Abstract
An algorithmic solution to the biological effective dose (BED) calculation from the Lea–Catcheside formula for a piecewise defined function is presented. Data from patients treated for metastatic thyroid cancer were used to illustrate the solution. The Lea–Catcheside formula for the G-factor of the BED is integrated numerically using a large number of small trapezoidal fits to each integral. The algorithmically calculated BED is compatible with an analytic calculation for a similarly valued exponentially fitted dose-rate plot and is the only resolution for piecewise defined dose-rate functions.
INTRODUCTION
The use of the biological effective dose (BED) has gained favor in dosimetry as it folds the biological effect of dose rate in with the standard absorbed dose. The success of the BED-response relationship established in the context of renal toxicity from radiopeptide therapy has given much credence to the formalism.1 The difficulty in applying the BED is that formulas only exist for analytic application for exponential based functional fits. In many instances the lack of a clear pattern for fitting leads to a piecewise trapezoidal fit of the dose-rate data for integration to absorbed dose. Recent attempts to extend the BED formalism to more general applications still depend on dose-rate contributions being analytically integrable.2, 3 Analytic calculation of the BED requires functional knowledge of the entire fit, which is not possible with a piecewise defined function such as trapezoidal fitting. We have devised an algorithmic resolution of the Lea–Catcheside G-factor formula to enable BED calculation for any function, including piecewise defined functions.
METHODS
The illustrative data comprises six PET image time points of the head and neck region at 0.5, 1, 2, 4, 24, and 48 h post 124I injection from patients treated at the University of Duisburg-Essen, Essen, Germany.4 The three-dimensional dosimetry package 3D-RD (Ref. 5) was used for the analysis and the regions of interest (salivary glands) were defined functionally from the PET images.
The BED is defined as
| (1) |
where α and β are the radiobiological parameters from the linear quadratic equation and D is the absorbed dose. The G-factor G(T) (Lea–Catcheside formula) is defined as6, 7
| (2) |
w and t are integration variables, while μ is the repair rate for DNA damage, assuming exponential repair. The algorithm will integrate G(T) numerically by approximation with a large number of small trapezoids; the integral over the second integration variable, w, in Eq. 2 is equal to
| (3) |
For the simple exponential fit of the form: , where λ is the effective clearance rate, Eq. 2 becomes
| (4) |
For large T values (T→∞) the exponentials disappear and the BED is reduced to the familiar8
| (5) |
For a piecewise defined function, an analytical formula cannot be established for the BED as it is time dependent and depends on full integral of the dose-rate function from time 0 to the desired time point. As such we have developed an algorithmic method for calculating the BED from the Lea–Catcheside equation.
-
(1)
Plot the dose-rate function. In the illustrative example, the results are from time point by time point Monte Carlo simulation and already in dose rates.4 A more conventional dosimetric approach consists of calculating absorbed dose using absorbed fractions from activity plots. A conversion from cumulated activity to dose is necessary to obtain the dose-rate plot first.
-
(2)
Bin the time range into small enough units (large number of bins, in our case 4000), from 0 to the desired maximum time (in our case four times 48 hours=192 hours).
-
(3)Iterate over the time bins for each n, with t(n)=nT. T is the time interval between bins.
-
(a)Determine the appropriate function piece and calculate the dose rate for each time bin, .
-
(b)Calculate the contribution to the integral in Eq. 3,
(6) -
(c)Calculate the Lea–Catcheside differential, ,
(7) -
(d)Calculate the contribution to the G-factor,
(8)
-
(a)
-
(4)
Substitute G(max time bin) into Eq. 1.
Application of the technique is illustrated in a clinical example of radioiodine uptake in the salivary glands, which is influenced by the timing of food intake such that the dose rate or activity kinetics are not amenable to simple exponential fits. Dose-rate values from 3D-RD calculations are plotted and fit with (A) a simple exponential function and (B) a hybrid trapezoid-partial exponential fit. The numeric integration algorithm is used to calculate the BED for both cases and the exponential result is compared to the analytic solution from Eq. 5.
Practical questions for the implementation of the algorithm are examined by considering the rate at which the G-factor converges, the variability as a function of binnumber and cutoff point for the simple exponential case. The function
| (9) |
where Gth is the analytical solution, is calculated for various cutoff points and total bin numbers as an assessment of accuracy.
RESULTS
The dose-rate plot and fits, both the simple exponential and the piecewise trapezoid-exponential hybrid, are shown in Fig. 1. Figures 23 show the G-factor as a function of time (or of number of iterations) for the piecewise defined function (Fig. 2) and the simple exponential (Fig. 3). The figures also show the dose-rate functions and the incremental change in the G-factor; these last two are scaled to fit on the same graph. Additionally in Fig. 3, the analytical value calculated from Eq. 5 is represented. Equation 4, the time-dependent analytical function, is not represented as it is virtually identical to the calculated values.
Figure 1.
Dose-rate values in the region of interest calculated from 3D-RD plotted as a function of time (triangles). The dotted line shows the exponential fit and the solid line shows the hybrid trapezoid-exponential fit.
Figure 2.
G-factor shown for the hybrid fit (thin line). The dotted line shows the hybrid functional fit to the dose rate, and the crosses (the thick line) show the incremental change in the G-factor (dG∕dt).
Figure 3.
G-factor plotted for the simple exponential fit (thin line). The dotted line shows the exponential fit, the thin dotted line represents the theoretical value, and the crosses (the thick line) show the incremental change in the G-factor (dG∕dt).
The graphs show results and calculations out to 48 h. The difference between the numeric and analytic G-factors [or accuracy A(n) of the numeric value] is 0.0017% for N=4000 bins (tcutoff=192 h) in the exponential case. For 35 other test cases the average difference was 0.0029%, with a standard deviation of 0.0018%. Table 1 shows the accuracy of the illustrated example calculated for different total numbers of bins (rows) and different cutoff points (columns).
Table 1.
Percent difference between analytic and numerical calculation for different bin numbers [Eq. 9]; negative values mean that the numerical calculation result is greater than the analytical result.
| No. of bins∕cutoff time | 48 h | 96 h | 144 h | 192 h |
|---|---|---|---|---|
| 40 | −13.1% | −14.2% | −14.2% | −14.2% |
| 400 | 0.352% | −0.169% | −0.171% | −0.171% |
| 4 000 | 0.495% | 1.67 10−4% | −1.71 10−3% | −1.71 10−3% |
| 40 000 | 0.494% | 1.84 10−3% | −1.01 10−5% | −1.71 10−5% |
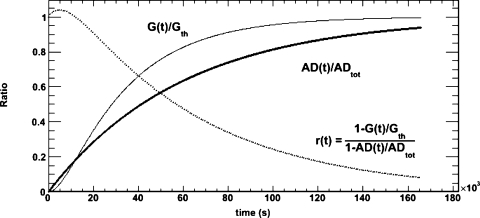
The rate of convergence is plotted in Fig. 4 and compared to the rate of convergence of the absorbed dose. The function r(t) is also plotted which compares the two functions graphically. For example, at 48 h r(t) has a value of ∼0.1, meaning that G(t) is approximately ten times closer to the theoretical value of G than AD(t) is to the total absorbed dose.
Figure 4.
convergence of the G-factor and the absorbed dose. The thin line shows the convergence of the G-factor, the thick line represents the convergence of the absorbed dose to its final value, and the dotted curve shows the ratio of the convergence of the two quantities.
DISCUSSION
The results show a great agreement between analytic and numeric solutions of G for the exponential case. There can be no analytic certainty of convergence for piece-wise defined functions that have exponential tails for the same reason that there is no analytic solution. However, for a numeric calculation, the G-factor converges rapidly (Table 1, Fig. 4) and a choice of several multiples of the last time point is sufficient to ensure precise results. Practically, precision can be increased and verified by showing that greater time does not change the calculated value; accuracy is improved by increasing the number of bins at a fixed time (e.g., as in Table 1).
CONCLUSION
The BED is widely regarded as a more biologically relevant quantity than absorbed dose in terms of establishing relationships with response or toxicity. This algorithm presents an important working solution in situations where the dose rate versus time curve cannot be expressed as an exponential function. By enabling a BED calculation for any dose-rate curve, we have practically extended the applicability of the BED and by extension, of reliable dose-response studies.
ACKNOWLEDGMENTS
The work presented here solves a problem which arose during a collaborative study with the University of Duisburg-Essen. The authors wish to express their thanks to Andreas Bockisch and Walter Jentzen for the data which illustrates the algorithm and for their support. This work supported by NIH∕NCI grant R01 CA116477.
References
- Barone R., Borson-Chazot F., Valkema R., Walrand S., Chauvin F., Gogou L., Kvols L. K., Krenning E. P., Jamar F., and Pauwels S., “Patient-specific dosimetry in predicting renal toxicity with (90)Y-DOTATOC: relevance of kidney volume and dose rate in finding a dose-effect relationship,” J. Nucl. Med. 46, 99S–106S (2005). [PubMed] [Google Scholar]
- Baechler S., Hobbs R. F., Prideaux A. R., Wahl R. L., and Sgouros G., “Extension of the biological effective dose to the MIRD schema and possible implications in radionuclide therapy dosimetry,” Med. Phys. 10.1118/1.2836421 35, 1123–1134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell R. W., Goddu M. S., and Rao D. V., “Application of the linear-quadratic model to radioimmunotherapy: Further support for the advantage of longer-lived radionuclides,” J. Nucl. Med. 35, 1861–1869 (1994). [PubMed] [Google Scholar]
- Jentzen W., “Prätherapeutische 124I-PET∕CT-Speicheldrüsendosimetrie bei der Radiojodtherapie differenzierter Schilddrüsenkarzinome und Untersuchung von wichtigen Einflussfaktoren auf die 124I-Quantifizierung,” Ph.D. thesis, University of Duisburg-Essen, 2009. [Google Scholar]
- Prideaux A. R., Song H., Hobbs R. F., He B., Frey E. C., Ladenson P. W., Wahl R. L., and Sgouros G., “Three-dimensional radiobiologic dosimetry: application of radiobiologic modeling to patient-specific 3-dimensional imaging-based internal dosimetry,” J. Nucl. Med. 10.2967/jnumed.106.038000 48, 1008–1016 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar W. T., “Application of the linear-quadratic model with incomplete repair to radionuclide directed therapy,” Br. J. Radiol. 64, 242–251 (1991). [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Hlatky L. R., Hahnfeldt P. J., Huang Y., and Sachs R. K., “The linear-quadratic model and most other common radiobiological models result in similar predictions of time-dose relationships,” Radiat. Res. 10.2307/3579648 150, 83–91 (1998). [DOI] [PubMed] [Google Scholar]
- Dale R. G., “The application of the linear-quadratic dose-effect equation to fractionated and protracted radiotherapy,” Br. J. Radiol. 10.1016/0167-8140(92)90244-O 58, 515–528 (1985). [DOI] [PubMed] [Google Scholar]