Abstract
We present an approximation to a molecule’s N-dimensional conformational probability density function (pdf) in terms of marginal pdfs of highest order l, where l is much less than N. The approximation is constructed as a product of conditional pdfs derived by recursive application of the generalized Kirkwood superposition approximation. Furthermore, an algorithm is presented to sample conformations from the approximate full-dimensional pdf based upon all input marginal pdfs. The sampling algorithm is tested for three small molecule systems by using the algorithm to sample conformations at levels l=1, 2, or 3 and comparing the distributions of sampled conformations with those from the molecular dynamics (MD) simulations. The distributions of conformations sampled at third (l=3) order resemble the MD distributions rather well and significantly better than those sampled at second (l=2) or first (l=1) order. In addition to highlighting the importance of correlations among internal degrees of freedom, these results suggest that low-order correlations suffice to describe most of the conformational fluctuations of molecules in a thermal environment.
INTRODUCTION
Molecules are flexible structures in a thermal environment, so their conformations fluctuate continuously. Characterization of molecular fluctuations is critical to understanding and ultimately controlling molecular properties and functions, such as binding and catalysis. Fluctuations also are of central importance in statistical thermodynamics, which links molecular motions to thermodynamic observables such as free energy, entropy, and enthalpy. A molecule’s configurational entropy, in particular, depends upon not only the magnitude of its conformational fluctuations along each degree of freedom but also the degree to which these fluctuations are mutually correlated.1 As a consequence, calculations of configurational entropy can shed light on correlations.
The relationship between correlation and entropy is especially clear in the mutual information expansion (MIE), a series expansion of the Gibbs entropy in terms of mutual information sums of increasing order.1, 2, 3 The mutual information at a given order goes to zero if there are no correlations at that order, and truncating the MIE series at a given order yields a value of the entropy under the approximation that higher-order correlations are absent. One may therefore assess the order of correlations in a given system by comparing the entropy from corresponding truncations of the MIE with the entropy from a second computational method that accounts for all physically relevant correlations. The mining minima (M2) method4 can serve as this second method for molecular systems of modest size (e.g., small molecules and host-guest systems). M2 obtains the Helmholtz free energy and average energy of a molecule by computing its configurational integral as a sum over local energy minima on an energy surface defined by an empirical force field and an implicit solvent model.4 The configurational entropy may then be computed as essentially the difference between the free energy and the mean energy. Since the M2 method uses the full configurational integral (subject to the approximation of summing over local energy wells) it effectively accounts for all physically relevant correlations at a given temperature.
It has been observed1 that molecular entropies estimated with MIE at the doublet and triplet levels—i.e., neglecting correlations above second and third orders, respectively—can agree well with independent M2 calculations for small molecules. Because M2 includes essentially all correlations, this observation suggests that most of the physically relevant fluctuations of these molecules involve only low-order correlations. Thus, it leads to the hypothesis that conformational probability distributions can be well described with a tractable set of low-order distribution functions. The present study describes a novel approach to test this hypothesis and provides results for molecular test systems.
THEORY
The conformational fluctuations of a molecule with M atoms at equilibrium are fully described by a Boltzmann probability distribution function (pdf) over its N=3M−6 internal degrees of freedom:
| (1) |
where β=(kT)−1, k being the Boltzmann constant and T the temperature, and E(X1,…,XN) is the energy as a function of internal coordinates . For a molecule in solution, the energy function comprises the molecule’s internal energy and an additional contribution from the solvent.5 The pdf, Eq. 1, can account for correlations of all orders up to N, but its high dimensionality makes it computationally unworkable for most molecules of interest.
The present study poses the following question: how accurately can the conformational fluctuations of a molecule in a thermal environment be described without accounting for high-order correlations? This is addressed by using a molecule’s low-order marginal pdfs, populated with well-converged simulation data, as a basis for sampling full molecular conformations. The resulting conformational distributions, which, by construction, account only for low-order correlations, are then evaluated by comparing them with those obtained from the simulations. The sampling is done with a novel sampling approximation in which the ancestral sampling algorithm6 is combined with a further generalization1 of the generalized Kirkwood superposition approximation3, 7 (GKSA) that allows the required high-order conditional pdfs to be estimated in terms of the available low-order marginals.
The following sections begin by introducing a notation enabling superposition approximations to be expressed compactly. The l-level superposition approximation (SA-l) is then described, followed by derivation of SA-l based conditional distributions in terms of given marginal pdfs. Finally, algorithms to sample conformations using the conditional distributions are presented.
Notation
Let be N random variables representing, for example, the N internal coordinates of a molecule. Specific numerical values of the random variables are denoted by lowercase letters, so that represents a particular conformation of the N-dimensional system. To make subsequent equations more readable, we introduce a compact notation for the pdf over a subset of k coordinates (Xi1,…,Xik), with i1,…,ik∊[1,N], in which the variables are denoted by just their index ik:
| (2) |
The order of the pdf is the total number of variable labels in the square bracket. For example, [3]≡p1(X3) denotes the one-dimensional (1D) marginal pdf, or singlet distribution, in X3; [3,5]≡p2(X3,X5) denotes the two-dimensional (2D) marginal pdf, or doublet distribution, in X3 and X5; and [3,5,6]≡p3(X3,X5,X6) denotes the three-dimensional (3D) marginal pdf, or triplet distribution, in variables X3, X5, and X6. Moreover, a specific value of the probability density pk(xi1,…,xik) at Xi1=xi1,Xi2=xi2,… is denoted by [xi1,…,xik], i.e.,
| (3) |
Specifying the values of a subset of variables effectively lowers the order of a distribution. For example, if variable X5 has a value of x5, the triplet distribution p3(X3,X5,X6)≡[3,5,6] becomes a 2D distribution p2(X3,X5=x5,X6), denoted by [3,x5,6].
The conditional distribution of variable Xl, given the condition that k other variables Xii,…,Xik have values xi1,…,xik, is denoted by
| (4) |
where l,i1,…,ik∊[1,N] and l≠i1≠⋯≠ik. Note that [l∣xi1,…,xik] is a 1D distribution of variable Xl, and that the product rule8 allows this distribution to be expressed as
| (5) |
Since xi1,…,xik are specific values of the variables, the denominator of Eq. 5 is just a number while the numerator is a 1D pdf.
We will be presenting an approximation of a k-dimensional pdf in terms of products of its lower-order marginal pdfs. The first-order (singlet), second-order (doublet), third-order (triplet), and fourth-order (quadruplet) marginals of a k-order distribution are given by
| (6) |
When the distributions are discretized, as in the present work, the integrals are replaced by summations. We furthermore denote the products of all unique singlet, doublet, triplet, and quadruplet distributions as
| (7) |
For N random variables, there are , , , and singlet, doublet, triplet, and quadruplet marginal distributions, respectively.
Superposition approximations
A superposition approximation approximates a higher-order pdf in terms of lower-order distributions. The original Kirkwood superposition approximation (KSA),9 which approximates a triplet correlation function in terms of singlet and doublet distribution functions,
| (8) |
was introduced in the distribution function theory of liquids as a closure equation to truncate the Bogoliubov-Born-Green-Kirkwood-Yvon (BBGYK) (Ref. 10) hierarchy of distribution functions and thereby enable calculation of the pair correlation function, which is equivalent to the doublet distribution here. The KSA was derived by approximating the three-particle potential of mean force as the sum of three two-particle potentials of mean force, one for each pair of the three particles. The Fisher–Kopelovich superposition approximation11 is an extension of the KSA, also introduced in theory of liquids, which approximates a four-particle distribution function in terms of three-particle marginals:
| (9) |
The GKSA,3, 7 introduced as a probabilistic closure equation, extends the KSA to a general order, approximating a k-order distribution function in terms of all lower-order (up to k−1) marginals:
| (10) |
Note that the KSA and its subsequent generalizations are not normalized pdfs.10
By recursive application of the GKSA, the k-order pdf can be approximated in terms of marginal pdfs of order l and lower to yield an l-level superposition approximation.1 For example, approximating the triplet distributions in the numerator of Eq. 9 by their KSAs according to Eq. 8 yields a second-level (l=2) approximation to a four-dimension (k=4) pdf:
| (11) |
Furthermore, approximating the doublets in the numerator of Eq. 11 as products of singlets yields the first-level (l=1) approximation to the four-dimension (k=4) pdf:
| (12) |
In general, approximations of a k-dimensional pdf at the singlet (SA-1), doublet (SA-2), and triplet (SA-3) levels are given by
| (13) |
| (14) |
and
| (15) |
where the symbols on the right-hand side are defined in Eq. 7. The level of approximation l may be much less than the dimensionality k of the full distribution. In this work, singlet (l=1), doublet (l=2), and triplet (l=3) level superposition approximations are used. Clearly a mixed level of approximation could also be written; for example, one could approximate only a subset of the triplets in Eq. 9 by their KSAs. As noted above, the SA-l pdfs are not normalized at any level except for the trivial singlet-level approximation.
Superposition approximations to conditional distributions
The SA-l approximation to the k-dimensional joint pdf can be used to derive approximate conditional distributions of a variable given values of all other variables. These approximate conditionals are obtained simply by substituting the SA-l approximations for the joint pdfs in Eq. 5. For example, using the SA-2 expression for [x1,x2,3] yields
| (16) |
and using the SA-2 expression for [x1,x2,x3,4] and [x1,x2,x3] gives
| (17) |
where N3 and N4 denote the normalization factor as a function of the fixed variables. In general, the conditional distributions are not normalized due to the use of superposition approximations for the pdfs in Eq. 5. The normalization factors may be obtained by using the standard procedure of summing over all values of the conditioned variable as follows:
| (18) |
and
| (19) |
The normalization factors can be computed efficiently as the summation is over values of a single variable.
More generally, the normalized conditional probability distribution of variable Xk (k>2), given values of the other k−1 variables, is approximated at the doublet level by
| (20) |
Similarly, Eq. 5 may be approximated at the triplet level by applying the SA-3, yielding the following conditional distribution:
| (21) |
For example, the conditional distributions at the triplet level for k=4 and k=5 are, respectively,
| (22) |
and
| (23) |
Algorithms for sampling based on low-order marginals
We wish to sample molecular conformations while accounting for the correlations represented in marginal pdfs of order l and below. This is easily done at the singlet level (l=1), at which all variables are assumed to be independent: one simply samples each variable from its 1D marginal distribution without reference to the other variables. Accounting for correlations by including higher-order marginals is more complicated. We now describe an approach that combines the ancestral sampling algorithm6 with superposition approximations of the required conditional pdfs.
Ancestral sampling algorithm
Any N-dimensional joint pdf can be factorized into a product of conditional distributions by repeated application of the product rule of probability theory:
| (24) |
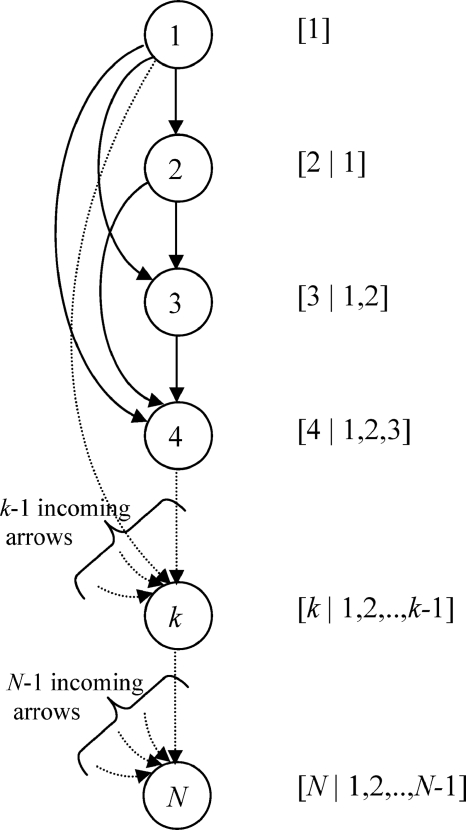
Equation 24 can be represented graphically by the directed, acyclic graph shown in Fig. 1, and the ancestral sampling algorithm6 can be used to sample from any pdf once it is represented by a directed acyclic graph. The following pseudocode explains how the ancestral algorithm samples a conformation from the N-dimensional pdf [1,2,…,N] expressed by the graph in Fig. 1:
Algorithm 1: Ancestral sampling algorithm.
| x1∼[1], | |
| x2∼[2∣x1], | |
| x3∼[3∣x1,x2], | |
| FOR k=4 to N | |
| xk∼[k∣x1,…,xk−1]. | |
| ENDFOR |
where “∼” means “sampled from.” The first variable is sampled from its 1D singlet distribution, and each subsequent variable is sampled from its 1D distribution conditioned upon the values of all the variables that have been sampled so far. Since the full N-dimensional pdf is known, all conditional distributions can be computed exactly and are normalized. The ancestral algorithm rigorously samples conformations from the full N-dimensional joint pdf. However, in many real-world applications, such as the present molecular studies, the required conditional distributions are not accessible because of the high dimensionality of the systems. This problem is addressed in the following subsection.
Figure 1.
Representation of an N-dimensional distribution function [1,2,…,N] as a directed graph. Variables are represented by circles containing the label of the respective variables. Solid arrows indicate the conditional dependencies of each variable as per the 1D conditional distribution indicated on the right of each node. The ancestral sampling uses exact conditional distributions for each variable. The l-level sampling algorithms presented in use approximations to conditional pdfs for (l+1)th and following variables. Doublet-level algorithm uses Eq. 20 while triplet-level algorithm uses Eq. 21. Dashed lines schematize elided portions of the graph.
Ancestral sampling with the superposition approximation
The present study uses an approximation of the standard ancestral sampling method wherein the exact conditional pdfs are replaced with superposition approximations of the conditional pdfs. Thus a doublet-level sampling algorithm is obtained by using conditional pdfs computed using only singlet and doublet distributions [Eq. 20]:
Algorithm 2: Doublet-level sampling algorithm.
| x1∼[1], | |
| FOR k=4 to N | |
| ENDFOR |
The triplet-level sampling algorithm is obtained simply by approximating the conditional distributions with the triplet-level superposition approximation [Eq. 21]:
Algorithm 3: Triplet-level sampling algorithm.
| x1∼[1], | |
| FOR k=5 to N | |
| ENDFOR |
These algorithms may be iterated to generate any desired number of sampled conformations, where the separate iterations are, of course, independent of each other and hence amenable to parallelization. These two specific algorithms also generalize to higher levels of approximation—i.e., to larger values of l—and the standard ancestral algorithm is recovered when l=N. Indeed, the l-level sampling algorithm is identical to ancestral sampling for the first l variables, because the conditional distributions become approximate only at the (l+1)th variable in the graph.
These approximate sampling algorithms require as input all marginals of the full pdf up to the level of the approximation, l; these input marginals are termed reference marginals. Thus, the doublet-level algorithm requires access to all singlet- and doublet-level reference marginals, and the triplet-level algorithm requires the triplet-level reference marginals as well. Importantly, these algorithms do not require computation or storage of the full N-level pdf or its N-dimensional superposition approximation. For the molecular systems studied here, the reference marginal pdfs may be computed from molecular simulations. For example, the doublet distribution of two torsion angles can be computed and stored simply as their normalized 2D histogram. When the distributions are represented as normalized histograms, as done in this work, then a conformational sample refers to a vector of bin numbers, one for each variable.
The distribution of sampled conformations obtained from these algorithms, unlike the standard ancestral algorithm, depends on the order in which the variables are sampled. This can be seen by recognizing that the first l variables are sampled directly from their reference marginals, whereas subsequent variables are sampled from approximations to their original, reference distributions. Note, too, that if the reference marginals have regions of zero probability, the present algorithms can fail to yield a complete set of variables for a conformation. This may be understood by considering the example of an SA-3 based conditional distribution:
| (25) |
After the first three variables have been sampled, the factors containing the next variable, X4, are essentially 1D histograms. When some bins contain values of zero, the product of two such histograms can have zero in all bins. This “null” condition makes it impossible to sample the fourth variable according to the algorithm. A related characteristic of the present approach is that the last variable is sampled from a conditional distribution constructed from the N-dimensional SA-l, which includes all reference marginal pdfs up to and including level l. Consequently, the populated regions of the marginal pdfs computed from the sampled conformations (termed the sampled marginals) will be contained within the populated regions of the corresponding reference marginal pdfs. Section 4D examines these issues by comparing sampled marginal pdfs with the corresponding reference marginal pdfs.
The probability of sampling each conformation with the present SA-l based sampling algorithms, , is readily obtained during the sampling procedure by substituting the conditional pdf used to sample each variable into the chain rule given by Eq. 24. The sampling probability thus obtained is automatically normalized since all conditional pdfs used in the chain rule are normalized. Note that, due to the normalization of the approximate conditional distributions, the sampling probability of a conformation with the present algorithm is different from the N-dimensional SA-l probability of that conformation, pSA-l. The Appendix0 provides an analytic expression for the probability of sampling a conformation in three dimensions using a doublet-level sampling algorithm, , and shows that it is normalized. The sampling distribution also is compared with the second-level superposition approximation, pSA-2(x1,x2,x3).
METHODS
Overview
We wish to determine whether the molecular conformations sampled with the algorithms described above are distributed similarly to those sampled by the molecular dynamics (MD) calculations from which the reference marginals used in the algorithms were obtained. The overall strategy is to
-
(1)
run a MD simulation of a molecule;
-
(2)
use the resulting trajectory to compute first-, second-, and third-order reference marginal pdfs—essentially normalized histograms—for internal coordinates of the molecule;
-
(3)
use these reference marginals in the sampling algorithms to sample molecular conformations in the absence of explicit high-order correlations;
-
(4)
reconstruct the 3D conformations of the molecule dictated by the sampled internal coordinates;
-
(5)
compare the distributions of internal coordinates, conformations, and energies obtained by sampling to those from the original MD run.
Two different systems of internal coordinates are considered, bond-angle-torsion (BAT) and anchored Cartesian (XYZ).12 For simplicity, distributions are not computed for some internal coordinates with very narrow distributions, such as dihedrals within a phenyl ring; instead, these coordinates are held fixed at their equilibrium values established by the force field. The coordinates which are sampled are termed “active.”
Molecular systems
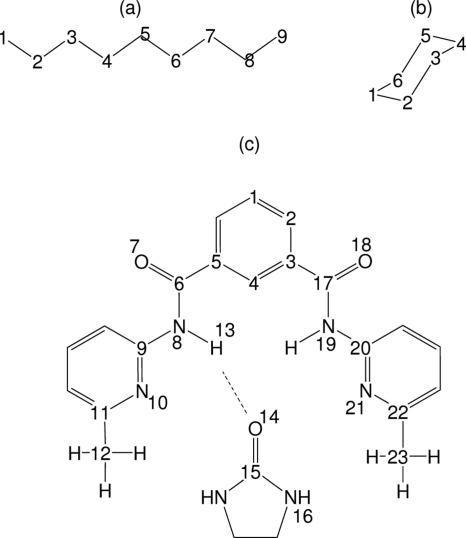
Three molecular systems were studied: linear nonane, cyclohexane, and a small host-guest complex.13 Figure 2 diagrams the chemical structures of these molecules and the subsets of atoms used for testing the sampling algorithms. For each system, 5×106 MD snapshots spanning 50 ns of a single MD simulation were processed. The MD trajectories are those used in a previous study of configurational entropy1 from our group. The MD simulations use an all-hydrogen energy model and approximate the effects of solvent with a simple distance-dependent dielectric model.
Figure 2.
Molecular systems used for testing the superposition approximation-based sampling algorithms: (a) nonane, (b) cyclohexane, and (c) host-guest complex (Ref. 13). Atoms included in the sampled structure are numbered. The internal coordinate system is set up such that atom 1 is on the origin, the bond between atoms 1 and 2 is along the x axis, and atom 3 is in the x-y plane. For nonane and cyclohexane, only the carbon chain is sampled, although the MD simulations included all hydrogens. For the host-guest complex, a dotted line represents the pseudobond used in defining the position and orientation of the guest relative to the host.
The analyses of nonane and cyclohexane were simplified by neglecting the coordinates of all-hydrogen atoms. This leaves 12 internal coordinates for the six carbons of cyclohexane (for BAT coordinates, these comprise 5 bond lengths, 4 bond angles, and 3 torsions) and 21 internal coordinates for the nine carbons of nonane (for BAT coordinates, these comprise 8 bond lengths, 7 bond angles, and 6 torsions). Conformations were sampled at the singlet level by independently sampling from the singlet marginal pdfs and at the doublet and triplet levels by using the algorithms described in Sec. 2D2 in both BAT and Cartesian coordinates.
The analysis of the host-guest system was simplified by limiting attention to a skeleton of 23 atoms (see Fig. 2) out of the simulated 56-atom complex. The retained atoms correspond to 63(3×23−6) internal degrees of freedom. Conformations were sampled at the singlet, doublet, and triplet levels in the BAT coordinate system. Out of the 63 BAT coordinates associated with the 23 atoms, 32 coordinates were treated as active: all bond-angle and torsional degrees of freedom except the ones in the rings, along with one pseudobond, two pseudoangles, and three pseudodihedrals4 that together specify the position and orientation of the guest with respect to the host. Table 1 lists the active degrees of freedom as well as the equilibrium values of the inactive ones. Three torsion angles (see Table 1) that might be viewed as flexible are treated as phase angles14 of flexible torsions that share the same rotatable bond; these phase angles have narrow distributions and are therefore treated as inactive.
Table 1.
List of BAT degrees of freedom corresponding to the 23-atom skeleton of the host-guest complex. All 22 bonds, 21 angles, and 20 torsions are listed using the atom numbers shown in Fig. 2. Active variables are indicated by A. For the inactive variables, the equilibrium values based on the force field are listed.
| Bond | Bond equilibrium value (Å) | Angle | Angle equilibrium value (rad) | Torsion | Torsion equilibrium value (rad) |
|---|---|---|---|---|---|
| 1-2 | 1.383 | 1-2-3 | 2.0944 | 1-2-3-4 | 0 |
| 2-3 | 1.383 | 2-3-4 | 2.0944 | 1-2-3-17 | A |
| 3-4 | 1.383 | 2-3-17 | A | 2-3-4-5 | 0 |
| 3-17 | 1.46 | 3-4-5 | 2.0944 | 2-3-17-18 | Phase of 2-3-17-19 |
| 4-5 | 1.383 | 3-17-19 | A | 2-3-17-19 | A |
| 5-6 | 1.46 | 3-17-18 | A | 3-17-19-20 | A |
| 6-8 | 1.345 | 4-5-6 | A | 3-4-5-6 | A |
| 6-7 | 1.225 | 5-6-8 | A | 4-5-6-7 | Phase of 4-5-6-8 |
| 8-9 | 1.355 | 5-6-7 | A | 4-5-6-8 | A |
| 8-13 | 1.0 | 6-8-9 | A | 5-6-8-13 | A |
| 9-10 | 1.327 | 6-8-13 | A | 5-6-8-9 | Phase of 5-6-8-13 |
| 10-11 | 1.327 | 8-9-10 | A | 6-8-9-10 | A |
| 11-12 | 1.5 | 8-13-14 | A | 6-8-13-14 | A |
| 13-14 | A | 9-10-11 | 2.0159 | 8-9-10-11 | A |
| 14-15 | 1.225 | 10-11-12 | A | 8-13-14-15 | A |
| 15-16 | 1.345 | 13-14-15 | A | 9-10-11-12 | A |
| 17-19 | 1.345 | 14-15-16 | A | 13-14-15-16 | A |
| 17-18 | 1.225 | 17-19-20 | A | 17-19-20-21 | A |
| 19-20 | 1.355 | 19-20-21 | A | 19-20-21-22 | A |
| 20-21 | 1.327 | 20-21-22 | 2.0159 | 20-21-22-23 | A |
| 21-22 | 1.327 | 21-22-23 | A | ⋯ | ⋯ |
| 22-23 | 1.5 | ⋯ | ⋯ | ⋯ | ⋯ |
Computing marginal and conditional distributions
The reference marginal pdfs required for the sampling algorithms were computed from MD trajectories by binning the active coordinates to create histograms and then normalizing the histograms. For example, the second-order marginal distribution of two torsion angles was obtained through a 2D histogram with square bins. For each variable, NB=30 equally spaced bins were set up between the minimum and maximum values of the data to be binned, except for the torsional coordinates of cyclohexane. To effectively capture the bimodal distribution of these variables, 15 equally spaced bins were used in the two intervals of 0°–110.2° and 248.7°–360°, for a total of 30 bins.
For N active coordinates with NB bins along each coordinate, the total numbers of elements in all matrices for the doublet (N2) and triplet (N3) distributions are, respectively,
| (26) |
These numbers can become large. For example, the largest molecular system considered here has N=32, giving N2=446 400 and N3=133 920 000, corresponding to about 1 Gbyte of memory at double precision (8 byte floats). However, we found that the storage requirements could be reduced about sixfold for all systems studied here by recognizing that many elements of the doublet and triplet distributions are zero and using sparse matrix storage techniques in the MATLAB® 7.5 (Ref. 15) programming environment.
The 1D conditional probability distribution constructed at each step of the SA-l based sampling algorithms consists of a product of distributions [Eqs. 20, 21], and the number of factors in this product becomes large as the number of variables increases. Direct multiplication of these factors can lead to underflow errors because the probabilities that are multiplied lie between 0 and 1. This problem was addressed by taking the logarithms of the factors and adding them instead of multiplying.
Evaluation of sampled conformations
In order to assess the contributions of successively higher-order correlations, distributions of conformations generated via sampling at the singlet, doublet, and triplet levels were compared to the distributions obtained directly from MD. Three types of comparisons were done.
First, the coordinates generated by the sampling algorithms were binned just as done for the MD snapshots, and the resulting 1D, 2D, and 3D sampled marginal pdfs were compared with the reference marginal pdfs obtained directly from MD. The difference between a sampled distribution and the corresponding reference distribution from MD is reported as the root mean square deviation (RMSD) across the bins of the stored distributions.
Second, the sampled distributions were compared with the reference MD distributions by comparing the distributions of energies and key intramolecular distances. Doing this requires reconstructing the molecular conformation associated with each set of internal coordinates generated by the sampling algorithm. Those internal coordinates not included in the sampling (Sec. 3B), such as bond stretches in the case of the host-guest complex, were set to their equilibrium values established by the force field. Because not all internal coordinates were allowed to vary in the sampling, it was necessary to modify the MD snapshots, in which all internal coordinates fluctuate, so that they would be on a comparable footing. This was done by extracting active internal coordinates from the MD snapshots and substituting the equilibrium values for the inactive coordinates, precisely as done for the sampled conformations. Then the MD conformations were reconstructed with these idealized coordinates and compared with the sampled conformations. For the host-guest complex, distributions were compared for multiple interatomic distances that characterize the conformations. For nonane, the distance between the terminal carbons was examined. For cyclohexane, the distance between carbons 1 and 6 was examined; this distance is not part of the BAT coordinate system and depends upon the values of the internal coordinates in the same way that nonane’s end-to-end distance depends upon its internal coordinates. Note that, in the XYZ coordinate system, the end-to-end distances for both nonane and cyclohexane are, in principle, functions of only the Cartesian coordinates of the last carbon. Nonetheless, since these coordinates are the last three variables to be sampled, the distributions of end-to-end distances in the sampled conformations depend on all sampled XYZ coordinates.
Third, the distributions of energy for the sampled conformations, computed with the same CHARMm16 force field model, were compared with those for the reconstructed (see above) MD conformations. Comparisons were made for both the total molecular energy and the separate terms provided by the force field. The separate terms provide additional physical insight; for example, if the sampled conformations were to yield more conformations with high Lennard-Jones energies, this would imply steric clashes.
RESULTS
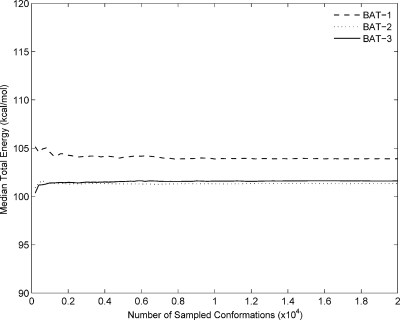
As detailed above, we (1) compute marginal probability distributions for the internal coordinates of nonane, cyclohexane, and a host-guest complex based upon MD simulations; (2) use these marginals to construct superposition approximations to conditional pdfs for the internal coordinates; (3) sample conformations with algorithms based upon these conditional distributions; and (4) compare the sampled conformations with the original MD conformations. For nonane and cyclohexane, 500 000 conformations were sampled, while for the host-guest complex, 200 000 conformations were sampled. Convergence was established based on the medians of energy and the sampled interatomic distances; a representative convergence plot of median total energy for the host-guest system is shown in Fig. 3.
Figure 3.
Convergence of median total energy as a function of the number of conformations sampled in the BAT coordinate system at the singlet (BAT-1), doublet (BAT-2), and triplet (BAT-3) levels for the host-guest complex. The means and standard deviations of the energy converge similarly, and similar results are obtained for the other molecular systems in this study.
The first three subsections here assess the accuracy of conformations sampled at the singlet, doublet, and triplet levels by studying the geometries and energies of the sampled conformations from the superposition approximations. The fourth subsection compares marginal distributions of the superposition approximations with the corresponding marginals of the original MD samples. Finally, the computational complexity of the sampling algorithms is analyzed.
Here BAT-1, BAT-2, and BAT-3 refer to conformational sampling in BAT coordinates at the singlet, doublet, and triplet levels, respectively, while XYZ-1, XYZ-2, and XYZ-3 refer to sampling in Cartesian coordinates at the corresponding levels of approximation. As noted in Sec. 2D, the distributions provided by the present algorithms depend to some degree upon the sequence in which the variables are sampled. Changing the sequence of sampling, such as by sampling bond lengths first versus sampling torsions first, when BAT coordinates are used, was found to alter the results in detail but had little effect on the overall accuracy of the sampled distributions. The sampling sequence in XYZ coordinates follows the atom numbering of Fig. 2 where, for each atom, the x coordinate is sampled first, followed by the y and z coordinates. In BAT coordinates, for nonane and cyclohexane the bond coordinates of all atoms in their indexed order were sampled first, followed by angles and torsions in the same order. Sampling for the host-guest system in BAT coordinates also followed the sequence of bonds, angles, and torsions, according to the order in Table 1.
As discussed in Sec. 2D2, the present algorithms can fail to yield a complete set of variables for a conformation. For the molecular systems studied here, such null samples never occurred for the doublet-level algorithm. At the triplet level, they occurred for <0.01% of the iterations for nonane and the complex and never occurred for cyclohexane.
Nonane
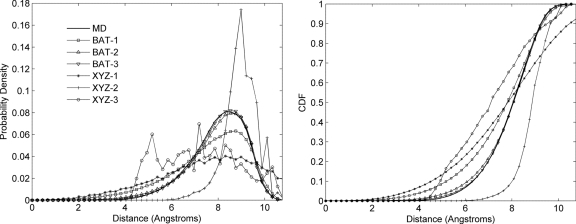
First-, second-, and third-order marginal distributions were computed for nonane in anchored Cartesian and BAT coordinates and used to sample conformations at the singlet, doublet, and triplet levels in both coordinate systems. Thus, conformations are sampled based on a total of 2×3=6 sampling approximations. The probability distributions of end-to-end distances (carbon 1 to carbon 9) from MD and from the six sampled sets are compared numerically in Table 2, and Fig. 4 graphs the corresponding distributions.
Table 2.
Statistics of end-to-end distances (Å) in nonane from MD and sampled conformations. See text for symbols.
| Median | Mean | Standard deviation | Minimum | Maximum | |
|---|---|---|---|---|---|
| MD | 8.16 | 8.01 | 1.14 | 2.99 | 10.98 |
| BAT-1 | 7.91 | 7.60 | 1.64 | 0.09 | 10.89 |
| BAT-2 | 8.18 | 7.96 | 1.29 | 0.24 | 10.80 |
| BAT-3 | 8.18 | 8.01 | 1.18 | 0.67 | 10.85 |
| XYZ-1 | 7.93 | 7.76 | 2.28 | 0.31 | 14.84 |
| XYZ-2 | 8.98 | 8.96 | 0.69 | 3.87 | 11.08 |
| XYZ-3 | 7.23 | 7.24 | 1.66 | 3.05 | 10.97 |
Figure 4.
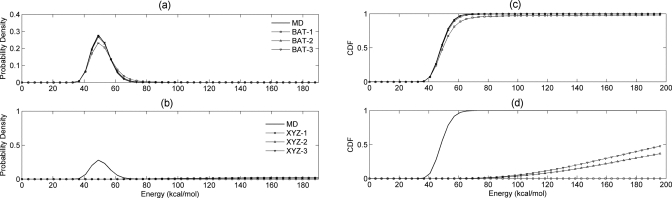
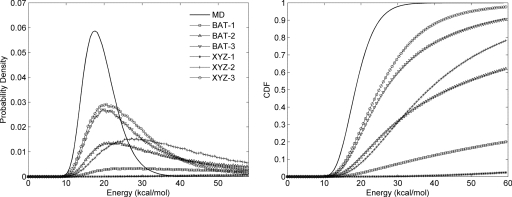
Probability distributions (left) and corresponding cumulative distributions functions (right) of end-to-end distances for nonane from MD conformations and sampling algorithms at singlet, doublet, and triplet levels in BAT and XYZ coordinates.
For BAT coordinates, both the doublet- and triplet-level superposition approximations provide excellent agreement with the MD results and are markedly more accurate than singlet-level sampling. In particular, Fig. 4 shows that doublet- and triplet-level samplings produce a substantially smaller fraction of conformations with excessively short end-to-end distances than does singlet-level sampling. This is also evident from the shorter minimum distance for singlet sampling (0.09 Å) as compared to doublet (0.24 Å) and triplet samplings (0.67 Å) (Table 2). The triplet level is slightly more accurate than the doublet, but the difference is less striking than that between the doublet and singlet.
For Cartesian coordinates, all three sampled cases yield distance distributions that deviate markedly from the MD distributions: although the numerical statistics in Table 2 look reasonable, Fig. 4 shows that the shapes of the distributions are poor. As with sampling in BAT coordinates, the population in the short-distance end of the distribution is notably higher for the singlet than for the doublet or triplet samples. Interestingly, in Cartesian coordinates the triplet-level approximation does not appear to yield greater accuracy than the doublet-level distribution, at least by the present measure.
The BAT coordinate samples of nonane yield distributions of total energy that agree very well with MD overall at the doublet and triplet levels, as shown in Fig. 5a. Interestingly, the total energy distribution at the singlet level is only slightly inferior to those at higher levels, indicating low correlations among various BAT internal coordinates. However, the energy distributions from superposition approximations in Cartesian coordinates disagree strongly with the reference MD distribution: they are wide and shifted to much higher energies, as shown in Fig. 5b. The singlet-level results are particularly poor, as the minimum total energy among all conformations is 307.1 kcal∕mol (see Table 3), which is outside the range of energies in Fig. 5b. The fraction of such high-energy conformations is lower for doublet- and triplet-level samples. It is not surprising that the energy distributions from Cartesian sampling are poor, given the similarly poor end-to-end distance distributions described above.
Figure 5.
Probability distributions and corresponding cumulative distributions functions (right) of total energy of nonane based upon MD conformations and conformations sampled in [(a) and (c)] BAT and [(b) and (d)] XYZ coordinate systems at singlet, doublet, and triplet levels.
Table 3.
Statistics of energy distributions (kcal∕mol) from MD and sampled conformations for nonane.
| Bond | Angle | Torsion | Coulomb | vdW | Total | |
|---|---|---|---|---|---|---|
| Median | ||||||
| MD | 7.7 | 4.4 | 3.23 | 34.5 | −0.63 | 50.2 |
| BAT-1 | 7.8 | 4.5 | 3.21 | 34.9 | −0.57 | 51.5 |
| BAT-2 | 7.8 | 4.5 | 3.22 | 34.4 | −0.62 | 50.6 |
| BAT-3 | 7.8 | 4.5 | 3.22 | 34.4 | −0.63 | 50.4 |
| XYZ-1 | 1.86×104 | 318.4 | 4.91 | 36.0 | 15.33 | 2.2×104 |
| XYZ-2 | 141.2 | 48.6 | 2.78 | 31.7 | −0.52 | 232.3 |
| XYZ-3 | 114.4 | 41.2 | 3.85 | 34.7 | −0.61 | 202.8 |
| Mean | ||||||
| MD | 8.34 | 4.9 | 3.22 | 34.8 | −0.6 | 50.7 |
| BAT-1 | 8.49 | 5.0 | 3.23 | 36.3 | 3.6×103 | 3.6×103 |
| BAT-2 | 8.48 | 5.0 | 3.23 | 35.0 | 182.6 | 234.3 |
| BAT-3 | 8.50 | 5.0 | 3.22 | 34.8 | 11.5 | 63.0 |
| XYZ-1 | 2.1×104 | 325.8 | 4.90 | 37.7 | 9.7×104 | 1.2×105 |
| XYZ-2 | 171.93 | 55.6 | 2.86 | 32.1 | 179.7 | 442.2 |
| XYZ-3 | 136.75 | 47.7 | 3.84 | 35.1 | 252.2 | 475.6 |
| Standard deviation | ||||||
| MD | 4.15 | 2.6 | 1.00 | 2.46 | 0.3 | 5.9 |
| BAT-1 | 4.22 | 2.7 | 1.11 | 5.59 | 1.24×105 | 1.24×105 |
| BAT-2 | 4.22 | 2.7 | 1.03 | 3.20 | 2.67×104 | 2.67×104 |
| BAT-3 | 4.26 | 2.7 | 1.00 | 2.66 | 4.62×103 | 4.62×103 |
| XYZ-1 | 1.21×104 | 120.0 | 1.14 | 7.94 | 6.51×105 | 6.50×105 |
| XYZ-2 | 120.9 | 36.2 | 0.92 | 1.85 | 2.41×104 | 2.41×104 |
| XYZ-3 | 93.4 | 31.9 | 1.10 | 3.22 | 3.33×104 | 3.33×104 |
| Minimum | ||||||
| MD | 0.04 | 0.02 | 0.01 | 28.1 | −1.49 | 31.7 |
| BAT-1 | 0.16 | 0.02 | 0.00 | 28.2 | −1.57 | 32.2 |
| BAT-2 | 0.17 | 0.02 | 0.00 | 28.4 | −1.51 | 32.0 |
| BAT-3 | 0.31 | 0.10 | 0.00 | 28.2 | −1.53 | 32.8 |
| XYZ-1 | 1.92×102 | 3.69 | 0.48 | 22.6 | −1.35 | 307.1 |
| XYZ-2 | 0.88 | 0.47 | 0.06 | 27.6 | −1.01 | 43.1 |
| XYZ-3 | 2.17 | 0.23 | 0.00 | 27.6 | −1.42 | 44.0 |
| Maximum | ||||||
| MD | 45.6 | 34.5 | 7.64 | 49.0 | 5.06 | 100.2 |
| BAT-1 | 49.1 | 28.1 | 7.88 | 244.4 | 9.98×106 | 9.99×106 |
| BAT-2 | 45.7 | 27.4 | 7.87 | 130.7 | 9.00×106 | 9.00×106 |
| BAT-3 | 42.0 | 23.7 | 7.22 | 80.5 | 2.54×106 | 2.54×106 |
| XYZ-1 | 1.16×105 | 955.5 | 9.56 | 352.4 | 9.99×106 | 1.00×107 |
| XYZ-2 | 1.55×103 | 452.4 | 7.44 | 84.7 | 9.96×106 | 9.97×106 |
| XYZ-3 | 1.27×103 | 386.5 | 8.52 | 80.6 | 8.95×106 | 8.95×106 |
The distributions of the total energy of nonane for the BAT samples show excellent agreement with the MD distributions especially at the doublet and triplet levels [in Figs. 5a, 5b]. This is consistent with the good agreement between the median total energy of the sampled conformations with the MD conformations (Table 3). The larger deviations for the mean energy and other statistics result from the small fraction of high-energy conformations among the sampled conformations, as evident from Fig. 5b and Table 4. Closer examination of Table 3 indicates that the bond-stretch and bond-angle energies are well behaved, but van der Waals energies are sometimes much too large. This indicates that the high-energy conformations among the BAT samples result from excessively close atom-atom contacts, consistent with the small values of the minimum end-to-end distances listed in Table 2. In contrast, the data in Table 3 show that the large errors in the energy distributions based upon XYZ coordinates result not only from van der Waals overlaps but also from severe errors in bond lengths and angles. The superior performance of the BAT coordinates, relative to Cartesians, is traceable to the fact that the BAT coordinates do an excellent job of capturing the marginal pdfs of those bond lengths and angles which are included in the coordinate set. Thus, the energetics of these stiff degrees of freedom are well reproduced.
Table 4.
Fraction of sampled conformations with energies greater than the maximum energy obtained in MD simulation of each test system.
| Number of conformations sampled | Number of high-energy conformations | Fraction of high-energy conformations | |
|---|---|---|---|
| Nonane | |||
| BAT-1 | 500 000 | 14 500 | 2.9×10−2 |
| BAT-2 | 500 000 | 1 450 | 2.9×10−3 |
| BAT-3 | 500 000 | 170 | 3.4×10−4 |
| XYZ-1 | 500 000 | 500 000 | 1.00 |
| XYZ-2 | 500 000 | 480 000 | 0.96 |
| XYZ-3 | 500 000 | 475 000 | 0.95 |
| Cyclohexane | |||
| BAT-1 | 500 000 | 401 539 | 0.80 |
| BAT-2 | 500 000 | 192 204 | 0.38 |
| BAT-3 | 500 000 | 48 680 | 0.09 |
| XYZ-1 | 500 000 | 488 561 | 0.98 |
| XYZ-2 | 500 000 | 112 324 | 0.22 |
| XYZ-3 | 500 000 | 12 392 | 0.02 |
| Host-guest complex | |||
| BAT-1 | 200 000 | 22 000 | 0.11 |
| BAT-2 | 200 000 | 2 000 | 0.01 |
| BAT-3 | 200 000 | 1 000 | 0.005 |
Cyclohexane
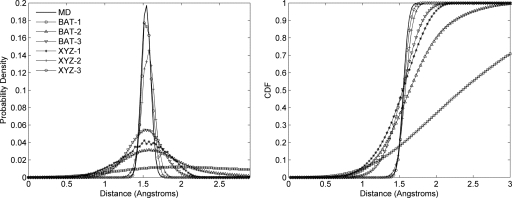
As for nonane, we studied the singlet-, doublet-, and triplet-level samples in Anchored Cartesian and BAT coordinates for a total of six superposition approximations. Results are assessed geometrically in terms of the distribution of distances between carbon 1 and carbon 6, the only two successive atoms in the ring whose bond length is not part of the BAT coordinate system. As shown in Fig. 6 and further detailed in Table 5, here the Cartesian coordinate system yields a more accurate distribution than does the BAT coordinate system, and the triplet-level distribution is more accurate than the doublet level, which in turn is substantially better than singlet-level distribution. The improvement upon including correlations is much more apparent in the distance distributions for samples in BAT coordinates. In BAT samples, the singlet-level distributions deviate drastically from the MD results, but the doublet- and triplet-level distributions are similar in structure to those from MD, being unimodal and centered at roughly the same bond length. Thus, for cyclohexane, higher-order correlations are required to accurately capture the geometry in both BAT and Cartesian coordinates. In absolute terms, the distribution of end-to-end distances from sampling in BAT coordinates for cyclohexane is more accurate than the distribution of end-to-end distances from sampling in Cartesian coordinates for nonane.
Figure 6.
Probability distributions (left) and corresponding cumulative distribution functions (right) of cyclohexane end-to-end distances computed from MD conformations and sampled conformations.
Table 5.
Statistics of end-to-end distances (Å) from MD and sampled conformations for cyclohexane.
| Median | Mean | Standard deviation | Minimum | Maximum | |
|---|---|---|---|---|---|
| MD | 1.54 | 1.54 | 0.06 | 1.24 | 1.83 |
| BAT-1 | 2.34 | 2.45 | 1.00 | 0.03 | 5.82 |
| BAT-2 | 1.63 | 1.66 | 0.43 | 0.09 | 3.74 |
| BAT-3 | 1.53 | 1.52 | 0.22 | 0.34 | 2.41 |
| XYZ-1 | 1.53 | 1.51 | 0.31 | 0.25 | 2.40 |
| XYZ-2 | 1.57 | 1.57 | 0.09 | 1.08 | 1.93 |
| XYZ-3 | 1.55 | 1.55 | 0.07 | 1.27 | 1.84 |
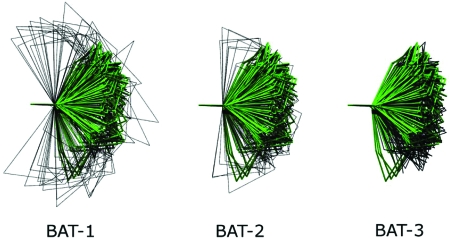
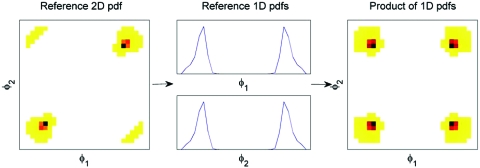
The MD trajectory used to compute the reference marginals includes chair, boat, and twist-boat conformations. In BAT coordinates, these conformations are established by the three internal torsions. Figure 7 compares representative conformations from MD and BAT-sampling at the three levels l=1, 2 and 3. The BAT-3 results resemble MD closely, and the BAT-2 results are similar, though somewhat less accurate. However, many of the BAT-1 conformations are quite distorted. The reason for the poor BAT-1 conformations has to do with the fact that singlet-level sampling completely ignores correlations, so that the effective 2D pdf linking each pair of torsions is just the product of their respective 1D pdfs. Figure 8 (left) shows that the correct 2D pdf of a pair of torsions has two maxima, corresponding to two different chair conformations. The corresponding singlet distributions (Fig. 8, middle) also have two maxima, so their product (Fig. 8, right) has four rather than two maxima. Two of the four maxima correspond to the correct maxima seen in the reference 2D pdf (Fig. 8, left); the other two are artifacts of singlet-level sampling and produce distorted conformations. Table 6 summarizes the statistics of energy components computed from sampled conformations and reference MD conformations. On the whole, in both BAT and XYZ coordinate systems, triplet-level samples are closer to MD values than doublet-level samples. At the triplet level, the statistics for the bond energies from Cartesian sampling match the reference MD values better than those from BAT sampling. However, the statistics are similar for the other energy components (angle, torsion, and van der Waals). This is generally consistent with the observation that sampling in BAT coordinates leads to an inappropriately wide distribution of the atom 1–atom 6 bond length, as noted in the previous paragraph. Overall, the distribution of total energy shown in Fig. 9 indicates that triplet-level samples yield the most accurate conformational distributions, Cartesian coordinates being slightly better than BAT, and that the results become progressively worse on going to doublet and then single-level sampling. The cumulative distribution functions in Fig. 9, as well as the data in Table 4, further document marked reductions in the numbers of abnormally high-energy conformations as more correlation is accounted for in both BAT and XYZ coordinates. Thus, the energy distributions, like the distance distributions, highlight the importance of including higher-order correlations for this constrained yet flexible chemical ring.
Figure 7.
Representative cyclohexane conformations from MD (in green) and from sampling (in black) in BAT coordinates at different levels. Each conformation is oriented such that atom 1 is at the origin, atom 2 is along the x axis, and atom 3 is in the x-y plane. Each set contains 100 conformations.
Figure 8.
The 2D pdf (right) obtained by multiplying 1D pdfs (middle) of cyclohexane has nonzero probability in regions not present in the true 2D pdf (left). Bins with zero probability in the 2D pdfs are colored white.
Table 6.
Statistics of energy (kcal∕mol) for cyclohexane computed from MD and sampled conformations. Coulombic term is not reported as it is always zero for this molecule.
| Bond | Angle | Torsion | vdW | Total | |
|---|---|---|---|---|---|
| Median | |||||
| MD | 5.3 | 3.9 | 7.9 | 1.0 | 18.8 |
| BAT-1 | 198.4 | 36.3 | 7.1 | 1.5 | 245.4 |
| BAT-2 | 24.4 | 8.4 | 7.6 | 1.1 | 44.1 |
| BAT-3 | 11.7 | 4.9 | 7.9 | 1.0 | 26.8 |
| XYZ-1 | 288.8 | 56.8 | 6.9 | 1.8 | 363.4 |
| XYZ-2 | 20.5 | 7.6 | 8.1 | 0.9 | 38.8 |
| XYZ-3 | 10.0 | 5.0 | 7.8 | 1.1 | 25.2 |
| Mean | |||||
| MD | 5.9 | 4.4 | 7.8 | 1.2 | 19.4 |
| BAT-1 | 498.7 | 53.8 | 7.1 | 3.4 | 563.0 |
| BAT-2 | 60.3 | 14.3 | 7.5 | 1.7 | 83.8 |
| BAT-3 | 18.5 | 5.8 | 7.8 | 1.4 | 33.5 |
| XYZ-1 | 448.8 | 67.6 | 7.0 | 8.1 | 531.5 |
| XYZ-2 | 27.7 | 9.2 | 8.0 | 1.2 | 46.1 |
| XYZ-3 | 12.7 | 6.1 | 7.8 | 1.3 | 27.9 |
| Standard deviation | |||||
| MD | 3.4 | 2.6 | 0.6 | 0.9 | 4.5 |
| BAT-1 | 669.9 | 50.4 | 0.8 | 8.4 | 713.0 |
| BAT-2 | 95.7 | 15.9 | 0.8 | 2.1 | 107.9 |
| BAT-3 | 19.9 | 4.1 | 0.7 | 1.2 | 21.5 |
| XYZ-1 | 444.0 | 48.4 | 0.9 | 90.2 | 485.9 |
| XYZ-2 | 24.2 | 6.5 | 0.7 | 0.9 | 27.0 |
| XYZ-3 | 10.2 | 4.3 | 0.6 | 0.9 | 12.1 |
| Minimum | |||||
| MD | 0.02 | 0.02 | 5.28 | −0.21 | 7.87 |
| BAT-1 | 0.16 | 0.09 | 4.71 | −0.30 | 9.09 |
| BAT-2 | 0.03 | 0.05 | 5.17 | −0.29 | 8.27 |
| BAT-3 | 0.01 | 0.04 | 5.32 | −0.26 | 8.26 |
| XYZ-1 | 0.32 | 0.15 | 3.99 | −0.30 | 12.44 |
| XYZ-2 | 0.08 | 0.07 | 5.37 | −0.16 | 8.63 |
| XYZ-3 | 0.08 | 0.04 | 5.66 | −0.12 | 8.47 |
| Maximum | |||||
| MD | 39.9 | 36.1 | 10.0 | 20.5 | 58.9 |
| BAT-1 | 4.9×103 | 292.4 | 9.9 | 1.6×103 | 5.2×103 |
| BAT-2 | 1.3×103 | 162.5 | 9.9 | 152.7 | 1.4×103 |
| BAT-3 | 379.3 | 79.8 | 9.9 | 28.4 | 412.8 |
| XYZ-1 | 4.3×103 | 353.3 | 10.0 | 2.8×104 | 2.8×104 |
| XYZ-2 | 369.1 | 108.0 | 10.0 | 30.4 | 415.8 |
| XYZ-3 | 180.1 | 76.0 | 10.0 | 18.1 | 245.2 |
Figure 9.
Probability distributions (left) and corresponding cumulative distribution functions (right) of the total energy of cyclohexane computed from MD conformations and sampled conformations using different sampling schemes.
Host-guest complex
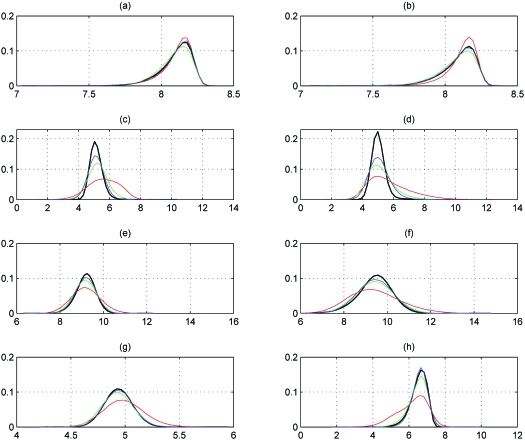
Conformations of the host-guest complex were sampled in BAT coordinates at the singlet, doublet, and triplet levels. Table 7 and Fig. 10 analyze the distances between eight atom pairs: 1-11, 1-22, 12-15, 15-23, 11-22, 12-23, 8-19, and 1-15. [See Fig. 2c for atom numbers.] Overall, the distance distributions from doublet- and triplet-level samplings agree well with the reference MD distributions, the triplet level being somewhat more accurate than doublet. The singlet-level samples give notably poorer distributions, especially for distances between host and guest atoms, as shown in Figs. 10c, 10d, 10h.
Table 7.
Statistics of seven key distances (Å) of host-guest conformations reconstructed from active MD and sampled BAT coordinates. Column headings give the atom pairs using atom numbers from Fig. 2c.
| 1-11 | 1-22 | 12-15 | 15-23 | 8-19 | 11-22 | 12-23 | 1-15 | |
|---|---|---|---|---|---|---|---|---|
| Median | ||||||||
| MD | 8.12 | 8.12 | 5.10 | 4.99 | 4.94 | 9.20 | 9.47 | 6.60 |
| BAT-1 | 8.13 | 8.14 | 5.63 | 5.47 | 4.98 | 9.17 | 9.35 | 6.31 |
| BAT-2 | 8.11 | 8.10 | 5.22 | 5.06 | 4.93 | 9.15 | 9.43 | 6.57 |
| BAT-3 | 8.13 | 8.12 | 5.11 | 5.04 | 4.94 | 9.18 | 9.43 | 6.62 |
| Mean | ||||||||
| MD | 8.10 | 8.09 | 5.14 | 5.03 | 4.94 | 9.17 | 9.43 | 6.54 |
| BAT-1 | 8.11 | 8.12 | 5.61 | 5.71 | 4.99 | 9.18 | 9.43 | 6.18 |
| BAT-2 | 8.09 | 8.08 | 5.24 | 5.17 | 4.93 | 9.15 | 9.43 | 6.50 |
| BAT-3 | 8.11 | 8.09 | 5.12 | 5.09 | 4.94 | 9.17 | 9.42 | 6.58 |
| Standard deviation | ||||||||
| MD | 0.11 | 0.12 | 0.43 | 0.45 | 0.13 | 0.46 | 0.79 | 0.45 |
| BAT-1 | 0.11 | 0.10 | 1.00 | 1.34 | 0.19 | 0.67 | 1.20 | 0.86 |
| BAT-2 | 0.11 | 0.13 | 0.64 | 0.88 | 0.14 | 0.52 | 0.88 | 0.54 |
| BAT-3 | 0.11 | 0.12 | 0.52 | 0.68 | 0.13 | 0.48 | 0.83 | 0.43 |
| Minimum | ||||||||
| MD | 7.06 | 7.07 | 3.42 | 3.12 | 4.27 | 6.31 | 4.95 | 3.22 |
| BAT-1 | 7.00 | 7.26 | 1.37 | 1.66 | 4.21 | 6.43 | 5.23 | 1.66 |
| BAT-2 | 7.19 | 7.16 | 2.20 | 2.13 | 4.30 | 6.58 | 5.22 | 2.20 |
| BAT-3 | 7.04 | 7.28 | 2.79 | 2.66 | 4.36 | 7.07 | 6.06 | 3.69 |
| Maximum | ||||||||
| MD | 8.37 | 8.38 | 10.34 | 10.33 | 5.92 | 12.17 | 14.50 | 7.92 |
| BAT-1 | 8.35 | 8.35 | 9.60 | 12.53 | 5.96 | 12.29 | 14.79 | 10.11 |
| BAT-2 | 8.37 | 8.36 | 9.00 | 11.58 | 5.68 | 11.45 | 13.56 | 8.41 |
| BAT-3 | 8.36 | 8.37 | 8.17 | 10.22 | 5.51 | 11.10 | 12.64 | 8.09 |
Figure 10.
Probability distributions of interatomic distances (Å) in the host-guest complex for atom pairs (a) 1-11, (b) 1-22, (c) 12-15, (d) 15-23, (e) 11-22, (f) 12-23, (g) 8-19, and (h) 1-15. Color code: MD in black, BAT-1 in red, BAT-2 in green, and BAT-3 in blue.
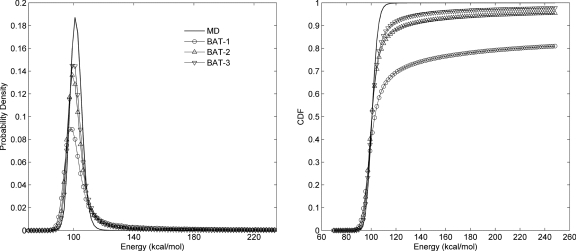
For both BAT-2 and BAT-3 samples, the median values of all energy components are in excellent agreement with MD, as are the mean values of all energy components other than van der Waals (Table 8). Although the tabulated statistics of BAT-1 samples are comparable to those of BAT-2 and BAT-3 samples, Fig. 11 shows that the distribution of total energies is substantially inferior with singlet-level sampling. It is also evident that the mean van der Waals energy of the BAT-3 samples is substantially better than that of BAT-1 and BAT-2 samples, though all are skewed toward higher values due to the presence of a few conformations with bad contacts. These lead to small tails of high-energy conformations for BAT-2 and BAT-3 samplings, as evident from the cumulative probability distributions of energy (Fig. 11). Table 4 furthermore documents sharp reductions in the number of conformations with abnormally high energy as more correlation is accounted for in the sampling.
Table 8.
Statistics of energy (kcal∕mol) of host-guest complex for conformations reconstructed from active MD and sampled BAT coordinates.
| Bond | Angle | Torsion | Improper | Coulomb | vdW | Total | |
|---|---|---|---|---|---|---|---|
| Median | |||||||
| MD | 0.0014 | 7.16 | 3.87 | 0.12 | 65.1 | 24.7 | 101.5 |
| BAT-1 | 0.0015 | 7.69 | 3.98 | 0.16 | 62.4 | 26.9 | 104.0 |
| BAT-2 | 0.0015 | 7.20 | 4.14 | 0.17 | 63.6 | 25.1 | 101.4 |
| BAT-3 | 0.0015 | 7.19 | 3.85 | 0.16 | 64.4 | 25.0 | 101.6 |
| Mean | |||||||
| MD | 0.0015 | 7.35 | 4.07 | 0.27 | 65.1 | 24.9 | 101.7 |
| BAT-1 | 0.0015 | 7.94 | 4.21 | 0.27 | 64.0 | 5.41×104 | 5.41×104 |
| BAT-2 | 0.0015 | 7.38 | 4.35 | 0.28 | 64.4 | 3.46×103 | 3.53×103 |
| BAT-3 | 0.0015 | 7.36 | 4.05 | 0.25 | 65.1 | 959.7 | 1.04×103 |
| Standard deviation | |||||||
| MD | 0.0004 | 1.89 | 1.53 | 0.38 | 2.97 | 1.66 | 4.26 |
| BAT-1 | 0.0004 | 2.19 | 1.65 | 0.39 | 11.3 | 4.82×105 | 4.82×105 |
| BAT-2 | 0.0004 | 1.90 | 1.72 | 0.40 | 5.49 | 1.15×105 | 1.15×105 |
| BAT-3 | 0.0004 | 1.87 | 1.50 | 0.34 | 4.75 | 6.20×104 | 6.20×104 |
| Minimum | |||||||
| MD | 0.0003 | 1.98 | 0.19 | 0.00 | 46.6 | 23.0 | 82.9 |
| BAT-1 | 0.0003 | 2.37 | 0.48 | 0.00 | −583.1 | 23.0 | 80.8 |
| BAT-2 | 0.0003 | 2.11 | 0.49 | 0.00 | −111.6 | 23.0 | 79.8 |
| BAT-3 | 0.0004 | 2.29 | 0.61 | 0.00 | 40.6 | 23.2 | 84.8 |
| Maximum | |||||||
| MD | 0.0044 | 24.1 | 17.2 | 8.23 | 84.0 | 2.51×103 | 2.62×103 |
| BAT-1 | 0.0042 | 23.1 | 16.4 | 7.48 | 1.22×103 | 9.99×106 | 9.99×106 |
| BAT-2 | 0.0041 | 19.4 | 16.9 | 4.54 | 238.7 | 9.20×106 | 9.20×106 |
| BAT-3 | 0.0040 | 19.0 | 12.5 | 2.97 | 262.9 | 8.97×106 | 8.97×106 |
Figure 11.
Probability distributions (left) and corresponding cumulative distribution functions (right) of total energy of the host-guest complex for MD and sampled conformations.
Comparing sampled and reference marginal pdfs
Another way to assess the accuracy of the sampled distributions is to compare their marginals with those of the original MD simulations. For singlet-level sampling, all 1D sampled marginal pdfs converge to the corresponding reference pdfs. For higher-level sampling, only the marginals of the first l variables converge to the reference marginals (data not shown), as expected based upon the structure of the sampling algorithm. The marginals of the subsequent (>l) variables are sampled from approximate conditional distribution and therefore are expected to deviate from the reference marginals.
The deviations were quantified by computing the RMSDs between all 1D, 2D, and 3D sampled and reference marginal pdfs for each sampling case. Table 9 reports the mean RMSD for the three marginal pdfs in each sampling case. As expected, for a specific molecule and coordinate system, the mean RMSD of singlet marginal pdfs is lower for singlet-level sampling than for higher-level sampling. However, the mean RMSDs of doublet and triplet marginal pdfs from doublet- or triplet-level samplings are, in general, lower than those from singlet-level sampling, indicating the presence of correlations similar to those in MD simulations. Sampling of nonane in XYZ coordinates does not follow this trend, indicating that triplet level is not sufficient to capture the correlations in this coordinate system. This observation is consistent with the analyses of distance and energy distributions above.
Table 9.
Accuracy of singlet, doublet, and triplet marginal distributions (columns) computed from conformations sampled at the singlet-, doublet-, and triplet-levels (rows) for the three molecular systems. Results are reported as mean RMSD across all marginals for a specific sampling case. Note that the table includes mean RMSDs of doublet and triplet marginal pdfs for conformations sampled at the singlet level, as well as mean RMSDs of triplet marginal pdfs for conformations sampled at the doublet level.
| Singlet | Doublet | Triplet | |
|---|---|---|---|
| Nonane | |||
| BAT-1 | 0.0013 | 0.0023 | 0.0020 |
| BAT-2 | 0.0013 | 0.0014 | 0.0015 |
| BAT-3 | 0.0020 | 0.0019 | 0.0019 |
| XYZ-1 | 0.0012 | 0.0267 | 0.0157 |
| XYZ-2 | 0.0915 | 0.0620 | 0.0336 |
| XYZ-3 | 0.0775 | 0.0530 | 0.0290 |
| Cyclohexane | |||
| BAT-1 | 0.0012 | 0.0062 | 0.0045 |
| BAT-2 | 0.0046 | 0.0037 | 0.0025 |
| BAT-3 | 0.0028 | 0.0022 | 0.0017 |
| XYZ-1 | 0.0012 | 0.0207 | 0.0129 |
| XYZ-2 | 0.0238 | 0.0180 | 0.0093 |
| XYZ-3 | 0.0195 | 0.0148 | 0.0083 |
| Host-guest complex | |||
| BAT-1 | 0.0021 | 0.0060 | 0.0050 |
| BAT-2 | 0.0098 | 0.0085 | 0.0055 |
| BAT-3 | 0.0063 | 0.0052 | 0.0039 |
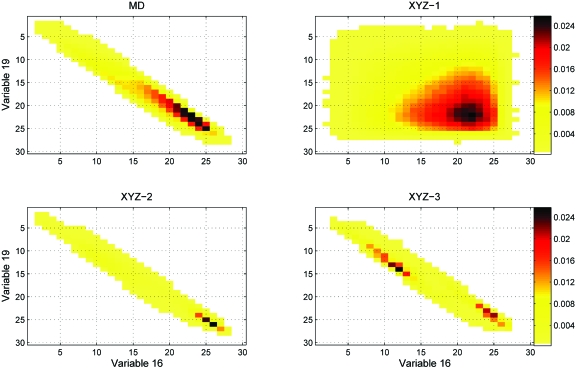
Marginal pdfs generated from sampled conformations are graphically compared with the corresponding reference marginals from the MD simulations in Figs. 1213. Figure 12 shows the doublet marginal pdf from the XYZ study of nonane which yielded the highest RMSD values at the singlet, doublet, and triplet levels, and Fig. 13 similarly analyzes the doublet marginal which yielded the highest RMSD in BAT-1 sampling. It is worth noting that the doublet marginal pdf from BAT-1 sampling [Fig. 13b] is much closer to its reference MD marginal than is the XYZ-1 result for nonane [Fig. 12b], confirming earlier results where BAT coordinate performed better than XYZ.
Figure 12.
Comparison of doublet marginals from MD with XYZ-1, XYZ-2, and XYZ-3 samplings of variables 16 and 19 of nonane, which correspond to the x coordinates of atoms 8 and 9 (Fig. 2). The RMSDs of the sampled doublet marginal pdfs are 0.0504, 0.0955, and 0.0692 for XYZ-1, XYZ-2, and XYZ-3, respectively. Cells are colored on a linear scale of probability, except that cells with identically zero probability are colored white.
Figure 13.
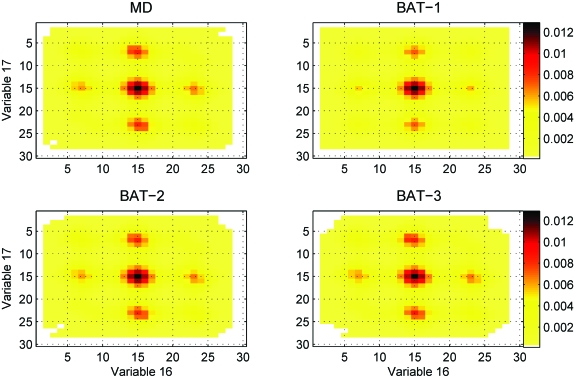
Comparison of doublet marginals from MD with BAT-1, BAT-2, and BAT-3 samplings of variables 16 and 17 of nonane, corresponding to torsions 1-2-3-4 and 2-3-4-5, respectively (Fig. 2). The RMSDs of the sampled doublet marginal pdfs are 0.0106, 0.0015, and 0.0018 for BAT-1, BAT-2, and BAT-3, respectively. Cells are colored on a linear scale of probability, except that cells with identically zero probability are colored white.
Range of configurational space sampled
The part of the N-dimensional configuration space accessible to the present sampling algorithms is largely dependent on the reference marginal pdfs used to construct the superposition approximations in the algorithms. In one extreme case, if the reference distribution sampled only a single conformation, then all the marginals of the reference distribution would be delta functions, and only the single reference conformation would be sampled by the present algorithm. More generally, as discussed in Sec. 2D2, l-level sampling will only generate conformations that have nonzero probability in all the reference marginals. This property is apparent in Figs. 1213, where the populated bins of a representative 2D marginal from doublet- and triplet-level samplings (right panel) are a subset of those sampled by the MD trajectories (left panel), unlike the marginal from singlet-level sampling. In this way, the reference marginals constrain the range of configuration space accessible to the sampling algorithm.
However, this constraint does not imply that the sampling algorithm generates only conformations identical to those used to compute the reference marginals. Rather, the omission of higher-level marginals from the superposition approximations leaves the sampling algorithm free to generate new conformations. In fact, for all of the present test systems, over 99% of the (2–5)×105 sampled conformations are new relative to the 5×106 conformations in the original MD trajectory used to generate the reference marginals. (Conformations were compared after binning their coordinates.) This small degree of overlap is intuitively reasonable, given the large number of potential conformations, ∼30ND, where 30 is the number of bins used to discretize each coordinate and ND is the dimensionality. Here ND is 12, 21, and 32 for cyclohexane, nonane, and host-guest complex, respectively, so the number of conformations ranges from ∼1017 to ∼1047. To better judge whether the high fraction of novel conformations generated by the sampling algorithm is reasonable, we did the following test on the MD trajectories of cyclohexane and nonane. For both molecules, we counted the number of conformations in the first 5×105 MD that were repeated in the following 4.5×106 conformations. We found no repeats for nonane, while <1% of the first 5×105 of cyclohexane were repeated, consistent with the low overlap between the sampled with corresponding MD conformations.
Computational complexity of the sampling algorithms
The computational cost of generating one conformation with the sampling algorithm (Sec. 2D2) depends mainly on the level of approximation (i.e., singlet, doublet, or triplet), the number of degrees of freedom sampled, and the number of bins used to represent the distribution functions. The computational cost of sampling at the singlet level is negligible as no conditional distributions need to be computed for each variable. Because the conditional distribution functions at the triplet level [Eq. 21] have more factors than at the doublet level [Eq. 20], sampling at the triplet level is computationally more expensive. The time complexities of the doublet- (Algorithm 2) and triplet-level (Algorithm 3) sampling algorithms are linear and quadratic, respectively, in the number of degrees of freedom, but both algorithms are linear in the number of bins used. The present MATLAB implementation took 0.0017, 0.04, and 0.6 s on a 3.8 GHz Pentium 4 personal computer to sample a conformation of the host-guest complex (32 degrees of freedom and 30 bins) at the singlet, doublet, and triplet levels, respectively. Computer time could be substantially reduced by implementation of the algorithm in a lower-level language, such as C or Fortran, and by straightforward code optimizations, such as substitution of an improved logarithm operation, since logarithms take 35% of the central processing unit time in the current implementation of the doublet- and triplet-level sampling algorithms.
DISCUSSION
The present paper introduces algorithms for sampling molecular conformations in a manner that includes correlations up to a desired order by means of superposition approximations and employs this algorithm to test the importance of correlations in capturing conformational fluctuations. We find that incorporating higher-order correlations systematically improves the distributions of sampled conformations as compared with the MD conformations, and that conformations sampled via superposition approximations at the doublet or triplet levels resemble reference conformations from MD simulations rather well for one or both of the BAT or Cartesian coordinate systems.
This observation supports the hypothesis that molecular fluctuations may be described to good approximation in the absence of high-order correlations. This assessment relies on the results obtained for the three molecular systems considered here, but we expect the picture to be similar for other molecular systems of similar size and type. It would clearly be of interest to know whether the same is true for larger systems, such as proteins.
This study was motivated by evidence that configurational entropy may be approximated to good accuracy without accounting for high-order mutual information terms (see Sec. 1). Comparing sampled conformations, as done here, provides a more stringent test of our hypothesis than merely comparing entropy values, because the same entropy could be obtained for two very different conformational distributions. The present study confirms that neglecting high-order correlations still allows generation of reasonably good conformations. As noted below, the present sampling algorithms can be generalized to use a selected set of higher-order reference pdfs (see also Sec. 2B). The present approach therefore provides a framework for investigating the contributions of selected correlations to fluctuations in a multidimensional system.
The present sampling algorithms are approximations to the well-known ancestral sampling algorithm.6 The ancestral sampling algorithm is a rigorous approach to sampling from a desired pdf, but it becomes difficult to apply to large, highly correlated systems because it requires access to all high-order marginals of the full pdf. This difficulty is addressed here by using approximate conditionals constructed from superposition approximations up to a selected order. This approach leads to some dependence upon sampling sequence, but this sequence dependence could readily be removed, if so desired, simply by randomizing the order in which the coordinates are sampled. The sequence dependence of the sampled conformations could also be removed by using the facts that we can readily compute the sampling probability to each conformation generated by the present algorithms, and that this distribution is expected to have good overlap with the SA-l approximation to the full distribution, which is symmetric in all variables. As a consequence, the present sampling algorithm could be used as a proposal distribution and corrected to the full-dimensional SA-l approximation by standard biased sampling methods.17
The generation of null samples (Sec. 2D2) represents a potential problem with the present approach, but this occurred very rarely in the present tests, and we have no reason to expect this to become more problematic for other molecular systems of similar size. A strength of the present approach is that using superposition approximations to approximate the conditional distributions limits the sampled conformations to regions of configuration space for which the reference marginal pdfs are populated. This helps avoid sampled conformations with grossly unrealistic conformations while still allowing construction of new conformations, i.e., ones not present in the MD samples used to construct the marginals.
Several avenues in accelerating and enhancing the present sampling algorithms can be discerned. Straightforward approaches to speeding the sampling are considered in Sec. 4F. It should also be noted that, unlike other methods of sampling from high-dimensional distributions, such as Gibbs sampling, the present approach generates successive samples that are uncorrelated with each other: in effect, the algorithm has no memory of prior conformations. As a result the sampling can be done in an embarrassingly parallel fashion on a distributed computing platform. As to improving accuracy, one obvious approach would be to construct higher-level (say, quadruplet) superposition approximations. However, this could lead to a combinatorial explosion in the number of higher-order marginals to be included. This explosion may be avoided by including only a few critically important high-level marginals in the superposition approximation. In addition, storage needs could be moderated by representing the higher-order marginals with a more sophisticated basis set, such as Gaussian or von Mises distributions, which require far fewer parameters than needed by the present histogram representation. Another approach in improving the accuracy of the sampled conformations is suggested by the dependence of the present results upon the choice of the coordinate system: BAT coordinates are clearly superior for nonane, while Cartesians appear to perform better for cyclohexane. More generally, different coordinate systems result in different degrees of correlation, or coupling, among coordinates. Accordingly, other coordinate systems, such as principal components of the MD trajectory in Cartesian or BAT (Ref. 18) coordinates, might better capture complex molecular fluctuations and other high-dimensional distributions in terms of tractable sets of low-order marginals.
The sampling algorithms introduced here may have practical uses. An immediate application would be to Ytreberg and Zuckerman’s reference system approach to computing absolute free energies.19 The approach entails computing energies of conformations sampled from an approximate reference distribution, and the convergence rate of the free energy depends critically on the degree of overlap between the reference distribution and the true physical distribution. Ytreberg and Zuckerman implemented the method with a reference distribution that neglects all correlations, as it uses only singlet marginal distributions computed from MD simulation data, but the authors noted that including correlations should improve convergence. The present sampling algorithm could represent an efficient means of introducing correlation into the reference distribution and thus speeding convergence. Another application of the present approach may be to characterize conformational uncertainty—rather than thermal fluctuations—and thus qualify molecular conformations inferred from experimental data, such as crystallography, NMR, or crosslinking studies.
Approximating high-dimensional distributions in terms of low-order marginals is a common theme in the many fields of inquiry that deal with high-dimensional systems.1, 2, 3, 20 Although one often has enough data to compute low-order marginals in such cases, the available data are typically too sparse to allow a full, high-dimensional distribution to be evaluated. The present paper describes a novel approach to approximate the high-dimensional distribution in terms of tractable low-order marginals and, furthermore, to sample from this approximate distribution. This work may have applications in fields beyond statistical mechanics, including bioinformatics, structural biology, data mining, and machine learning.
ACKNOWLEDGMENTS
This publication was made possible by Grant No. GM61300 from the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
APPENDIX: DERIVATION OF ANALYTIC EXPRESSION OF SAMPLING PROBABILITY
The l-level sampling algorithms of Sec. 2D2 sample conformations from a distribution, , which is different from the corresponding l-level superposition approximation pSA-l(X1,…,XN). In this section, we derive an analytic expression for the sampling distribution corresponding to doublet-level (l=2) sampling of 3D conformations, , and compare it with the corresponding superposition approximation, pSA-2(X1,X2,X3). We show that , unlike pSA-2, is identically normalized. Additional differences between and pSA-l will also be highlighted.
To sample a conformation in three dimensions (x1,x2,x3), the doublet-level sampling algorithm requires the following normalized singlet and doublet marginal pdfs as input: [1], [2], [3], [1,2], [1,3], and [2,3]. Let variable X1 be sampled first, followed in order by X2 and X3. The variables are sampled from the following 1D pdfs;
| (A1) |
| (A2) |
and
| (A3) |
Substituting N3(x1,x2) from Eq. 18 into Eq. A3 gives
| (A4) |
Using the chain rule of Eq. 24, the probability is given by the product of the above 1D pdfs,
| (A5) |
or
| (A6) |
where is used to distinguish the summation label from the value of the variable X3. Next we show that is normalized, i.e.,
| (A7) |
Substituting from Eq. A6 in the left hand side of Eq. A7 and extracting the terms independent of x3 from the summation over x3 gives
| (A8) |
The factors involving summations over x3 and cancel, giving
| (A9) |
The last equality uses the fact that all input marginals are normalized. Thus is identically normalized. The above procedure can be generalized to distributions in higher dimensions and to higher-level sampling.
Next we point out some general differences between the sampling distribution and pSA-l by comparing the two distributions for the above case. The pSA-2 in three dimensions is given by setting k=3 in Eq. 14 which gives the KSA of Eq. 8. For easy comparison, we rewrite the two distributions,
| (A10) |
and
| (A11) |
The factors [x1] and [x2] in pSA-2, which are absent from , are guaranteed to be nonzero for any sampled conformation. As a result, any conformation with a nonzero probability will also have a nonzero pSA-2 probability. In this sense, the two distributions overlap. Also, while pSA-2 is symmetric in all three variables, is symmetric in only x1 and x2. More generally, the sampling probability is symmetric in only the first l sampled variables in the present sampling algorithms; the remaining asymmetry accounts for the dependence of on the sequence in which the variables are sampled.
References
- Killian B. J., Kravitz J. Y., and Gilson M. K., J. Chem. Phys. 10.1063/1.2746329 127, 024107 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda H., Phys. Rev. E 10.1103/PhysRevE.62.3096 62, 3096 (2000). [DOI] [PubMed] [Google Scholar]
- Attard P., Jepps O. G., and Marcelja S., Phys. Rev. E 10.1103/PhysRevE.56.4052 56, 4052 (1997). [DOI] [Google Scholar]
- Chang C. E. and Gilson M. K., J. Am. Chem. Soc. 10.1021/ja047115d 126, 13156 (2004). [DOI] [PubMed] [Google Scholar]
- Gilson M. K., Given J. A., Bush B. L., and McCammon J. A., Biophys. J. 72, 1047 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C., Pattern Recognition and Machine Learning (Information Science and Statistics) (Springer, New York, 2006), p. 365. [Google Scholar]
- Reiss H., J. Stat. Phys. 10.1007/BF01060200 6, 39 (1972) [DOI] [Google Scholar]; Singer A., J. Chem. Phys. 10.1063/1.1776552 121, 3657 (2004) [DOI] [PubMed] [Google Scholar]; Stell G., in The Equilibrium Theory of Classical Fluids, edited by Frisch H. L. and Lebowitz J. L. (Benjamin, New York, 1964). [Google Scholar]
- Jaynes E. T., Probability Theory: The Logic of Science (Cambridge University Press, Cambridge, 2003). [Google Scholar]
- Kirkwood J. G. and Boggs E. M., J. Chem. Phys. 10.1063/1.1723737 10, 394 (1942). [DOI] [Google Scholar]
- Hansen J. -P. and McDonald I. R., Theory of Simple Liquids, 3rd ed. (Academic, New York, 2006), pp. 83–85. [Google Scholar]
- Fisher I. Z. and Kopeliovich B. L., Sov. Phys. Dokl. 5, 761 (1960). [Google Scholar]
- Potter M. J. and Gilson M. K., J. Phys. Chem. A 10.1021/jp0135407 106, 563 (2002). [DOI] [Google Scholar]
- Chang S. K. and Hamilton A. D., J. Am. Chem. Soc. 10.1021/ja00212a065 110, 1318 (1988) [DOI] [Google Scholar]; Goswami S. and Mukherjee R., Tetrahedron Lett. 10.1016/S0040-4039(97)00103-2 38, 1619 (1997). [DOI] [Google Scholar]
- Abagyan R., Totrov M., and Kuznetsov D., J. Comput. Chem. 10.1002/jcc.540150503 15, 488 (1994). [DOI] [Google Scholar]
- MATLAB, The MathWorks, Inc., 2007. (http://www.mathworks.com/products/matlab/).
- A. D.MacKerell, Jr., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-Mccarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., and Karplus M., J. Phys. Chem. B 10.1021/jp973084f 102, 3586 (1998). [DOI] [PubMed] [Google Scholar]
- Press W., Flannery B., Teukolsky S., and Vetterling W., Numerical Recipes in C: The Art of Scientific Computing (Cambridge University Press, New York, 1992), pp. 316–328. [Google Scholar]
- Altis A., Nguyen P., Hegger R., and Stock G., J. Chem. Phys. 126, 244111 (2007). [DOI] [PubMed] [Google Scholar]
- Ytreberg F. M. and Zuckerman D. M., J. Chem. Phys. 10.1063/1.2174008 124, 104105 (2006). [DOI] [PubMed] [Google Scholar]
- Bishop C., Pattern Recognition and Machine Learning (Information Science and Statistics) (Springer, New York, 2006) [Google Scholar]; Daub C., Steuer R., Selbig J., and Kloska S., BMC Bioinf. 10.1186/1471-2105-5-118 5, 118 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]; Deshpande A., Garofalakis M., and Rastogi R., Bell Labs Technical Report, 2001; Stephens G. J. and Bialek W., http://arxiv.org/abs/8081.0253.