Abstract
Background
Neurocognitive deficits in patients with hepatitis C virus (HCV) infection prompted a search for HCV in brain.
Results
HCV was present in the brains of 7 (54%) of 13 patients with viremia, as determined by 5' UTR and E1 (envelope 1) gene analysis. Brain HCV RNA consensus sequences differed from those in plasma and liver in 4 (57%) of 7 patients. The quality ofHCVRNA from postmortem brain and liver was assessed and demonstrated to be suitable for sequence analysis. Quasispecies analysis revealed that several mutations present in clones from >1 brain region were absent in clones from liver and plasma. Brain-specific mutations defined several families of related sequences. The patterns of brain-specific mutations in these families were consistent with the evolution of HCV RNA from a common ancestor. Single-nucleotide-polymorphism analysis confirmed that a prominent brain-specific mutation constituted ~10% of HCV RNA in cerebellum and medulla but that this mutation was undetectable in the liver and plasma of the same patient.
Conclusions
This study introduces novel methods for assessing RNA from postmortem samples. It increases the reported cases of HCV in the brain, provides the first E1 sequences from the brain, and contributes to the growing evidence that HCV replicates and evolves within the brain.
Hepatitis C virus (HCV) causes chronic infection in ~3% of the world's population. A search for HCV in brain was prompted by complaints of fatigue and cognitive dysfunction made by patients with HCV infection [1-7]. Specific cognitive deficits and nuclear magnetic resonance abnormalities have been reported in HCV-infected patients [1, 4, 8-10]. These observations, along with increasing evidence of HCV infection and replication in peripheral blood mononuclear cells (PBMCs) [11-16], suggest that the brain may also be a compartment for extrahepatic HCV replication.
To date,HCVRNAhas been amplified and sequenced from brain tissue of a limited number of subjects [17-21]. Detection of antigenomic HCV RNA has been reported in various brain regions [19]. The population of brain HCV contains sequence variants that are absent in serum. Both the presence of antigenomic HCV RNA and the presence of brain-specific variants suggest that HCV infection and replication may occur in the brain.
Despite the importance of sequence analysis for investigations of brain HCV, no study has examined the suitability of postmortem (PM) material for sequence analysis, although, of necessity, such studies are almost universally performed on PM material. We developed a bioinformatic method to assess the quality of RNA templates from PM tissue. This method demonstrated that brain HCV RNA is a suitable template for sequence analysis. Sequence analysis was then conducted on portions of the 5' untranslated region (UTR) and the E1 (envelope 1) gene. Results of both direct sequencing and quasispecies analysis support the hypothesis that HCV replicates and evolves within the brain.
PATIENTS, MATERIALS, AND METHODS
Details on sample and clinical data collection and sequence preparation can be found in appendix A, which appears only in the electronic edition of the Journal, and in tables 1 and 2. Sequences have been deposited in GenBank under accession numbers EU165716-EU166327.
Table 1.
RT and PCR primers.
| Region | Primer Set | Sequence1 |
|---|---|---|
| 5'UTR | Reverse (-) | 184 - CGTCCTGGCAATTCCGGTGT - 165 |
| Forward (+) | 59 - TGTCTTCACGCAGAAAGCGTCTAGC - 83 | |
| E1 external | Reverse (-) | 1328 - CGTAGGGGACCAGTTCATCATCAT - 1305 |
| Forward (+) | 834 - GCAACAGGGAACCTTCCTGGTTGCTC - 859 | |
| E1 internal | Reverse (-) | 1316 - GTTCATCATCATATCCCATGCCAT - 1293 |
| Forward (+) | 843 - AACCTTCCTGGTTGCTCTTTCTCTAT - 868 | |
| GAPDH | Reverse (-) | 750 - TTCTAGACGGCAGGTCAGGT - 731 |
| Forward (+) | 542 - TCACTGCCACCCAGAAGACT - 561 |
Table 2.
PCR Conditions
| Regiona | Forward Primer | Reverse Primer | Cycling Conditions | Expected Fragment |
|---|---|---|---|---|
| 5'UTRb | 5'UTR | 5'UTR | 98C 10 sec | 128 bp |
| forward | reverse | 35 X | ||
| 98 10 sec | ||||
| 71 30 sec | ||||
| 72 4 sec | ||||
| 72 10 min | ||||
| E1 outerb | E1 | E1 | 98 30 sec | 495 bp |
| external | external | 35 X | ||
| forward | reverse | 98 10 sec | ||
| 72 20 sec | ||||
| 72 10 min | ||||
| E1 innerb | E1 inner | E1 inner | 98 30 sec | 474 bp |
| forward | reverse | 35 X | ||
| 98 10 sec | ||||
| 69 2 sec | ||||
| 72 18 sec | ||||
| 72 10 min | ||||
| GAPDHc | GAPDH | GAPDH | 94 C 2 min | 208 bp |
| forward | reverse | 35 X | ||
| 94 30 sec | ||||
| 60 30 sec | ||||
| 72 45 sec | ||||
| 72 C 7 min |
All reactions were performed in 20 μl volume with μl template, 1X Phusion HF buffer, 200μM of each dNTP, 0.5uM primers, 3% DMSO, and 0.4U Phusion (Finnzymes).
The negative control for this PCR assay was a reaction with no cDNA and water to volume, the positive control was a reaction containing sequence verified HCV cDNA
Primer design for GAPDH was performed by Dr. D. Zhang (The Mount Sinai Medical Center).
Sequence and statistical analysis
Sequences were aligned using the ClustalW algorithm in MegAlign (DNAStar). E1 sequences were compared with 783 1a and 373 1b sequences downloaded from http://hcv.lanl.gov/components/hcv-db/combined_search/searchi.html on 19 December 2005. Substitution frequencies were tallied using custom Perl scripts. Comparisons of the average number of mutations in sequence sets were performed using 2-sample t tests. P < .05 was considered to indicate statistical significance.
Single-nucleotide polymorphism (SNP) analysis
Amplicons from patient 20024 were either analyzed directly or digested with Alw261 (Fermentas) in a 20-μL reaction containing 12.5 μL of gel-purified DNA. Restriction fragments were fractionated in 1.3% aga-rose 1× TBE (Tris-borate-EDTA) gels and subsequently stained with Syto 60 (Invitrogen). Gels were scanned by use of an Odyssey scanner (Licor). Bands were quantified using accompanying software.
RESULTS
Prevalence of HCV in PM brain
The study population was comprised of 27 subjects (table 3). On the basis of testing of stored premortem plasma, 13 had HCV viremia, and 14 did not have HCV viremia. None received HCV-targeted therapies between determination of HCV load and death. Ten of the 13 patients with HCV viremia were positive for HIV antibodies, as were 10 of the 14 patients without HCV viremia.
Table 3.
Characteristics of patients and polymerase chain reaction (PCR) results.
| PCR results |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma |
Liver |
Cortex |
GAPDH |
||||||||||
| Patient | HIV Ab | Sex | PMI, h | VLI, days | Serum HCV loada | 5' UTR | E1 | 5' UTR | E1 | 5' UTR | E1 | Liver | Cortex |
| 10001 | + | F | 4.5 | 108 | 7.67 | + | + | + | + | - | - | + | + |
| 10016 | + | F | 17 | 186 | 6.82 | + | + | + | + | - | + | + | + |
| 10027 | + | M | 6 | 120 | 6.33 | + | + | + | + | - | - | + | + |
| 10066 | + | F | 7 | 102 | 3.42 | - | + | + | + | - | + | + | + |
| 10086 | + | F | 7.5 | 3 | 6.39 | + | + | + | + | - | + | + | + |
| 20015 | + | F | 6.5 | 150 | 6.03 | + | + | + | + | - | - | + | + |
| 20024 | + | M | 4 | 2 | 7.88 | + | + | + | + | + | + | + | + |
| 20028 | + | M | 6 | 285 | 6.29 | + | + | + | + | + | + | + | + |
| 20053 | + | M | 29 | 66 | 6.91 | + | + | + | + | - | - | + | + |
| 10034 | + | M | 5 | 36 | 7.09 | + | + | + | + | + | + | + | + |
| 40003 | - | M | 14.5 | 15 | 2.80 | - | - | + | + | - | - | + | + |
| 40007 | - | M | 11.5 | 108 | 6.35 | - | +b | - | + | + | [+] | + | + |
| 553 | - | M | 16 | 195 | 5.40 | NA | NA | + | + | - | - | + | + |
| 20016 | + | M | 8 | 2 | UN | - | - | - | - | - | - | + | + |
| 20025 | + | M | 25.5 | 2 | UN | - | - | - | - | - | - | + | + |
| 10018 | + | M | 3 | 0 | UN | - | - | - | - | - | - | + | + |
| 10025 | + | M | 7.5 | 39 | UN | - | - | - | - | - | - | + | + |
| 10043 | + | F | 17 | 90 | UN | - | - | - | - | - | - | + | + |
| 10095 | + | M | 5 | 309 | UN | - | - | - | - | - | - | + | + |
| 10065 | + | M | 7 | 99 | UN | - | - | - | - | - | - | + | + |
| 10133 | + | M | 7.5 | 345 | UN | - | - | - | - | - | - | + | + |
| 10105 | + | M | 12 | 90 | UN | - | - | - | - | - | - | + | + |
| 10162 | + | M | 16.5 | 120 | UN | - | - | - | - | - | - | + | + |
| 555 | - | M | 8.5 | 48 | UN | - | - | - | - | - | - | + | + |
| 568 | - | F | 16 | NA | UN | - | - | - | - | - | - | + | + |
| 556 | - | F | 18.5 | 15 | UN | - | - | - | - | - | - | + | + |
| 584 | - | M | 22.5 | NA | UN | - | - | - | - | - | - | + | + |
NOTE. [+], PCR product obtained but material was insufficient for accurate sequencing; Ab, antibody; F, female; M, male; NA, not available; PCR, polymerase chain reaction; PMI, postmortem interval; VLI, viral load interval; UN, undetectable; UTR, untranslated region.
Log10 IU/mL.
Forward and reverse sequences were not identical.
In the first set of experiments, HCV was amplified from brain (frontal cortex), liver, and plasma. Amplicons were fractionated by gel electrophoresis. Specimens were scored positive if a 5' UTR and/or an E1 amplicon were present. For the 13 HCV-infected patients, 5' UTR amplicons were obtained from 12 of 13 liver, 9 of 12 plasma, and 4 of 13 brain/frontal cortex specimens (table 3); E1 amplicons were obtained from 13 of 13 liver, 11 of 12 plasma, and 7 (54%) of 13 brain/frontal cortex specimens. The average PM interval (PMI; the time between death and sample collection) was 6.6 h for brain specimens that yielded amplicons in the nonnested 5' UTR assay, whereas the PMI was 12.75 h for specimens that yielded neither 5' UTR nor E1 amplicons (P = .12). Thus, HCV RNA content may decrease during the PMI. Four of 8 brain specimens from men were positive in the 5' UTR assay, versus 0 of 5 brain specimens from women—a trend that may accord with the observation that women are more likely to clear HCV spontaneously [22]. Brain HCV was present in 6 of 10 patients with HIV/HCV coinfection and in 1 of 3 patients with HCV monoinfection. Every amplicon except brain/frontal cortex from 40007 (denoted 40007B) was analyzed by direct sequencing. No E1 amplicon had the sequence of a laboratory strain (such as H77), ruling out plasmid contamination. No appropriately sized amplicons were obtained from 84 HCV reverse-transcription polymerase chain reactions (RT-PCRs) performed on 42 specimens from 14 control subjects without viremia; in all cases, however, a GAPDH amplicon was produced (table 3).
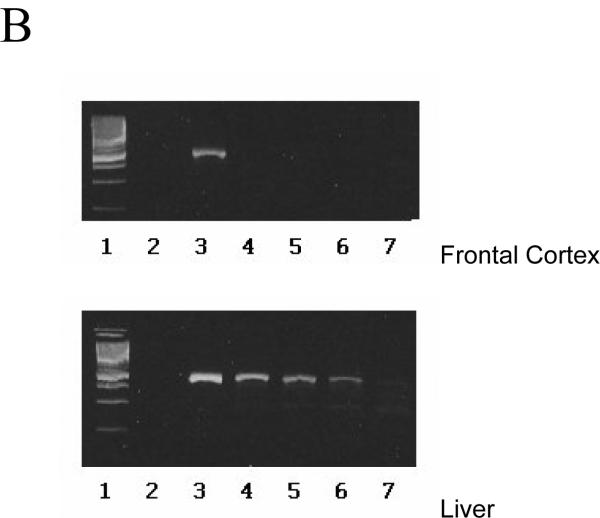
End-point dilution was used to compare the amount of liver and brain/frontal cortex RNA in patients 10086 and 20024. The greatest dilutions yielding E1 amplicons from 10086L and 20024L were 1:10,000 and 1:1000, respectively, whereas the greatest dilutions yielding amplicons from 10086B and 20024B were 1:10 and no dilution, respectively (figure 1). These data suggest that, at PMIs of 4 to 7.5 h, liver HCV was 1000-fold more abundant than brain/frontal cortex HCV.
Figure 1. Relative concentration of HCV RNA in frontal cortex vs. liver, as assessed by end point dilution.

RNA extracted from frontal cortex or liver was either added to an RT/PCR at its initial concentration or it was serially diluted 10 fold and then the E1 region was amplified as described in the text. (A) Patient 10086. The top panel shows the amplicons from frontal cortex RNA used without dilution (lane 3), diluted 1:10 (lane 4), 1:100 (lane 5), 1:1000 (lane 6), and 1:10,000 (lane 7). The bottom panel shows amplicons of liver RNA used without dilution (lane 2), and diluted 1:10 (lane 3), 1:100 (lane 4), 1:1000 (lane 5), 1:10,000 (lane 6), and 1:100,000(lane 7). Size markers are shown in lane 1. In the top panel, lane 2 is a no RNA contamination control. (B) Patient 20024. The top panel shows the amplicons from frontal cortex RNA used without dilution (lane 3), diluted 1:10 (lane 4), 1:100 (lane 5), 1:1000 (lane 6), and 1:10,000 (lane 7). The bottom panel shows amplicons of liver RNA used without dilution (lane 3), and diluted 1:10 (lane 4), 1:100 (lane 5), 1:1000 (lane 6), and 1:10,000 (lane 7). In both panels, size markers are shown in lane 1; lane 2 is a no RNA contamination control.
A bioinformatic approach to assessing the quality of RNA in PM tissues
Detailed analysis of brain HCV sequences would be warranted only if the extracted brain RNAs are reflective of the sequences present at the time of death. We reasoned that PM degradation might alter the template properties of the RNA (e.g., by deamination). A multicomponent bioinformatic assay was developed to assess the quality of PM RNA. This assay takes advantage of the fact that naturally occurring mutations are subject to natural selection, whereas artifactual mutations introduced after death are not. In naturally occurring sequences, natural selection reduces the ratio of nonsynonymous to synonymous (dN/dS) codon substitutions far below the ratio produced by chance. More generally, selection pressures eliminate sequences that contain lethal mutations, causing the frequency distribution of naturally occurring and artifactual mutations to differ. Sequences with artifactual mutations created by degradative processes will have an excess of mutations because they will contain both the mutations that were present at the time of death and any mutations introduced during the PMI.
For the first part of the bioinformatic assay, a 423-base-long region of the E1-encoding sequences (bases 871-1293) from our amplicons were compared with 1156 sequences from GenBank, which were primarily from premortem serum and plasma samples. The dN/dS ratio forGenBanksequences was 0.49,comparedwith 0.39 for (premortem) plasma sequences, 0.32 for PM brain/frontal cortex sequences, and 0.44 for PM liver sequences (table 4A). Thus, the proportion of nonsynonymous amino acid substitutions was actually lower in brain/frontal cortex HCV than in GenBank sequences (P = .045). By this measure, brain HCV was certainly no less constrained than GenBank HCV. This result provides evidence that brain HCV is a suitable template for sequence analysis.
Table 4.
Mutational analysis.
| Set | Average (SD) | P | Average (SD) | P | Average (SD) | P |
|---|---|---|---|---|---|---|
| A. dN/dS substitution analysis | ||||||
| GenBank | 0.49 (0.21) | |||||
| Cortex | 0.32 (0.12) | .04 | ||||
| Liver | 0.44 (0.18) | .39 | ||||
| Plasma | 0.39 (0.16) | .12 |
| B. Nucleotide mutational frequency analysis | Nucleotide | Amino acid | ||||
|---|---|---|---|---|---|---|
| GenBank | 20.55 (6.88) | 6.32 (2.46) | ||||
| Cortex | 23.00 (5.40) | .38 | 5.83 (2.22) | .62 | ||
| Liver | 20.08 (4.33) | .814 | 5.91 (2.31) | .57 | ||
| Plasma | 20.51 (4.42) | .84 | 5.90 (2.42) | .59 | ||
| C. Mutational frequency analysis | Very rare | Rare | Common | |||
|---|---|---|---|---|---|---|
| Nucleotide | ||||||
| GenBank | 1.78 (2.70) | 4.68 (2.94) | 14.08 (5.35) | |||
| Cortex | 3.00 (2.96) | .27 | 4.66 (1.36) | .98 | 15.33 (3.72) | .56 |
| Liver | 2.00 (2.21) | .78 | 4.58 (2.35) | .90 | 13.50 (4.52) | .70 |
| Plasma | 1.78 (2.54) | .81 | 4.68 (1.85) | .58 | 14.05 (3.72) | .50 |
| Amino acid | ||||||
| GenBank | 1.10 (1.55) | 1.46 (1.57) | 3.75 (1.88) | |||
| Cortex | 1.00 (1.26) | .87 | 1.00 (0.63) | .47 | 3.83 (1.72) | .92 |
| Liver | 1.00 (0.85) | .81 | 1.41 (1.37) | .91 | 3.50 (1.56) | .64 |
| Plasma | 0.90 (1.10) | .68 | 1.40 (1.43) | .89 | 3.60 (1.71) | .80 |
| D. BLOSUM score substitution analysis | Very rare (BLOSUM < 0) | Rare (BLOSUM = 0) | Common (BLOSUM > 0) | |||
|---|---|---|---|---|---|---|
| GenBank | 1.63 (1.35) | 2.25 (1.43) | 2.56 (1.48) | |||
| Cortex | 2.00 (1.67) | .50 | 1.83 (1.32) | .48 | 2.00 (1.54) | .35 |
| Liver | 1.83 (1.46) | .60 | 2.09 (1.57) | .71 | 2.09 (1.44) | .29 |
| Plasma | 1.60 (1.26) | .94 | 2.10 (1.52) | .74 | 2.20 (1.54) | .43 |
NOTE. P values are for the comparison with GenBank sequences. Boldface indicates a statistically significant difference. dN/dS, ratio of nonsynonymous to synonymous substitutions.
Second, individual genotype 1a brain sequences and 783 genotype 1a GenBank sequences were compared with the consensus of the GenBank genotype 1a sequences, and individual genotype 1b brain/frontal cortex sequences and 373 1b GenBank sequences were compared with the consensus of the genotype 1b GenBank sequences. The average number of nucleotide mutations was 20.55 in GenBank sequences, compared with 23.0 in PM brain/frontal cortex sequences, 20.08 in PM liver sequences, and 20.51 in premortem plasma sequences (no significant difference for all comparisons) (table 4B). The average number of amino acid mutations was 6.32 in GenBank sequences, 5.83 in brain/frontal cortex sequences, 5.91 in liver sequences, and 5.90 in plasma sequences (no significant difference for all comparisons) (table 4B).
Third, mutations were scored as very rare (present in 1% or fewer GenBank sequences), rare (1%-10%), or common (10%-50%). This approach, which is an alternative to the BLOSUM score analysis [23], can be applied to both nucleic acid and amino acid sequences. The frequency distribution of nucleotide and amino acid mutations and a parallel analysis of BLOSUM scores are provided in table 4C and 4D. Neither PM brain/frontal cortex sequences nor PM liver HCV sequences differed significantly from GenBank sequences in any comparison. These results indicate that HCV RNA extracted from PM tissues did not contain a significantly increased proportion of unusual mutations. On the basis of results from all 3 components of the bioinformatic analysis, PM brain/frontal cortex and liver specimens are appropriate for HCV-sequencing studies.
Direct sequencing analysis of the 5' UTR and the E1 region
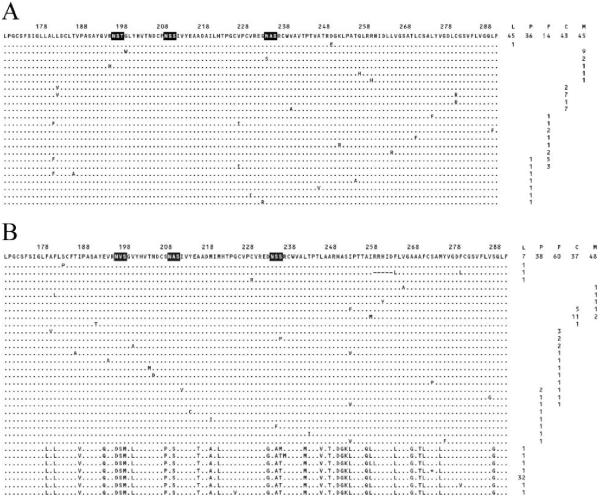
Amplicons of the 5' UTR from 9 plasma, 12 liver, and 4 brain/frontal cortex specimens were analyzed. Five sequences contained mutations at the positions indicated in figure 2, which depicts the HCV internal ribosome entry site [24, 25]. In patients 10034, 20024, and 20028, the 5' UTR sequence from the brain did not match that from the liver or the plasma. E1 region amplicons were sequenced from 12 plasma, 13 liver, and 7 brain/frontal cortex specimens. On the basis of these sequences, most specimens contained genotype 1 HCV. Specimens 40007L and 40007P contained genotype 3a. It is likely that 40007B also contained genotype 3a HCV; however, the sequence could not be determined. No sequence was obtained from >1 patient. This result shows that specimens did not cross-contaminate each other. Sequences of the E1 protein and signal sequence appear in figure 3A-3C. Mutations in the 1a and 1b sequences are marked to indicate their abundance relative to GenBank sequences. Four putative N-linked glycosylation sites, which have the general sequence N-X-S/T [26], occur in this region of E1 and are indicated in the alignment. The E1 signal sequences were compared with 420 genotype 1a (figure 3A) or 629 genotype 1b sequences (figure 3B). Amplicon 20028P had an I186T substitution, whereas 20028L had an I187T substitution. Amplicon 20024P had an I187S substitution. Extensive changes in this region are known to inhibit cleavage by the signal peptide peptidase and alter localization of the core protein [27].
Figure 2.
Diagram of the hepatitis C virus (HCV) internal ribosome entry site (IRES), with the mutations found in patient sequences. The IRES backbone is adapted from Lukavsky et al. [25]. Shading indicates the region amplified in the reverse-transcription polymerase chain reaction. The asterisk indicates that this mutation was found in 1 of 2 extracts of brain/frontal cortex tissue from this patient.
Figure 3.
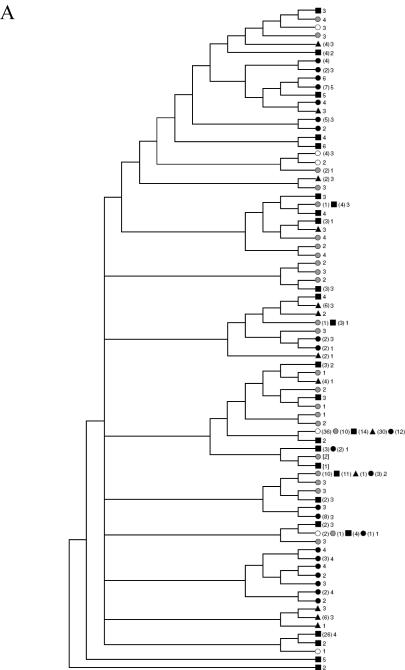
Dendogram and alignment of E1 amino acid sequences from plasma (P), brain/frontal cortex (B), and liver (L). The genotype 1a consensus amino acid sequence appears at the top of the alignment in panel A, and the 1b and 3a consensus sequences are shown at the top of panels B and C, respectively. Black regions in the consensus sequence are potential N-linked glycosylation sites. Very rare mutations (shown in squares) are present in <1% of GenBank sequences of the same genotype, rare mutations (shown in circles) are present in 1%-10% of GenBank sequences, and common mutations (unadorned) are present in >10% of GenBank sequences. Note that the brain/frontal cortex and plasma sequences of patient 20028 are 1a, whereas the liver sequence is 1b. Amino acid position numbering is based on the numbering of the H77 clone. In panel D, samples from the 13 patients with hepatitis C virus viremia yielded 30 specimens with E1 amplicons that could be unambiguously sequenced: Sequences marked with an asterisk differed from the liver and plasma sequences from the same patient. The sequences from patient 20028 (shown in black) did not cluster together.
The phylogenetic relationships of the E1 nucleotide sequences are presented in figure 3D. The RNA sequences of different specimens from the same patient were more closely related to each other than to any other sequence in the set, with the exception of sequences from patient 20028. In the 2 cases indicated by asterisks—10086B and 20028B—the consensus sequence of brain HCV differed from that of liver and plasma HCV. In total, the consensus sequence from brain did not match the plasma and liver sequence in 4 of 7 patients. In the E1 region, HCV RNA from 10086 and 20028 in brain differed from that from plasma and liver. In the 5' UTR, HCV RNA from 10034, 20024, and 20028 in brain did not match that in liver and plasma.
Quasispecies analysis of the E1 region in 2 patients
If HCV is produced in the brain, it might be possible to find supporting evidence by cloning individual sequence variants and comparing the pattern of mutations in brain clones with that in liver and plasma clones. Quasispecies analysis was performed on a portion of the E1 gene inHCVRNAfrom 2 patients, 20024 and 10086. Plasma samples from these patients were collected 2 and 3 days before death (table 3). Three brain regions (frontal cortex, medulla, and cerebellum) were analyzed. Three separate RT-PCRs were performed on each brain region, producing 3 amplicons per region. At least 15 clones of each brain amplicon were amplified, and at least 43 clones of liver and plasma amplicons were sequenced. The mutational patterns of brain and liver sequences were compared with those of plasma sequences in a quality-control analysis similar to the one presented in table 4. Clones from PM tissues did not contain a greater number of mutations or a higher percentage of rare mutations than those from plasma (data not shown), confirming the suitability of the PM tissues for quasispecies analysis. dN/dS ratios for the quasispecies were not significantly different between compartments (table 5).
Table 5.
Quasispecies statistics: Numbers of variants and Dn/Ds ratios.
| Patient and Compartmenta | N | Distinct Variants (Nuc) | Distinct Variants (AA) | Ds | Dn | Dn/Ds |
|---|---|---|---|---|---|---|
| 10086 | 278 | 79 | 24 | 0.002 (0.001) | 0.034 (0.009) | 0.058 |
| Brain total | 189 | 55 | 18 | 0.003 (0.001) | 0.036 (0.01) | 0.083 |
| Cerebellum | 60 | 15 | 5 | 0.004 (0.002) | 0.031 (0.01) | 0.129 |
| Cortex | 70 | 24 | 9 | 0.002 (0.001) | 0.033 (0.011) | 0.06 |
| Medulla | 59 | 22 | 6 | 0.002 (0.001) | 0.033 (0.011) | 0.06 |
| Liver | 46 | 7 | 2 | 0.001 (0.001) | 0.02 (0.01) | 0.05 |
| Plasma | 43 | 25 | 8 | 0.002 (0.001) | 0.035 (0.01) | 0.057 |
| 20024 | 278 | 78 | 34 | 0.453 (0.117) | 0.053 (0.09) | 0.116 |
| Brain total | 178 | 33 | 18 | 0.014 (0.004) | 0.004 (0.001) | 0.285 |
| Cerebellum | 50 | 6 | 4 | 0.004 (0.004) | 0.004 (0.002) | 1 |
| Cortex | 75 | 22 | 12 | 0.016 (0.005) | 0.004 (0.01) | 0.25 |
| Medulla | 53 | 11 | 6 | 0.01 (0.005) | 0.003 (0.002) | 0.3 |
| Liver | 48 | 28 | 11 | 0.521 (0.146) | 0.058 (0.012) | 0.111 |
| 1a | 38 | 22 | 7 | 0.024 (0.01) | 0.002 (0.001) | 0.083 |
| 1b | 10 | 6 | 4 | 0.007 (0.005) | 0.003 (0.002) | 0.428 |
| Plasma | 47 | 19 | 9 | 0.012 (0.004) | 0.004 (0.001) | 0.333 |
The sequences from patient 10086 were all genotype 1a, while the sequences from 20024 were all genotype 1b in the plasma and brain, but were a mixture of genotype 1a and 1b in the liver.
Synonymous substitutions per synonymous site (Ds) and non synonymous substitutions per nonsynonymus site (Dn) were calculated by the MEGA package using the algorithm of Nei and Gojobori with the Jukes-Cantor correction.
A total of 24 distinct amino acid sequence variants were cloned from patient 10086, and 34 were cloned from patient 20024 (figure 4). In both patients, a dominant sequence was shared by the brain (frontal cortex, medulla, and cerebellum combined) and plasma. In patient 10086, the topmost variant in figure 4A was dominant in all tissues; in patient 20024, the topmost variant in figure 4B was dominant in brain and plasma, whereas the 32nd variant, which is a different subgenotype, was dominant in liver.
Figure 4.
Alignment of distinct amino acid sequence variants in patients 10086 and 20024. The quantity of clones that were obtained and the tissue(s) in which the variant was observed are listed on the right. Potential N-linked glycosylation sites are shown in black. Amino acid position numbering is based on the numbering of the H77 clone. A, Variants from patient 10086. B, Variants from patient 20024. C, cerebellum; F, frontal cortex; L, liver; M, medulla; P, plasma.
Brain-specific mutations
Subsequent quasispecies analysis was focused on mutations present in clones from brain and absent in clones from plasma and liver. Mutations that were found only in clones from brain and that were present in at least 2 brain clones—but only if they did not represent the same template (i.e., the mutations were present in the products of at least 2 RT-PCRs and/or occurred in variants that differed from each other at 1 or more secondary positions)—were considered to be brain specific. These restrictions were imposed to counter the skewing of the data that can occur whenever a tiny fraction of the RNA population is cloned and template concentrations are low.
Analysis of brain-specific mutations revealed that brain HCV contained several families of related sequences (figures 5 and 6). Figure 5A presents a family with 14 members from the brain of patient 10086. All members of the family share at least 1 brain-specific mutation with another member of the family. All 10 of the nucleotide substitutions highlighted in figure 5A occurred only in brain HCV and were not present in liver or plasma HCV. Variant 1 can be considered the nexus of the family. It has 3 brain-specific mutations: U930C, A1062G, G1118A, and C1152U. The U930C mutation is shared by variants 2-5; the A1062G mutation is shared by variants 2, 5, 6, and 7. The C1152U mutation is shared by 6, 7, 13, and 14. The family has multiple subcomponents that form branches when phylogenetic analysis is performed (figure 5B). An explanation for the presence of families is that a sequence with a brain-specific mutation replicated locally to yield progeny that retained the original mutation and accumulated additional brain-specific mutations over time. Three additional families occurred in the clones of patient 10086, and 6 families occurred in the clones of 20024 (figure 6).
Figure 5.
A 14-member family of sequences with shared brain-specific mutations. A, Positions of the brain-specific mutations in the largest sequence family from patient 10086. Mutations in the sequences that were not found exclusively in brain are not shown. The nos. at the top of the figure indicate the nucleotide position according to the H77 genotype 1a sequence. The dominant sequence in the brain from patient 10086 is considered to be wild type (WT) and is presented at the top for comparison. The nos. on the left identify the variants, and the letters on the right identify the tissues from which the variant was isolated (C, cerebellum; F, frontal cortex; M, medulla). Repeated letters indicate that the variant was found in multiple reverse-transcription polymerase chain reactions for the same tissue. Starred mutations are not brain specific (as defined in the text) but were observed only in brain-derived clones. Variant 1 shares the U930C mutation with variants 2, 3, 4, and 10; variant 10 has an additional brain-specific mutation, U1128C, which is shared by variants 11-14. The A1062G mutation is shared by variants 1, 2, 4, 5, and 6, and the C1152U mutation is shared by 1, 4, 5, 8, and 9. B, Phylogenetic analysis of all brain-specific variants from patient 10086. Variants are labeled to indicate their position in the tree.
Figure 6. Additional families of sequences with shared brain-specific mutations.
(A-F) Six families from the quasispecies of patient 20024. The numbers on the left identify the variants in the family, and the letter(s) on the right identify the tissue(s) from which the variant was isolated: F (frontal cortex) C (cerebellum), M (medulla). Repeated letters indicate that the variant was found in multiple RT/PCR reactions from the same tissue. The numbers at the top indicate the nucleotide position according to the numbering of H77. Brain-specific nucleotide mutations are boxed. Family A centers at three mutations: C880U, C1001U, and C1157U. Variant 2 in this family contains all three mutations, and shares C880U with variant 1 and C1001U and C1157U with variant 3. Family B consists of two mutations, G887A and U940C which are both found in variant 1. The U940C mutation is also found in variant 2. In the family shown in Panel C there are four shared brain-specific mutations: G897A, C965U, A1098G, A1113G. Sequence 1 shares the C965U mutation with sequences 2, 3, and 4. Sequence 4 is related to sequence 5 via the A1113G mutation. Each section of the brain analyzed is represented in this family, and variants 3 and 5 were found in two different brain regions. Similar to family B, family D has two members, one mutation is present in both and a second mutation is present in one of the members. Families E and F differ from the other families in that they consist of only one variant. This variant creates a family on its own because it was identified in multiple RT reactions and contains brain specific mutations. In the case of family E, the variant was identified in two separate RT-PCR reactions from the frontal cortex. In family F, the variant was identified in the medulla and cerebellum. (G-I) Three additional families were constructed from the quasispecies of patient 10086. Family G contains three mutations C885G, U804C, and U1182C. Variant 1 contains all three mutations. The remaining variants contain subsets of the three mutations. Interestingly, variant two was found in multiple RT/PCR reactions from the cerebellum. Families H and I resemble families B and D of patient 20024. They consist of two mutations which appear together in one variant, and individually in additional variants.
SNP analysis of a prominent brain-specific mutation
To validate the cloning and sequencing results, we used SNP analysis to measure the prevalence of one of the brain-specific mutations. The brain-specific mutation A1113G found in patient 20024 creates a novel restriction site. The amplicons of this patient were incubated with a restriction enzyme specific for this site, and the resulting restriction fragments were analyzed by gel electrophoresis (selected samples are shown in figure 7). The band resulting from cleavage at the site of the mutation (arrow C) constituted an average of 10% over the 3 cerebellum amplicons and a similar percentage of the 3 medulla amplicons. In contrast, this band was not present in digests of liver and plasma amplicons (figure 7). This SNP analysis indicates that the A113G mutation is, indeed, more prevalent in brain than in liver or plasma.
Figure 7.
Single-nucleotide polymorphism analysis of a prominent brain-specific mutation: Amplicons from all tissues from patient 20024 were digested with restriction enzyme Alw261. The A1113G mutation observed in clones from cerebellum and medulla created a new restriction site, allowing the prevalence of this mutation in the hepatitis C virus (HCV) population amplified from each tissue to be estimated. The plus sign indicates that enzyme was included in the digestion reaction; parallel no-enzyme reactions were performed and are shown in the lanes indicated by minus signs. Arrow A indicates the position of uncut amplicon. Arrow B indicates the position of the fragments containing the wild-type sequence. Arrow C indicates the position of the fragments containing the mutant sequence. Seven percent of the cut amplicon in the cortex 4 reaction contained the mutation, and 22% of the cut amplicon in the cerebellum 1 reaction contained the mutation.
Statistical evaluation of compartmentalization
To statistically analyze the extent of sequence compartmentalization, we examined correlations between the genetic distance of the sequences and their body compartment of origin using Mantel's test. As a control for the validity of this approach, we compared sequences obtained from 2 separate RT-PCRs on the same brain region. Statistically significant compartmentalization was observed for sequences from 2 separate RT-PCRs on the cerebellum from patient 20024. Because these sequences were obtained from the same RNA population, this result demonstrates that Mantel's test cannot be used to test for sequence compartmentalization on the basis of genetic distance. For each patient, phylogenetic trees of all variants were constructed (figure 8). For patient 10086, medulla variants partially clustered. For patient 20024, most liver sequences segregated from brain and plasma. This reflects the difference in the predominant subgenotype between these compartments.
Figure 8. Phylogenetic trees of distinct nucleotide sequences amplified from patients 10086 and 20024 and application of Mantel's test.
Bootstrapped phylogenetic trees of the distinct variants suggested that brain variants clustered separately from liver variants. The symbols next to the branches indicate the tissue(s) in which the sequence was observed. Open circles are liver, grey circles are plasma, filled circle are medulla, filled triangles are cerebellum, and filled squares are from frontal cortex. The numbers in parentheses indicate the number of clones of the variant observed if greater than one. The number of nucleotide changes relative to the dominant sequence is noted alongside each sequence branch.
A) 10086. Medulla variants of patient 10086 appeared to cluster together in two main groups. B)20024. Most liver sequences amplified from this patient were genotype 1a, while plasma and brain variants were uniformly from genotype 1b.
DISCUSSION
The most basic question in HCV neurobiology is whether HCV occurs in living brain outside the vasculature. An affirmative answer would provide an impetus for investigating the clinical significance of the invasion of the brain by HCV. Our investigation advances HCV brain research by presenting evidence that mutations in brain HCV do not have the aberrant features expected of mutations produced by PM degradation, supporting the use of PM brain in HCV sequencing studies. Our study also shows that the consensus sequence of HCV RNA in brain often differs from that in liver and plasma and demonstrates that families of brain sequences have shared brain-specific mutations that are consistent with HCV RNA evolution occurring within the brain. Particularly in light of the bioinformatic data demonstrating that brain HCV did not contain artifactually introduced mutations, the disparity between brain and plasma sequences effectively rules out blood contamination as the source of the brain HCV and focuses attention on the possibility that brain RNA was synthesized locally within the brain.
Our analysis also revealed a nonsignificant trend toward an inverse relationship between PMI and the detection of brain HCV. This trend suggests that the actual prevalence may exceed our observed frequency, which was 54% (7/13). A single 35-cycle protocol was used to amplify the 5' UTR. The ability to detect HCV without a “nested” PCR assay suggests that brain HCV is relatively abundant, although it is 1000-fold lower than liver HCV in PM specimens.
Direct sequencing identified mutations in the first or third putative N-linked glycosylation sites [28] in the brain E1 amplicon from 4 patients (10066, 20024, 20028, 10034) and in 2 clones from the medulla of patient 10086 (figure 3B and 3C). These mutations are of interest because glycans at these sites are thought to play a role in viral entry [28]. Like the E1 amplicons, the 5' UTR amplicons of 10034B and 20028B had unusual substitutions, suggesting that in these patients both the UTR and the envelope protein may have adaptations that enhance entrance into and expression within the brain. Of the 7 patients with brain HCV, the consensus sequence did not match liver and plasma HCV in 4, contributing to the growing evidence that the brain supports replication of a separate and distinctive subpopulation of HCV.
To obtain further evidence of HCV replication and evolution within the brain, quasispecies analysis was done on HCV RNA from plasma, liver, and 3 noncontiguous brain regions of 2 patients. Specific nucleotide and amino acid mutations (i.e., brain-specific mutations) were found in clones from multiple brain regions and were not found in clones from plasma or liver of the same patient. The repeated detection of the same mutations suggests that they were not the result of polymerase or sequencing errors. Moreover, brain-specific nucleotide mutations present in clones from 2 RT-PCRs on the frontal cortex of patient 10086 were missing from the clones from cerebellum and medulla. This finding further suggests that blood contamination was not likely to be the source of brain HCV RNA, because the viral population from contaminating blood would be the same throughout the brain and would be identical to that in plasma.
Among brain sequences, several families of variants were identified that shared brain-specific mutations. Some families consisted of variants from >1 brain region and were supported by cluster analysis. To investigate the total population of molecules in amplicons (not only those that were individually cloned and sequenced), SNP analysis was performed using a restriction enzyme specific for the A1113G mutation present in the brain quasispecies of patient 20024. This analysis confirmed that the A1113G mutation was, indeed, brain specific. This finding and the presence of sequence families whose members are related to each other through shared brain-specific mutations support the hypothesis that HCV replicates and evolves within the brain.
Our results add to previously reported evidence of brain compartmentalization. The quasispecies in the gray matter of 1 (of 2) liver-transplant recipients did not match that in serum, although the consensus sequences were identical [17]; brain and serum 5' UTR sequences differed in 2 of 6 patients at the genotype level [18]; and the brain hypervariable region (HVR) 1 master sequence did not match that in serum in 1 of 2 patients [19]. The mutations we observed in the 5' UTR of brain HCV are an interesting and nonrandom group of substitutions. The U147C mutation in 10034B has been reported in lymph node HCV [19], and the insertion at position 126 in 20024B and 20028B* has been reported in lymph node [18, 19] and cerebellum [18] HCV, suggesting that these substitutions might represent adaptations to replication in extrahepatic compartments. The insertion, however, has also been reported in serum HCV [29, 30].
Sequence analysis is currently the mainstay of molecular HCV brain research. Complementary methods, such as immunohisto-chemistry and in situ hybridization, may become more widely available in the future, but despite notable successes [20, 31-33] these techniques are extremely difficult to apply effectively and accurately to HCV [34, 35]. Minus-strand detection has often been used to identify sites of viral replication, but in the case of brain HCV this method can never be definitive because of the potential for PBMCs to portage inert HCV minus strands in and out of the brain. Proof of HCV replication in brain will probably await rigorous and unequivocal demonstration that the population of HCV in 1 or more parts of the brain is distinct from HCV in the periphery.
In conclusion, the detection of HCV in brain is not sufficient to implicate HCV in brain dysfunction; however, it is a necessary first step. The substitution patterns we detected in brain HCV provide a basis for further investigation. Occult kidney damage in patients with HCV was recently found to be prevalent and extensive [36], suggesting that HCV may cause more extrahepatic damage than has been appreciated. The bioinformatic approach introduced here will facilitate future studies of HCV in brain and other extrahepatic sites.
Acknowledgments
Financial support: National Institutes of Health (grants DA016156 and DK066939 to A.D.B. and grant R24MH59724 to S.M.); American Liver Foundation Scholar's Award (to F.J.E.); National Center for Research Resources (grant MO1-RR-00071).
Footnotes
Potential conflicts of interest: none reported
References
- 1.Ryan EL, Morgello S, Isaacs K, Naseer M, Gerits P. Neuropsychiatric impact of hepatitis C on advanced HIV. Neurology. 2004;62:957–62. doi: 10.1212/01.wnl.0000115177.74976.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soogoor M, Lynn HS, Donfield SM, Gomperts E, Bell TS, Daar ES. Hepatitis C virus infection and neurocognitive function. Neurology. 2006;67:1482–5. doi: 10.1212/01.wnl.0000240255.42608.99. [DOI] [PubMed] [Google Scholar]
- 3.Hilsabeck RC, Perry W, Hassanein TI. Neuropsychological impairment in patients with chronic hepatitis C. Hepatology. 2002;35:440–6. doi: 10.1053/jhep.2002.31257. [DOI] [PubMed] [Google Scholar]
- 4.McAndrews MP, Farcnik K, Carlen P, et al. Prevalence and significance of neurocognitive dysfunction in hepatitis C in the absence of correlated risk factors. Hepatology. 2005;41:801–8. doi: 10.1002/hep.20635. [DOI] [PubMed] [Google Scholar]
- 5.Forton DM, Taylor-Robinson SD, Thomas HC. Cerebral dysfunction in chronic hepatitis C infection. J Viral Hepat. 2003;10:81–6. doi: 10.1046/j.1365-2893.2003.00416.x. [DOI] [PubMed] [Google Scholar]
- 6.Kramer L, Bauer E, Funk G, et al. Subclinical impairment of brain function in chronic hepatitis C infection. J Hepatol. 2002;37:349–54. doi: 10.1016/s0168-8278(02)00172-1. [DOI] [PubMed] [Google Scholar]
- 7.Laskus T, Radkowski M, Adair DM, Wilkinson J, Scheck AC, Rakela J. Emerging evidence of hepatitis C virus neuroinvasion. AIDS. 2005;19(Suppl 3):S140–4. doi: 10.1097/01.aids.0000192083.41561.00. [DOI] [PubMed] [Google Scholar]
- 8.Forton DM, Thomas HC, Murphy CA, et al. Hepatitis C and cognitive impairment in a cohort of patients with mild liver disease. Hepatology. 2002;35:433–9. doi: 10.1053/jhep.2002.30688. [DOI] [PubMed] [Google Scholar]
- 9.Forton DM, Allsop JM, Main J, Foster GR, Thomas HC, Taylor-Robinson SD. Evidence for a cerebral effect of the hepatitis C virus. Lancet. 2001;358:38–9. doi: 10.1016/S0140-6736(00)05270-3. [DOI] [PubMed] [Google Scholar]
- 10.Weissenborn K, Ennen JC, Bokemeyer M, et al. Monoaminergic neuro-transmission is altered in hepatitis C virus infected patients with chronic fatigue and cognitive impairment. Gut. 2006;55:1624–30. doi: 10.1136/gut.2005.080267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu YK, Igarashi H, Kanematu T, et al. Sequence analysis of the hepatitis C virus genome recovered from serum, liver, and peripheral blood mononuclear cells of infected chimpanzees. J Virol. 1997;71:5769–73. doi: 10.1128/jvi.71.8.5769-5773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laskus T, Radkowski M, Wang LF, Jang SJ, Vargas H, Rakela J. Hepatitis C virus quasispecies in patients infected with HIV-1: correlation with extrahepatic viral replication. Virology. 1998;248:164–71. doi: 10.1006/viro.1998.9269. [DOI] [PubMed] [Google Scholar]
- 13.Navas S, Martin J, Quiroga JA, Castillo I, Carreno V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–6. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roque-Afonso AM, Ducoulombier D, Di Liberto G, et al. Compartmentalization of hepatitis C virus genotypes between plasma and peripheral blood mononuclear cells. J Virol. 2005;79:6349–57. doi: 10.1128/JVI.79.10.6349-6357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lerat H, Berby F, Trabaud MA, et al. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Invest. 1996;97:845–51. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dammacco F, Sansonno D, Piccoli C, Racanelli V, D'Amore FP, Lauletta G. The lymphoid system in hepatitis C virus infection: autoimmunity, mixed cryoglobulinemia, and overt B-cell malignancy. Semin Liver Dis. 2000;20:143–57. doi: 10.1055/s-2000-9613. [DOI] [PubMed] [Google Scholar]
- 17.Vargas HE, Laskus T, Radkowski M, et al. Detection of hepatitis C virus sequences in brain tissue obtained in recurrent hepatitis C after liver transplantation. Liver Transpl. 2002;8:1014–9. doi: 10.1053/jlts.2002.36393. [DOI] [PubMed] [Google Scholar]
- 18.Radkowski M, Wilkinson J, Nowicki M, et al. Search for hepatitis C virus negative-strand RNA sequences and analysis of viral sequences in the central nervous system: evidence of replication. J Virol. 2002;76:600–8. doi: 10.1128/JVI.76.2.600-608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forton DM, Karayiannis P, Mahmud N, Taylor-Robinson SD, Thomas HC. Identification of unique hepatitis C virus quasispecies in the central nervous system and comparative analysis of internal translational efficiency of brain, liver, and serum variants. J Virol. 2004;78:5170–83. doi: 10.1128/JVI.78.10.5170-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letendre S, Paulino AD, Rockenstein E, et al. Pathogenesis of hepatitis C virus coinfection in the brains of patients infected with HIV. J Infect Dis. 2007;196:361–70. doi: 10.1086/519285. [DOI] [PubMed] [Google Scholar]
- 21.Bolay H, Soylemezoglu F, Nurlu G, Tuncer S, Varli K. PCR detected hepatitis C virus genome in the brain of a case with progressive encephalomyelitis with rigidity. Clin Neurol Neurosurg. 1996;98:305–8. doi: 10.1016/0303-8467(96)00040-6. [DOI] [PubMed] [Google Scholar]
- 22.Bakr I, Rekacewicz C, El Hosseiny M, et al. Higher clearance of hepatitis C virus infection in females compared with males. Gut. 2006;55:1183–7. doi: 10.1136/gut.2005.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–9. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown EA, Zhang H, Ping LH, Lemon SM. Secondary structure of the 5' nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7:1105–10. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 26.Marshall RD. The nature and metabolism of the carbohydrate-peptide linkages of glycoproteins. Biochem Soc Symp. 1974:17–26. [PubMed] [Google Scholar]
- 27.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980–8. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goffard A, Callens N, Bartosch B, et al. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol. 2005;79:8400–9. doi: 10.1128/JVI.79.13.8400-8409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto H, Okada S, Sugiyama Y, et al. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991;72:2697–704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- 30.van Leeuwen HC, Reusken CB, Roeten M, et al. Evolution of naturally occurring 5' non-translated region variants of hepatitis C virus genotype 1b in selectable replicons. J Gen Virol. 2004;85:1859–66. doi: 10.1099/vir.0.79924-0. [DOI] [PubMed] [Google Scholar]
- 31.Pal S, Shuhart MC, Thomassen L, et al. Intrahepatic hepatitis C virus replication correlates with chronic hepatitis C disease severity in vivo. J Virol. 2006;80:2280–90. doi: 10.1128/JVI.80.5.2280-2290.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ballardini G, De Raffele E, Groff P, et al. Timing of reinfection and mechanisms of hepatocellular damage in transplanted hepatitis C virus-reinfected liver. Liver Transpl. 2002;8:10–20. doi: 10.1053/jlts.2002.30141. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Inigo E, Bartolome J, de Lucas S, et al. Histological damage in chronic hepatitis C is not related to the extent of infection in the liver. Am J Pathol. 1999;154:1877–81. doi: 10.1016/S0002-9440(10)65445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gowans EJ. Distribution of markers of hepatitis C virus infection throughout the body. Semin Liver Dis. 2000;20:85–102. doi: 10.1055/s-2000-9503. [DOI] [PubMed] [Google Scholar]
- 35.Fenwick F, Bassendine MF, Agarwal K, et al. Immunohistochemical assessment of hepatitis C virus antigen in cholestatic hepatitis after liver transplantation. J Clin Pathol. 2006;59:174–8. doi: 10.1136/jcp.2005.028126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGuire BM, Julian BA, Bynon JS, Jr, et al. Brief communication: glomerulonephritis in patients with hepatitis C cirrhosis undergoing liver transplantation. Ann Intern Med. 2006;144:735–41. doi: 10.7326/0003-4819-144-10-200605160-00007. [DOI] [PubMed] [Google Scholar]
- 37.Murray J, Fishman SL, Ryan E. Clinicopathologic correlates of hepatitis C virus in brain: a pilot study. J Neurovirol. doi: 10.1080/13550280701708427. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corbet S, Bukh J, Heinsen A, Fomsgaard A. Hepatitis C virus subtyping by a core-envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J Clin Microbiol. 2003;41:1091–100. doi: 10.1128/JCM.41.3.1091-1100.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]