Abstract
Visualizing developing organ formation as well as progession and treatment of disease often heavily relies on the ability to optically interrogate molecular and functional changes in intact living organisms. Most existing optical imaging methods are inadequate for imaging at dimensions that lie between the penetration limits of modern optical microscopy (0.5-1mm) and the diffusion-imposed limits of optical macroscopy (>1cm) [1]. Thus, many important model organisms, e.g. insects, animal embryos or small animal extremities, remain inaccessible for in-vivo optical imaging.
Although there is increasing interest towards the development of nanometer-resolution optical imaging methods, there have not been many successful efforts in improving the imaging penetration depth. The ability to perform in-vivo imaging beyond microscopy limits is in fact met with the difficulties associated with photon scattering present in tissues. Recent efforts to image entire embryos for example [2,3] require special chemical treatment of the specimen, to clear them from scattering, a procedure that makes them suitable only for post-mortem imaging. These methods however evidence the need for imaging larger specimens than the ones usually allowed by two-photon or confocal microscopy, especially in developmental biology and in drug discovery.
We have developed a new optical imaging technique named Mesoscopic Fluorescence Tomography [4], which appropriate for non-invasive in-vivo imaging at dimensions of 1mm-5mm. The method exchanges resolution for penetration depth, but offers unprecedented tomographic imaging performance and it has been developed to add time as a new dimension in developmental biology observations (and possibly other areas of biological research) by imparting the ability to image the evolution of fluorescence-tagged responses over time. As such it can accelerate studies of morphological or functional dependencies on gene mutations or external stimuli, and can importantly, capture the complete picture of development or tissue function by allowing longitudinal time-lapse visualization of the same, developing organism.
The technique utilizes a modified laboratory microscope and multi-projection illumination to collect data at 360-degree projections. It applies the Fermi simplification to Fokker-Plank solution of the photon transport equation, combined with geometrical optics principles in order to build a realistic inversion scheme suitable for mesoscopic range. This allows in-vivo whole-body visualization of non-transparent three-dimensional structures in samples up to several millimeters in size.
We have demonstrated the in-vivo performance of the technique by imaging three-dimensional structures of developing Drosophila tissues in-vivo and by following the morphogenesis of the wings in the opaque Drosophila pupae in real time over six consecutive hours.
Protocol
The Drosophila melanogaster stocks used in this experiment are the following:
elav-Gal4 (FBti0002575)
ap-Gal4md544 (FBal0051787)
UAS-srcEGFP (FBti0013990)
w1118 (FBal0018186)
For all experiments, the UAS-Gal4 system was used to overexpress GFP in the tissue of interest (i.e the salivary glands or the wing discs). More specifically, female flies transgenic with the appropriate Gal4 were crossed to UAS-EGFP transgenic mates. The crosses were reared at 25°C in a humidified incubator. When pupae start populating the vials (approximately 5-6 days after initiation of the cross), they were selected for GFP fluorescence at the white prepupal stage (0-1 hr after puparium formation) and collected for tomography imaging.
Set up appropriate crosses and rear at 25°C
5-6 days after the initiation of the cross, select fluorescent progeny for imaging. Use a wet paintbrush for gentle removal of the white prepupae from the sides of the vial.
Put prepupae in a drop of water or PBS placed in a petri dish to remove food particles and the glue that covers the pupal case and makes the specimen autofluorescent. Gently clean with a paintbrush. If aging of the pupae is necessary, place selected white prepupae in a petri dish with a wet paper towel (a humid chamber) or at the sides of a clean vial with fly food. This will ensure that there is enough humidity, to allow proper development of the animal.
Drosophila Mounting
Glue the bottom part of the pupal case into a capillary glass tube (800 microns diameter) such that the fly’s anterior/posterior axis is oriented parallel to the tube.
Fix the capillary tube vertically on a rotational stage.
Adjust the tilting axis of the rotational stage in order for the rotational axis to be parallel to the pixel columns of the imaging CCD.
Determination Forward Model
Fix a pupa in a capillary glass and mount as described before
Fill a silica capillary tube (100 microns, OD) with a fluorescent dye (Cy5.5)
Insert the capillary tube inside the pupa at 45 degrees with respect to the anterior/posterior axis
Illuminate the pupa with an excitation laser beam with a low numerical aperture lens. Collect fluorescence transillumination images over 360 degrees rotating the pupa along its vertical axis.
Fit the experimental data with an appropriate forward model
Imaging Salivary Glands
Mount a pupa expressing GFP in the salivary glands as described before
Illuminate with an excitation beam and collect in transillumination the GFP fluorescence signal over 360 degrees rotating the pupa along its vertical axis.
Reconstruct the acquired data utilizing the forward model
Time-Lapse Imaging
Mount a white prepupa expressing GFP in the wing imaginal discs
Keep the imaging environment humid and at room temperature to avoid dehydration of the pupa.
Place a shutter to avoid continuous illumination
Illuminate with an excitation beam and collect in transillumination the GFP fluorescence signal over 360 degrees rotating the pupa along its vertical axis.
Reconstruct the acquired data utilizing the forward model.
Histology
Dissect salivary glands CNS/eye discs in 1x PBS.
Fix tissues in 4% Formaldehyde, 1x PEM (100mM Pipes, 1mM EGTA, 1mM MgCl2) for 20min at room temperature.
Remove fixation and mount in Vectashield with dapi (Vector)
Image with a fluorescent microscope.
Time-Lapse Histology
Collect white prepupae and stage them in a humid chamber.
Collect pupae at different developmental stages
Prick the pupal case twice (0 hrs After Puparium Formation) with a fine needle (insect pin 000, FST) before fixation.
Fix pupae in 8% Formaldehyde, 1x PEM for 6-8 hrs at room temperature with gentle agitation.
After removal of fixation and rinsing with 1x PBS, cut the fixed pupa with a sharp blade in two pieces perpendicular to the A/P axis;
Submerge the two pieces in Vectashield with dapi (Vector) and mount on coverglass slides (Lab-Tek) for imaging.
Acquire images with a fluorescence confocal microscope.
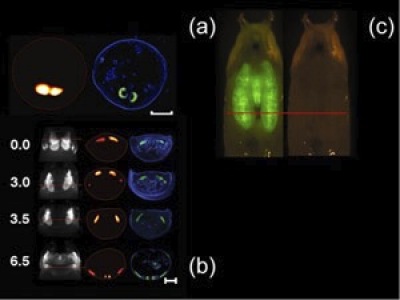 Figure 1. Please click here to see a larger version of figure 1.
Figure 1. Please click here to see a larger version of figure 1.
Discussion
In-vivo reconstructions of the pupal case and the GFP-expressing salivary glands of a D. melanogaster prepupa (Scale bar, 500 microns) with the corresponding histology (blue, dapi staining; green, GFP fluorescence) is shown in Fig 1(a). Time-lapse series imaging of D. melanogaster wing imaginal discs is shown in Fig.1(b). The images are acquired from a single live specimen, at four different time points (0,3.0, 3.5, and 6.5 hours). In the first column one projection at 0 degrees with respect to the pupa's dorsal view is shown. In the second column the reconstructions correspond to the sections indicated by the red lines. Finally, a comparison with the histology is shown the third column. Clearly, the histological preparations correlate well with the tomographic reconstructions. In (c) planar image of a Drosophila’s pupal case and the GFP-expressing salivary glands.
Acknowledgments
C. Vinegoni acknowledges support from National Institutes of Health (NIH) grant 1-RO1-EB006432.
References
- Ntziachristos V. Looking and listening to light: the evolution of whole-body photonic imaging. Nat. Biotechnol. 2005;23:313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- J Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Dodt HU. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Vinegoni C. In vivo imaging of Drosophila melanogaster pupae with mesoscopic fluorescence tomography. Nat. Methods. 2008;5:45–47. doi: 10.1038/nmeth1149. [DOI] [PubMed] [Google Scholar]


