicating that these residues play critical roles in RacE catalysis. Interestingly, these mutants were capable of stereospecifically deprotonating opposite enantiomers of threo-3-chloroglutamate 20. This data, coupled with the results from asymmetric, kinetic-isotope effects performed using the C73S and C184S RacE mutants 15, indicate that the α-proton of D-glutamate is abstracted by C73, whereas C184 is responsible for the deprotonation of L-glutamate. Kinetic analysis of mutations of other highly conserved residues, specifically D10N, D36N, E152Q, and H186N showed significant changes in either kcat, Km, or both, indicating that these residues are involved in substrate binding or that they play important roles in catalysis, 21; 22.
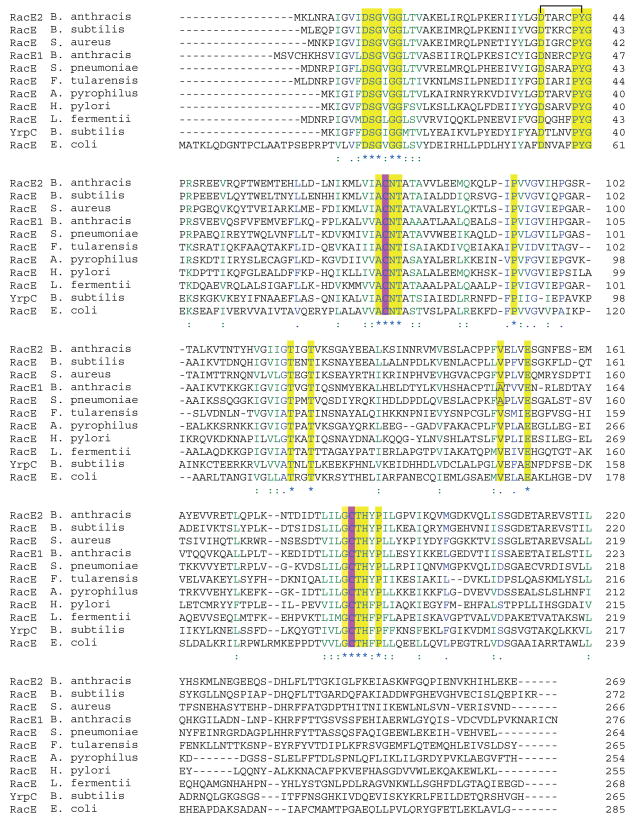
The two putative isozymes of glutamate racemase from B. anthracis, RacE1 and RacE2, share 67% homology in their amino acid sequences, and significant homology to other RacE enzymes including the well-studied enzyme from Lactobacillus fermenti (Figure 1). All of the aforementioned amino acid residues are 100% conserved and are deemed to be critical for RacE catalysis. The location of these amino acids within the active site has been confirmed via the x-ray structure of glutamate racemase from the thermophile Aquifex pyrophilus 21 and more recently from the RacE structure from B. subtilis 23. The A. pyrophilus structure provided the first detailed structural information on the active site of glutamate racemase which is formed, in part, by the “head-to-head” juxtaposition of two RacE monomers, with the active sites facing each other along the dimer interface 21. In contrast, the RacE structure from Bacillus subtilis has been reported to form a ‘tail-to-tail’ dimer, with the catalytic sites of the two monomers facing outward into solution, and with a somewhat different arrangement of residues within the catalytic site 23. A recent conference report indicates that RacE isozymes isolated from different bacterial species have different functional oligomers, with RacE from E. coli functioning as a monomer, H. pylori RacE functioning as a head-to-head dimer, and RacE from both E. faecalis and S. aureus functioning as tail-to-tail dimers with somewhat differing dimeric interfaces 9.
Figure 1.
Multiple sequence alignments of RacE. The catalytic cysteines are highlighted in purple and all other strictly conserved residues are highlighted in yellow. The black bar connecting D37 and Y43 (B. anthracis RacE2 sequence numbering) denotes a hydrogen bond that is responsible for maintaining the positioning of loop 4, which is important for substrate recognition.
To more accurately define the positions of amino acids within the active site and to determine the quaternary structure arrangements of the two RacE isozymes from B. anthracis, we co-crystallized them in the presence of the product, D-glutamate, and determined their x-ray structures to a resolution of 1.75 Å (RacE1) and 1.99 Å (RacE2). The quaternary structures of RacE1 and RacE2 are similar to the quaternary structure of the RacE isozyme from B. subtilis 23. We have also characterized the oligomeric and kinetic properties of both RacE1 and RacE2 to more clearly understand their functional roles in B. anthracis. Moreover we also propose a strategy for structure-based drug design to obtain inhibitors that may have a broader spectrum than ones reported to date 8; 9; 10.
Results
Purification of RacE1, RacE2 and R2-V149A mutant Enzymes
The two glutamate racemase isozymes, RacE1 and RacE2, and the R2-V149A mutant enzyme from Bacillus anthracis were overexpressed in E. coli BL21(DE3) and purified to homogeneity in a single step via a Co2+-charged Hi-Trap affinity column. The final purification yields of RacE1, RacE2, and R2-V149A were 75 mg, 100 mg and 70 mg per liter of TB culture respectively, with a final purity of over 95% based on SDS-PAGE analysis (data not shown).
Confirmation of RacE1 and RacE2 as glutamate racemase enzymes
To confirm that the cloned genes are indeed glutamate racemases, the activities of RacE1 and RacE2 toward other L-amino acids were determined using the CD assay. When the change in molar ellipticity of high concentrations of the amino acids glutamine, aspartate, asparagine, serine, alanine, proline or lysine were monitored upon the addition of high concentrations of B. anthracis RacE1 or RacE2, only the ellipticity of D- or L-glutamate changed (data not shown). These results, and a comparison of the kinetic properties of RacE1 and RacE2 with those of other glutamate racemases (Table 2), confirm that RacE1 and RacE2 are bona fide glutamate racemase isozymes.
Table 2.
Comparison of the quaternary structure and kinetic parameters of Bacillus anthracis glutamate racemase isozymes RacE1 & RacE2 to enzymes isolated from other organisms.
| a Organism | Isozyme | b Oligomer Structure | c Assay & Direction |
kcat (s−1) |
kcat e ratio |
Km (mM) |
Kme ratio |
kcat/Km (s−1M−1) |
kcat/Kme ratio |
|---|---|---|---|---|---|---|---|---|---|
| B. anthracis | RacE2 | Dimer |
L to D | 38 ± 5 | 24 | 3.7 ± 0.8 | 19 | (10.3 ± 2.6) × 103 | 1.3 |
| D to L | 1.6 ± 0.05 | 0.2 ± 0.03 | (8.0± 1.2) × 103 | ||||||
| d D to L | 1.3 ± 0.02 | 0.2 ± 0.01 | (6.5 ± 0.34) × 103 | ||||||
| RacE1 | M ↔ D |
L to D | 17.7 ± 0.3 | 4.5 | 8.0 ± 0.4 | 9 | (2.2± 0.12) × 103 | 0.5 | |
| D to L | 3.9 ± 0.07 | 0.9± 0.1 | (4.3± 0.91) × 103 | ||||||
| d D to L | 2.8 ± 0.08 | 1.9 ± 0.2 | (1.5± 0.16) × 103 | ||||||
| B. subtilis | RacE | M ↔ D |
L to D | 580 | 21 | 50 | 20 | 11.6 × 103 | 1.0 |
| D to L | 28 | 2.5 | 11.2 × 103 | ||||||
| f YrpC | Monomer |
L to D | .02 | 0.13 | 0.18 | 0.15 | 1.1 × 102 | 0.8 | |
| D to L | 0.16 | 1.2 | 1.3 × 102 | ||||||
| L. fermenti | RacE | Monomer |
L to D | 69 | 1.0 | 0.33 | 1.3 | 2.1 × 105 | 0.8 |
| D to L | 68 | 0.26 | 2.6 × 105 | ||||||
| d D to L | 76 | 0.24 | 3.2 × 105 | ||||||
| A. pyrophilus | RacE | M ↔ D |
L to D | 0.25 | 3.5 | 3.3 | 6 | 63 ± 28 | 0.6 |
| D to L | 0.06 | 0.55 | 102 ± 13 | ||||||
| E. coli | f RacE |
a Monomer |
g L to D | 3.1 | 0.31 | 4 | 0.28 | 7.8 × 102 | 1.0 |
| g D to L | 9.8 | 14 | 7.0 × 102 | ||||||
| S. pneumoniae | RacE | h NR | L to D | NR | - | 18 | 6.2 | - | - |
| D to L | NR | - | 2.9 | - | - | ||||
| H. pylori | RacE |
a Dimer |
L to D | NR | - | NR | - | - | - |
| D to L | NR | - | NR | - | - | - | |||
| E. faecalis | RacE |
a Dimer |
L to D | NR | - | NR | - | - | - |
| D to L | NR | - | NR | - | - | - | |||
| S. aureus | RacE |
a Dimer |
L to D | NR | - | NR | - | - | - |
| D to L | NR | - | NR | - | - | - |
The references for the other organism are; L. fermenti (20, 21, B. subtilis (36), A. pyrophilus, E. coli (11, 34), S. pneumoniae (10), H. pylori (11), E. faecalis (11), and S. aureus (11).
The quaternary structure for each isozyme is listed as either a monomer (M), dimer (D) or as a monomer-dimer equilibrium (M ↔ D).
RacE1 and RacE2 from B. anthracis were assayed using two different assay methods as described in materials and methods. The forward (L to D) and reverse (D to L) rates were both measured by the circular dichroism assay. The reverse rate was also measured by the coupled-enzyme assay. For the CD assay, the buffer contained 10 mM potassium phosphate, pH 8.2, and 0.2 mM DTT.
Rate constants determined via the coupled-enzyme assay. For B. anthracis, the assay contained 50 mM CHES buffer, pH 9.2, 5 mM NAD+, 37.5 units of bovine L-glutamate adehydrogenase, 2.5 mM ADP, 0.65 mM INT, 2 units of diaphorase, and D-glutamic acid at variable concentrations.
The ratios are calculated from the kinetic parameters determined from the circular dichroism assay in the forward (L to D) direction divided by the values for the reverse (D to L) direction.
B. subtilis YrpC and E.coli RacE also function as DNA gyrase inhibitors (40, 41).
The enzymatic activity of E.coli RacE in both directions is absolutely dependent on the binding of the peptidoglycan precursor UDP-N-acetylmuramoyl-L-alanine (34).
NR Not Reported
Oligomeric State of RacE1 and RacE2 Enzymes in Solution
The quaternary structures of glutamate racemase from a variety of organisms are reported to be monomers in solution in the absence of substrates 8; 9; 18; 24; 25; 26. However, RacE from B. subtilis forms a dimer in solution in the presence of saturating concentrations of either D- or L-glutamate or D- or L-glutamine, but it remains a monomer in the presence of aspartate 26. The RacE enzyme from A. pyrophilus, a monomer under low salt concentrations, has also been reported to form a dimer under high salt concentrations 25. The formation of a dimeric structure for the A. pyrophilus enzyme was also inferred from the x-ray crystal structure 21. Since the oligomeric state of glutamate racemase is seemingly variable among species and isozymes, and since equilibrium between a monomer and a dimer has been reported for B. subtilis and A. pyrophilus RacE enzymes, the quaternary structures of B. anthracis RacE1 and RacE2 were determined in the absence and presence of saturating concentrations of D,L-glutamate by analytical, size-exclusion chromatography. In the absence of substrates and at a concentration of 10 mg/mL, B. anthracis RacE1 exists almost exclusively as a monomer with an apparent molecular weight of 37.5 ± 2.0 kDa. However, when RacE1 is saturated with the substrate-product, L,D-glutamate, there is an equilibrium shift from a monomer to almost exclusively a dimer with an apparent molecular weight of 62.9 ± 0.3 kDa. In contrast, RacE2 exists entirely as a dimer both in the absence (62.4 ± 1.8 kDa) and presence (62.6 ± 0.4 kDa) of saturating concentrations of L,D-glutamate. A summary of the results for RacE1 and RacE2 from B. anthracis is presented in Table 2 along with the results available for glutamate racemase from other organisms.
Kinetic Properties of RacE1 and RacE2 Enzymes
The steady-state kinetic parameters, kcat and Km, for the forward (L to D) and reverse (D to L) reactions catalyzed by B. anthracis RacE1 and RacE2 are summarized in Table 2 along with the kinetic parameters determined for glutamate racemase isozymes from other organisms. The steady-state kinetic parameters were determined using two different kinetic assays. The circular dichroism assay was used to determine the kinetic parameters for the forward and reverse reactions. For the forward direction, RacE2 has a two-fold higher kcat than RacE1 (38 sec−1 versus 17.7 sec−1), and a two-fold lower Km value for L-glutamate than RacE1 (3.7 mM vs. 8.0 mM). In contrast, for the reverse reaction, RacE1 has an approximately 2.5-fold higher kcat than RacE2 (3.9 sec−1 versus 1.6 sec−1 respectively), and a Km value for D-glutamate that is approximately 4.5-fold higher than that for for RacE2 (0.9 mM versus 0.2 mM). A similar trend in the kinetic parameters for the reverse reaction is also observed using the coupled-enzyme assay, which was performed at a slightly different pH value (Table 2). The ratios of the kinetic parameters, kcat/Km, for the forward and reverse reactions of RacE1 and RacE2 are near unity as would be predicted from the Briggs-Haldane relationship 27.
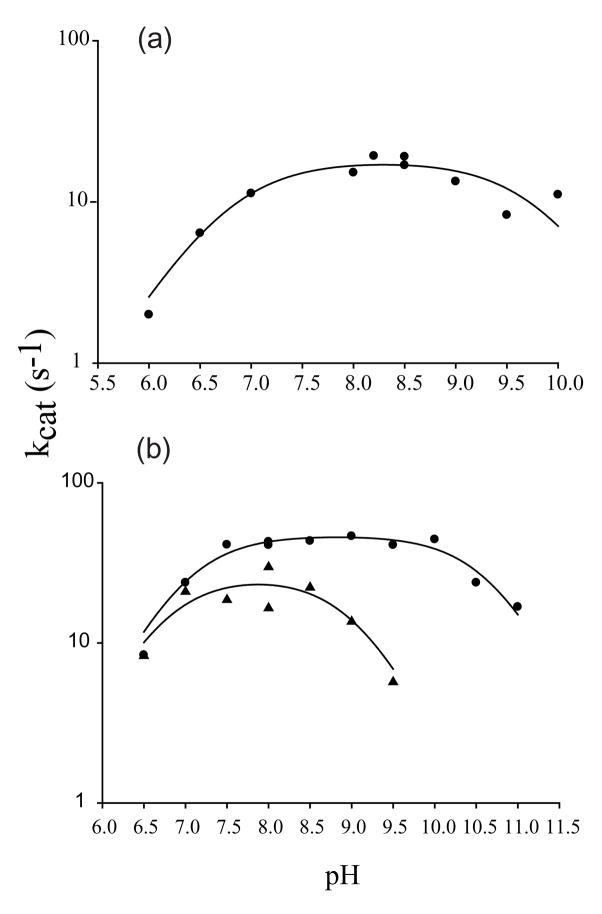
The pH rate profiles were determined for the forward reactions catalyzed by RacE1 and RacE2 using the CD assay. The rates of the reaction were determined at saturating concentrations of L-glutamate over the pH range of 6.0 to 11.0. Plots of kcat versus pH follow typical bell-shaped curves with two ionizable groups in the ES complex (Figure 2). The pKa values in the ES complex were determined by fitting the data for RacE1 and RacE2 to Equation 2. The resulting pKa values for RacE1 are pKa1 = 6.8 ± 0.2, and pKa2 = 9.8 ± 0.2 (Figure 2(a)); and for RacE2 are pKa1 = 7.0 ± 0.1 and pKa2 = 10.7 ± 0.1 (Figure 2(b)). From these data, the pH optima were calculated from Equation 3 to be 8.3 for RacE1 and 8.8 for RacE2.
Figure 2.
The pH dependence of RacE1 and RacE2 isozyme reactions using L-glutamate as a substrate. Steady-state kinsetic measurments were made using the circular dichroism assay. (a) The kcat values of the RacE1 catalysed reaction, tested under conditions of saturating L-glutamate at 25°C, are plotted as a function of pH to determine the pH optimum. (b) The kcat values of the RacE2 reaction are plotted as a function of pH at both saturating (●) and subsaturating (▲) concentrations of L-glutamate. Data were fit to equation 2 to obtain kcat,max, pKa1 and pKa2 values (see text).
We also determined the pH rate profile for RacE2 at subsaturating concentrations of L-glutamate where the observed rate approximates a first-order dependence on L-glutamate concentration with kobs = [E]o•kcat/Km, and titrations reflect pKa values of the free enzyme 28. The resulting pH rate profile is also bell-shaped but with a notable difference of approximately 1.7 pH units in the value of pKa2 (Figure 2(b)). The pKa values obtained under subsaturating concentrations of L-glutamate are 6.7 ± 0.3 for pKa1, and 9.0 ± 0.3 for pKa2 (Figure 2(b)). The shift of pKa2 from 9.0 in the free enzyme or substrate to 10.9 in the enzyme-substrate complex suggests that the substrate plays a role in altering the pKa of a sidechain in the active site of RacE2, or the alpha-amino group of glutamate, upon binding.
X-ray Structure of RacE1 and RacE2
Numerous crystallization trials were performed in an attempt to crystallize the apoenzymes of RacE1 and RacE2. Unfortunately, crystals of the apoenzymes could not be obtained from commercial screens. Therefore, screens of both isozymes in the presence of saturating concentrations of L-glutamate were performed. Crystals of B. anthracis RacE1 and RacE2 grew as a complex with only the product D-glutamate bound in the active site; this is supported by the well-resolved and unambiguous electron density for D-glutamate (Figure 3). The presence of D-glutamate in the active sites indicates that RacE1 and RacE2 are active under crystallization conditions.
Figure 3.
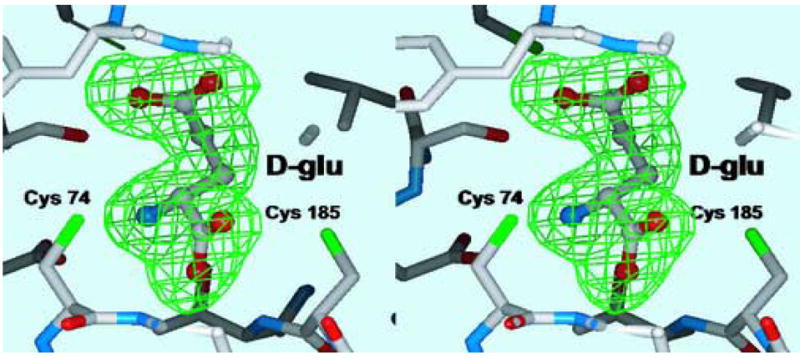
Stereoview of the final electron density (Fo-Fc) omit map in the active site surrounding the substrate D-glutamate. The electron density is contoured at 3 σ (green) and the product D-glutamate was omitted from the Difference-Fourier calculations.
For the RacE2 isozyme, four monomers are found in the asymmetric unit. The average, root-mean-square deviation (rmsd) between the Cα atoms of the four monomers is at most 0.46 Å, indicating that they have nearly identical structures. For simplicity, all distances stated hereafter are for the A chain of B. anthracis RacE2 since the distances in all chains are nearly identical. Each monomer of RacE2 is comprised of two domains with an overall α/β structure. Domain I consists of residues 1-95 and 208-270 which form five α-helices that encircle a six-stranded parallel β-sheet (Figure 4(a)). Domain II includes residues 96-207, and is comprised of six α-helices that surround a four-stranded parallel β-sheet. RacE2 crystallizes as two homodimers, with each dimer formed by the “tail-to-tail” association of two monomers in which the active site of each monomer is located at opposite ends of the dimer (Figure 4(b)). One dimer, chains A & B, in the asymmetric unit is stabilized by 19 inter-chain hydrogen bonds, whereas the other dimer, chains C & D, is stabilized by 25 hydrogen bonds (Supporting Information, Figure 1
Figure 4.
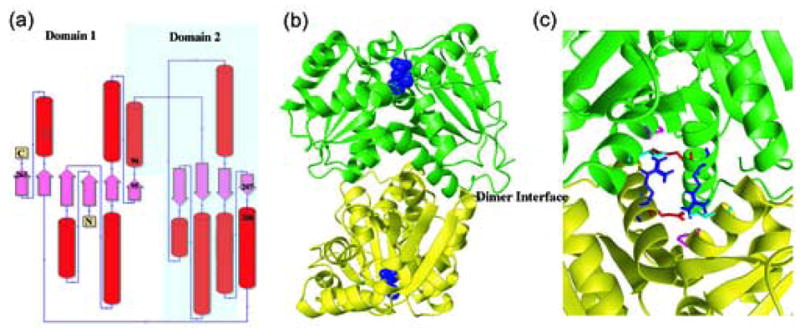
X-ray structures of RacE isozymes from Bacillus anthracis. (a) Topology diagram for the secondary structure elements of RacE1 and RacE2. Domain 1 is composed of residues 1-95 & 208-270 for RacE2, and 1-98 & 211-276 for RacE1. Domain 2 is composed of residues 96-207 for RacE2, and 99-210 for RacE1. (b) The homodimers are shown with the two chains colored green and yellow. The substrate D-glutamate is shown as space fill model (blue) and is located in the active site opposite the dimer interface (labeled). (c) Close-up view of the dimer interface of RacE2. The two monomers are shown in green and yellow. Amino acid R214 (blue), located in the middle of a helix in one monomer, is hydrogen bonded to the amino acids E215 (red), P99 (magenta) and T103 (cyan) in the other monomer. In RacE1, the corresponding residue R214 is replaced with Ile (I217). This alteration disrupts a total of 6 hydrogen bonds that stabilize the dimer interface between the two monomers.
For RacE1, the model contains three monomers, chains A, B and C, in the asymmetric unit. Each monomer is almost identical in topology to RacE2 (Figure 4(a)). Chains A and B were determined to be the biologically-active, dimeric unit by studying the interface area between the two chains. Similar to RacE1, the homodimer is comprised of two monomers associating in a “tail-to tail” orientation with the active sites of each monomer located opposite of the dimer interface (Figure 4(b)). The dimer interface is stabilized by approximately 22 hydrogen bonds (Supporting Information, Figure 1).
Comparison of RacE1 and RacE2 X-ray Structures
The amino acid sequences of RacE1 and RacE2 from B. anthracis are 48% identical and 67% homologous (Figure 1). The crystal structures of the two isozymes are remarkably similar. An rmsd of 0.81 Å for 256 Cα-atoms was obtained when the A-chains from both isozymes were superimposed. Similarly, when the dimers of each isozymes are superimposed, an rmsd of 0.89 Å was obtained for Cα of 557 atoms. Most of the homologous regions overlap perfectly between the two crystal structures.
Although the crystal structures of these two isoforms are very similar, their dimerization properties and structures at the interface are quite different. RacE1 exists almost exclusively as a monomer in the absence of substrate. In the presence of saturating amounts of L-glutamate, RacE1 exists as a dimer. In contrast, RacE2 is a dimer irrespective of the presence of substrate. Analysis of the dimer interface revealed that R214 in RacE2 is hydrogen bonded to P99, T103 and E215 (Figure 4(c)). In RacE1, the corresponding R214 residue is replaced with Ile (I217) which eliminates a key hydrogen bond between the two monomers. To test this importance of the hydrogen bond contributed by R214 in RacE2, this residue was mutated to an alanine residue. Size exclusion chromatography showed that removal of this hydrogen bond upon mutation altered the equilibrium between the monomer and dimer form of RacE2 in favor of the monomer (data not shown). Therefore, the hydrogen bond provided by R214 contributes a significant interaction at the dimer interface of RacE2 and is absent at the dimer interface of RacE1. For simplicity, the RacE2 isoform will henceforth be used for further detailed discussions
Active Sites of Glutamate Racemases
Mutagenesis and isotope effect studies on the L. fermenti RacE enzyme revealed that the amino acid residues that act directly in acid-base catalysis are C73 and C184 15; 20. The equivalent residues in the B. anthracis RacE1 are C77 and C188, and C74 and C185 in RacE2. These cysteine residues are also located in the active-site pocket with their sulfur atoms separated by approximately 7.4 Å (Figure 5(a)). Based on the crystal structures and orientation of D-glutamate in the active site of RacE1 and RacE2, C74 is proposed to serve as the catalytic base in abstracting the C-2 hydrogen from D-glutamate in the reverse reaction, and C185 is proposed to be responsible for abstraction of the C-2 hydrogen from the substrate L-glutamate in the forward reaction 20; 22. The only amino acid located within close proximity to C74 that could potentially act to deprotonate the sulfur atom in the reverse reaction is an aspartic acid residue, D11, which is approximately 3.8 to 4.0 Å (Sγ-Oε1) away (Figures 5(a) & 6(a)). D11 is 100% conserved in all glutamate racemases (Figure 1), and is held in position via direct hydrogen bonds (~2.9 Å) between one of its side-chain carboxylate oxygens and the backbone amide hydrogens of G13 and G16 (not shown). These two glycines form part of a strictly conserved loop (h10-D11-S12-G13-h14-G15-G16-h17) at the end of the first helix where h is any hydrophobic amino acid (Figure 1). The other side-chain oxygen atom of D11 comes within close contact (~3.0Å) to the amide nitrogen of the D-glutamate substrate (Figure 6(a)). D11 is observed in a similar conformation in the active site of the A. pyrophilus RacE (Figure 5(b)), although its distance from the cysteine sulfur is more than 1 Å further away than in B. anthracis RacE2.
Figure 5.
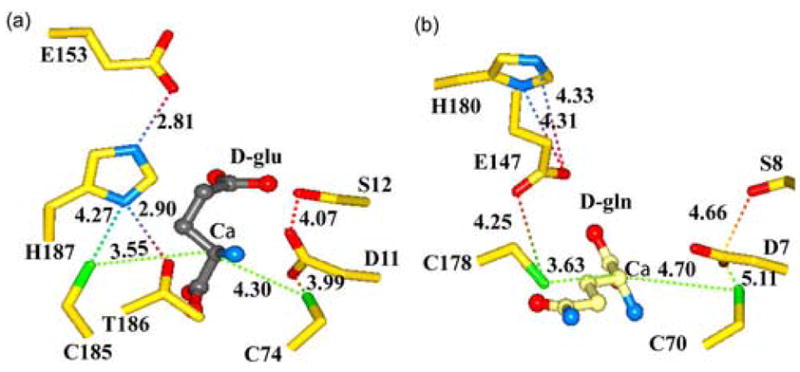
Comparison of the active sites of B. anthracis RacE2 with that of RacE from A. pyrophilus. (a) RacE2 active site with D-glutamate bound (grey). The conformation of residues H187 and G153 in the RacE2 structure is notably different from that of A. pyrophilus RacE. The possible interactions between the catalytic residues and other residues in the immediate vicinity are shown by dotted lines with distances given in Angstroms. (b) Active site of A. pyrophilus RacE with D-glutamine bound in the reverse orientation with its side chain pointing away from the active site pocket.
Figure 6.
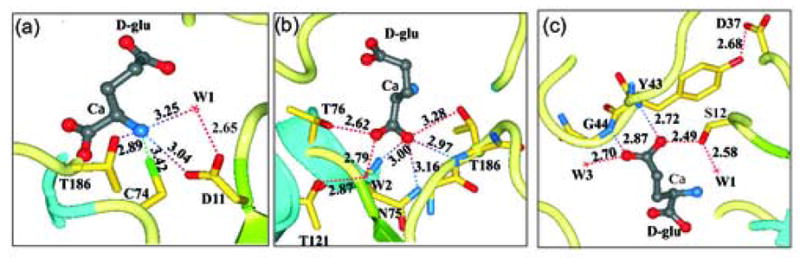
Interactions of amino acids within the RacE2 catalytic site with D-glutamate. (a) Potential interactions between the active site residues and the α amino group of D-glutamate. (b) Interactions involved in recognition and stabilization of the Cα-carboxyl group. (c) Interactions likely involved in substrate/product recognition and stabilization of the D-glutamate side chain carboxyl group. A highly conserved loop forms the floor of the active site with D37 forming a hydrogen bond with Y43 that is significant and results in orienting this loop in a conformation that favors substrate binding thereby maintaining an important secondary structure conformation that is involved in substrate recognition. The dashed lines in all panels represent distances between atoms in Å.
Analysis of the active site region surrounding C185 reveals that Y188 and H187 are the only two residues within proximity of the side chain that would be capable of deprotonating the sulfur atom (Figure 5(a)). The C185 sulfur atom is fairly restricted by the surrounding residues. The side chain hydroxyl oxygen of Y188 is located 4.1 Å from the C185 sulfur atom (not shown), and the Nδ1 atom of H187 is located 4.3 Å away (Figure 5(a)). Interestingly, the second nitrogen atom (Nδ2) of H187 forms a hydrogen bond (2.8 Å) with one of the carboxylate oxygens of E153 within the same monomer (Figure 5(a)). The hydrogen-bonding pattern observed is reminiscent of the catalytic triad of papain-like cysteine proteases with a slightly altered geometry 29. In contrast, in A. pyrophilus the geometry in the active site region surrounding this cysteine (C178) is completely different, and the catalytic-triad is not observed (Figure 5(b)). Interestingly, although located within the active site, the glutamate residue (E147) of A. pyrophilus that would be part of the triad is not within the same monomer, but instead enters the active site from another monomer as a result of dimerization (20). E147 in A. pyrophilus is actually in a position to directly deprotonate the cysteine, although it is over 4.2 Å away. Similarly, the histidine (H180) side chain in the A. pyrophilus structure is too far away from E147 to be involved in hydrogen-bonding to either E147 or C178 (Figure 5(b)). It is noteworthy that the equivalent active site residues of the B. subtilis RacE structure, S11, A12, C74, G153, C185 and H187, are in nearly identical positions in the active site relative to D-glutamate as observed in the B. anthracis RacE1 and RacE2 structures 23.
The structure of RacE2 also reveals a number of other important contacts between the enzyme and D-glutamate that are not observed in the x-ray structure of A. pyrophilus bound with the weak inhibitor glutamine. For instance, the α-amine- nitrogen of D-glutamate is within hydrogen-bonding distance to a number of residues including D11 (3.0 Å), S12 (3.2 Å) and T186 (2.9 Å), and is also positioned only 3.4 to 3.5 Å from the sulfur atom of C74 (Figure 6(a)). In addition, it is hydrogen bonded to a water molecule (W1). In contrast to the more hindered sulfur atom of C185, with a small rotation about the Cβ dihedral axis of C74, the sulfur atom can come within hydrogen bonding distance of the α-amine nitrogen.
Recognition of the α-carboxylate of D-glutamate by the enzyme is achieved via a series of hydrogen bonds with the side chains of T76 (2.6 Å), N75 (3.2 Å) and T186 (3.3 Å), the backbone amides of N75 (3.0 Å) and T186 (3.0 Å), and the side chain of T121 through a water molecule (W2) (Figure 6(b)). These residues form a highly polar “threonine pocket”, and the hydrogen bonds formed between the α-carboxylate oxygens and these residues may serve to position the substrate for catalysis as well as in stabilizing any developing negative charge on the α-carboxylate that may form during the reaction.
The γ-carboxylate group of D-glutamate is recognized and stabilized via hydrogen-bonding interactions with the side-chain hydroxyl group of S12 (2.5 Å), and with the backbone amide hydrogens of residues Y43 (2.7 Å) and G44 (2.9 Å) (Figure 6(c)). These latter two residues form part of a highly conserved loop in racemases with the consensus sequence D37-X-X-X-X-P-Y-G44, where X is any amino acid (Figure 1). This loop, which is stabilized by the hydrogen bond formed between the side chain carboxylate of D37 and the side chain hydroxyl of Y43, forms the floor of the active site with the Y43 and G44 backbone nitrogens directed toward the γ-carboxylate group (Figure 6(c)). Mutation of D37 to an asparagine residue in the L. fermenti RacE produced an enzyme that has a 100-fold increase in Km 22. Tanner et al. were unable to interpret the results since the A. pyrophilus crystal structure did not show this residue in the active site. Thus, the hydrogen bond shared by D37 and Y43 is likely to play an important role in orienting the γ-carboxylate recognition loop to a conformation that favors substrate binding.
Kinetic Properties of a RacE2-V149A Mutant and Inhibition by D-glutamate Analogs
Based on our analysis of the active-site structures of RacE1 and RacE2 in combination with the primary sequence alignment presented in Figure 1 and inhibitor data against S. pneumoniea RacE, we hypothesized that the valine residue at position 149 may partially or fully occlude a hydrophobic pocket that could be exploited for inhibitor design. Therefore, we mutated this valine residue to the corresponding alanine residue in B. anthracis RacE1 and S. Pneumoniae. The steady-state kinetic parameters, kcat and Km, for the forward (L to D ) and reverse (D to L) reactions catalyzed by B. anthracis RacE2-V149A were determined using the circular dichroism assay. In the forward direction, RacE2-V149A has about a two-fold higher kcat than wild type RacE2 (67 sec−1 versus 38 sec−1), and has a Km value for L-glutamate similar to that of RacE2 (4.6 mM versus 3.7 mM). In the reverse reaction, RacE2-V149A has a higher kcat than RacE2 (4.9 sec−1 versus 1.6 sec−1 respectively), and its Km value for D-glutamate is the same as that of RacE2 (0.2 mM).
Next, we tested the inhibition of RacE1, RacE2 and RacE2-V149A by three different D-glutamate analogs, 4, 8, and 9. Inhibition constants were determined by assays conducted in the reverse (D- to L-) direction using the coupled-enzyme assay. All kinetic parameters and inhibition constants, along with their standard errors, are summarized in Table 3. Compounds 4, 8, and 9 were observed to be good competitive inhibitors of RacE1 with Ki values of 4.6 μM, 3.5 μM, and 1.5 μM. In contrast, these compounds were found to be much weaker competitive inhibitors of RacE2 with Ki values of 289 μM, 64 μM, and 73 μM. Finally, for the mutant RacE2 enzyme R2-V149A, 4, 8, and 9 were shown to be potent competitive inhibitors with Ki values of 0.3 μM, 0.2 μM, and 0.2 μM respectively.
Table 3.
Ki values for the D-glutamate analogs, 4, 8 and 9, against RacE1, RacE2 and R2-V149A.
| Enzyme | 4 Ki (μM) |
8 Ki (μM) |
9 Ki (μM) |
|---|---|---|---|
| RacE1 | 4.6 ± 0.4a | 3.5 ± 0.3 | 1.5 ± 0.1 |
| RacE2 | 289 ± 51 | 64 ± 15 | 73 ± 20 |
| R2-V149A | 0.30 ± 0.05 | 0.20 ± 0.05 | 0.20 ± 0.01 |
Standard error in the Ki value resulting from a fit of the data to equation 4 describing competitive inhibition.
Computational docking of the D-glutamate analogs using GOLD
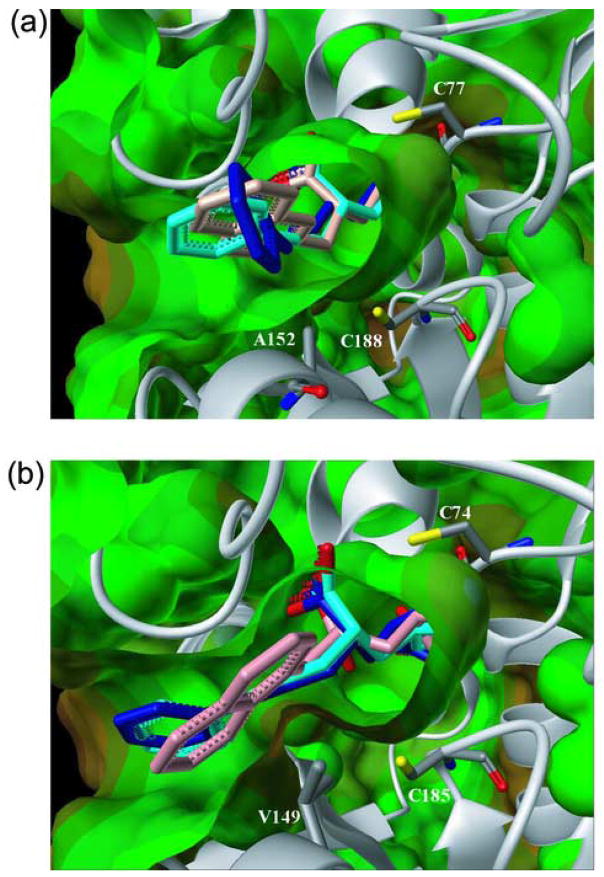
To better understand the binding geometry of 4, it was docked into the RacE2 active site with and without the V149A mutation using the program GOLD (Cambridge Crystallographic Data Centre, Cambridge, UK). It was found that the tunnel, which is constricted by valine 149 in native RacE2, is much less accommodating of the large naphthyl ring of 4 (Figure 7(b)), whereas the more open tunnel formed by an alanine in RacE1 at this position creates more space to accommodate this moiety (Figure 7(a)). Compounds 8 and 9, which differ only in that the α-carboxylate moiety in 8 is reduced to a hydroxyl in 9, were also docked into the RacE2 and R2-V149A active sites to assess the potential that a longer carbon-linker between D-glutamate and the hydrophobic substituents could lead to a more potent RacE2 inhibitor. The docking results demonstrate that the 3-C linker of compounds 8 and 9, when compared to the 1-C linker of 4, allow the full hydrophobic moiety to extend out of the tunnel leading to the catalytic site (Figure 7). The docking results also indicate that modification of the α-carboxyl group (8) of the glutamate core structure to a hydroxyl (9) retains the hydrogen bonding capabilities in this region, and is thus likely to retain activity, consistent with the assay results. Combined with the kinetic results indicating minor differences between 8 and 9, this suggests that minor modification of the glutamate core structure may be a complementary design strategy.
Figure 7.
Compounds 4 (pink), 8 (blue) and 9 (cyan), docked into the active sites of RacE1 (a) and RacE2 (b) using GOLD. A152 in RacE1 causes the opening to the active site to be less sterically hindered than RacE2, which has V149 at the corresponding position.
Discussion
Kinetic characterization of the Bacillus anthracis gene products (racE1 and racE2) demonstrates that both genes produce bona fide glutamate racemase isozymes designated RacE1 and RacE2. Both isozymes are highly specific for the amino acid glutamate, and each catalyzes the racemization of D- or L- glutamate with high catalytic efficiency, comparable to RacE isozymes isolated from other bacteria (Table 2). For both RacE1 and RacE2 from B. anthracis, the ratios of the forward and reverse rate constant, kcat, are 5-fold and 24-fold in favor of the formation of D-glutamate. A comparison of the Km values for the interaction D-glutamate with both isozymes reveal that D-glutamate binds RacE1 and RacE2 approximately 9 and 19 times more tightly than L-glutamate assuming that these Km values reflect the affinity of the enzymes for the substrate and product (Table 2).
The trends in the kinetic parameters observed for RacE1 and RacE2 are not specific to B. anthracis, but appear to be a more general characteristic of glutamate racemases isolated from organisms where the Km and kcat values have been reported (Table 2). With the exception of E. coli RacE and B. subtilis YrpC (discussed below), the forward rate constant, kcat, is typically 1 to 24 times higher than the reverse rate constant. The observation that the Km values for the D-glutamate product are typically 1.3 to 20 times lower than those for the L-glutamate substrate, indicates that D-glutamate is likely bound and stabilized more tightly than L-glutamate. Another general observation is that the ratios of the kinetic parameter, kcat/Km, for the forward and reverse reactions are all near unity (0.5 to 1.3) for these enzymes, as would be predicted from the Briggs-Haldane relationship 27. Together, the trends in the kinetic parameters for glutamate racemase from a number of organisms suggest that the enzymes have evolved to kinetically favor the formation of D-glutamate within the cell. This is to be expected since during cell wall and capsule biosynthesis any D-glutamate formed would likely be rapidly incorporated into the peptidoglycan precursors.
Two exceptions to these general observations are YrpC from B. subtilis and RacE from E. coli. Both enzymes are homologous to glutamate racemase (Figure 1), yet YrpC has poor catalytic activity for L-glutamate, and both enzymes are less efficient when it comes to catalyzing the forward reaction. Notably, these two enzymes have opposite trends in their kcat and Km values, as compared to the other racemases. Both YrpC and E.coli RacE enzymes have a higher catalytic rate for the reverse reaction, and appear to bind L-glutamate more tightly than D-glutamate. Interestingly, YrpC and E.coli RacE are unique among the racemases in Table 2 in having an alternate function within the cell where they also function as inhibitors of DNA gyrase (DNA topoisomerase II) 30; 31. Phylogenetic analyis of bacterial glutamate racemases shows that B. anthracis RacE1, RacE2, and B. subtilis RacE are closely related, whereas B. subtilis YrpC is distantly related to these isozymes and appears to be distinct from that of other Bacillus racemase genes 32.
A model of the active site structure of B. anthracis RacE2, based upon the four-location model for enantiomeric specificity of proteins 33; 34, is illustrated in Figure 8. This model encompasses the x-ray structural data on glutamate racemases from B. anthracis and B. subtilis, as well as the available data from kinetic and mutagenesis studies on the enzymes listed in Table 2. We propose that three of the subsituent groups, –NH3+, –COO− and –(CH2)2COO−, attached to the alpha-carbon of glutamate, interact at the three locations of the enzyme identified in Figure 8. Moreover, we propose that these interactions are likely maintained with slight geometric alterations throughout the catalytic process, and that racemization occurs via an antarafacial transfer of the protons from one of the “fourth” location positions, either the C185-E153-H187 system, or C74, depending on the enantiomeric substrate that binds. The model in Figure 8 is consistent with the x-ray structure and modeling studies of aspartate racemase 35, and is supported by the recent computational QM/MM simulation studies on the glutamate racemase reaction 36.
Figure 8.
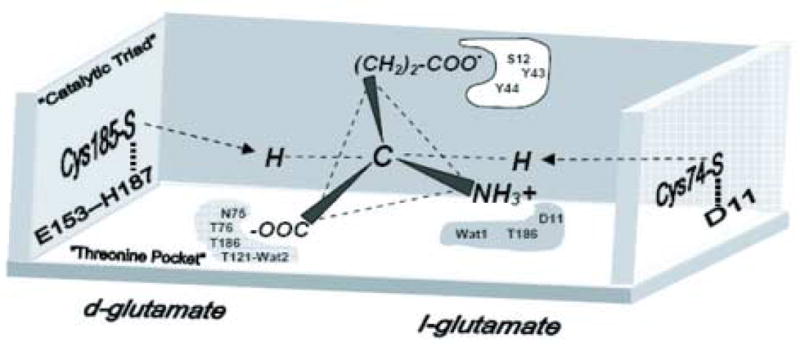
Proposed active site model for the racemization of glutamate by B. anthracis RacE isozymes. The illustration is based on the four-location model for protein stereospecificity 33; 34. Three locations including the “threonine pocket” (N75,T76,T186, T121 & Wat6), and the α-NH3+ binding site (D11, T186 and Wat 1), and the R-group –(CH2)2-COO− binding site (S12, Y43 & Y44), which help to anchor the substrate, are shown and shaded. The “fourth location” or binding site(s) that are responsible for protonation of a possible ylide-intermediate and hence formation of D-glutamate (Cys185-H187-E153), or L-glutamate (Cys74-D11), are shown.
Comparison of the crystal structures of RacE1 and RacE2 reveals that the three dimensional structures of these two isozymes are remarkably similar. Over 90% of the hydrogen bonding patterns are the same. Minor differences are seen in each monomer as well as at the dimer interface. The question as to why the organism expresses two enzymes from two different genes that have the same structure and function remains unanswered. Ongoing studies focusing on the gene expression patterns of these enzymes at different stages of growth and under different growth conditions may provide us with an understanding of how this organism utilizes two isozymes of glutamate racemase.
Comparison of the x-ray structures of RacE1 and RacE2 and the sequence alignments of other racemases in Figure 1 provides new insight into the possibility of developing compounds that would selectively inhibit these enzymes. Specifically, the active site pocket near the 4-methylene carbon of the substrate is wider for RacE1 than RacE2 (Figure 9). The widening of the pocket stems from a single amino acid difference, V149A, that removes two methyl groups that sterically block access to an additional hydrophobic pocket Thus, it should be possible to selectively target and inhibit RacE1 without inhibiting RacE2. For the organisms shown in Figure 1, only RacE1 of B. anthracis and the RacE of S. pneumoniae have Ala at this position. All other RacE forms have Val at this position. Therefore, a focus on inhibiting RacE2 has two advantages. First, RacE2 has a smaller catalytic site opening due to the presence of a valine at the mouth of the active site versus an alanine. Our data in Table 3 indicate that a RacE2 inhibitor should also bind to and inhibit RacE1, which has a larger entrance to the catalytic site, and should accommodate an inhibitor of RacE2 with smaller groups. Second, the activity of RacE2 was shown to be essential for the viability of B. anthracis 13, so inhibiting RacE2 alone should be effective in inhibiting the growth of B. anthracis. The RacE2 structure shows that the active site is constricted and would best accommodate analogs substituted at the 4-position of D-glutamate.
Figure 9.
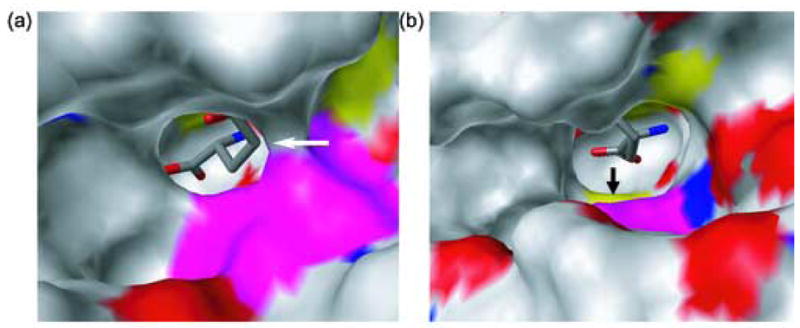
X-ray Structures of RacE2 (a) and RacE1 (b) with D-glutamate bound in the active site depicted with a solid surface showing the active site pockets. RacE2 has Val at position 149 (shown in magenta) while RacE1 has Ala at the corresponding position. These figures demonstrate the effect that the Val and Ala residues have on the opening and accessibility of the active site pocket, which may have important implications in the design of inhibitors for glutamate racemase enzymes. RacE1 has a more open structure, with the active site Cys more visible (shown by the black arrow) as opposed to the more occluded active site of RacE2 (shown by the white arrow).
In recent years, several inhibitors of glutamate racemase have been developed 8; 9; 10. De Dios and coworkers successfully designed a series of 4-substituted D-glutamate analogs that function as potent competitive inhibitors against RacE from S. pneumoniae, with IC50 values ranging from 0.04 to 9.8 μg/mL 8. For instance, a 2-napthyl and 2-benzothienyl substituent inhibits RacE from S. pneumonia with IC50 values of 0.1 and 0.036 μg/mL, respectively, with corresponding MIC values against S. pneumoniae of 0.25 μg/mL for both compounds. However, neither of these compounds inhibit glutamate racemase from S. aureus, which has a valine in this position (IC50 >250 μg/mL), and are absolutely ineffective in inhibiting the organism (MIC >250 μg/mL). Since most pathogens contain a valine at the equivalent position, the most viable strategy for developing a broad-spectrum inhibitor that targets glutamate racemase would be to design compounds against the more restricted active sites of glutamate racemases such as RacE2.
The structure-activity relationship (SAR) for these compounds is consistent with the hypothesis that a large, lipophilic binding pocket exists in close proximity to the RacE active site, and that the addition of aromatic, nonpolar substituents to D-glutamate, if displayed in the correct stereochemical position, can be accommodated by this pocket. Activity profiles against different bacterial species demonstrated that these D-glutamate derivatives lack broad-spectrum activity, most probably due to steric bumping of the aromatic substituent with the V149 that is present in most organisms. Additionally, different bacterial cell permeability may result in differential uptake of compounds. Second, an alternate supply of D-glutamate, possibly via the d-amino acid transferase (DAT) pathway, may be available. Finally, the primary amino acid sequence of glutamate racemase enzymes differs from species to species (Figure 1). These differences may evoke changes in the active site geometry and/or electrostatics that would determine the binding, or lack thereof, of this class of inhibitors.
We chose the most potent of these inhibitors, (2R, 4S)-2-Amino-4-(2-napthyl) methyl pentanedioic acid (4), and tested its inhibitory activity against RacE1 and RacE2. Compound 4 inhibits RacE1 with a 65-fold higher efficacy than RacE2. Analysis of the active site surroundings suggests that V149 produces a partial occlusion of the tunnel connecting the active site and the hydrophobic pocket as compared to an alanine occupying the same position. A less sterically hindered tunnel when alanine occupies this position, appears to be the principal source for the difference in inhibitory specificity of 4 between RacE1 and RacE2.
We suggest that by altering compounds similar to 4 by increasing the length of the carbon chain that links the hydrophobic moiety to the D-glutamate skeleton, that more potent RacE2 inhibitors could be designed that would have the potential to also inhibit other RacE enzymes that have alanine at this position. Targeting enzymes with a valine at position 149, or equivalent, would seem to be the most sensible approach for inhibitor design since a molecule that will fit the more constricted area created by the valine is likely to be easily accommodated by the more open, less sterically hindered tunnel created by the alanine.
To investigate this hypothesis, we engineered a RacE2-V149A mutant enzyme, characterized it kinetically, and tested the ability of this mutant to be inhibited by three different D-glutamate analogs, 4, 8, and 9. The inhibition constants for these three compounds were determined for RacE1, RacE2 and R2-V149A via the coupled enzyme assay. Compound 4 had about a 1000-fold lower Ki for R2-V149A when compared to the native RacE2, and compounds 8 and 9 were approximately 35-fold lower. The remarkable increase in affinity of 4 for R2-V149A lends strong support for our hypothesis.
The docking results demonstrate that the 3-C linker of compounds 8 and 9, when compared to the 1-C linker of 4, allows the bulkier hydrophobic moiety more extension out of the tunnel leading to the catalytic site (Figure 7). We experimentally tested the effectiveness of 8 and 9 to serve as more potent RacE2 inhibitors and found that both compounds exhibit an approximate 4-fold decrease in their Ki value against wild type RacE2, 64 μM and 73 μM, when compared with shorter linker of 4 (Table 3). These values, along with the docking studies, demonstrate that strategies for bypassing the constrictive V149 side chain may be effective in decreasing the inhibition constant and making this class of compounds more potent inhibitors for RacE2 and other glutamate racemases with valine at this position.
From this data, we conclude that designing a small molecule around the constraints imposed by a valine in the connecting tunnel may be the best strategy for broad-spectrum inhibitor design against glutamate racemases. Efforts are underway for optimizing the length of the carbon linker as well as optimizing the hydrophobic moiety. Minor modifications to the glutamate core structure may also prove to be successful, as demonstrated by 9.
Materials and Methods
Cloning and Expression of Bacillus anthracis Glutamate Racemase Isozymes RacE1 and RacE2
The genes encoding RacE1 and RacE2 were PCR-amplified from genomic DNA isolated from Bacillus anthracis Sterne 34F2. A 5′-primer complementary to the beginning of the RacE1 gene generated a SalI restriction site upstream of the start codon, and a 3′-primer complementary to the end of the RacE2 gene created a BamHI site directly downstream of the termination codon. The PCR amplification of the RacE2 gene was performed using the same strategy as RacE1. Qiagen synthesized all primers. The primers used for PCR amplification of the RacE1 and RacE2 genes were: RacE1 N-terminal primer: 5′GTGATGTCGACATGTCTGTATGTCATAAACAT; C-terminal primer: 5′GCTAGGATCCTAA TTACAGATGCGAGCATTCTT; RacE2 N-terminal primer: 5′GTGATGTCGACATGAAGT TGAATAGAGCAATC, C-terminal primer: 5′GCTAGGATCCTATTCTTTTTCTAAATGAATATG.
The resulting PCR products were digested with SalI and BamHI, gel purified, and then ligated into a XhoI and BamHI digested pET15b vector carrying a 23 amino acid N-terminal extension encoding a (His)6-tag (Novagen). The ligation products were transformed into E. coli (JM109), and the resulting clones were screened for the presence of inserts by PCR amplification using the complementary primers. The resulting expression plasmids for RacE1 and RacE2 are designated as pRacE1-15b and pRacE2-15b. The full gene sequences were verified by dideoxy sequencing at the UIC DNA core facility.
E. coli BL21(DE3) was transformed with pRacE1-15b, plated onto LB-ampicillin plates, and a single colony was used to inoculate 10 mL of a starter culture that was grown overnight at 37 °C. 1 L of Terrific Broth (TB) media, supplemented with 100 μg/mL ampicillin, was inoculated with 10 mL of the starter culture and was grown at 37 °C until an optical density of 0.6 at 600 nm was reached. The same procedures were implemented for the pRacE2-15b construct. Expression of the RacE1 and RacE2 enzymes was induced by the addition of 1 mM IPTG into the media, and the culture was grown for an additional 4 hours.
Cloning and Expression of Bacillus anthracis RacE2-V149A mutant enzyme
The protocol published by J.L. Reymond et al. 37 was used to generate the V149A RacE2 mutant enzyme (R2-V149A). The primers used for PCR amplification of the mutant pET-15b-RacE2 vectors are as follows with the mutations underlined; direct primer: 5′ CCT TTC GCT GAA CTT GTA GAG AGT GGC 3′, and reverse primer: 5′ AAG TTC AGC GAA AGG CGG ACA CGC TAA 3″. Integrated DNA Technologies Inc. synthesized all primers. The PCR reaction was performed in a 25 μL reaction volume and included 4% DMSO. The reaction contained 63 ng of purified, pET15b-RacE2 DNA, 1 μM of primer pairs, 300 μM dNTP’s, and 2 U of DNA polymerase (Expand High Fidelity PCR System from Roche). The amplification reaction was initiated by pre-heating the reaction mixture to 94 °C for 3 min and then continued for 16 cycles at 94 °C for 1 min, 50 °C or 56 °C for 1 min, and 68 °C for 13 min. A final incubation at 68 °C for 30 min was used to ensure complete elongation of the amplified vectors. Agarose gel electrophoresis was used to evaluate the PCR-amplification products, which were subsequently treated with the restriction enzyme DpnI. 1 μL of PCR product was used to transform XL1 Blue supercompetent E. coli cells, which were then plated on Luria-Bertani (LB) media supplemented with 100 μg/mL ampicillin. The plasmid DNA was purified, and the full gene sequence was determined by dideoxy sequencing at the UIC DNA core facility to verify the correct mutation. The resulting mutated expression plasmid was designated pR2-V149A.
E. coli BL21(DE3) were transformed with mutant vector pR2-V149A and plated onto LB-ampicillin plates. The RacE2 expression protocol was successfully employed to overexpress the mutant protein R2-V149A.
Purification of RacE1, RacE2, and R2-V149A
The purification procedures are the same for RacE1, RacE2, and R2-V149A. E. coli BL21(DE3) cells containing overexpressed RacE1, RacE2, or R2-V149A were harvested by centrifuging at 5,000 rpm in 1 L, centrifuge tubes using a Sorvall RC5-B centrifuge equipped with an SLC-4000 rotor. The cell pellets were suspended in lysis buffer [1x PBS, 1% Triton, DNase I, protease inhibitors cocktail, and lysozyme] 3 ml per gram of cell pasted, and were then lysed using a French press (SLM-Aminco). The lysate was centrifuged at 18,000 rpm using a Sorvall SA-600 rotor, and the supernate was removed and loaded onto a 5 mL, HiTrap affinity column (Amersham Biosciences) charged with Co2+ and equilibrated with HiTrap equilibration buffer (50 mM Tris pH 8.0, 500 mM NaCl and 10 mM imidazole). The RacE1, RacE2, or R2-V149A enzyme was eluted with a linear gradient of 0–50% elution buffer (50 mM Tris pH 8.0, 500 mM NaCl and 500 mM imidazole). Fractions were collected and assayed for glutamate racemase activity using the coupled-enzyme assay. The fractions with the highest specific activity, and judged to be pure by SDS-PAGE analysis, were then pooled and concentrated using a 15 mL, centrifugal ultra-filtration device with a 10,000 MWC (Amicon-Millipore). The His tags were not removed as the enzyme activity with and without these tags were not significantly different. For RacE1, the concentrated enzyme was then buffer exchanged into 50 mM Tris pH 8.0, 100 mM NaCl, and 0.1 mM DTT using the same concentrator. RacE2 and R2-V149A were prepared in a similar fashion, except that the storage buffer was 50 mM Tris pH 8.0, 250 mM NaCl, and 0.1 mM DTT. For long-term storage, glycerol was added to the enzymes to a final concentration of 10%, and the samples were frozen using liquid nitrogen and stored at −80 °C for up to three months.
Size-Exclusion Chromatography
The oligomeric states of RacE1 and RacE2 were determined using a Superdex 200 HR 10/30 (Amersham Biosciences) high-performance, gel-filtration column. The column was equilibrated with equilibration buffer, 50 mM Tris (pH 8.0), 250 mM NaCl, and 2 mM DTT, and was calibrated by running a set of protein standards (with molecular weights and elution volumes: ribonuclease A, 13,700 Da (16.26 mL); chymotrypsinogen, 25,000 Da (15.69 mL); ovalbumin, 43,000 Da (13.76 mL); albumin, 67,000 Da (12.46 mL); catalase, 232,000 Da (10.93 mL); ferritin, 440,000 Da (9.06 mL). A 10 mg/mL sample of blue dextrin was used to determine the void volume. Blue dextrin and all molecular weight standards were prepared by reconstituting lyophilized samples in the equilibration buffer, and centrifuging at 13,000 g for 5 min to remove any particulates.
To determine the quaternary structure of the apoenzymes, a 50 μL sample of purified, recombinant RacE1 or RacE2 at a concentration of 10 mg/mL was applied to the column, and the elution volume of the enzyme peak was determined using the AKTA FPLC software (Amersham Biosciences). To determine the quaternary structure of RacE1 and RacE2 in the presence of L-glutamic acid, a 50 μL sample of each enzyme was exchanged into the above equilibration buffer that was supplemented with 50 mM L-glutamic acid. The column was equilibrated with the supplemented equilibration buffer, and the enzymes at 10 mg/mL were run over the column as described for the apoenzyme. The apparent molecular weights for RacE1 and RacE2 in the absence and presence of 50 mM L-glutamic acid were estimated by comparing their relative mobility (Kav) to those of the standard enzymes via a plot of the log of the molecular weights of the standards vs. the Kav. The value of Kav is equal to (VR − VO)/(VC − VO), where VR is the retention volume, VO is the void volume, and VC is the bed volume of the column.
Glutamate Racemase Activity Assays
The enzymatic activities of RacE1, RacE2, and R2-V149A in the forward and reverse directions were measured by a continuous, circular dichroism (CD) assay based on methods described previously 18. Ellipticity was monitored at 25 °C using a Jasco J-710 circular dichroism spectrometer equipped with a thermostated cuvette holder. The molar ellipticity of L-glutamic acid at 204 nm and pH 8.2 was determined to be 32.4 mdeg•cm−1•mM−1, in reasonable agreement with the previously reported values of 31.0 mdeg•cm−1•mM−1 18 and 33.7 mdeg•cm−1•mM−1 38. This wavelength was used to determine the reaction rates of the enzymes at concentrations of D- or L-glutamate below 8 mM. To measure the reaction rates at higher concentrations, above 8 mM and up to 30 mM, a wavelength of 220 nm was utilized in order to attenuate the CD signal and thus avoid saturating the photomultiplier. The molar ellipticity of L-glutamate at 220 nm and pH 8.2 was determined to be 7.53 mdeg•cm−1•mM−1.
The enzymatic activity of RacE1 and RacE2 in the reverse direction with D-glutamate as the substrate was also measured using a continuous, coupled-enzyme assay based on the method described previously 18. Briefly, the product of the reverse reaction, L-glutamate, serves as the substrate for L-glutamate dehydrogenase which converts NAD to NADH. This reaction is coupled to diaphorase, which uses the generated NADH to reduce p-iodonitrotetrazolium violet, which has an absorbance maximum at 500 nm. The initial rates of the D-glutamate conversion were measured by monitoring the change in absorbance at 500 nm over time using a Varian/Cary 50 UV/Vis spectrometer equipped with a thermostated, 18-position, cuvette holder. The reaction mixture (1.00 mL) contained 50 mM CHES buffer, pH 9.2, 5 mM NAD+, 37.5 units of bovine L-glutamate dehydrogenase (Sigma), 2.5 mM ADP, 0.65 mM INT, 2 units of diaphorase, and D-glutamic acid at variable concentrations. After preincubation of the reaction mixture for 10 minutes at 25 °C to achieve thermal equilibration, the reaction was initiated by the addition of recombinant RacE1 or RacE2 to give a final enzyme concentration of approximately 0.3 μM. The initial velocity data were then plotted against substrate concentration and fit to the Michaelis-Menten equation as described below.
Steady-State Kinetic Parameters
The steady-state kinetic parameters for the forward and reverse reactions catalyzed by RacE1, RacE2, and R2-V149A, using L- and D-glutamate as substrates, were determined in an assay buffer consisting of 10 mM potassium phosphate, pH 8.2, and 0.2 mM DTT. For RacE1, the concentration of L- or D-glutamic acid was varied between 0.5 and 30.0 mM with a final enzyme concentration of 0.34 μM RacE1. For RacE2, the concentration of L- or D-glutamic acid was varied between 0.1 and 20.0 mM with a final enzyme concentration of 0.22 μM RacE2. The assay volumes for both enzymes were 3.00 mL, and the pathlength of the cuvettes was 1.00 cm. Initial velocity data at each substrate concentration were measured in triplicate or quadruplicate and averaged. Initial velocity data were plotted against substrate concentration and the data were fit to the Michaelis-Menten equation (Eq. 1) using non-linear regression and the Enzyme Kinetics Module of the software program SigmaPlot (Systat Software, Inc.).
| (Eq. 1) |
The resulting kinetic parameters kcat and Km are reported with their standard error values, which are determined from the residual values at the 95% confidence interval.
pH Rate Profiles and Optima
The pH rate profiles and optima of purified, recombinant RacE1 and RacE2 were determined in the forward direction using the circular dichroism assay at wavelengths of 220 nm and 204 nm for high and low concentrations of substrate as described above. The L-glutamate concentrations were fixed at 22 mM for RacE1, and at 25 mM and 4 mM for RacE2. The pH range covered was 6.0 to 11.0. Assays over the pH range of 6.0 to 8.0 were conducted using 10 mM potassium phosphate buffer containing 0.2 mM DTT. For the pH range of 8.5 to 11.0, a 10 mM borate buffer containing 0.2 mM DTT was used. The reactions were incubated at 25 °C for 10 min and were then initiated by the addition of enzyme to give a final enzyme concentration of 0.34 μM for RacE1, and 0.07 μM for RacE2. pH rate data were fit to Equation 2 which describes a bell-shaped pH curve with two pKa values, pKa1 and pKa2, and a maximum catalytic rate, kcat,max.
| (Eq. 2) |
The values for kcat,app were determined at each pH value, and the pH optimum was calculated from equation 3.
| (Eq. 3) |
RacE1, RacE2 and R2-V149A Ki determination using D-glutamate analogs
The enzymatic activity of RacE1 and R2-V149A in the reverse direction with D-glutamate as the substrate was measured in the presence of three D-glutamate analogs, 4, 8, or 9 using the coupled enzyme assay described previously. For a control assay, identical conditions were used except water was used in place of the compounds. Wild-type RacE2 activity was assayed under the same conditions. A Tecan Freedom Evo 200 liquid handling robot was used to pipette the assay components to a final volume of 200 μL in a 96 well plate format. The pathlength of the solution in a well of a 96-well plate containing 200 μL liquid was determined to be 0.44 cm. The initial rates of the D-glutamate conversion in the presence of compound were measured by monitoring the change in absorbance at 500 nm over time at 25 °C using a SPECTRAmax Plus 384 spectrometer. For these experiments, D-glutamate concentration ranged from 0.05 to 4.0 mM. The test compounds, 4, 8, and 9, were initially dissolved in 100% DMSO and then diluted in water to a final stock concentration of 10, 100 and 1000 μM and to a final DMSO concentration of 10%. DMSO concentrations in the assay never exceeded 2% to avoid inactivation of the enzymes. The final concentration of compounds 4, 8, and 9 ranged from 0.5 to 100 μM. Reactions were initiated with the addition of enzyme. A final concentration of 0.37 μM for R2-V149A and RacE2 or 0.20 μM for RacE1 was used. Initial velocity data at each substrate/inhibitor concentration were measured in triplicate or quadruplicate and were averaged. The data were fit to the equation 4 that describes pure competitive inhibitioin using non-linear regression and the Enzyme Kinetics Module of the software program SigmaPlot (Systat Software, Inc.).
| (Eq. 4) |
Kinetic parameters including the resulting inhibition constant (Ki) are reported with standard error values, which were determined from the residual values at the 95% confidence interval.
Crystallization and X-ray Data Collection for RacE2
Crystallization trials were performed by the hanging-drop, vapor-diffusion method, and initial conditions were found from the Hampton Research PEG 6000 grid screen. Crystals were obtained by mixing 1 μL protein solution at concentrations of 7.5 to 11.0 mg/mL in the presence of 50 mM L-glutamic acid (sodium salt), pH 7.0 and 2 mM DTT with 1 μL well buffer solution consisting of 100 mM MES, pH 6.0 and 5% PEG 6000. Thin crystal plates appeared after 24 h and were stable at room temperature for up to six months. We were unable to consistently grow RacE2 crystals directly from these starting conditions, so we routinely grew crystals by adding RacE2 microcrystals to drops containing 1 μL RacE2 solution and 1 μL well solution as described above. Diffraction intensities were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, using a MAR300 CCD detector. The enzyme crystallizes in space group P21, based upon the symmetry of the diffraction pattern and the systematic absences, and has four molecules in the asymmetric unit. X-ray data were processed to 1.99 Å resolution using XDS 39.
Structure Solution and Refinement for RacE2
The structure was solved by molecular replacement using Phaser 40 with data from 20 to 2.5 Å. The coordinates of RacE from A. pyrophilus (PDB entry 1B74) were modified to the B. anthracis RacE2 sequence. The initial search model included only residues 46-134 in one chain, and led to a reasonable solution for two chains. Refinement of the coordinates of the partial structure was then performed from 20 to 2.5 Å using the program CNS 41 and a maximum-likelihood target function. Overlap of the structure with MurI in a sigma-A weighted electron density map allowed for the addition of residues 198-210 to one chain, yielding residues 46-134 and 198-210, which was then used as a search model for molecular replacement using the program Phaser. Four chains were then evident from the solution with a Log Likelihood Gain (LLG) of 548. Iterative rounds of positional and B-factor refinement and electron density map generation with model building using O 42 led to generation of a model with four complete chains. At this stage, a ligand identified as D-glutamate was evident in all four active sites and was introduced into the model. Subsequent rounds of refinement and model building led to the addition of 487 water molecules and dual conformations for 11 residues. The final model has an Rcryst value of 0.203, an Rfree value of 0.241, and a map figure-of-merit (FOM) of 0.833 (Table 1).
Table 1.
Data Collection and Refinement Statistics
| RacE1 | RacE2 | |
|---|---|---|
| Data Collection | ||
| Space group | C2 | P21 |
| Cell parameters: | ||
| a,b,c (Å) | 85.78(1)a, 49.66(1), 198.25(3) | 56.35(5), 90.52(6), 116.80(9) |
| β (deg) | 90.06 (7) | 100.02 (2) |
| Resolution (Å) | 20.0–1.75 | 20.0–2.11 |
| No. reflections recorded | 561,856 | 289,750b |
| No. averaged reflections | 82,804 | 77,916b |
| R merge (%) | 6.4 (15.3)c | 12.7 (44.0) |
| I/σI | 23.0 (11.8) | 7.15 (3.01) |
| % completeness | 97.5 (93.0) | 99.0 (100.0) |
| Refinement | ||
| Resolution range | 20.0 – 1.75 | 20.0 – 2.11 |
| no. reflections in working set | 78,689 (93.0%) | 73,972 (93.4%) |
| no. reflections in test set | 4178 (4.9%) | 3930 (5.0%) |
| Rcrys (%) | 15.9 | 20.3 |
| Rfree (%) | 19.4 | 24.1 |
| Wilson B | 19.1 | 35.0 |
| average B-factor (Å2) | ||
| protein | 15.0 | 39.9 |
| water | 37.8 | 46.1 |
| D-glutamate | 9.8 | 26.4 |
| no. water molecules | 1167 | 486 |
| no. of protein molecules in asymmetric unit | 3 | 4 |
| No. of protein atoms in final model | 6542 | 8357 |
| rmsd from ideal geometry: | ||
| Bond lengths (Å) | 0.009 | 0.009 |
| Bond angles (deg) | 1.48 | 1.41 |
| Ramachandran plot | ||
| allowed (%) | 99.6 | 99.1 |
| generous (%) | 0.4 | 0.9 |
| disallowed (%) | 0.0 | 0.0 |
Estimated standard error in unit cell edge lengths and angles are given in parentheses.
Number of reflections recoreded are for data to 1.99 Å resolution. Cuttoff resolution is 2.11 Å.
Statistics for highest resolution shell are given in parentheses.
Crystallization and X-ray Data Collection for RacE1
Initial crystallization conditions were found from the Hampton Research PEG/Ion grid screen by the hanging–drop vapor diffusion method. RacE1 was crystallized in 0.2M KF with 20% PEG3350 at pH 7.2. 50mM D-glutamate was added to the enzyme at 10 mg/mL and 2 μL of the protein solution was mixed with 2 μL of the well solution. Crystals were obtained in 48 hours at room temperature and grew in the form of rod-like clusters, however they diffracted poorly. The best crystals were obtained when yttrium chloride (from the Hampton Research Additive Screen) was added into the protein drop to a final concentration of 10 mM. Diffraction intensities were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. To reduce radiation damage, crystals were frozen at 100K, with 20% (v/v) glycerol added as a cryoprotectant. X-ray data were processed to 1.75 Å resolution using XDS 39. The enzyme crystallizes in space group C2, and has three chains in the asymmetric unit.
Structure Solution and Refinement for RacE1
The structure was solved by molecular replacement using Phaser with data from 20 to 2.5Å. The coordinates of RacE2 (PDB ID code 2GZM) were used for the molecular replacement. Refinement of the coordinates of the partial structure was then performed using the program CNS 41, and a randomly selected 5% of the total observed unique reflections were used as a test set to calculate the free R value. The model was manually built using the program ‘O’ 42 and CCP4. A total of 1167 water molecules and a D-glutamate for each active site were added to the model. The final model has an Rcryst value of 0.159, an Rfree value of 0.194, and a map figure-of-merit (FOM) of 0.901 (Table 1).
Both models were validated using Molprobity 43, What-If 44, and Procheck 45. Molecular graphics figures were prepared using the program NOC 46 and Lithium (Tripos, Inc., St. Louis, MO). The coordinates and structure factors for the final model have been deposited with the PDB with ID codes 2DWU for RacE1 and 2GZM for RacE2.
Synthesis of D-Glu analogs
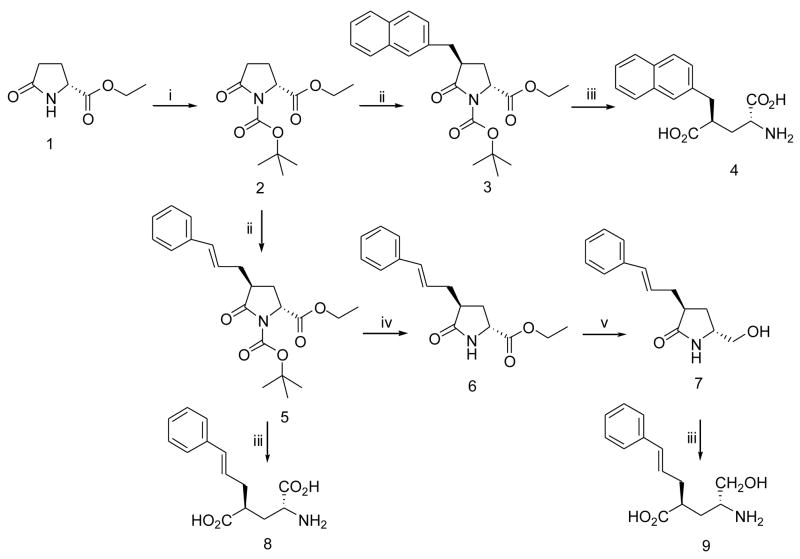
Synthesis of D-Glu analogs, 4, 8 and 9 followed the general strategy shown in Scheme 1. The commercially available starting material 1, protected as the t-butoxycarbonyl N-BOC derivative, 2, was converted to the pyroglutamate lactam enolate using LiHMDS in THF at −78 °C and reaction with electrophiles by minor modifications of literature procedures 8; 47. Major diastereomers (3 or 5) were isolated in moderate yields, without affecting the pyroglutamate stereogenic center and were deprotected directly with 6N hydrochloric acid for 8, or in 2 steps for 4, giving products in good yields. Treatment of 5 with trifluoroacetic acid in methylene chloride selectively removed the N-BOC protecting group to give 6, allowing ester reduction with LiBH4 in THF and subsequent 2 step depreotection to give the desired product 9, in good yield.
Scheme 1.
Conditions:(i) Di-tert-butyl dicarbonate, DMAP, Et3N, CH2Cl2. (ii) LHDMS, THF, −78 °C, and then ArCH2Br. (iii) 2.5 LiOH, THF. (iv) CF3COOH, CH2Cl2, rt. (v) LiBH4, THF.
General Experimental
All solvents and synthetic reagents were purchased from Aldrich (WI) and were used without further purification unless otherwise noted. Melting points were measured on a Thomas-Hoover melting point apparatus without correction. 1H NMR spectra were recorded on a Bruker Avance 300 spectrometer at 300 MHz. Chemical shift are reported in ppm (δ) using tetramethylsilane (TMS) as the internal reference. Electrospray mass spectra (ESMS) were recorded on an Agilent 1100 LC/MSD ion trap and HRMS were collected with a Waters Micromass Q-TOF2 mass spectrometer. Flash column chromatography was performed using silica gel 60 (particle size 200–400 mesh) purchased from Aldrich (WI). Thin layer chromatography (TLC) was performed on aluminum-backed silica gel plates (Aldrich 60 F254. 200 μm in thickness).
Ethyl (2R)-(−)-N-(tert-butoxycarbonyl)-D-pyroglutamate (2)
m.p 54–5 °C. 1H NMR (CDCl3): δ 4.56 (dd, 1H), 4.20 (q, 2H), 2.66-2.22 (m, 3H), 2.03-1.96 (m, 1H), 1.46 (s, 9H), 1.26 (t, 3H). ESMS: m/e 280.0 [M+Na]+, 296.0 [M+K] +.
Ethyl (2R,4S)-N-(tert-butoxycarbonyl)-4-(2-naphthyl)methyl-D-pyroglutamate (3)
m.p. 162–4 °C. 1H NMR (CDCl3): δ 7.82-7.77 (m, 3H), 7.61 (s, 1H), 7.49-7.42 (m, 2H), 7.30 (dd, 1H), 4,46 (dd, 1H), 4.17 (q, 2H), 3.46 (dd, 1H), 3.06 (dq, 1H), 2.84 (dd, 1H), 2.11-2.00 (m, 2H), 1.49 (s, 9H), 1.23 (t, 3H). ESMS: m/e 420.2 [M+Na]+, 436.1 [M+K]+.
(2R,4S)-2-Amino-4-(2-naphthyl)methyl pentanedioic acid (4)
m.p. 178–9 °C (lit 8: m.p. 144 °C). 1H NMR (DMSO-d6): δ 8.44 (bs, 2H), 7.87-7.81 (m, 3H), 7.68 (s, 1H), 7.47-7.44 (m, 2H), 7.38 (dd, 1H), 3.83 (dd, 1H), 3.06-3.01 (m, 3H), 2.14-2.08 (m, 1H), 1.89-1.83 (m, 1H). 13C NMR (DMSO-d6): δ 176.5, 170.6, 136.9, 133.8, 132.7, 128.2, 127.7, 127.5, 127.3, 126.0, 125.6, 51.5, 43.4, 38.3, 31.8. ESMS: m/e 288.1 [M+H]+. HRMS calcd for [C16H17NO4−H]− 286.10793. Found 286.10873.
Ethyl (2R,4S)-N-(tert-butoxycarbonyl)-4-cinnamyl-D-pyroglutamate (5)
m.p. 96–7 °C. 1H NMR (CDCl3): δ 7.35-7.19 (m, 5H), 6.35 (d, 1H), 6.15 (ddd, 1H), 4,54 (dd, 1H), 4.22 (q, 2H), 2.85-2.71 (m, 2H), 2.43-2.36 (m, 1H), 2.24-2.01 (m, 2H), 1.50 (s, 9H), 1.28 (t, 3H). ESMS: m/e 396.2 [M+Na]+, 412.2 [M+K]+.
Ethyl (2R,4S)-4-cinnamyl-D-pyroglutamate (6)
To a solution of ethyl (2R,4S)-N-(tert-butoxycarbonyl)-4-cinnamyl-D-pyroglutamate (5) (680 mg, 1.82 mmol) in methylene chloride (68 mL) was added trifluoroacetic acid (1.05 g, 9.15 mmol). The reaction mixture was stirred at room temperature for 3 h and then poured in a mixture of ice (20 g) and saturated sodium bicarbonate (10 mL). The organic layer was separated and water layer was extracted with methylene chloride (20 mL × 3). The organic layers were combined, washed with brine (50 mL), dried over sodium sulfate, and evaporated under reduced pressure to dryness. Chromatography of the residue on a silica gel column (hexane/methylene chloride/ethyl acetate 1:1:1) gave 350 mg (70.3 %) of 6 which was crystallized in ethyl acetate/hexane to yield while fluffy crystals. m.p. 100–1°C. 1H NMR (CDCl3): δ 7.36-7.19 (m, 5H), 6.47 (d, 1H), 6.16 (ddd, 1H), 5.95 (bs, 1H, N-H), 4.20 (q, 2H), 4.14 (dd, 1H), 2.73-2.61 (m, 2H), 2.45-2.36 (m, 2H), 2.29-2.21 (m, 1H), 1.28 (s, 3H). 13C NMR (CDCl3): δ 179.1, 172.3, 137.2, 132.9, 128.7, 127.5, 126.5, 126.3, 61.8, 53.9, 39.7, 34.2, 30.6, 14.3. ESMS: m/e 274.3 (M+H+), 296.2 [M+Na]+, 312.1 [M+K]+. HRMS calcd for [C16H19NO3+H]+ 274.14439. Found 274.14353.
(5R,3S)-3-cinnamyl-5-hydroxylmethl-2-prrolidinone (7)
Under a positive pressure of nitrogen, a 2.0 M solution of lithium borohydride in THF (1.37 mL, 2.74 mmol) was dropwise added to a solution of ethyl (2R,4S)-4-cinnamyl-D-pyroglutamate (6) (750 mg, 2.74 mmol) in anhydrous THF (20 mL). The resulting reaction mixture was stirred at room temperature overnight (about 16 hours). The mixture was cooled in an ice-bath and then quenched with ice (25 g). The THF was removed under reduce pressure and the remaining was extracted with ethyl acetate (50 mL × 3). The ethyl acetate layers were washed with sodium bicarbonate (50 mL), brine (50 ml × 2), dried over sodium sulfate and evaporated to dryness. Chromatography of the residue on a silica gel column (acetone/methylene chloride 2:1) gave 610 mg (96.2 %) of 7 as colorless oil, which was solidified while standing on bench and recrystallized in ethyl acetate/hexane to yield white crystals. m.p. 102–3°C. 1H NMR (CDCl3): δ 7.35-7.18 (m, 5H), 6.5 (d, 1H), 6.16 (ddd, 1H), 4.04 (m, 1H), 3.69-3.63 (m, 2H), 3.48-3.44 (m, 1H), 2.69-2.61 (m, 2H), 2.36-2.31 (m, 1H), 1.99-1.94 (m, 2H). 13C NMR (CDCl3): δ 180.7, 137.6, 132.8, 128.9, 127.6, 127.1, 126.5, 66.3, 54.9, 41.2, 35.1, 28.8. ESMS: m/e 232.4 [M+H]+, 254.2 [M+Na]+, 270.1 [M+K]+. HRMS calcd for [C14H17NO2+H]+ 232.13387. Found 232.13302.
(2R,4S,E)-2-Amino-4-(3-phenylprop-2-enyl)pentanedioic acid (8)
m.p. 161–4°C. 1H NMR (DMSO-d6): δ 7.26-7.09 (m, 5H), 6.40 (d, 1H), 6.06 (ddd, 1H), 4.40-4.33 (qq, 2H), 2.62-2.37 (m, 3H), 2.29-2.18 (m, 1H). 13C NMR (DMSO-d6): δ 178.7, 174.6, 137.6, 132.8, 128.9, 127.6, 127.1, 126.5, 53.2, 46.8, 37.1, 33.4. ESMS: m/e 264.1 [M+H]+. HRMS calcd for [C14H17NO4−H]− 262.10793. Found 262.10846.
(4R,2S)-2-cinnamyl-4-amino-5-hydroxypentanoic acid (9)
(5R,3S)-3-cinnamyl-5-hydroxylmethl-2-prrolidinone (7) (150 mg, 0.648 mmol) was dissolved in THF (5 mL) and then 2.5 N lithium hydroxide solution (5 mL, 12.5 mmol) was added to the above solution. The resulting mixture was heated to gent reflux under nitrogen and the reflux lasted for 3 hours. The mixture was cooled to room temperature and acidified with 1 N HCl to pH = 2, extracted with ethyl acetate (30 mL × 3). The combined organic layers were dried over sodium sulfate and evaporated to dryness. The residue was treated with 2 N HCl by refluxing gently for 1 hour. The oily residue was gradually turned to while solid. The solid was filtered, triturated with ethyl ether, filtered off and dried under vacuum to give 111 mg (75.1 %) of 9 as white solid. m.p. 110–2°C. 1H NMR (Methanol-d4): δ 7.19-6.99 (m, 5H), 6.32 (d, 1H), 6.05 (ddd, 1H), 3.66-3.58 (m, 1H), 3.48-3.36 (m, 1H), 3.16-3.14 (m, 1H), 2.61-2.52 (m, 1H), 2.42-2.304 (m, 2H), 1.85 (s, 1H), 1.83-1.77 (m, 1H), 1.70-1.62 (m, 1H). 13C NMR (Methanol-d4): δ 178.8, 138.6, 134.2, 129.6, 128.4, 127.3, 126.5, 62.7, 53.3, 43.2, 37.1, 32.1. ESMS: m/e 250.1 [M+H]+. HRMS calcd for [C14H19NO3−H]− 248.12867. Found 248.12941.
Supplementary Material
Dimer interfaces of RacE2 and RacE1. Interactions between residues are shown with dashed lines. Residues are colored according to their side chain properties.
Acknowledgments
We gratefully acknowledge the synchrotron beamline personnel at the Advanced Photon Source (APS). All data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID (or 22-BM) beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
This work was supported by a grant from the National Institutes of Health (AI056575).
Coordinates and structure factors for the final model have been deposited in the Protein Data Bank with RCSB ID code rcsb025941 and PDB ID code 2DWU for RacE1 and RCSB ID code rcsb037744 and PDB ID code 2GZM for RacE2.
Abbreviations: RacE1, glutamate racemase isozyme 1 from B. anthracis; RacE2, glutamate racemase isozyme 2 from B. anthracis; RacE, glutamate racemase; R2-V149A, V149A mutant of RacE2; ΔANR, (pX01-, pX02-) B. anthracis; DTT, dithiothreitol; TB, Terrific Broth; CD, circular dichroism; ES, enzyme/substrate; FOM, figure-of-merit; LLG, least-likelihood gain; rms, root mean square; PDB, Protein Data Bank; GOLD, Genetic Algorithm for Protein Ligand Docking.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Atlas RM. Bioterriorism: from threat to reality. Annu Rev Microbiol. 2002;56:167–85. doi: 10.1146/annurev.micro.56.012302.160616. [DOI] [PubMed] [Google Scholar]
- 2.Athamna A, Athamna M, Abu-Rashed N, Medlej B, Bast DJ, Rubinstein E. Selection of Bacillus anthracis isolates resistant to antibiotics. J Antimicrob Chemother. 2004;54:424–8. doi: 10.1093/jac/dkh258. [DOI] [PubMed] [Google Scholar]
- 3.Bast DJ, Athamna A, Duncan CL, de Azavedo JC, Low DE, Rahav G, Farrell D, Rubinstein E. Type II topoisomerase mutations in Bacillus anthracis associated with high-level fluoroquinolone resistance. J Antimicrob Chemother. 2004;54:90–4. doi: 10.1093/jac/dkh294. [DOI] [PubMed] [Google Scholar]
- 4.Grohs P, Podglajen I, Gutmann L. Activities of different fluoroquinolones against Bacillus anthracis mutants selected in vitro and harboring topoisomerase mutations. Antimicrob Agents Chemother. 2004;48:3024–7. doi: 10.1128/AAC.48.8.3024-3027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price LB, Vogler A, Pearson T, Busch JD, Schupp JM, Keim P. In vitro selection and characterization of Bacillus anthracis mutants with high-level resistance to ciprofloxacin. Antimicrob Agents Chemother. 2003;47:2362–5. doi: 10.1128/AAC.47.7.2362-2365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep. 2001;18:503–19. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 7.Mock M, Fouet A. Anthrax. Annu Rev Microbiol. 2001;55:647–71. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- 8.de Dios A, Prieto L, Martin JA, Rubio A, Ezquerra J, Tebbe M, Lopez de Uralde B, Martin J, Sanchez A, LeTourneau DL, McGee JE, Boylan C, Parr TR, Jr, Smith MC. 4-Substituted D-glutamic acid analogues: the first potent inhibitors of glutamate racemase (MurI) enzyme with antibacterial activity. J Med Chem. 2002;45:4559–70. doi: 10.1021/jm020901d. [DOI] [PubMed] [Google Scholar]
- 9.Fisher SL, Kern G, Newton T, Lundqvist T, Folmer R, Xue Y. 45th Annual ICAAC, American Society for Microbiology 2005 [Google Scholar]
- 10.Keating TA, Fisher SL, Lundqvist T. Interscience Conference on Antimicrobial Agents and Chemotherapy. San Francisco, CA: 2006. [Google Scholar]
- 11.Doublet P, van Heijenoort J, Bohin JP, Mengin-Lecreulx D. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J Bacteriol. 1993;175:2970–9. doi: 10.1128/jb.175.10.2970-2979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnard JP, Friedlander AM. Vaccination against anthrax with attenuated recombinant strains of Bacillus anthracis that produce protective antigen. Infect Immun. 1999;67:562–7. doi: 10.1128/iai.67.2.562-567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shatalin KY, Neyfakh AA. Efficient gene inactivation in Bacillus anthracis. FEMS Microbiol Lett. 2005;245:315–9. doi: 10.1016/j.femsle.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 14.Tanner ME. Understanding nature’s strategies for enzyme-catalyzed racemization and epimerization. Acc Chem Res. 2002;35:237–46. doi: 10.1021/ar000056y. [DOI] [PubMed] [Google Scholar]
- 15.Glavas S, Tanner ME. Catalytic acid/base residues of glutamate racemase. Biochemistry. 1999;38:4106–13. doi: 10.1021/bi982663n. [DOI] [PubMed] [Google Scholar]
- 16.Rios A, Richard JP. Biological Enolates: Generation and Stability of the Enolate of N-Protonated Glycine Methyl Ester in Water. J Am Chem Soc. 1997;119:8375–8376. [Google Scholar]
- 17.Shaw JP, Petsko GA, Ringe D. Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution. Biochemistry. 1997;36:1329–42. doi: 10.1021/bi961856c. [DOI] [PubMed] [Google Scholar]
- 18.Gallo KA, Knowles JR. Purification, cloning, and cofactor independence of glutamate racemase from Lactobacillus. Biochemistry. 1993;32:3981–90. doi: 10.1021/bi00066a019. [DOI] [PubMed] [Google Scholar]
- 19.Gallo KA, Tanner ME, Knowles JR. Mechanism of the reaction catalyzed by glutamate racemase. Biochemistry. 1993;32:3991–7. doi: 10.1021/bi00066a020. [DOI] [PubMed] [Google Scholar]
- 20.Tanner ME, Gallo KA, Knowles JR. Isotope effects and the identification of catalytic residues in the reaction catalyzed by glutamate racemase. Biochemistry. 1993;32:3998–4006. doi: 10.1021/bi00066a021. [DOI] [PubMed] [Google Scholar]
- 21.Hwang KY, Cho CS, Kim SS, Sung HC, Yu YG, Cho Y. Structure and mechanism of glutamate racemase from Aquifex pyrophilus. Nat Struct Biol. 1999;6:422–6. doi: 10.1038/8223. [DOI] [PubMed] [Google Scholar]
- 22.Glavas S, Tanner ME. Active site residues of glutamate racemase. Biochemistry. 2001;40:6199–204. doi: 10.1021/bi002703z. [DOI] [PubMed] [Google Scholar]
- 23.Ruzheinikov SN, Taal MA, Sedelnikova SE, Baker PJ, Rice DW. Substrate-induced conformational changes in Bacillus subtilis glutamate racemase and their implications for drug discovery. Structure (Camb) 2005;13:1707–13. doi: 10.1016/j.str.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 24.Doublet P, van Heijenoort J, Mengin-Lecreulx D. The glutamate racemase activity from Escherichia coli is regulated by peptidoglycan precursor UDP-N-acetylmuramoyl-L-alanine. Biochemistry. 1994;33:5285–90. doi: 10.1021/bi00183a035. [DOI] [PubMed] [Google Scholar]
- 25.Kim SS, Choi IG, Kim SH, Yu YG. Molecular cloning, expression, and characterization of a thermostable glutamate racemase from a hyperthermophilic bacterium, Aquifex pyrophilus. Extremophiles. 1999;3:175–83. doi: 10.1007/s007920050114. [DOI] [PubMed] [Google Scholar]
- 26.Taal MA, Sedelnikova SE, Ruzheinikov SN, Baker PJ, Rice DW. Expression, purification and preliminary X-ray analysis of crystals of Bacillus subtilis glutamate racemase. Acta Crystallogr D Biol Crystallogr. 2004;60:2031–4. doi: 10.1107/S0907444904021134. [DOI] [PubMed] [Google Scholar]
- 27.Briggs GE, Haldane JB. A Note on the Kinetics of Enzyme Action. Biochem J. 1925;19:338–9. doi: 10.1042/bj0190338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleland WW. The use of pH studies to determine chemical mechanisms of enzyme-catalyzed reactions. Methods Enzymol. 1982;87:390–405. doi: 10.1016/s0076-6879(82)87024-9. [DOI] [PubMed] [Google Scholar]
- 29.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. Embo J. 1997;16:3787–96. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashiuchi M, Kuwana E, Komatsu K, Soda K, Misono H. Differences in effects on DNA gyrase activity between two glutamate racemases of Bacillus subtilis, the poly-gamma-glutamate synthesis-linking Glr enzyme and the YrpC (MurI) isozyme. FEMS Microbiol Lett. 2003;223:221–5. doi: 10.1016/S0378-1097(03)00381-1. [DOI] [PubMed] [Google Scholar]
- 31.Ashiuchi M, Kuwana E, Yamamoto T, Komatsu K, Soda K, Misono H. Glutamate racemase is an endogenous DNA gyrase inhibitor. J Biol Chem. 2002;277:39070–3. doi: 10.1074/jbc.C200253200. [DOI] [PubMed] [Google Scholar]
- 32.Kimura K, Tran LS, Itoh Y. Roles and regulation of the glutamate racemase isogenes, racE and yrpC, in Bacillus subtilis. Microbiology. 2004;150:2911–20. doi: 10.1099/mic.0.27045-0. [DOI] [PubMed] [Google Scholar]
- 33.Mesecar AD, Koshland DE., Jr Sites of binding and orientation in a four-location model for protein stereospecificity. IUBMB Life. 2000;49:457–66. doi: 10.1080/152165400410326. [DOI] [PubMed] [Google Scholar]
- 34.Mesecar AD, Koshland DE., Jr A new model for protein stereospecificity. Nature. 2000;403:614–5. doi: 10.1038/35001144. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Iwata K, Kita A, Kawarabayasi Y, Yohda M, Miki K. Crystal structure of aspartate racemase from Pyrococcus horikoshii OT3 and its implications for molecular mechanism of PLP-independent racemization. J Mol Biol. 2002;319:479–89. doi: 10.1016/S0022-2836(02)00296-6. [DOI] [PubMed] [Google Scholar]
- 36.Puig E, Garcia-Viloca M, Gonzalez-Lafont A, Lluch JM. On the ionization state of the substrate in the active site of glutamate racemase. A QM/MM study about the importance of being zwitterionic. J Phys Chem A Mol Spectrosc Kinet Environ Gen Theory. 2006;110:717–25. doi: 10.1021/jp054555y. [DOI] [PubMed] [Google Scholar]
- 37.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katzin LI, Gulyas E. Absorption, rotatory dispersion, and circular dichroism studies on some hydroxy and amino acids. J Am Chem Soc. 1968;90:247–51. doi: 10.1021/ja01004a006. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch WJ. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 (Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 43.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 44.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–6. 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A Program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 46.Chen ME. Nitrogen, Oxygen, Carbon 2.0.1.12 edit. Beijing: 2005. [Google Scholar]
- 47.Pedregal C, Collado I, Escribano A, Ezquerra J, Dominguez C, Mateo AI, Rubio A, Baker SR, Goldsworthy J, Kamboj RK, Ballyk BA, Hoo K, Bleakman D. 4-Alkyl- and 4-cinnamylglutamic acid analogues are potent GluR5 kainate receptor agonists. J Med Chem. 2000;43:1958–68. doi: 10.1021/jm9911682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dimer interfaces of RacE2 and RacE1. Interactions between residues are shown with dashed lines. Residues are colored according to their side chain properties.