Abstract
Epilepsy is a complex neurological disease. Currently ~20 genetic variants are known to cause Mendelian forms of human epilepsy, leaving a vast heritability undefined with future hopes resting on candidate gene resequencing and/or large scale genome-wide association studies. Rodent models for genetically complex epilepsy have been studied for many years, but only recently have strong candidate genes emerged, including Cacna1g in the GAERS rat model of absence epilepsy and Kcnj10 in the low seizure threshold of DBA/2 mice. In parallel, a growing number of mouse mutations studied on multiple strain backgrounds reveal how genetic modifiers have an enormous impact on seizure severity, incidence or form - perhaps mimicking the complexity seen in humans. The field of experimental genetics in rodents is now poised to study discrete epilepsy mutations on a diverse choice of strain backgrounds to develop better models and identify modifiers. But it must find the right balance between embracing the strain diversity available, with the ability to detect and characterize genetic effects. In practice, the use of alternate strain backgrounds when studying epilepsy mutations will enhance the modeling of epilepsy as a complex genetic disease.
Genetically complex epilepsy and animal models
Epilepsy is a complex disease that is defined primarily by recurrent seizures. It affects approximately 1% of the population, presenting a serious burden to society - $15.5B annually in the US (see www.cdc.gov/epilepsy). Epilepsy has both genetic and environmental causes. The triggering of a seizure might be a specific sensory stimulus (e.g. strobe flash, stress, sound), or might appear random. Antiepileptic drug therapies work for many patients, but can have adverse side effects and fail to achieve complete seizure control in about one-quarter of all patients 1. In addition, the mortality rate associated with epilepsy is at least 2–3 times higher than in the general population 2. But even isolated seizures can be fatal. One advocacy group estimates that over 50,000 people die annually in the US directly or indirectly as a result of seizures (http://www.cureepilepsy.org/about/epilepsy_facts.asp) – more than other causes that are more prominent in the media, such as breast, colon or prostate cancer.
Most idiopathic epilepsies – those without any known or obvious organic cause or lesion – are thought to have an appreciable genetic component. However, from casewise concordance in twins and comparative risk to first-degree relatives, it is clear that most heritable epilepsies are genetically complex 3. Thus, while over 20 pathological genetic variants have been identified in human families 4,these tend to represent rare, Mendelian forms of disease. As might be expected for a neuroexcitability disorder, at least two thirds of the genes involved encode ion channels directly. Nevertheless, for the vast majority of idiopathic epilepsy, the underlying genetic causes are not yet known. While genome-wide association studies (GWAS) for epilepsy have not yet been described, it difficult to know whether they will be any more successful than those tried for other complex functional diseases of the brain, such as schizophrenia 5.
Idiopathic epilepsy is an accessible and potentially very rewarding disease for geneticists to study. When compared with other brain disorders that lack an obvious anatomical focus, such as schizophrenia, autism or depression, the phenotypic endpoint of idiopathic epilepsy – a seizure - is a relatively straightforward and robust entity to measure, especially in the laboratory. It should also be very powerful to study as a “model” complex trait in experimental animals. Indeed it has been known for many years that laboratory mice and rats can model complex heritable seizure disorders – from the DBA inbred mouse strain discovered in 1947 to experience severe tonic-clonic seizures in response to audiogenic stimuli 6, to the genetic absence epilepsy rat strain (GAERS), with its absence seizures first described in 1982 7, 8, to the EL mouse strain which has limbic and tonic-clonic convulsions that are precipitated by routine animal handling (i.e. reflex epilepsy) 9, 10 in addition to several other strains that model either convulsive or absence epilepsy 11–16.
The genetic basis of disease in DBA, GAERS, EL and those other epileptic rodent strains is usually multifactorial, due to a combination of additive and epistatic genetic variables, along with environmental and even random, stochastic effects. From genetic mapping approaches, various candidate genes have been nominated to account for portions of the phenotype in these and other strains. The most promising candidate was put forward in at least one seizure threshold model, DBA/2 mice – Kcnj10, encoding an inwardly rectifying K+ channel 17, 18. Interestingly, a KCNJ10 variant was also associated with human idiopathic 19 and syndromic epilepsy 20, 21. In general, however, progress in gene discovery in these models has been slow – in part, because idiopathic epilepsy is a complex trait and difficult to study as such, and partly because there are not many laboratories working in this field.
Keeping your candidate genes close, and your mating partners closer: Genetic background matters
When studying complex traits, whether in humans or in animal models, the “signal-to-noise ratio” is the major challenge: a given genetic signal is diluted by many other effects (e.g. other variants and environmental factors) that interfere with the ability to follow it in a population. Although non-genetic factors can sometimes be controlled by experimental procedures or overcome by increasing sample size, it is genetic complexity itself that presents the greatest obstacle. In humans this is compounded by the fact that most variants are not homozygous but are segregating within families or populations. In laboratory rodents, this complexity can be fixed on inbred strain backgrounds that can be manipulated in a more controlled fashion. In principle, this level of control provides an opportunity to better follow the sought-after genetic signal down to the molecular level.
Recently, progress has been made in understanding the genetic basis of epilepsy in the GAERS rat – an absence epilepsy model that has been studied for many years but for which definitive mutations were not known until recently. By examining only a single candidate gene, Powell and colleagues found that GAERS has a functional mutation in the Cacna1h gene encoding the Cav3.2 low voltage activated Ca2+ channel (LVAC) 22. The study showed that the effect was due to a gain-of-function splice variant mutation, and that it was semidominant, explaining at least 30% of the phenotypic variance in the cross (N.b. it is possible that it could explain more, due to untested epistatic interaction between Cacna1h and other variants in the genome). In heterologous expression studies the study showed that the GAERS splice variant allele on Cav3.2 conferred faster recovery from channel inactivation and greater charge transference during high-frequency bursts 22 – consistent with what one might expect given the role of the LVAC in thalamic burst firing. The LVAC is a critical component of the thalamocortical rhythm that generates absence seizure activity. CACNA1H and its related gene CACNA1G are also the only genes in which sequence variants have been consistently found in human childhood absence epilepsy (e.g. see 23–25). CACNA1H is indeed the strongest a priori candidate gene for this disease.
Although this recent finding might have made sense to ion channel aficionados, it might have comes as a surprise to those who followed the GAERS story from the forward genetics perspective. This is because in 2004, another group had employed quantitative trait locus (QTL) mapping to identify genetic variants in absence seizures of GAERS 26. The approach was unbiased and systematic– examining the F2 (second intercross) generation between GAERS and an unrelated, non-epileptic rat strain – scoring individual F2 rats for seizure incidence, duration and amplitude, and examining the genome-wide correlation between seizure phenotypes and genetic markers. The study reported several chromosomal regions that each accounted for a fraction (12%–14%) of the total variance in one or more of the seizure phenotypes – some of these correlations reached statistical significance, whereas others were only suggestive. As is often the case for QTL with large critical intervals, several interesting candidate genes were suggested for future study, including Cacng2 (“stargazin”), Kcnj4 and Scn2b. However, given the results of the recent Powell study, it was surprising that there was no signal in the chromosomal region where Cacna1h resides - not even a suggestive “blip!”
How was such a confounding result possible? While it is difficult to exclude technical oversight, it is much more likely that genetic background is to blame. Indeed, each study utilized a different non-epileptic partner strain to generate the segregating population in which association was determined between seizures and genotype. The QTL study utilized a common non-epileptic rat strain, “brown Norway” (BN), which is not so closely related to GAERS, whereas the Cacna1h candidate gene study used the “non-epileptic control” (NEC) rat, strain, selectively bred from the same stock from which GAERS arose. In other words, many fewer genes would be expected to differ between GAERS and NEC, giving more pronounced effect to the Cacna1h splice variant. Although the GAERS-NEC difference is still complex (with some of the genetic contribution not yet explained), presumably many additional variants differed between GAERS and BN strains, diluting the ability to detect the Cacna1h main effect. It is important to note that just because Cacna1h was not detected in the QTL study, does not mean that it was not a factor. The nature of its effect might be much more complex – involving multiple interactions between it and other segregating variants– going beyond the ability to detect and model it in the necessarily modest sampling of a conventional QTL study that involves a laborious phenotyping method - EEG. In addition to Cacna1h potentially being hidden behind these interactions, it is also possible that some of the 25% of the individuals that did not inherit the bad allele had independent susceptibility alleles that phenocopied the Cacna1h effect. This illustrates the principle of locus heterogeneity (identical phenotype due to different genetic variants from one family to the next) – a hallmark of some complex traits and one that is recognized to be a major obstacle for progress in genetic mapping of common human epilepsy 27.
The study by Powell et al while important for understanding GAERS and for the functional significance of alternative splicing, resulted from an “obvious” candidate gene approach. Despite this success, clearly we cannot limit inquiries to such candidate genes; we must push ahead with forward genetics or other discovery-driven approaches to better understand function in these complex biological systems. However, it is troubling when discovery-driven efforts, such as the GAERS QTL study, can miss critical genes like Cacna1h. Clearly there is a balance that needs to be achieved, between the “potency” of a genetic effect in a population and the sought-after allelic diversity that defines a given complex genetic system.
103 mutations: might may already know more than we think we know…
Although discovery-driven progress on the genetics of epilepsy remains slow, we might know more than we thought we did. Mutant mice studied or created for other purposes, but which happen to have recurrent seizures as the prominent - and in some cases, the only - abnormal phenotype can reveal useful information that will help us to better understand the underlying genetics of epilepsy.
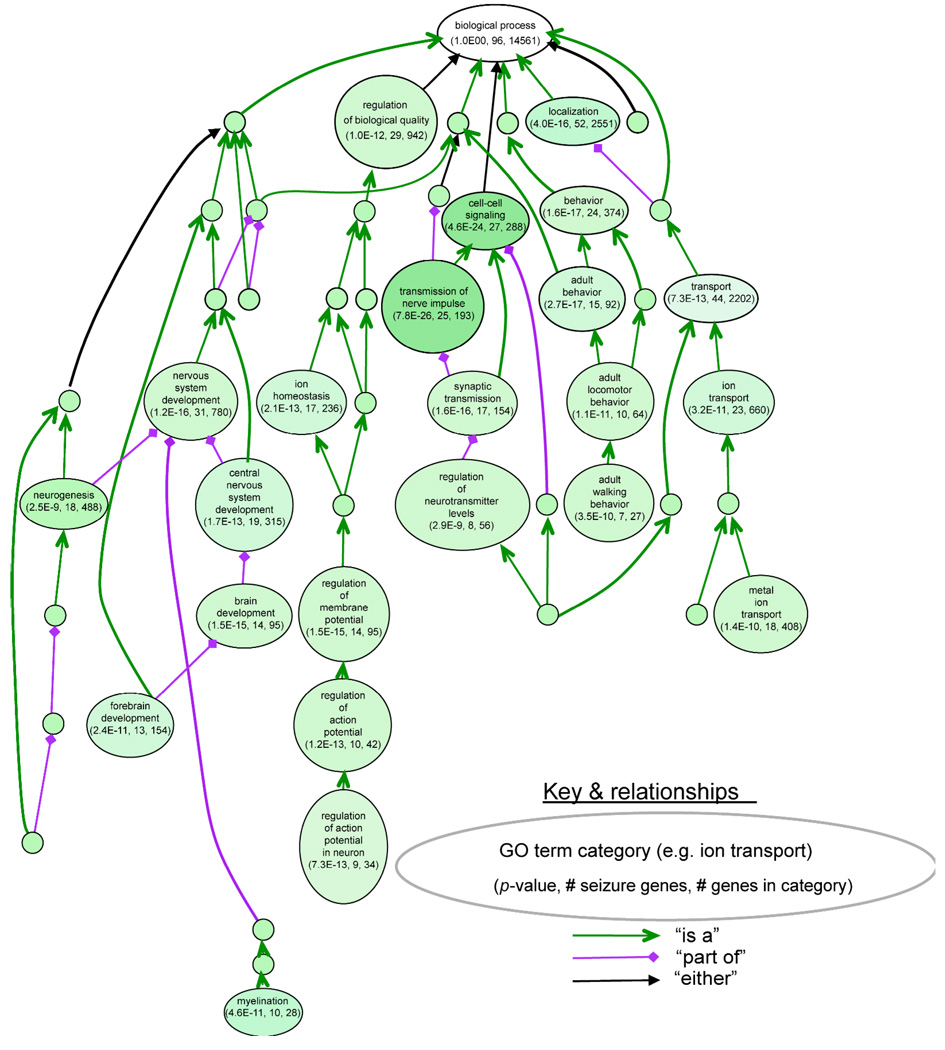
All genetic and phenotypic information for mouse mutations is summarized in the world’s largest mammalian genetics database, published by the Mouse Genome Informatics (MGI) group (www.informatics.jax.org). The searchable resource summarizes results from published articles according to mammalian phenotype ontology, incorporating a community effort to provide standard terms for annotating mammalian phenotypic data. Using “seizure” as a search term, the database lists almost 400 genes which, when mutated, are associated with a variety of seizure-related phenotypes– including relative sensitivity to chemically-induced seizures. When limiting this set to spontaneous or more readily-evoked seizures (such as after routine animal handling), perhaps better approximating idiopathic epilepsy, still 103 genes remain (Table 1). This group represents a rather wide array of molecules, ranging from cytostructural (e.g., myosin), to neurotransmitter receptor (GABAB), to synaptic vesicle functions (e.g., synaptojanin-1), to cellular signaling (Nr4a3), to ion channels (Kcna1, Scn8a). Using a prototype visualization tool called VLAD (VisuaL Annotation Display), we can observe how these 103 genes cluster according to gene ontology (GO) functional annotation of biological process (Figure 1). These systematic analyses together support the casual observation that recurrent seizures in mice can be associated with a wide variety of molecules and biological processes.
Table 1.
Genes associated with spontaneous or handling-associated seizures in mutant micea
| Gene | Encoded protein | Gene | Encoded protein |
|---|---|---|---|
| Acp2 | acid phosphatase 2, lysosomal | Htr2c | 5-hydroxytryptamine (serotonin) receptor 2C |
| Adam22 | a disintegrin and metallopeptidase domain 22 | Htra2 | HtrA serine peptidase 2 |
| Aldh5a1 | aldhehyde dehydrogenase family 5, subfamily A1 | Itpr1 | inositol 1,4,5-triphosphate receptor 1 |
| Amph | amphiphysin | Jrk | jerky |
| potassium voltage-gated channel, shaker-related | |||
| Ank3 | ankyrin 3, epithelial | Kcna1 | subfamily, member 1 |
| potassium voltage-gated channel, shaker-related | |||
| Ap3b2 | adaptor-related protein complex 3, beta 2 subunit | Kcna2 | subfamily, member 2 |
| potassium inwardly-rectifying channel, subfamily | |||
| Ap3m2 | adaptor-related protein complex 3, mu 2 subunit | Kcnj6 | J, member 6 |
| potassium inwardly-rectifying channel, subfamily | |||
| Asc1 | anterior suture cataract 1 | Kcnj9 | J, member 9 |
| potassium inwardly-rectifying channel, subfamily | |||
| Atcay | ataxia, cerebellar, Cayman type homolog (human) | Kcnj10 | J, member 10 |
| potassium voltage-gated channel, subfamily Q, | |||
| Atxn7 | ataxin 7 | Kcnq2 | member 2 |
| Bax | Bcl2-associated X protein branched chain ketoacid dehydrogenase E1, alpha |
Myo5a | myosin Va |
| Bckdha | polypeptide | Neurod2 | neurogenic differentiation 2 |
| Bmi1 | Bmi1 polycomb ring finger oncogene | Npy | neuropeptide Y |
| Brunol4 | bruno-like 4, RNA binding protein | Nr4a3 | nuclear receptor subfamily 4, group A, member 3 |
| Bsn | bassoon | Numb | numb gene homolog (Drosophila) |
| calcium channel, voltage-dependent, P/Q type, alpha | |||
| Cacna1a | 1A subunit | Numbl | numb-like |
| calcium channel, voltage-dependent, alpha 2/delta | |||
| Cacna2d2 | subunit 2 | Otx1 | orthodenticle homolog 1 (Drosophila) |
| Cacnb4 | calcium channel, voltage-dependent, beta 4 subunit | Pcmt1 | protein-L-isoaspartate (D-aspartate) O- methyltransferase 1 |
| Cacng2 | calcium channel, voltage-dependent, gamma subunit | Pitpna | phosphatidylinositol transfer protein, alpha |
| Cdk5r1 | cyclin-dependent kinase 5, regulatory subunit (p35) 1 | Plaur | plasminogen activator, urokinase receptor |
| Chrna4 | cholinergic receptor, nicotinic, alpha polypeptide 4 | Plcb1 | phospholipase C, beta 1 |
| Clcn3 | chloride channel 3 | Plp1 | proteolipid protein (myelin) 1 |
| Cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase | Pmp22 | peripheral myelin protein |
| Cplx1 | complexin 1 | Ppt1 | palmitoyl-protein thioesterase 1 |
| Cstb | cystatin B | Psap | prosaposin |
| Ctnnb1 | catenin (cadherin associated protein), beta 1 | Pten | phosphatase and tensin homolog |
| Ctsd | cathepsin D | Pura | purine rich element binding protein A |
| Dbnl | drebrin-like | Qk | quaking |
| Dbp | D site albumin promoter binding protein | Scg5 | secretogranin V |
| Drd1a | dopamine receptor D1A | Serpine2 | serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| Emx1 | empty spiracles homolog 1 (Drosophila) epilepsy, progressive myoclonic epilepsy, type 2 gene |
Sgce | sarcoglycan, epsilon |
| Epm2a | alpha | Scn1a | sodium channel, voltage-gated, type I, alpha |
| Fgf14 | fibroblast growth factor 14 | Scn1b | sodium channel, voltage-gated, type I, beta |
| Fyn | Fyn proto-oncogene | Scn2a | sodium channel, voltage-gated, type-2 alpha |
| Gabbr1 | gamma-aminobutyric acid (GABA-B) receptor, 1 | Scn8a | sodium channel, voltage-gated, type VIII, alpha |
| Gabbr2 | gamma-aminobutyric acid (GABA-B) receptor 2 | Shh | sonic hedgehog |
| Gabrb3 | gamma-aminobutyric acid (GABA) B receptor 3 | Slc12a5 | solute carrier family 12, member 5 |
| Gabrg2 | gamma-aminobutyric acid (GABA-A) receptor gamma | Slc13a1 | solute carrier family 13 (sodium/sulphate symporters), member 1 |
| Gad2 | glutamic acid decarboxylase 2 | Slc1a2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| Gjb1 | gap junction protein, beta 1 | Slc25a12 | solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| Gjc2 | gap junction protein, gamma 2 | Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 |
| Gnao1 | guanine nucleotide binding protein, alpha O | Slc9a1 | solute carrier family 9 (sodium/hydrogen exchanger), member 1 |
| Gng3 | guanine nucleotide binding protein (G protein), gamma 3 |
Snap25 | synaptosomal-associated protein 25 |
| Gpr98 | G-protein coupled receptor, 98 (a.k.a. Mass1) | Sod2 | superoxide dismutase 2, mitochondrial |
| Gria2 | glutamate receptor, ionotropic, AMPA2 (alpha 2) | Sox1 | SRY-box containing gene 1 |
| Gria3 | glutamate receptor, ionotropic, AMPA2 (alpha 3) | Syn1 | synapsin I |
| Grm7 | glutamate receptor, metabotropic 7 | Syn2 | synapsin II |
| Helt | Hey-like transcription factor (zebrafish) | Synj1 | synaptojanin 1 |
| Hexa | hexosaminidase A | Tal2 | T-cell acute lymphocytic leukemia 2 |
| Hexb | hexosaminidase B | Tef | thyrotroph embryonic factor |
| Hlf | hepatic leukemia factor | Ube3a | ubiquitin protein ligase E3A |
| Usp18 | ubiquitin specific peptidase 18 |
Results were tabulated from a standard phenotype query of the Mouse Genome Informatics database (www.informatics.jax.org)
Figure 1. Classification of 101 mouse seizure genes using gene-ontology functional annotation.
This analysis was done using VLAD, a multipurpose prototype tool for visualizing and analyzing gene-ontology (GO) annotation data for sets of genes (http://proto.informatics.jax.org/prototypes/vlad-1.0.3/). Here VLAD was used to determine the overrepresentation of the 101 mouse seizure genes relative to total number of genes in each GO class for a biological process. Each large bubble shows a specific GO class, with the number of seizure genes in the class, the number of total genes in the class (from the Mouse Genome Informatics, MGI, database), and a P-value to estimate the relative overrepresentation of seizure genes in that class (see http://proto.informatics.jax.org/prototypes/vlad-1.0.3/ for methods). The arrows indicate the “parent-child” relationship of GO classes to one another; small, unlabelled bubbles are classes that were not significantly overrepresented. The existence of multiple larger bubbles containing significant class overrepresentations is indicative of multiple etiologies for seizure disorders in mice.
Only a modest subset (25%) of these 103 mouse genes encodes ion channels, compared with 66% for currently known Mendelian human epilepsy genes (P<0.0002, Fisher exact test). Is this difference the result of ascertainment bias in the Mendelian form of the disease? Perhaps the 103 mouse genes are more representative of the broader spectrum of molecules involved in genetically complex epilepsies. Alternatively, ion channel variants might be underrepresented by the set of 103 genes because many of the phenotypes in the database result from knockout (null) mutations, resulting in little or no normal protein and some of which might be embryonically lethal. In nature, one finds a good mix of null, reduced and altered function. For some ion channels (e.g. Scn8a 28) seizures might occur regardless of the type of allele. Therefore, the difference seems noteworthy
But aren’t those 101 mouse mutations all single-gene mutations, i.e. Mendelian? How can we claim their phenotypes are genetically complex? To be sure, because most of these mutants were not studied for their seizure disorder in any depth, one cannot know whether the inheritance of recurrent seizures in them is genetically complex; most were only ever studied on a single genetic background (e.g. C57BL/6J – B6 or 129S1/SvImJ -129). However, at least a few were studied on multiple strain backgrounds and each showed appreciable variation in seizure penetrance, incidence, severity or form (e.g. Slc9a1 29, Gad65 30, Scn8a 28, 31, Scn2a 31, 32, Brunol4 33; interestingly, often showing more severe seizures in strains other than B6). Therefore, it is plausible that some or many of these 103 genes can participate in a genetically complex seizrue disorder.
Extensive mouse strain diversity in seizure phenotypes
The recognized strain background influence on seizure mutations is just the tip of the iceberg. Although only a few strains experience spontaneous seizures, experimentally provoked seizures are typically used to estimate seizure threshold. Seizure threshold is a theoretical “set-point” below which an individual has a seizure, and has been examined in mouse strains for many years. Strain diversity in seizure threshold was indeed evident from the earliest strain surveys, which examined sensitivity to audiogenic seizures 34, to more recent ones examining sensitivity to electroconvulsion 35, and from many smaller studies of chemically-induced seizures (e.g. using proconvulsants such as pentylenetetrazole, kainic acid, even cocaine 36) and recently flurothyl-induced kindling 37. Depending upon the stimulus type (which alone can affect the results) and the seizure endpoint measured,C57BL/6 (B6) mice is often one of the most seizure-resistant and DBA/2 (D2) mice one of the most seizure-sensitive strains – hence, much focus in the past 20 years has been in efforts to understand the genetic basis of seizure susceptibility between B6 and D2 mice, resulting in strong candidates such as Kcnj10.
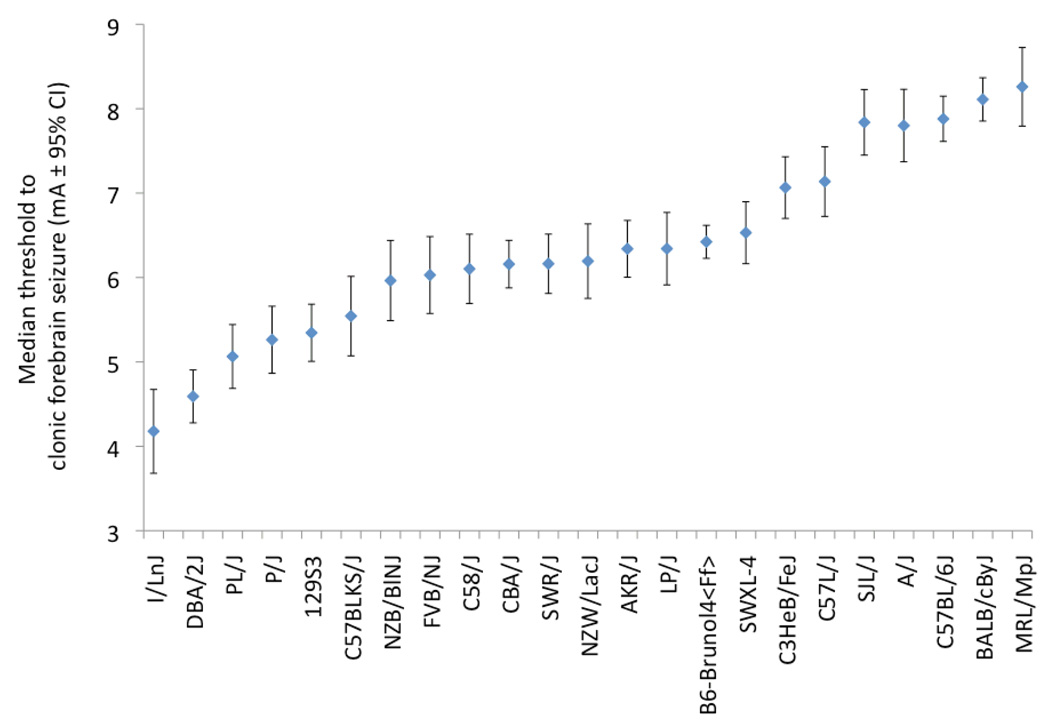
While differences between B6 and D2 are well established, surveys that involve a wider variety of strains have revealed extensive seizure threshold variation among the less-often used mouse strains. For example, in one survey 35 electroconvulsive threshold was examined for several seizure endpoints in 16 inbred strains: the FVB/NJ strain (a popular strain for creating transgenic mice) and CBA/J (a general purpose strain) had clonic seizure thresholds almost as low as the D2. By contrast, these two strains diverged greatly in their threshold to the more severe, tonic seizure endpoint– with FVB/NJ even more sensitive than D2, and CBA/J almost as resistant as C57BL/6J (at the far right end of the spectrum). Recently we have extended this strain survey to less commonly used mouse strains, and have found several additional strains that have very low seizure thresholds – including I/Ln, P/J and PL/J (N.b. PL/J also has handling-associated seizures 13) and an additional high seizure threshold strain – MRL/MpJ (Figure 2). These studies show that there are several other strains in addition to B6, D2 and 129 (the strain commonly used for gene-targeting), to consider for studies of seizure predisposition in mice. They also suggest that for seizure susceptibility phenotypes, one does not need to venture into wild-derived mouse strains or elaborate genetic designs (such as the Collaborative Cross 38); the old inbred mouse strains derived 100 years ago from “fancy” mice 39 seem to have plenty of phenotypic diversity to be useful…
Figure 2. Electroconvulsive threshold variation in 23 mouse strains.
The median thresholds (mA ± 95% CI) to clonic forebrain seizures are shown. Thresholds for I/LnJ, P/J, and MRL/MpJ have not been previously published. The remaining values were recalculated and re-plotted from previous studies 13, 33, 35; the new data were collected and analyzed as described in those studies. For simplicity of presentation, only female data are shown, but the relative strain thresholds are similar for male. A four mA range of median seizure threshold amongst the different inbred strains is suggestive of a very appreciable amount of genetic variation.
One interesting corollary of the strain surveys is that the relationship between seizure threshold and epilepsy is not one-to-one. Certainly, some spontaneously epileptic strains can also have the lowest seizure thresholds – PL/J, for example (Figure 2 13). But others, for example, the epileptic SWXL-4 (originally derived from normal progenitor strains – SWR/J and C57L/J 11) do not; several normal inbred strains have even lower thresholds than SWXL-4 35. Conversely, some mutations may express seizures preferentially on high threshold strains, for example, recurrent limbic and tonic-clonic seizures of frequent-flyer mutant mice on the B6 background (B6-Brunol4Ff, Figure 2). Thus, while low seizure threshold is a component of epilepsy, it is not an absolute indicator. However, the imperfect relationship does not necessarily detract from the utility and pragmatism of measuring induced seizure threshold. Also, the fact that many effective antiepileptic drugs were selected from seizure threshold screens in rodents proves that it is very relevant to epilepsy and a useful indicator. The salient point is that specific interactions between a mutation and genetic variants in the strain background are equally important factors.
Change partners for success
Seizure disorders of mouse and rat strains - whether spontaneous or experimentally induced - are clearly complex, non-linear entities with no shortage of endogenous genetic variants to influence phenotypic severity, incidence and/or form. What is the most effective way ahead to exploit these resources to better understand genetic mechanisms that underlie epilepsy as a complex trait?
Whether looking for susceptibility genes in epileptic strains (e.g. GAERS, PL/J), or trying to discover modifier genes of human mutations made transgenic or targeted in mice, it may be self-defeating restrict the study to one or two strain backgrounds; some strains may even be the wrong choice for certain mutations. And we have already established that seizure threshold is not the sole criterion. But since there are so many strains, and since specific genetic interactions are unknowable in advance of their discovery, which strains to chose? In mice it been common practice to involve a very distantly related strain partner derived from a different species such as Mus spretus or subspecies such as Mus musculus castaneus; assuring significant genetic distance. For seizure research, this might be a good approach if one is merely trying model diverse phenotypes without care to identifying underlying genes. But if the goal is gene identification, it might be counterproductive to venture too far afield. For example, wild-derived strains derived from different subspecies, or elaborate genetic designs such as multi-way crosses to introduce maximum diversity, may introduce “dilution” like that which rendered Cacna1h in GAERS undetectable in the original QTL study. While elaborate populations might reflect a heterogeneous genome structure that resembles in at least superficial ways a human population, what is the advantage of using them if progress towards detecting or identifying the underlying genes is impeded? Studying very diverse populations might not even be necessary in epilepsy, given the apparent diversity of seizure susceptibility in common mouse strains,.
A reasonable compromise is to produce several pilot crosses each with a strain partner of different genealogical origin (avoiding cross-species or sub-species combinations), and pursue only those that show notable phenotypic modification or for which modifier loci can be detected. For some laboratories, even this might seem easier said than done; if genome scans are not too expensive, animal housing costs might be prohibitive to do multiple crosses (normal mouse cage per diem costs range from $0.15 to $1.00 depending upon the institution and the level of care) or space-limited. However it is important to consider that even pilot, small-scale crosses of 40–50 individuals each will suffice to determine whether there is significant phenotypic modification, and suffice to detect loci that explain more than 25% of the phenotypic variance (as was the case for Cacna1h in GAERS). Successful crosses can then be expanded for deeper analysis and candidate gene evaluation.
New hope for the rat…
Between mice and rats, mice have been the more powerful genetic model due to the relative ease of manipulating the genome via gene targeting and transgenesis. But the rat might be catching-up. While it is not yet possible to successfully create rat mutants by homologous recombination in ES cells, the ability to combine transposon mutagenesis (e.g. L1, Sleeping Beauty) with various gene-trap and reporter strategies, facilitates effective conditional gene-targeting in rat 40, 41. In the coming years will be fascinating to compare genetic effects of epilepsy mutations between these two species as more rat mutants and strains are used in preclinical research.
Acknowledgements
The author is grateful to Dr. Verity Letts for having read a draft of this manuscript. This research was supported by a grant from the NIH-NINDS.
References
- 1.Kwan P, Brodie MJ. Early identification of refractory epilepsy. N Engl J Med. 2000;342:314–319. doi: 10.1056/NEJM200002033420503. [DOI] [PubMed] [Google Scholar]
- 2.Nei M, Bagla R. Seizure-related injury and death. Curr Neurol Neurosci Rep. 2007;7:335–341. doi: 10.1007/s11910-007-0051-1. [DOI] [PubMed] [Google Scholar]
- 3.Ottman R. Analysis of genetically complex epilepsies. Epilepsia. 2005;46 Suppl 10:7–14. doi: 10.1111/j.1528-1167.2005.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucarini N, et al. Genetic polymorphisms and idiopathic generalized epilepsies. Pediatr Neurol. 2007;37:157–164. doi: 10.1016/j.pediatrneurol.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Need AC, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000373. e1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall CS. Genetic differences in fatal audiogenic seizures between two inbred strains of mouse mice. J. Hered. 1947;38:2–6. [PubMed] [Google Scholar]
- 7.Vergnes M, et al. Spontaneous paroxysmal electroclinical patterns in rat: a model of generalized non-convulsive epilepsy. Neurosci Lett. 1982;33:97–101. doi: 10.1016/0304-3940(82)90136-7. [DOI] [PubMed] [Google Scholar]
- 8.Marescaux C, et al. Genetic absence epilepsy in rats from Strasbourg--a review. J Neural Transm Suppl. 1992;35:37–69. doi: 10.1007/978-3-7091-9206-1_4. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki J, Nakamoto Y. El mouse: a model of sensory precipitating epilepsy. Excerpta Medica. 1977;427:81–82. [Google Scholar]
- 10.Rise ML, et al. Genes for epilepsy mapped in the mouse. Science. 1991;253:669–673. doi: 10.1126/science.1871601. [DOI] [PubMed] [Google Scholar]
- 11.Frankel WN, et al. Genetic epilepsy model derived from common inbred mouse strains. Genetics. 1994;138:481–489. doi: 10.1093/genetics/138.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coenen AM, Van Luijtelaar EL. Genetic animal models for absence epilepsy: a review of the WAG/Rij strain of rats. Behav Genet. 2003;33:635–655. doi: 10.1023/a:1026179013847. [DOI] [PubMed] [Google Scholar]
- 13.Kitami T, et al. Genetic and phenotypic analysis of seizure susceptibility in PL/J mice. Mamm Genome. 2004;15:698–703. doi: 10.1007/s00335-004-3007-7. [DOI] [PubMed] [Google Scholar]
- 14.Strohl KP, et al. Sleep-related epilepsy in the A/J mouse. Sleep. 2007;30:169–176. doi: 10.1093/sleep/30.2.169. [DOI] [PubMed] [Google Scholar]
- 15.Beyer B, et al. Absence seizures in C3H/HeJ and knockout mice caused by mutation of the AMPA receptor subunit Gria4. Hum Mol Genet. 2008;17:1738–1749. doi: 10.1093/hmg/ddn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokuda S, et al. Genetic complexity of absence seizures in substrains of C3H mice. Genes Brain Behav. 2009;8:283–289. doi: 10.1111/j.1601-183X.2008.00472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferraro TN, et al. Fine mapping of a seizure susceptibility locus on mouse Chromosome 1: nomination of Kcnj10 as a causative gene. Mamm Genome. 2004;15:239–251. doi: 10.1007/s00335-003-2270-3. [DOI] [PubMed] [Google Scholar]
- 18.Ferraro TN, et al. Analysis of a quantitative trait locus for seizure susceptibility in mice using bacterial artificial chromosome-mediated gene transfer. Epilepsia. 2007;48:1667–1677. doi: 10.1111/j.1528-1167.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- 19.Lenzen KP, et al. Supportive evidence for an allelic association of the human KCNJ10 potassium channel gene with idiopathic generalized epilepsy. Epilepsy Res. 2005;63:113–118. doi: 10.1016/j.eplepsyres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Bockenhauer D, et al. Epilepsy, ataxia, sensorineural deafness, tubulopathy, and KCNJ10 mutations. N Engl J Med. 2009;360:1960–1970. doi: 10.1056/NEJMoa0810276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholl UI, et al. Seizures, sensorineural deafness, ataxia, mental retardation, and electrolyte imbalance (SeSAME syndrome) caused by mutations in KCNJ10. Proc Natl Acad Sci U S A. 2009;106:5842–5847. doi: 10.1073/pnas.0901749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell KL, et al. A Cav3.2 T-type calcium channel point mutation has splice-variant-specific effects on function and segregates with seizure expression in a polygenic rat model of absence epilepsy. J Neurosci. 2009;29:371–380. doi: 10.1523/JNEUROSCI.5295-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, et al. Association between genetic variation of CACNA1H and childhood absence epilepsy. Ann Neurol. 2003;54:239–243. doi: 10.1002/ana.10607. [DOI] [PubMed] [Google Scholar]
- 24.Singh B, et al. Mutational analysis of CACNA1G in idiopathic generalized epilepsy. Mutation in brief #962. Online. Hum Mutat. 2007;28:524–525. doi: 10.1002/humu.9491. [DOI] [PubMed] [Google Scholar]
- 25.Heron SE, et al. Extended spectrum of idiopathic generalized epilepsies associated with CACNA1H functional variants. Ann Neurol. 2007;62:560–568. doi: 10.1002/ana.21169. [DOI] [PubMed] [Google Scholar]
- 26.Rudolf G, et al. Polygenic control of idiopathic generalized epilepsy phenotypes in the genetic absence rats from Strasbourg (GAERS) Epilepsia. 2004;45:301–308. doi: 10.1111/j.0013-9580.2004.50303.x. [DOI] [PubMed] [Google Scholar]
- 27.Dibbens LM, et al. A polygenic heterogeneity model for common epilepsies with complex genetics. Genes Brain Behav. 2007;6:593–597. doi: 10.1111/j.1601-183X.2007.00333.x. [DOI] [PubMed] [Google Scholar]
- 28.Papale LA, et al. Heterozygous mutations of the voltage-gated sodium channel SCN8A are associated with spike-wave discharges and absence epilepsy in mice. Hum Mol Genet. 2009;18:1633–1641. doi: 10.1093/hmg/ddp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox GA, et al. Sodium/hydrogen exchanger gene defect in slow-wave epilepsy mutant mice. Cell. 1997;91:139–148. doi: 10.1016/s0092-8674(01)80016-7. [DOI] [PubMed] [Google Scholar]
- 30.Kash SF, et al. Epilepsy in mice deficient in the 65-kDa isoform of glutamic acid decarboxylase. Proc. Natl. Acad. Sci. USA. 1997;94:14060–14065. doi: 10.1073/pnas.94.25.14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearney JA, et al. Molecular and pathological effects of a modifier gene on deficiency of the sodium channel Scn8a (Na(v)1.6) Hum Mol Genet. 2002;11:2765–2775. doi: 10.1093/hmg/11.22.2765. [DOI] [PubMed] [Google Scholar]
- 32.Bergren SK, et al. Fine mapping of an epilepsy modifier gene on mouse Chromosome 19. Mamm Genome. 2009 doi: 10.1007/s00335-009-9193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, et al. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS Genet. 2007;3:e124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuller JL, Sjursen FH. Audiogenic seizures in eleven mouse strains. J. Hered. 1967;58:135–140. doi: 10.1093/oxfordjournals.jhered.a107565. [DOI] [PubMed] [Google Scholar]
- 35.Frankel WN, et al. Electroconvulsive thresholds of inbred mouse strains. Genomics. 2001;74:306–312. doi: 10.1006/geno.2001.6564. [DOI] [PubMed] [Google Scholar]
- 36.Golden GT, et al. Acute cocaine-induced seizures: differential sensitivity of six inbred mouse strains. Neuropsychopharmacology. 2001;24:291–299. doi: 10.1016/S0893-133X(00)00204-9. [DOI] [PubMed] [Google Scholar]
- 37.Papandrea D, et al. Dissociation of seizure traits in inbred strains of mice using the flurothyl kindling model of epileptogenesis. Exp Neurol. 2009;215:60–68. doi: 10.1016/j.expneurol.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Churchill GA, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 39.Silver LM. Mouse genetics: concepts and practice. Oxford University Press; 1995. [Google Scholar]
- 40.Kitada K, et al. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007;4:131–133. doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- 41.Lu B, et al. Generation of rat mutants using a coat color-tagged SleepingBeauty ransposon system. Mamm Genome. 2007;18:338–346. doi: 10.1007/s00335-007-9025-5. [DOI] [PubMed] [Google Scholar]