Abstract
One third of patients with type 1 diabetes and microalbuminuria experience an early, progressive decline in renal function that leads to advanced stages of chronic kidney disease and ESRD. We hypothesized that the urinary proteome may distinguish between stable renal function and early renal function decline among patients with type 1 diabetes and microalbuminuria. We followed patients with normal renal function and microalbuminuria for 10 to 12 yr and classified them into case patients (n = 21) with progressive early renal function decline and control subjects (n = 40) with stable renal function. Using liquid chromatography matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, we identified three peptides that decreased in the urine of patients with early renal function decline [fragments of α1(IV) and α1(V) collagens and tenascin-X] and three peptides that increased (fragments of inositol pentakisphosphate 2-kinase, zona occludens 3, and FAT tumor suppressor 2). In renal biopsies from patients with early nephropathy from type 1 diabetes, we observed increased expression of inositol pentakisphosphate 2-kinase, which was present in granule-like cytoplasmic structures, and zona occludens 3. These results indicate that urinary peptide fragments reflect changes in expression of intact protein in the kidney, suggesting new potential mediators of diabetic nephropathy and candidate biomarkers for progressive renal function decline.
Microalbuminuria (MA) has been the primary diagnostic test to identify patients who have type 1 diabetes and are at risk for overt proteinuria and subsequent declining renal function leading to ESRD1,2; however, the predictive value of MA is now questioned. First, a large proportion of patients with MA can revert to normoalbuminuria3–5; second, only a minority of patients with MA progress to proteinuria3,6; and, third, in one third of patients with MA, progressive renal function decline starts already at the onset of MA, not proteinuria. The last process we refer to as early renal function decline (ERFD).7 Our understanding of the disease process underlying ERFD is limited. The existing hypotheses about the cause of diabetic nephropathy (DN) are in question given that progressive ERFD is initiated while there is only minimal elevation of urinary albumin excretion and renal function is still in the normal or even elevated range.
In this report, we investigated the hypothesis that qualitative differences in urine proteins might provide insight into the mechanisms underlying progressive ERFD and may serve as candidate biomarkers to diagnose ERFD. To address this hypothesis, we analyzed urine specimens obtained during the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes.3,7–9 In this study, the low molecular weight protein (<10 kD), or peptidomic, fraction in urine was analyzed. Our data demonstrate that the urinary expression of several protein fragments differs significantly between patients who had MA and diabetes with early ERFD compared with patients who had MA and diabetes and in whom renal function remained stable for 10 to 12 yr. Three peptides, fragments of extracellular matrix proteins [α-I (IV) and α-I (V) collagens and tenascin-X] had diminished urinary excretion in patients with early ERFD. Three peptides, fragments of inositol pentakisphosphate 2-kinase (IPP2K) and zona occludens 3 (ZO-3) and cadherin-like protein FAT tumor suppressor 2, had increased urinary excretion in patients with early ERFD. The excretion of cadherin-like protein FAT tumor suppressor 2 was completely correlated with ZO-3. IPP2K and ZO-3 were further studied in human renal biopsies to determine whether urine peptide fragments correlate with renal protein expression. Our study demonstrated that urine peptide analysis can define renal signaling pathways active in DN and that the urine peptidome contains putative biomarkers that correlate with the future development of diabetic renal disease.
Results
Characteristics of the Study Groups
The study groups were derived from a population composed of patients whose onset of MA was documented in the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes.9 Additional eligibility criteria included follow-up examinations spanning at least 10 to 12 yr after MA onset for estimating the rate of GFR decline (slopes of cC-GFR) and availability of a 6-ml aliquot of stored urine for peptide analysis. Of 86 patients who had new-onset MA and were followed, 61 met eligibility criteria. They were subdivided into 21 case patients with early renal function decline (decliners), defined as a decline of cC-GFR slope of ≥3.3%/yr (range −18.0 to −3.3%/yr), and 40 control subjects with stable renal function (range −3.2 to 1.9%/yr).
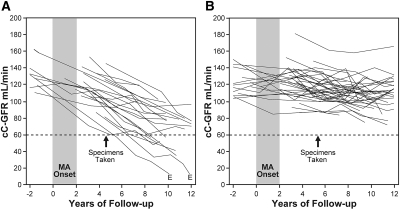
Longitudinal changes in renal function in the study groups are shown in Figure 1A for case patient (decliners) and Figure 1B for control subjects (nondecliners). Both groups had new-onset MA diagnosed during the 2-yr interval of observation. The beginning of this interval has been considered as the start of the 10- to 12-yr follow-up. The time point when the urine specimens for the proteomic analysis were collected is indicated by arrows in Figure 1.
Figure 1.
Patterns of changes in estimates of cC-GFR in patients with type 1 diabetes and new-onset MA during 10 to 12 yr of follow-up. cC-GFR was estimated using serum concentration of cystatin C and the formula of Macisaac et al.,26 and changes in renal function were estimated by slopes. (A and B) Patients were divided into two study groups: those with early renal function decline (decliners) when cC-GFR loss was ≥3.3%yr (A) and stable renal function (nondecliners) when cC-GFR loss was <3.3%yr (B). E, ESRD. Arrows indicate examinations from which urine samples were used for peptide component analysis.
Characteristics of the case and control groups are summarized in Table 1. All patients were white, and the groups had a similar number of men and women. At the onset of MA, both groups had similar duration of diabetes, similar age, and similarly elevated urinary albumin excretion. Case patients had higher average glycated hemoglobin (HbA1c) than did control subjects. At the time of urine sampling for peptide analysis, the groups had similar MA duration, BP, urinary albumin excretion, and medications. HbA1c continued to be significantly higher in case patients than in control subjects. The cC-GFR was nonsignificantly lower in case patients than in control subjects but still remained in the normal range in both groups.
Table 1.
Clinical characteristics of case patients with ERFD and control subjects without ERFD at three points in time (onset of MA, collection of urine sample, and end of follow-up)a
| Characteristic | Patients with New Onset of MA Grouped by Changes in Renal Function during 10 to 12 yr of Follow-up |
||
|---|---|---|---|
| Case Patients with ERFD (Decliners; n = 21) | Control Subjects without ERFD (Nondecliners; n = 40) | P | |
| At onset of MA | |||
| duration of diabetes (yr; mean ± SD) | 20.0 ± 8.5 | 18.0 ± 8.2 | NS |
| age (yr; mean ± SD) | 35.0 ± 8.7 | 31.0 ± 9.5 | NS |
| HbA1c (%; mean ± SD)b | 9.5 ± 1.4 | 8.7 ± 1.3 | 0.02 |
| urinary albumin excretion (μg/min; median [25th, 75th percentiles])b | 45 (38, 55) | 52 (40, 76) | NS |
| At collection of urine sample for peptide analysis | |||
| duration of MA (yr; mean ± SD)c | 4.6 ± 1.7 | 5.4 ± 1.6 | NS |
| HbA1c (%; mean ± SD) | 9.6 ± 1.3 | 8.3 ± 1.5 | 0.004 |
| SBP (mean ± SD) | 127 ± 21 | 121 ± 14 | NS |
| urinary albumin excretion (μg/min; median [25th, 75th percentiles])d | 40 (26, 150) | 33 (21, 68) | NS |
| use of ACEIs (%) | 74 | 51 | NS |
| cC-GFR (ml/min; mean ± SD) | 103 ± 25 | 117 ± 21 | NS |
| At end of follow-up | |||
| duration of follow-up (yr; mean ± SD)e | 11.1 ± 2.3 | 12.0 ± 2.1 | NS |
| cC-GFR slope (%/yr; median [range]) | −4.9 (−18.0 to −3.3) | −1.3 (−3.2 to 1.0) | By design |
| CKD stage ≥3 (%) | 40g | 0 | By design |
| urinary albumin excretion (μg/min; median [25th, 75th percentiles])f | 212 (35, 380) | 29 (18, 71) | 0.001 |
aChanges in cC-GFR during the follow-up in the study groups are shown in Figure 1. ACEI, angiotensin-converting enzyme inhibitor; SBP, systolic BP.
bAll measurements of urinary albumin excretion were used for the 2-yr interval when MA was diagnosed.
cTime interval between the beginning of the 2-yr interval when MA was diagnosed and the collection of urine samples.
dSingle measurement of urinary albumin excretion was used
eTime interval between the beginning of the 2-yr interval when MA was diagnosed and the last determination of cC-GFR.
fAll measurements of urinary albumin excretion were used for the last 2 yr of follow-up.
gDuring follow-up, eight patients developed stage 3 CKD or higher. ESRD developed in two of these eight (see also Figure 1).
At the end of follow-up (Table 1), the mean duration of observation was approximately 11 yr. By design, the rate of renal function loss (cC-GFR slope) was much greater in cases of ERFD (median slope −4.9%/yr) than in control subjects (median slope −1.4%/yr). As a result of this difference, at the end of the 10 to 12 yr of follow-up, 40% of case patients had reached advanced chronic kidney disease (CKD; i.e., stage 3 CKD or higher, including two case patients with ESRD). Assuming that same renal function decline continues, one may expect that the rest of the decliners would reach stage 3 CKD during the subsequent 10 yr of follow-up (Figure 1). In contrast, all control subjects had normal renal function at the end of 10 to 12 yr of follow-up and most likely will remain in the same stage if followed for another 10 yr (Figure 1). Regarding abnormalities in urinary albumin excretion at the end of the follow-up, half of case patients had proteinuria and the rest had MA or normoalbuminuria. In control subjects, one third had either MA or proteinuria and the rest had normoalbuminuria.
Urine Peptide Analysis
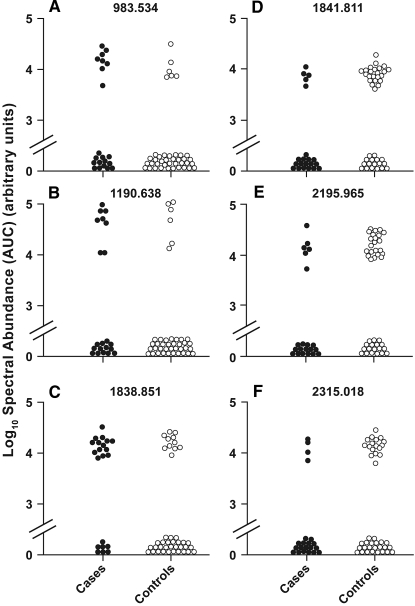
As pointed out in previous studies, analysis of mass spectral data derived from urine may be challenging.10 To focus effort on the most promising peptides, we imposed several stringent criteria for selecting peptides for further analysis. These criteria were chosen to decrease the number of false-positive associations between peptide expression and ERFD; however, they may also exclude some true-positive associations. First, we eliminated 3364 peptides that were detected in <20% of the specimens. Second, using a modification of the approach of Rossing et al.,10 we required that there be at least a 50% difference in the frequency of a peptide between case patients and control subjects and that this difference be statistically significant. This reduced the number of peptides from 825 to six. For these peptides, the differences in the distributions of their urinary abundances, which were derived from peptide peak's characteristics using the integrated signal area for the peptide isotopic series, was determined and their distributions are presented as one-dimensional scatter plots in Figure 2.
Figure 2.
Distribution of peptide abundance in case and control urine. Aligned MS data sets were constructed from peptide mass and retention time, and peptide spectral abundance was calculated from the MS ion cluster area. Peptide abundance in all samples is presented. The difference in frequency of urine peptides between cases and controls was determined using Fisher exact test. All peptides shown were significantly different between case and control groups.
Three peptides were present more frequently in urine of case patients in comparison with urine of control subjects, and three were present less frequently in urine of case patients in comparison with urine of control subjects. Interesting, the abundance and detection frequency of two peptides 983.534 and 1190.638 m/z were completely correlated whereas the others were less correlated or not correlated at all among themselves.
The association of these peptides with ERFD was studied further using logistic regression analysis controlling for the effects of other covariates such as HbA1c and albumin excretion rates. Table 2 shows the frequency of the peptides in urine specimens of case patients and control subjects with P values obtained from Fisher exact test together with the results of logistic analysis. Urinary presence of peptides 983.534, 1190.638, and 1838.851 m/z was strongly and independently associated with presence of ERFD. Adjusted odds ratios (ORs) varied from 4.4 to 4.9 (95% confidence interval [CI] 1.2 to 20.0). Conversely, urinary presence of peptides (i.e., 1841.811, 2195.965, and 2315.018 m/z) was protective against ERFD. Adjusted ORs varied from 0.2 to 0.4. 95% CI for the 1841.811 m/z was <1.0 but for the two others slightly above 1, indicating that after adjustment for other covariates, the negative association of these peptides with ERFD had only borderline significance.
Table 2.
Comparison of frequency of specific peptides in urine specimens of case patients and control subjects and ORs for the development of ERFD according to urinary presence of specific peptidesa
| Peptide Mass (m/z) | Frequency of Peptides (% with Peptide) |
Adjusted OR (95% CI)c | ||
|---|---|---|---|---|
| Case Patients (Decliners; n = 21) | Control Subjects (Nondecliners; n = 40) | Pb | ||
| 983.534 | 38 | 15 | 0.03 | 4.4 (1.1 to 20.0) |
| 1190.638 | 38 | 15 | 0.03 | 4.4 (1.1 to 19.0) |
| 1838.851 | 67 | 28 | 0.005 | 4.9 (1.4 to 17.0) |
| 1841.811 | 24 | 58 | 0.01 | 0.2 (0.1 to 0.8) |
| 2195.965 | 29 | 59 | 0.03 | 0.4 (0.1 to 1.1) |
| 2315.018 | 19 | 43 | 0.04 | 0.3 (0.1 to 1.1) |
aBinary (0,1) peptide abundance data were used to calculate the frequency (in %) of specific peptides in urine specimens. Distribution of peptide abundances in case patients and control subjects are shown in Figure 2.
bFisher exact test.
cLogistic regression was used to adjust for HbA1c at the time of urine specimen collection. Other covariates albumin excretion rate and angiotensin-converting enzyme treatment did not have an impact on the association of the examined peptides with presence or absence of ERFD.
Urine Peptides that Identify Patients with ERFD Are not Present in Plasma
The discriminating peptides present in urine had a molecular weight (m/z) ranging approximately from 842 to 1838 Da and therefore might be present in the urine after being freely filtered from the plasma by the glomerulus. To investigate the source of the selected peptides, we examined the plasma obtained with the urine specimens we had analyzed. Liquid chromatography matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (LC-MALDI-TOF MS) analysis of plasma peptides was carried out in 18 case and 18 control patients. The isolated plasma peptides were analyzed using identical LC-MALDI-TOF MS methods described for the urine analyses. One plasma peptide (1190.638 m/z) seen in the urine was detected in plasma; however, there was no difference in this peptide's plasma abundance between case and control patients. Although three of the four urine peptides could not be detected in plasma, it does not necessarily mean that they are not there. Differences in ion suppression between plasma and urine specimens as well as the relative low peptide abundance in urine compared with plasma could also account for the difference.
Identification of ERFD Discriminating Urine Peptide Amino Acid Sequence by MS
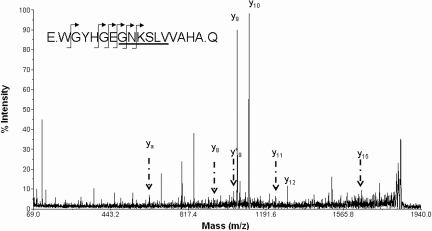
To understand better ERFD mechanisms, we sought to identify the protein source for the discriminating urinary peptides. We performed tandem MS analysis identify the amino acid sequence of the discriminating urine peptides. Because the Mascot scoring algorithms we used are biased against scoring the single noncanonical tryptic peptides we were studying, all Mascot results were manually reviewed as in the Concise Methods section. As shown in Table 3, the parent peptide mass and the partial amino acid sequence information gained from tandem MS experiments allowed us to identify six peptides with increased expression in ERFD specimens. These peptides were fragments of the cadherin-like protein FAT tumor suppressor 2, ZO-3, and IPP2K. Three peptides decreased in ERFD specimens were fragments of extracellular matrix proteins tenascin-X, α-I (IV) collagen, and α-I (V) collagen. The candidate peptide sequences are offset from the preceding and subsequent amino acids by periods as specified by the Paris Guidelines for reporting protein identification data.11 Analysis of the tandem MS data for the 1838.851 m/z peptide was consistent with a glycyl-glycyl posttranslational modification to the ε amino group of the internal lysine, which would be presumed to result from ubiquitination of the parent protein IPP2K. Figure 3 shows a representative LC-MS/MS spectra used to identify the 1838.851 m/z peptide as IPP2K. As noted already, the urinary presence of peptide 983.534 (ZO-3) and 1838.851 (IPP2K) was strongly and independently associated with the presence of ERFD. Adjusted ORs varied from 4.4 to 4.9 (95% CI 1.2 to 20.0).
Table 3.
Assignment of peptide sequence to differentially expressed urinary peptidesa
| Mass (m/z) | Parent Protein | Abundance Change in Cases | Peptide |
|---|---|---|---|
| 983.534 | ZO-3 | Increased | G.VSSQNLSLN.D |
| 1190.638 | FAT tumor suppressor 2 | Increased | S.PEFQQHLYE.A |
| 1838.851 | IPP2K | Increased | E.WGYHGEGNKSLVVAHA.Q + GlyGly (K) |
| 1841.811 | Tenascin-X | Decreased | Y.RNGDGQPK(OH)AVRGPGHEE.G |
| 2195.965 | α-I (IV) collagen | Decreased | S.PGP(OH)WGAP(OH)GLPGEK(OH)GDHGFPGSS.G |
| 2315.018 | α-I (V) collagen | Decreased | G.PKGRGGPNGDPGPLGPP(OH)GEKGKLG.V |
aThe candidate peptide sequences are offset within the parent protein sequence by bracketing periods with preceding and subsequent amino acids as per the Paris Guidelines for reporting protein identification data.11 The glycyl-glycyl posttranslational modification to ϵ amino group of the internal lysine.
Figure 3.
Analysis of peptide fragmentation spectra and assignment to candidate proteins. The fragmentation spectrum for 1838.851 m/z is assigned to a 16–amino acid fragment of IPP2K after analysis by Matrix Science Mascot with subsequent manual review. The candidate peptide sequence is shown (inset) with arrows marking observed y and y* ions. The underlined sequence is inferred by prominent ya internal fragment. No b ions were observed. Mass data for observed y, y*, and ya (internal) ions are inclusive of a glycyl-glycyl modification to the ε amino group of the internal lysine amino acid.
Changes in IPP2K and T Cell Intracellular Antigen 1 Expression in the Intact Kidney
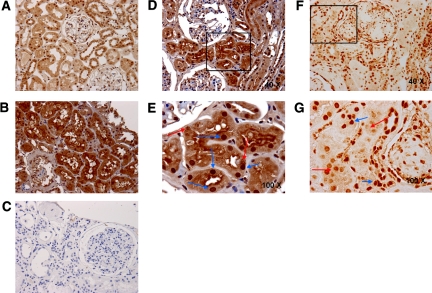
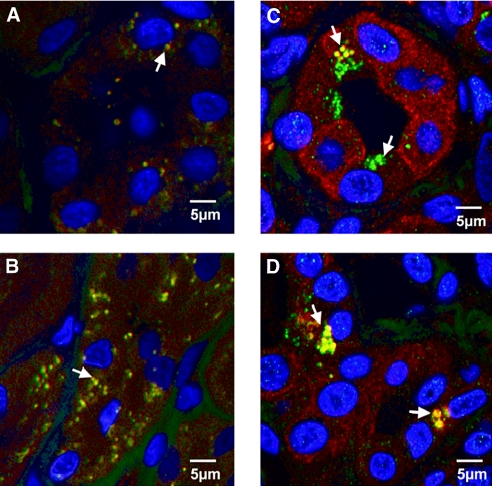
We then examined whether the candidate urinary peptides might provide insight into disease-induced changes in renal protein expression. We analyzed IPP2K expression in kidneys from normal individuals and patients with diabetes by immunohistochemistry. In keeping with our focus on ERFD, the biopsies were from patients with diabetes, minimal albuminuria, and serum creatinine levels of 1.2 to 1.9 mg/dl. Shown in Figure 4, A through C, shows representative images from six pairs of experiments that examined IPP2K expression in kidneys from normal patients (Figure 4A), a patient with early DN (Figure 4B), and negative control for nonspecific binding (Figure 4C). Patients with DN had increased IPP2K expression in renal tubules and glomeruli. IPP2K was shown to be a constituent of mRNA-containing granules responsible for protein translation arrest in stressed cells.12 These cytoplasmic inclusions, referred to as stress granules, are observed in cells subjected to environmental stress, including heat, irradiation, oxidative conditions, and hyperosmolarity.13 Because we observed increased intact IPP2K in the kidney and IPP2K urinary fragments, we examined whether stress granules are present in DN. We then determined the expression of another stress granule protein, T cell intracellular antigen 1 (TIA1). Shown in Figure 4, D and E, are representative sections from four pairs of normal and diabetic kidney biopsies stained for TIA1 from normal kidney (Figure 4D) or early DN (Figure 4E). We observed increased TIA1 staining in DN. TIA1 was primarily localized to the cytoplasm and in a granular pattern, indicative of stress granules. To confirm the presence of stress granules in DN, we determined whether TIA1 and IPP2K co-localize renal tissue from patients with diabetes. Renal biopsies from normal donors and patients with diabetes were stained with TIA1 and IPP2K antibodies. Shown in Figure 5, A and B, are representative sections from normal kidney biopsies. Distinct cytoplasmic expression and co-localization of TIA1 and IPP2K is present in normal kidneys as indicated by punctate red and green staining and yellow when the proteins are co-localized. These structures are indicative of cytoplasmic distribution but are far smaller than stress granules. In Figure 5, C and D, TIA1 co-localization with IPP2K is evident as indicated by the yellow pseudocolor fluorescence and its presence in structures the size of stress granules as reported in the literature.14
Figure 4.
IPP2K renal expression is increased in the renal parenchyma of patients with type 1 diabetes and minimal nephropathy. (A through C) Immunohistochemical localization of IPP2K in a control kidney biopsy specimen (A); type 1 diabetes biopsy specimen (B); and secondary antibody negative control (C) demonstrates increased renal tubular cytoplasmic, nuclear, and perinuclear IPP2K staining within the diabetic phenotype. (D and E) Further experiments for IPP2K (D) and TIA1 (E) were conducted in type 1 diabetes biopsy specimens. Blue arrows identify regions of nuclear staining; red arrows identify regions of punctate cytoplasmic staining. Unless otherwise noted, the presented image is at a . Images are representative of experiments performed in four sets of normal and diabetic kidney. Magnifications: ×40 in A, B, C, D, and F; ×100 in E and G.
Figure 5.
Co-localization of stress granule proteins IPPK and TIA1 in proximal tubules of human renal biopsies. Co-localization of IPP2K and TIA1 in proximal tubules of human renal biopsies. (A through D) Representative fluorescence images of control (A and B) and early type 1 diabetic (C and D) renal biopsies stained for IPP2K (red fluorescence) and TIA1 (green fluorescence). Nuclei are stained with DAPI. Arrows indicate structures staining for co-localized TIA1 and IPP2K and appear as a yellow pseudocolor. Diabetic sections stain more intensely for cytoplasmic IPP2K. IPPK in diabetic sections is co-localized with TIA1 in larger granular structures. Images are representative of three sets of normal and diabetic renal biopsies.
Changes in ZO-3 Protein Expression in the Kidney
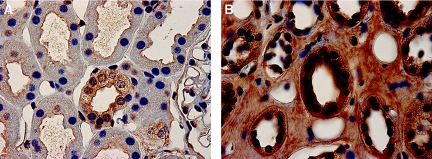
To examine further whether urine peptide changes may reflect alterations in intact kidney proteins, we analyzed ZO-3 expression in kidneys from normal individuals (n = 4) and patients with diabetes (n = 4) by immunohistochemistry. We used the same biopsies studied already, from patients with diabetes, minimal albuminuria, and serum creatinine levels of 1.2 to 1.9 mg/dl. As shown in Figure 6, ZO-3 expression was increased in biopsies from patients with DN (Figure 6B) when compared with control subjects (Figure 6A). We also observed that ZO-3 staining was less linear, not confined to the cell periphery, and increased in the cytoplasm when compared with normal biopsy sections. This distribution of ZO-3 is similar to that of ZO-1 in cultured podocytes incubated in high-glucose medium.15
Figure 6.
Cytoplasmic ZO-3 expression and cell junction staining are increased in renal biopsies of patients with diabetes and minimal nephropathy. (A and B) Control (A) and diabetic (B) renal biopsy sections stained for ZO-3 demonstrated enhanced staining for ZO-3 in the cytoplasm of diabetic biopsy sections and prominent, dense staining in the apical membrane of renal proximal tubular cells and in sites of cell–cell interaction within renal tubules and were taken to be diagnostic of tubular adherens junctions. Images are representative of experiments performed in four sets of normal and diabetic kidney.
Discussion
Our goal in this study was to identify urinary peptides that are associated specifically with the risk for progressive ERFD. ERFD is indicative of the initiation of renal function loss in patients with type 1 diabetes and MA while GFR is still in the normal range.7
The urine specimens we studied were obtained soon after the development of MA and could be classified into cases and controls according to their future changes in renal function; therefore, the specimens could be used to measure peptides that may discriminate patients who will retain stable renal function from those with ERFD and therefore are at high risk for subsequent development of ESRD. We reasoned whether such discriminating peptides that might provide new insights into the pathophysiology of the initiation of renal function loss and possibly serve as candidate biomarkers in future studies could be identified. Using stringent selection criteria, we identified six peptides whose urinary excretion significantly varied between case patients as compared with control subjects. Further MS analysis provided partial amino acid sequence information for the six peptides. From the bioinformatic analysis of the MS data, the three peptides more prevalent in the urine of case patients were concluded to be derived from the ZO-3, cadherin-like protein FAT tumor suppressor 2, and IPP2K. The three peptides more prevalent in the urine of control subjects were determined to be derived from the extracellular matrix proteins tenascin-X, α-I (IV) collagen, and α-I (V) collagen.
We wished to examine whether urine peptide fragment differences reflected changes of intact proteins in the kidney. On the basis of the differential expression of an IPP2K urinary fragment in case and control specimens, we examined whether this protein was differentially regulated in normal kidneys and those from patients with type 1 diabetes. We observed increased expression of IPP2K in renal tubules of patients with type 1 diabetes compared with normal kidney. Increased IPP2K expression was observed both in nuclei and in the cytoplasm. The role in renal cell function of IPP2K is incompletely understood. One major role for IPP2K has been in the regulation of mature messenger RNA nuclear export and mRNA translation arrest during cellular stress.16 Most recently, cytoplasmic IPP2K has been associated with stress granule formation. Stress granules are phase-dense cytoplasmic particles that appear in eukaryotic cells exposed to environmental stress (e.g., oxidative conditions, hyperosmolarity, nutrient deprivation, ultraviolet irradiation). Stress granule assembly is a consequence of abortive translational initiation. If the stress is relieved, then the stress granules disassemble and translation resumes. The persistence of stress granules is considered a reflection of ongoing insults that, if left unchecked, may lead to apoptotic cell death. We observed structures consistent with stress granule formation in kidneys with early DN, as well as in other renal diseases (data not shown), suggesting that stress-induced changes in translation had occurred. Because stress granule formation may result from endoplasmic reticulum stress, our findings are consistent with those of Lindenmeyer et al.,17 who reported that endoplasmic reticulum stress occurs in kidneys with DN.
We also observed that increased amounts of urine peptides derived from the ZO-3 protein reflected changes in the diabetic kidney. Diabetic kidneys had increased tubule cell ZO-3 expression and apparent translocation of the protein from the cell membrane to cytoplasm. ZO-3 is a member of the ZO protein family found in epithelial cell apical junction complexes.18 ZO proteins play an important role in maintaining barrier integrity formed by epithelial cells, and loss of ZO-3 in particular results in increased permeability of cell layers.19 Recently, Sharma et al.15 showed glucose causes translocation of ZO-1 from the cell membrane to the cytoplasm in cultured podocytes. We observed similarly increased cytoplasmic and decreased membrane ZO-3 staining in renal tubules in human diabetic kidneys. These data and the literature suggest that increased ZO-3 fragments in the urine may be a result of the diabetes-induced change in ZO-3 cellular distribution. Considered together, our ZO-3 and IPP2K data indicate that urine peptidomic analysis can reflect protein expression changes in the renal parenchyma.
Our finding of decreased amounts of urinary collagen fragments in patients with ERFD is consistent with the work of Rossing et al.,10 who observed decreased abundance of several collagen types in the urine of patients with DN. Several explanations are possible for decreased urinary collagen excretion in patients at risk for ERFD. First, the accumulation of collagen in advanced DN is well described and may result from both increased synthesis and decreased proteolysis.20,21 Similarly, we showed in an animal model of DN that the matrix protein elastin accumulates as a result of decreased elastase activity leading to diminished elastase turnover.22 Another factor influencing collagen proteolysis of collagen in diabetes is the action of advanced glycation end products. Advanced glycation end products promote collagen synthesis and inhibit collagen proteolysis, thereby linking glucose control to diminished collagen degradation.23,24
One of the goals of these studies was to identify urine peptides that predicted progressive ERFD. The confirmation of these peptides as biomarkers of ERFD in patients with type 1 diabetes will require validation studies using large patient populations, and their predictive value may also be strengthened by inclusion in multiple marker panels.25 A second goal was to determine whether urinary peptides associated with ERFD reflected changes in the renal parenchyma and thereby provide insight into the mechanisms of diabetic kidney disease. In biopsies from patients with type 1 diabetes and early signs of DN, ZO-3 expression in renal tubular epithelial cells was significantly increased compared with that in control subjects and ZO-3 translocated to the cytoplasm. IPP2K expression was increased and co-localized in renal tubules with granule-like cytoplasmic structures. TIA1, another protein associated with stress granules, was also increased in diabetic kidney biopsies. These results indicate that urine peptide analysis can define renal signaling pathways active in DN and that the urine peptidome contains putative biomarkers that correlate with the future development of diabetic renal disease.
In summary, we have identified peptides in the low molecular weight fraction of urine that correlate with early renal function decline in type 1 diabetes. These peptides reflect changes in both tubular and glomerular protein expression that are associated with the formation of stress granules and may define a new cellular mechanism by which DN is initiated or progresses. Thus, urine peptide expression may provide insight into renal pathophysiologic mechanisms. The usefulness of these discriminating peptides as biomarkers of diabetes-associated renal function decline should be determined in additional rigorous studies in a larger patient population.
Concise Methods
Patient Selection and Case Definition for ERFD
The protocol and consent procedures were approved by the Committee on Human Studies of the Joslin Diabetes Center, and use of the de-identified specimens was approved by the University of Louisville Human Subjects Review Committee. Among the 943 patients with normoalbuminuria enrolled in the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes, MA developed in 109 during the first 4 yr of observation, it is this cohort of 109 patients with a documented onset of MA that is the population eligible for this study.9
Eighty-six patients were followed until 2007, and 61 met the following additional criteria for inclusion in the analysis: (1) Follow-up examinations spanning at least 10 yr after the onset of MA for the estimation of the slope of GFR over time, and (2) at least one 6-ml aliquot of stored urine from an examination within the early years of follow-up was available for peptidomic analysis. Specimens were collected without protease inhibitors and centrifuged immediately, and aliquots of the supernatant were removed for clinical and proteomic analysis. Samples used for proteomic analysis had been bioreposited at −80°C and had not been thawed before proteomic analysis. Among available urine specimens, the earliest after MA onset was selected.
The 61 patients were divided into cases of ERFD and control subjects according to the regression slope fitted to their follow-up estimates of cC-GFR. The protocol used for assessment of early progressive renal function decline has been described previously.7 In brief, GFR in ml/min was approximated using concentration of serum cystatin C (in mg/L) and application of formula recently developed by Macissec et al.26 for patients with diabetes. This estimate is referred to as cC-GFR. A regression slope fitted to serial measurements of cC-GFR accurately tracks the trend in renal function during that time. Patients had, on average, 4.8 cC-GFR estimates. As previously published, a negative slope or trend in renal function change >3.2%/yr qualified as an abnormal rate of decline (designated ERFD).7
Sample Handling and Peptide Isolation
The order of case or control sample handling during peptide isolation was randomized to minimize technical or methodologic bias during data collection. Urinary peptides were isolated from the urine using Millipore Amicon Ultra-4 spin concentrators (Millipore, Billerica, MA) having a 10,000 molecular weight cutoff membrane made of regenerated cellulose. The Ultra-4 concentrators were spun at 3220 × g for 15 min at 4°C.
The peptides were isolated and desalted from the urine 10-kD ultrafiltrate by solid-phase extraction methods adapted from a published method,27 and all co-purifying material was assumed from this point on to be peptide unless proved otherwise. Briefly, 1 ml of Oasis HLB (Waters Corp., Milford, MA) solid-phase extraction cartridges was equilibrated with sequential 1-ml washes of 100% methanol and distilled water. Urine ultrafiltrate containing urinary peptides (<10 kD) was acidified and diluted 1:1 with 0.5% triflouroacetic acid (TFA) and applied to the 1-ml HLB cartridges. The ultrafiltrate was pulled through the HLB cartridges with constant vacuum using a vacuum manifold. The loaded HLB cartridges were desalted with three sequential 1-ml washes of 5% methanol. The bound peptides were eluted using a single 1-ml wash of 80% methanol/0.1% TFA, and the eluted samples were immediately chilled to −80°C overnight. The next day, the eluted samples containing the urinary peptides were lyophilized to dryness. The dried peptides were resuspended using 0.01% TFA, and the peptide concentration was determined using a μBCA kit (Pierce, Rockford, IL). Before injection into the LC system, the samples were filtered through regenerated cellulose syringe (0.2 μm; National Scientific Co., Rockwood, TN) to remove particulate matter. After filtration, the samples (50 to 200 μl) were transferred to autosampler vials in a final concentration of 0.1 μg/μl. Autosampler vials were arranged in a randomized manner so as not to bias the order of sample selection for chromatographic selects (i.e., samples were randomly loaded to avoid the introduction of propagating bias or error).
Peptides (5 μg) were separated using a reversed-phase capillary scale column (Dionex 75 μm I.D. Nano Series Column, C18 PepMap 100, 3 μm, 100 Å) with 45 min of Solvent A (0.05% TFA) to Solvent B (80% acetonitrile/0.05% TFA) gradient at a 0.300-μl/min flow rate, and 45 1-min fractions were collected with a robotic fraction collector (Accuspot; Shimadzu Corp., Kyoto, Japan). MALDI matrix (5 mg/ml α-Cyano-4-hydroxycinnamic acid) and a 5.33-fmol/μl angiotensin I standard dissolved in 50% acetonitrile/0.1% TFA was overlaid onto dried LC fractions collected on archivable MALDI sample plates (ABI Opti-TOF plates; ABI, Foster City, CA) after run using the Accuspot to include an internal mass calibration standard at 4 fmol of angiotensin I per spot.
Positive ion MALDI-TOF mass spectra were acquired using an Applied Biosystems (Foster City, CA) AB4700 Proteomics Analyzer operating in reflectron mode and with ion source pressure approximately 0.5 μTorr. After a 400-ns time-delayed ion extraction period, the ions were accelerated to 20 kV for TOF mass spectrometric analysis. A total of 1000 laser shots (355 nm of Nd:YAG solid-state laser operating at 200 Hz) were acquired, and the signal was averaged. MALDI-TOF spectra were exported as.t2d files as well as peak list files for chromatographic data alignment and for analysis by MarkerView (MDS Sciex, Concord, ON, Canada).
Analysis of MALDI-TOF MS Data Sets
Aligned MS data sets were constructed (peptide mass [m/z; +1] and retention time pairs) with peptide abundance based on the MS ion cluster area. The prevalence of peptide abundance was determined. Cases of early renal function decline were defined by loss in GFR that exceeded −3.3%/yr, a threshold that corresponds to the 2.5th percentile of the distribution of GFR slopes in an independent nondiabetic normotensive population. Observed peptide masses were assigned an intensity of 1, and unobserved masses were assigned an intensity of 0. The fractional abundance (f) of each case and control peptide was used to compute a fractional abundance ratio (fcase/fcontrol). Fractional abundance ratios >1.5 or <0.5 in case-to-control ratios were considered significantly differentially expressed within the two populations. The fractional abundance ratios for these peptides were reconsidered using only the patient samples found in the first and third tertiles. Peptides having a ratio that became more extreme (>1.5 or <0.5) were tabulated for further MS/MS analyses. For determination of the correlation of peptide expression with ERFD, the slope of the decline in renal function (slope glm) was regressed on the s/n three peak intensities with natural log scaling, and significance was assessed using simple linear regression.
MALDI-TOF/TOF MS Data Analysis and Assignment of Peptide Amino Acid Sequence
Both postsource decay and collision-induced dissociation fragmentation methods were used. These data sets for each tabulated peptide were acquired using the AB4700 MALDI TOF/TOF using 1KeV collision energy, atmospheric gases (medium pressure), and collection averaged data from 1500 laser shots. Concatenated lists of averaged fragmentation spectra (postsource decay and collision-induced dissociation) were submitted for computer-assisted data analysis. Analyses were conducted in an iterative manner with the concatenated lists starting with search parameters of a mass accuracy of 0.15 Da for peptides and 0.3 Da for fragment masses. All MS/MS data were analyzed first by scoring algorithms (Global Protein Server software [Applied Biosystems] and Matrix Science Mascot 2.1). Because the Mascot algorithm is biased against assigning significant protein MOWSE scores on the basis of single peptides, all results were subjected to a manual data review step. Peptide fragmentation peak lists were searched against the unconstrained (nonenzyme) NCBInr 20060712 database (378,3042 sequences, 1,304,471,729 residues) with Homo sapiens taxonomy (111,934 sequences) initially with no post-translational modifications. The urine is known to contain fragments of collagen. Because collagen is known to be frequently hydroxylated at proline and less frequently at asparagine and lysine, the TOF/TOF data were analyzed in an iterative manner proceeding sequentially with addition of possible post-translational modifications sodiation and hydroxylation. Mascot search of the MALDI TOF-TOF data proceeded with assuming possible hydroxylation of lysine (K), asparagine (N), proline (P), and possible sodiation of asp or glu side chains or carboxy termini. Returned Mascot probabilistic scores for peptides of ≥67 indicate identity or extensive homology (P < 0.05). Because Mascot scoring is biased against single-peptide identification, manual review of returned Mascot fragmentation data analyses was conducted and used to make sequence tagging assignments. Data were then re-searched using a NCBInr Homo sapiens decoy database constructed using the online tool from Matrix Science MASCOT for randomization of database protein sequences. The top peptide assignments by the Homo sapiens and the Homo sapiens decoy databases were rank ordered, and masses having a higher Homo sapiens decoy rank order were discarded and not used for further analyses. Fragmentation spectra assigned a MASCOT score of ≥20 for m/z <3000 were manually reviewed using acceptance criterion including y, b, a, ya, and yb ion coverage, percentage of fragmentation spectra assigned, root mean square (rms) mass accuracy errors for precursor (rms <50 ppm) and precursor fragmentation products (rms <500 ppm).
Immunohistochemistry
A modified version of a previously described IHC protocol was used.28 Formalin-fixed, paraffin-embedded kidney sections from human biopsy specimens were obtained, and sections were dewaxed and treated with antigen-unmasking solution (H-3300; Vector Laboratories, Burlingame CA), and endogenous peroxidase activity was blocked. Kidney sections were selected from three different normal, disease control, and disease biopsy specimens. Kidney sections were blocked, probed with primary antibodies (1) 1:50 dilution mouse polyclonal antibody to IPP2K (cat. no. H00064768-A01; Novus Biologicals, Littleton, CO), (2) 1:50 dilution of goat polyclonal antibody to ZO-3 (cat. no. sc11478; Santa Cruz Biotechnology, Santa Cruz, CA), or (3) 1:50 dilution of rabbit polyclonal antibody to TIA1 (cat ab40693; Abcam, Cambridge, UK). The primary antibodies were probed with secondary antibody and treated with ABC reagent. For negative controls, serial sections were stained as before with the exception of no primary antibody. Processed sections were scored by three blinded individuals using a scoring rubric for staining to nuclei, cytoplasm, basement membrane, and interstitium. Negative control and deletion control experiments were conducted to optimize for antibody combination, antibody concentration, incubation times, and order of antibody visualization.
Fluorescence Immunohistochemistry
Sections were cleared of paraffin with xylene and rehydrated by incubation in graded ethanol, followed by water and Tris-buffered saline (TTBS). Antigen retrieval was performed with DIVA decloaking agent at 95 to 99°C for 20 min in a water bath, followed by an additional 20 min after removal of the coplin jar from the water bath. Tissue sections were washed with water and incubated with blocking solution (2% goat serum, 1% BSA, 0.05% Triton X-100, and 0.05% Tween-20) for 1 h at room temperature. The primary antibodies used were a mouse anti-human IPP2K (Abnova, Taipei, Taiwan) and rabbit anti-human TIA1 (Abcam). IPPK and TIA antibodies were diluted at a concentration of 1:50 in antibody dilution buffer (1% BSA, 0.05% Triton X-100, and 0.05% Tween-20), and sections were incubated for 1 h at room temperature. After three 5-min washes in TTBS, sections were incubated in secondary antibodies (goat anti-rabbit Alexa Fluor-488, and goat anti-mouse Alexa Fluor-545; Invitrogen, Carlsbad, CA) at a concentration of 2 μg/ml in antibody dilution buffer for 1 h at room temperature. Tissue sections were washed in TTBS three times, 5 min each, and incubated with 300 nM DAPI for 5 min. Negative control for staining was performed by incubation of sections only with secondary antibody. Slides were mounted with a coverslip using Vectashield hard mount mounting medium (Vector Laboratories). Images were acquired by laser scanning confocal microscopy using a Zeiss Axiovert 100 microscope and LSM 510 6.2 software. A multitrack scanning configuration for sequential activation of the Enterprise 364 nm, Argon 488 nm, and HeNe1 543 nm lasers was used.
Statistical Analysis
The difference in frequency of urine peptides between cases and controls was determined using Fisher exact test. Logistic regression analysis was used to examine statistical significance between specific urinary peptides and ERFD controlling for other covariates.
Disclosures
None.
Acknowledgments
This work was supported by National Institutes of Health grants R01 DK067638 (A.S.K. and J.B.K.), R01 DK041526 (A.S.K.), and DOE DE-FG02-05ER6406 (J.B.K. and M.L.M.) and the Department of Veterans Affairs (J.B.K.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Viberti GC, Hill RD, Jarrett RJ, Argyropoulos A, Mahmud U, Keen H:Microalbuminuria as a predictor of clinical nephropathy in insulin-dependent diabetes mellitus. Lancet 1: 1430–1432, 1982 [DOI] [PubMed] [Google Scholar]
- 2.Mogensen CE, Keane WF, Bennett PH, Jerums G, Parving HH, Passa P, Steffes MW, Striker GE, Viberti GC:Prevention of diabetic renal disease with special reference to microalbuminuria. Lancet 346: 1080–1084, 1995 [DOI] [PubMed] [Google Scholar]
- 3.Perkins BA, Ficociello LH, Silva KH, Finkelstein DM, Warram JH, Krolewski AS:Regression of microalbuminuria in type 1 diabetes. N Engl J Med 348: 2285–2293, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Giorgino F, Laviola L, Cavallo Perin P, Solnica B, Fuller J, Chaturvedi N:Factors associated with progression to macroalbuminuria in microalbuminuric type 1 diabetic patients: The EURODIAB Prospective Complications Study. Diabetologia 47: 1020–1028, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Araki S, Haneda M, Sugimoto T, Isono M, Isshiki K, Kashiwagi A, Koya D:Factors associated with frequent remission of microalbuminuria in patients with type 2 diabetes. Diabetes 54: 2983–2987, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Caramori ML, Fioretto P, Mauer M:The need for early predictors of diabetic nephropathy risk: Is albumin excretion rate sufficient? Diabetes 49: 1399–1408, 2000 [DOI] [PubMed] [Google Scholar]
- 7.Perkins BA, Ficociello LH, Ostrander BE, Silva KH, Weinberg J, Warram JH, Krolewski AS:Microalbuminuria and the risk for early progressive renal function decline in type 1 diabetes. J Am Soc Nephrol 18: 1353–1361, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Krolewski AS, Laffel LM, Krolewski M, Quinn M, Warram JH:Glycosylated hemoglobin and the risk of microalbuminuria in patients with insulin-dependent diabetes mellitus. N Engl J Med 332: 1251–1255, 1995 [DOI] [PubMed] [Google Scholar]
- 9.Scott LJ, Warram JH, Hanna LS, Laffel LM, Ryan L, Krolewski AS:A nonlinear effect of hyperglycemia and current cigarette smoking are major determinants of the onset of microalbuminuria in type 1 diabetes. Diabetes 50: 2842–2849, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Rossing K, Mischak H, Dakna M, Zurbig P, Novak J, Julian BA, Good DM, Coon JJ, Tarnow L, Rossing P:Urinary proteomics in diabetes and CKD. J Am Soc Nephrol 19: 1283–1290, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradshaw RA, Burlingame AL, Carr S, Aebersold R:Reporting protein identification data: The next generation of guidelines. Mol Cell Proteomics 5: 787–788, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Brehm MA, Schenk TM, Zhou X, Fanick W, Lin H, Windhorst S, Nalaskowski MM, Kobras M, Shears SB, Mayr GW:Intracellular localization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. Biochem J 408: 335–345, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson P, Kedersha N:RNA granules. J Cell Biol 172: 803–808, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson P, Kedersha N:Stress granules: The Tao of RNA triage. Trends Biochem Sci 33: 141–150, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Sharma K, Ramachandrarao S, Qiu G, Usui HK, Zhu Y, Dunn SR, Ouedraogo R, Hough K, McCue P, Chan L, Falkner B, Goldstein BJ:Adiponectin regulates albuminuria and podocyte function in mice. J Clin Invest 118: 1645–1656, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR:Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol 8: 711–716, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Lindenmeyer MT, Rastaldi MP, Ikehata M, Neusser MA, Kretzler M, Cohen CD, Schlondorff D:Proteinuria and hyperglycemia induce endoplasmic reticulum stress. J Am Soc Nephrol 19: 2225–2236, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Margolis B:Apical junctional complexes and cell polarity. Kidney Int 72: 1448–1458, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Kiener TK, Selptsova-Friedrich I, Hunziker W:Tjp3/zo-3 is critical for epidermal barrier function in zebrafish embryos. Dev Biol 316: 36–49, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Umezono T, Toyoda M, Kato M, Miyauchi M, Kimura M, Maruyama M, Honma M, Yagame M, Suzuki D:Glomerular expression of CTGF, TGF-beta 1 and type IV collagen in diabetic nephropathy. J Nephrol 19: 751–757, 2006 [PubMed] [Google Scholar]
- 21.Mohan PS, Carter WG, Spiro RG:Occurrence of type VI collagen in extracellular matrix of renal glomeruli and its increase in diabetes. Diabetes 39: 31–37, 1990 [DOI] [PubMed] [Google Scholar]
- 22.Thongboonkerd V, Barati MT, McLeish KR, Benarafa C, Remold-O'Donnell E, Zheng S, Rovin BH, Pierce WM, Epstein PN, Klein JB:Alterations in the renal elastin-elastase system in type 1 diabetic nephropathy identified by proteomic analysis. J Am Soc Nephrol 15: 650–662, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Lee CI, Guh JY, Chen HC, Hung WC, Yang YL, Chuang LY:Advanced glycation end-product-induced mitogenesis and collagen production are dependent on angiotensin II and connective tissue growth factor in NRK-49F cells. J Cell Biochem 95: 281–292, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Bakris GL, Bank AJ, Kass DA, Neutel JM, Preston RA, Oparil S:Advanced glycation end-product cross-link breakers: A novel approach to cardiovascular pathologies related to the aging process. Am J Hypertens 17: 23S–30S, 2004 [DOI] [PubMed] [Google Scholar]
- 25.Hewitt SM, Dear J, Star RA:Discovery of protein biomarkers for renal diseases. J Am Soc Nephrol 15: 1677–1689, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Macisaac RJ, Tsalamandris C, Thomas MC, Premaratne E, Panagiotopoulos S, Smith TJ, Poon A, Jenkins MA, Ratnaike SI, Power DA, Jerums G:Estimating glomerular filtration rate in diabetes: A comparison of cystatin-C- and creatinine-based methods. Diabetologia 49: 1686–1689, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Cutillas PR, Norden AG, Cramer R, Burlingame AL, Unwin RJ:Urinary proteomics of renal Fanconi syndrome. Contrib Nephrol 141: 155–169, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Barati MT, Merchant ML, Kain AB, Jevans AW, McLeish KR, Klein JB:Proteomic analysis defines altered cellular redox pathways and advanced glycation end-product metabolism in glomeruli of db/db diabetic mice. Am J Physiol Renal Physiol 293: F1157–F1165, 2007 [DOI] [PubMed] [Google Scholar]