Abstract
Heparan sulfate proteoglycans are essential components of the cell-surface and extracellular matrix which provide structural integrity and act as storage depots for growth factors and chemokines, through their heparan sulfate (HS) side chains. Heparanase is the only mammalian endoglycosidase known that cleaves HS, thus contributing to matrix degradation and cell invasion. The enzyme acts as an endo-β-D-glucuronidase resulting in HS fragments of discrete molecular weight size. Cell-surface HS is known to inhibit or stimulate tumorigenesis depending upon size and composition. We hypothesized that heparanase contributes to melanoma metastasis by generating bioactive HS from the cell-surface to facilitate biological activities of tumor cells as well as tumor microenvironment. We removed cell-surface HS from melanoma (B16B15b) by HPSE treatment and resulting fragments were isolated. Purified cell-surface HS stimulated in vitro B16B15b cell migration but not proliferation, and importantly, enhanced in vivo angiogenesis. Furthermore, melanoma cell-surface HS did not affect in vitro endothelioma cell (b.End3) migration. Our results provide direct evidence that, in addition to remodeling extracellular matrix and releasing growth factors and chemokines, HPSE contributes to aggressive phenotype of melanoma by releasing bioactive cell-surface HS fragments which can stimulate melanoma cell migration in vitro and angiogenesis in vivo.
Keywords: Heparanase, heparan-sulfate proteoglycans, melanoma, migration, angiogenesis
Introduction
Enzymatic remodeling of heparan sulfate proteoglycans (HSPG) within the tumor microenvironment is emerging as an important mechanism for the dynamic regulation of tumorigenesis [1-3]. HSPG are a family of glycoproteins that are distinguished by the covalent attachment of one or more HS chains to their protein core. HS are directly involved with the angiogenic process by acting as co-receptors with angiogenic growth factors [3]. As the interface between tumor cells and host cells, HS mediate cellular interactions. HS also influence tumor metastasis to sites such as the brain by arbitrating interactions between cancer cells, platelets, endothelial cells, and host organ cells. Intact HS prevent metastasis by acting as a physical barrier in the extracellular matrix (ECM). Enzymes that cleave HS may release fragments that can either support or inhibit tumorigenesis [4, 5].
HPSE is the only mammalian endo-β-D-glucuronidase which cleaves HS at specific intrachain sites, resulting in fragments of appreciable size (10–20 sugar units) [7]. Numerous in vitro and in vivo studies have asserted a role for HPSE in tumor invasion and metastasis [6-12]. In addition, HPSE activity is upregulated in human cancers; a number of studies using clinical samples demonstrated a correlation between tumor malignancy and HPSE levels [13-18]. In vivo animal studies have also indicated that changes in the fine structure of tumor-cell-surface insoluble HS or soluble HS in the ECM have profound effects on tumor-cell growth kinetics, angiogenesis and metastasis formation [4, 5, 19-22]. Depending upon HS composition and/or sequence involved in the process of tumorigenesis, they can either act as pro-tumorigenic, or anti-tumorigenic agents [4, 5, 19].
Soluble or ECM HS can also differentially regulate fibroblast growth factor-2 (FGF2) binding and signaling leading to modification of angiogenesis of brain-metastatic melanoma cells depending upon its concentration [19]. Amongst additional signaling molecules, some of HS-binding growth factors important for angiogenesis and tumor development are vascular endothelial growth factor (VEGF), hepatocyte growth factor (HGF), transforming growth factor-β (TGF-β), and platelet-derived growth factor (PDGF) [23]. VEGF or vascular permeability factor (VPF) [24] was initially described as a highly specific mitogen for endothelial cells [25] and as a potent inducing agent for angiogenesis and vasculogenesis in a variety of physiological and pathological conditions [26]. Inhibition of VEGF signaling by VEGF antagonists, antisense VEGF, or dominant negative VEGF receptor (VEGFR) impaired tumorigenesis in vivo [27, 28]. The VEGF family of proteins includes VEGF A through E and placenta growth factor [26]. VEGF A is the most studied and one of its splice variants, VEGF165, is the most potent and widely expressed isoform known [29]. VEGF165 is secreted in the ECM as a disulfide-linked homodimer with two identical heparin-binding sites. HS, by binding to VEGF, regulate the diffusion, half-life, and affinity of VEGF165 to its cognate receptors [3, 30].
We have shown previously that HPSE treatment or HPSE-generated bovine kidney HS products increase FGF2 binding and signaling in melanoma cells in vitro and FGF2-medited angiogenesis in vivo. In this report, we have extended our study using cell surface HS from murine brain-metastatic melanoma cells (B16B15b) to investigate their effect on melanoma biology in respect to VEGF signaling. We sought to assess the role of HPSE-degraded cell-surface HS in VEGF-mediated activity in brain-metastatic melanoma cells since VEGF is known to be essential for brain metastasis in melanoma [28]. We hypothesized that HPSE contributes to melanoma metastasis by generating bioactive HS from the cell-surface that stimulate biological activities associated with metastatic cascade. We also examined if these fragments could differentially affect VEGF165-mediated biological activities of melanoma and endothelioma. We demonstrate that the isolated cell-surface HS stimulate in vitro migration but not proliferation of melanoma cells (B16B15b). Furthermore, they also promote in vivo angiogenesis by Matrigel™ plug assays. Interestingly, VEGF165 does not affect melanoma migration or angiogenesis alone or together with the cell-surface HS in these experiments. Finally, melanoma cell-surface HS do not stimulate in vitro migration of murine brain endothelioma cells (b.End3). Our results suggest that, in addition to remodeling the ECM and releasing ECM-based growth factors and chemokines, HPSE can contribute to aggressive phenotype of melanoma by releasing bioactive cell-surface HS which in turn stimulate melanoma cell migration and angiogenesis.
Experimental Procedures
Materials
Heparan sulfate (HS) from bovine kidney was purchased from Sigma Chemical Company (St. Louis, MO). Heparin-lyase III (heparitinase, EC 4.2.2.8) from Flavobacterium heparinium was obtained from Seikagaku (Seikagaku America, Falmouth, MA). DMEM and Ham's F-12 nutrient medium and trypsin-EDTA were purchased from Gibco (Grand Island, New York, NY), and FBS from Hyclone Laboratories (Logan, UT). Reduced-growth factor Matrigel™ was obtained from BD Biosciences Discovery Labware (Bedford, MA). All other chemicals used were reagent grade or better.
Cells and Tissue Culture Conditions
Early-passage, Mycoplasma-negative, murine melanoma (B16B15b) cells with high metastatic capabilities [12, 31] were maintained as monolayer cultures in a 1:1 (v/v) mixture of Dulbecco's modified Eagle's medium/F-12 (DMEM/F12) supplemented with 5% (v/v) fetal bovine serum. Murine brain endothelioma cells (b.End3) [32] were passaged in DMEM/F12 supplemented with 4 mM L-glutamine adjusted to contain 1.5 g/L sodium bicarbonate and 4.5 g/L glucose supplemented with 10% (v/v) fetal bovine serum. Cells were maintained at 37°C in a humidified 5% CO2/95% air (v/v) atmosphere and passaged using 2 mM EDTA (B16B15b) or trypsin-EDTA (b.End3) before reaching confluence.
For enzymatic treatment with HPSE, cells were washed 3 times in DMEM/F-12 containing 0.1% (w/v) BSA, penicillin (100 U/ml) and streptomycin (100 μg/ml), then incubated with indicated concentrations (0-10 μg/ml) of recombinant HPSE in 50 mM HEPES-buffered DMEM/F-12 (pH 6.8) containing penicillin (100 U/ml) and streptomycin (100 μg/ml) for 18 h at 37°C in a shaker incubator at 50 rpm.
HPSE Isolation and Activity
Recombinant human HPSE was purified as previously described [19, 33]. Briefly, Sf9 insect cells, transfected with baculovirus transfer vectors containing HPSE subunits, were grown in SF900II serum-free medium (Gibco BRL, Grand Island, NY) for high-titer stocks. Tni cells cultured in suspension using ExCell405 serum-free medium (JRH Bioscience, Lenexa, KS) were infected with high-titer stock for 48 h, and cells were subsequently removed by centrifugation. The supernatant was then tested for HPSE activity, filtered through a 0.45 μm filter, and loaded on a HiTrap heparin column (Amersham Biosciences, Piscataway, NJ). The column was subsequently washed in TBS, and then eluted using a 100 ml gradient of 0.15-1.0 M NaCl in 25 mM Tris-HCl (pH 7.5). Collected fractions (1 ml) were screened for HPSE activity (Heparan Degrading Enzyme Assay Kit; Takara Mirus, Madison, WI) [19, 33]. HPSE eluted at 0.67 M NaCl, as expected [19, 33].
Flow Cytometric Analysis
Degradation of cell-surface HS was confirmed by flow cytometric analyses. Briefly, B16B15b metastatic melanoma cells were treated with different concentrations of HPSE (0-10 μg/ml) for 18 h at 37°C using a shaker incubator. The medium was removed at the end of treatment and cells were collected by PBS-EDTA. Cells (5 × 105) were then incubated with HS mAb 10E4 (Seikagaku) followed by incubations with super sensitive biotin-goat anti-mouse IgM (BioGenex, San Ramon, CA) and PE streptavidin (Molecular Probes, Eugene, OR) respectively. Cells were then fixed in 200 μl cold 1% (v/v) paraformaldehyde and stored at 4°C until analysis. Samples were analyzed for cell-surface HS staining using FACScan Flowcytometer. Data were analyzed with WinMDI. Appropriate control samples without the primary and/or the secondary antibody (anti-IgM) were run to subtract background staining.
Isolation of cell surface HS
B16B15b metastatic melanoma cells were treated with or without HPSE (10 μg/ml) overnight at 37°C using a shaker incubator. Conditioned medium was collected and pH was adjusted to 6.0. The medium was then centrifuged at 5000 rpm for 10 minutes, filtered through 0.22 μm filters and boiled for 10 minutes. The medium was incubated with DEAE-Sephacel (Sigma) for 18 h and subsequently poured onto column. HS fragments bound to DEAE were washed with 0.1M sodium phosphate and 0.15M NaCl, pH 6.0. The fragments were gradient eluted with 0.5M, 1.0M, and 2.0M NaCl in 0.1M sodium phosphate buffer pH 6.0. The fragments were then concentrated and buffer-exchanged into ultra-pure water by application to a Centricon filter (Millipore Corporation, Bedford, MA). Finally, isolated fragments were treated with pronase (Sigma; 58) to remove protein core from isolated HS.
HPSE-degraded HS used in biological experiments was analyzed by separating HS on a Criterion 4-20% TBE gel (Bio-Rad Laboratories, Hercules, CA) for 20 min at 60 mAmp. Bands were visualized with alcian blue 8GX (Sigma-Aldrich) followed by silver staining (Pierce Endogen, Rockford, IL) [34]. Densitometric analyses were performed using a Versadoc imaging system (Bio-Rad Laboratories) to determine profiles leading edge. Non-treated commercial HS from bovine kidney were electrophoresed at various concentrations to obtain quantitative analysis.
Wound Healing Assay
Migratory properties of melanoma cells were analyzed by a standard wound healing assay. Briefly, cells were plated in 12-well plates at a high density and allowed to grow to confluence. Cells were washed 3 times in DMEM/F-12 containing 0.1% (w/v) BSA, penicillin (100 U/ml) and streptomycin (100 μg/ml) and then incubated with the same medium for one hour. Using a sterile 100 μl tip, a single scratch was made through the middle of each well. The medium was subsequently removed and the wells were rinsed three times with DMEM/F-12 containing 0.1% (w/v) BSA and penicillin/streptomycin to remove the detached cells. Cell-surface HS (1 ng/ml) or recombinant VEGF165 (10-50 ng/ml) were added to cells in DMEM/F-12 containing 0.1% (w/v) BSA, 4mM Hepes, penicillin (100 U/ml), and streptomycin (100 μg/ml) for 8 h at 37°C in a humidified 5% CO2/95% air (v/v) atmosphere. Photomicrographs were taken at 0 hour (T0) and at the end of the experiment (T8) using identical conditions to calculate percent relative gap closure. Relative gap closure was measured as [1-(T8/T0)]. Migration assays for endothelioma were incubated for 24 h (T24).
Proliferation Assay
Proliferation of melanoma cells were assayed by using alamarBlue™ (BioSource International, Camarillo, CA, USA) [35], a non-toxic dye which monitors the reducing environment of the proliferating cell, as per manufacturer's instructions. Briefly, 1 × 104 cells/ml were plated into 24-well plates and incubated for 24 h. At the start of the proliferation assay, cells were washed 3 times in DMEM/F-12 containing 0.1% (w/v) BSA and penicillin (100 U/ml) and streptomycin (100 μg/ml), and then incubated with the same medium for one hour. Indicated concentrations of cell-surface HS or recombinant VEGF165 were added to cells in triplicates in DMEM/F-12 containing 0.1% (w/v) BSA, 4mM Hepes, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a humidified 5% CO2/95% air (v/v) atmosphere. For indicated time points alamarBlue™ (10%, v/v) was added per well and were incubated for 4 h at 37°C in a humidified 5% CO2/95% air (v/v) atmosphere. Cell proliferation was measured by monitoring the fluorescence of alamarBlue™ supplemented cell culture media at excitation and emission wavelengths of 540 and 630 nm respectively. The greater is the percentage of reduction (fluorescent count), the higher is the proliferative activity.
In Vivo Angiogenic Assay
B16B15b cells were released with PBS-EDTA, washed two times in DMEM/F-12, and resuspended at 1×107 cells/ml in 50% (v/v) reduced-growth factor Matrigel™ (Becton Dickinson, Labware, Bedford, MA) in DMEM/F-12 at 4°C. HS fragments and VEGF165 were added accordingly. Cells (2 × 106) were injected using a 25-gauge needle to the left and right abdominal subcutaneous tissue of female C57BL6 (Harlan Teklan, Madison, WI) mice (n=6-9). B16B15b cells, in reduced-growth factor Matrigel along with HPSE-degraded melanoma cell-surface HS or without, as mock control, were injected into the right (with VEGF) and left (without VEGF) abdominal subcutaneous tissue of female C57BL6 mice (n = 6-9). Animals were divided in three groups in a split-plot arrangement. Group A received HPSE-treated HS with VEGF (right) or without VEGF (left). Group B received HPSE-treated HS that were further treated with Hep III to cleave them into inactive disaccharide fragments with VEGF (right) or without VEGF (left). Group C received melanoma cells in mock-buffer with VEGF (right) or without VEGF (left). Mice were sacrificed on the 10th day; tumors were excised, fixed in 10% (v/v) formalin, and embedded in paraffin. Tumor sections (7 μm thick) were then stained with haematoxylene and eosin (H&E) and examined under the microscope. Blood vessel density was assessed by counting vessels within the tumor region in five sections in each tumor. Tumor sections were photographed using Olympus DP70 camera, Olympus BX45 microscope and saved in JPEG format using DP Manager (Olympus America Inc., Center Valley, PA). Tumor areas were measured by counting pixels on ImageJ software (NIH). Pixel counts were converted to mm2 to present the number of vessels per unit area. Statistical analyses were done using SAS (Version 9.1.3) in an analysis of variance in a split-plot arrangement of treatments. Main plot effects included Group and Animal Id within Group; subplot effects included Side and Group*Side interaction. Pairwise comparisons of main effects were conducted with Tukey's HSD test. When appropriate, interaction effect comparisons were performed with t-tests of least-square means. All comparisons were considered significant at p<0.05. Prior to analysis, the data were natural log-transformed to stabilize variance terms.
Results
Removal and Isolation of Cell-Surface HS by HPSE Treatment
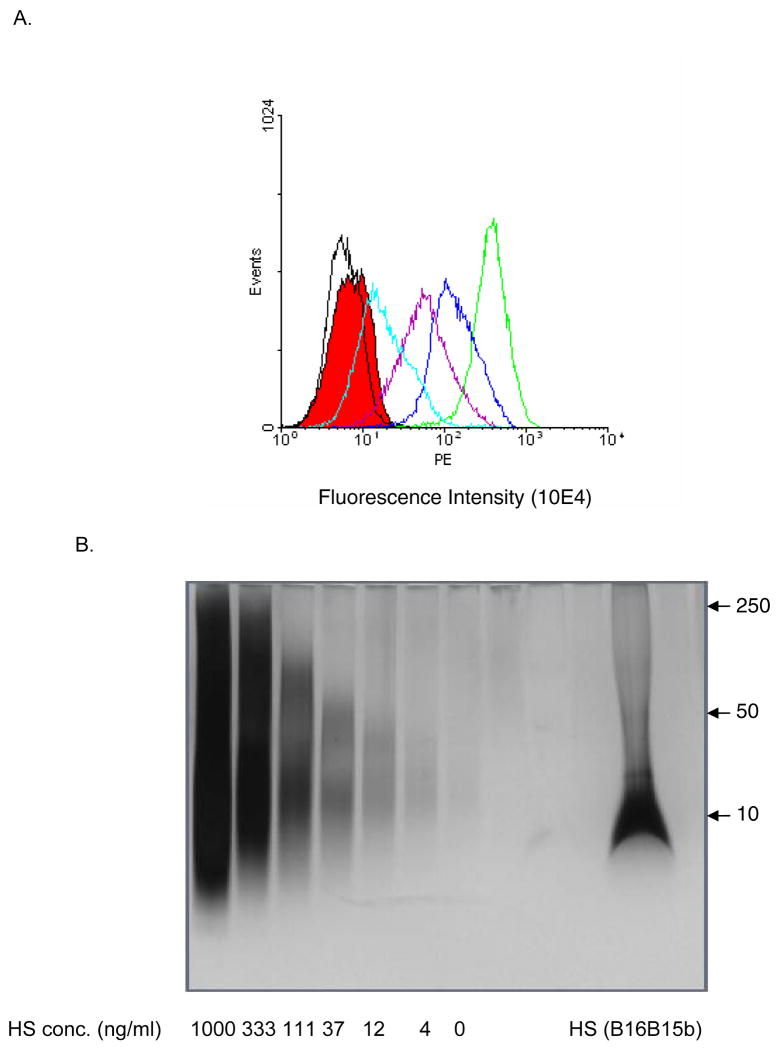
Highly brain-metastatic B16B15b melanoma cells were chosen as a source of HS since they possess high HSPG expression on the cell-surface [36]. The extent of HS degradation by HPSE was assessed by detection of cell-surface HS on FACS analysis. Detectable reduction in HS expression levels was seen with as low as 5.0 ng/ml HPSE compared to no HPSE treatment (data not shown). To optimize isolation of cell surface HS, when cells were treated with higher HPSE concentrations (100-10,000 ng/ml), a dose-dependent decrease in cell-surface HS expression was observed (Fig. 1A).
Figure 1. Dose-dependent reduction of cell surface HSGAG with HPSE.
A. Murine B16B15b melanoma cells were treated with 0-10,000 ng/ml HPSE. Cell surface HS level was detected by flowcytometry using mAb10E4. HPSE removes cell surface HSGAG in a dose-dependent manner, (green) 0 ng/ml HPSE, (blue) 100 ng/ml HPSE, (purple) 1,000 ng/ml HPSE, (light blue) 10,000 ng/ml HPSE. Appropriate controls were run to account for background staining, (black) no primary antibody control, (solid red) no primary and no secondary antibody control.
B. HPSE-degraded cell surface HSGAG profile on silver stain. As expected, HPSE digestion generated HS fragments of about 10 kDa. Various concentrations of untreated HS was run to generate a standard curve for determination of concentration of HSGAG after densitometric analysis. Numbers on the right hand of figure refer to M.W. standards (kDa).
Conditioned medium following HPSE treatment (10 μg/ml) from B16B15b melanoma cells was collected and HS were isolated by ion-exchange column chromatography. Assessment of HPSE-mediated HS degradation was determined by gel electrophoresis of isolated fragments (Fig. 1B). Since the isolated HS were a heterogeneous mixture of oligosaccharides due to HPSE digestion, it migrated as a broad band during gel electrophoresis (Fig. 1B). The leading edge of HS profiles was determined after densitometric analysis and concentration was determined by generating a curve with known standards (Fig. 1B).
Effects of HPSE-Degraded HS on Endothelioma In Vitro
Angiogenesis is an important step in solid tumor growth beyond a certain dimension (0.2-2.0 mm or about 105-106 cells) that requires formation of new blood vessels from the preexisting vascular network [37]. Endothelial cells migrate and proliferate during angiogenesis and are influenced by the tumor microenvironment including heparin/HS-binding growth factors secreted by the tumors such as FGF2 and VEGF [38]. Therefore, we decided to study effects of HPSE-degraded HS in an endothelial system. We investigated changes in migratory properties in murine brain endothelioma cellline b.End3, since migration is a critical event in angiogenesis [38].
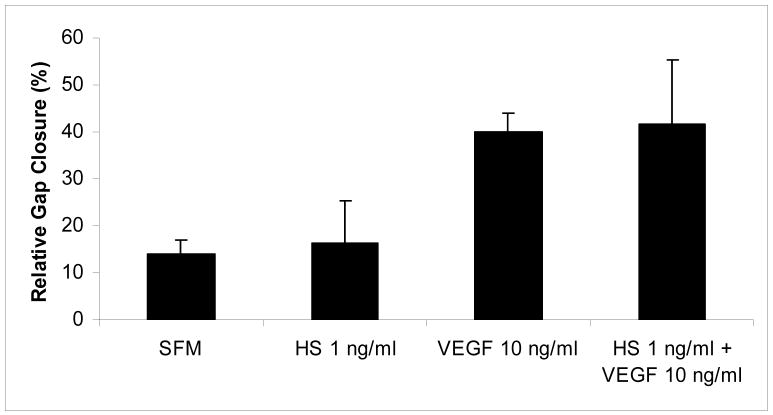
To study how exogenous addition of HS will influence endothelioma (b.End3) biological activity, we added HPSE-digested melanoma cell surface HS to serum-free endothelioma medium. HS did not have any effect on endothelioma cell migration (Fig. 2). We also used VEGF165, a known mitogenic factor for endothelial cells, which stimulated endothelioma cell migration compared to no VEGF treatment (p< 0.05, Fig. 2). Moreover, addition of melanoma cell surface HS to VEGF165 treatment did not augment this response (Fig. 2).
Figure 2.
Endothelioma cell migration is not influenced by HPSE-degraded HS but VEGF stimulates wound healing. To study how exogenous addition of HS will influence endothelioma (b.End3) biological activity, we added HPSE-digested melanoma cell surface HS to serum-free endothelioma medium. HS treatment did not stimulate endothelioma cell migration while VEGF did (p< 0.05), as expected. HS, when added along with VEGF, did not augment VEGF response.
HPSE-Degraded Cell Surface HS Modulates Melanoma Cell Migration
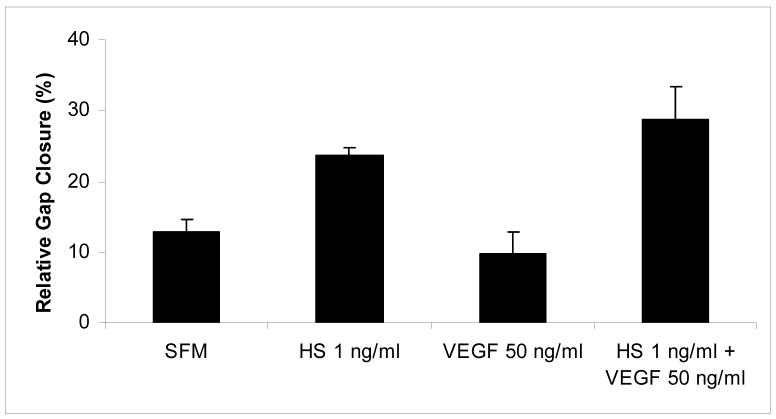
To directly test whether the B16B15b cell-surface HS were biologically active, we tested their effects on melanoma cell migration. B16B15b cells possess an aggressive migratory behavior, express VEGF receptors, and respond to VEGF. Interestingly, when HS (1 ng/ml) were added externally, there was a 30% up to a 2-fold increase in cell migration compared to no treatment (p< 0.05, Fig. 3). Addition of VEGF165 (0-100 ng/ml) did not augment this effect compared to no VEGF165 control (data not shown). Interestingly, VEGF165 did not effect migration even when added along with HS compared to HS alone (Fig. 3). One possibility could be that melanoma cells tested secrete autocrine growth factors [39] even after serum starvation, and thus do not respond to exogenously added stimuli. This was unexpected since VEGF165 is known to require HS to exert its biological effects. However, changes in cell migratory properties following addition of melanoma HS isolated by HPSE-digestion suggested that the HPSE-degraded HS fragments are bio-active and possess tumor stimulatory activity.
Figure 3.
HPSE-degraded cell surface HS modulate melanoma cell migration. When HS (1 ng/ml) were added externally to melanoma cells in wound healing assays, there was increased migration compared to control (p< 0.05). Addition of VEGF (50 ng/ml) did not affect migration with or without HS.
HPSE-Degraded Cell-Surface HS does not Influence Melanoma Cell Proliferation
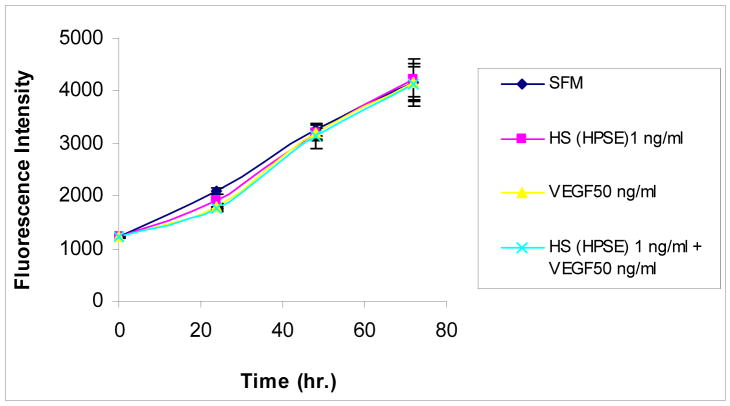
We next explored effects of B16B15b cell-surface HS on melanoma cell proliferation to test if similar conditions used in our wound healing assays also affect cell proliferation. Melanoma cell proliferation was assayed by alamarBlue™ (see “Experimental Procedures”), cell proliferation was monitored every 24 h for 72 h. The basal cell proliferation rate is high in these cells; we did not observe any change in proliferative properties of the cells either by HS or by VEGF treatment over a period of 72 hours (Fig. 4). Thus, exogenous addition of melanoma cell-surface HS isolated by HPSE treatment influences melanoma cell migration without affecting proliferation.
Figure 4.
HPSE-degraded cell surface HSGAG does not influence melanoma cell proliferation. Proliferation of melanoma cells were assayed by alamarBlue™, a non-toxic dye that monitors the reducing environment of the proliferating cell. Melanoma cell proliferation was monitored every 24 h for 72 h. Exogenous addition of VEGF (50 ng/ml) or melanoma cell surface HS (1 ng/ml) isolated by HPSE treatment did not influence cell proliferation.
HPSE-Degraded Cell-Surface HS Promotes In Vivo Angiogenesis
To investigate effects of HPSE-derived cell-surface HS on in vivo angiogenesis, Matrigel plug assays were performed. B16B15b cells, in reduced-growth factor Matrigel along with HPSE-degraded melanoma cell-surface HS (1 ng/ml) or without, as mock control, were injected into the right (with 50 ng/ml VEGF) and left (without VEGF) abdominal subcutaneous tissue of female C57BL6 mice (n = 6-9). Mice were sacrificed on the 10th day post-injection and Matrigel plugs were removed. Sections (7 μm thick) were then H&E-stained to examine for blood vessel formation. Blood vessel density was assessed by counting vessels within the Matrigel plug region in five sections in each plug.
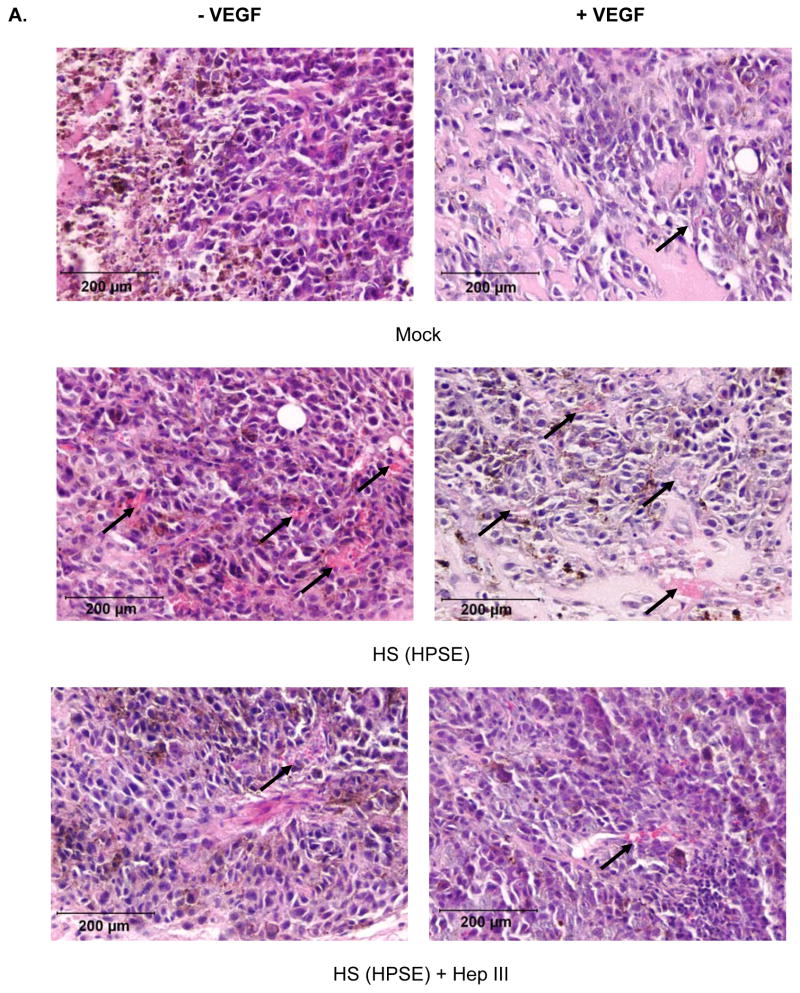
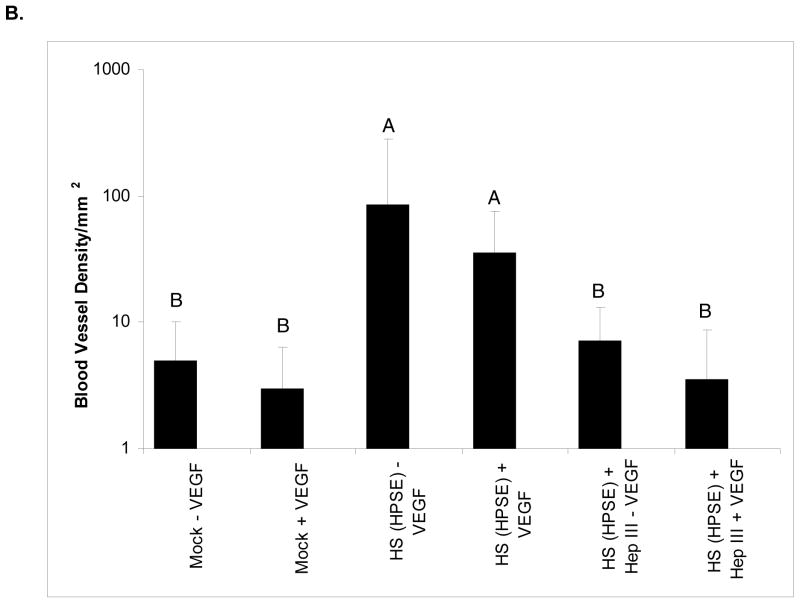
HPSE-treated cell surface HS induced a significant increase in intratumor blood vessel formation (Fig. 5A and 5B) in animals in group A compared to mock (group C) or Hep III treatment of HPSE-derived cell-surface HS (group B) (p < 0.0001). Interestingly, the increased numbers of tumor vessel formation were VEGF-independent. Presence of VEGF did not affect angiogenesis in all three groups (p = 0.2-0.8). Notably, the absence of blood vessels inside the tumors led to areas of necrosis due to lack of nutrients and oxygen (Fig. 5A). Importantly, the Matrigel plugs in all groups seemed to be approximately the same size and this observation was confirmed following excision and weighing of the plugs (not shown). Therefore, the differences in vascularity between the mock control and/or Hep III control vs. the HPSE-generated cell-surface HS tumors are not due to differences in tumor size, rather it is due to the tumor promoting effects of cell-surface HS fragments on tumor microenvironment.
Figure 5.
HPSE-degraded cell surface HSGAG promotes angiogenesis in vivo. Blood vessel density was assessed by counting vessels within the tumor region in five different sections in each tumor. Tumor sections were photographed using Olympus DP70 camera, Olympus BX45 microscope and saved as JEPG format using DP Manager (Olympus). Tumor areas were measured by counting pixels on ImageJ software (NIH). Pixel counts were converted to mm2 to present the number of vessels per unit area. Statistical analyses were done using SAS (Version 9.1.3) in an analysis of variance in a split-plot arrangement of treatments.
A. Representative tumor sections from each treatment group (H & E). HPSE-treated cell surface HSGAG induced a significant increase in intratumor blood vessel (arrow) formation in animals compared to mock or HepIII treatment of HPSE-treated cell surface HSGAG (p<0.0001). Hep III treatment renders the HPSE-degraded fragments inactive, hence abolishes their biological activity. Presence of VEGF did not affect angiogenesis in all three groups (p=0.2-0.8). Notably, inside the tumors, absence of blood vessels, thereby lack of nutrition and oxygen led to areas of necrosis.
B. Bar graph representation of mean blood vessel density with standard deviation plotted on a log scale. Bars with the same letter are not significantly different from each other.
Discussion
In the present study, we have investigated roles of HPSE-degraded cell-surface HS in melanoma tumorigenesis and possible effects on host endothelial system. Our findings suggest that melanoma cell-surface HS isolated by HPSE treatment promotes 1) melanoma migration, and 2) angiogenesis independent of VEGF activity. These results also provide evidence that, in addition to remodeling the ECM and releasing growth factors and chemokines, HPSE contributes to the aggressive phenotype of melanoma by releasing bioactive HS which stimulate melanoma tumorigenesis.
HSPG are recognized as key cell-surface/ECM active biological modulators [3-5, 23, 40]. Their degradation at the level of HS chains by glycosidases has significant regulatory consequences in cancer metastasis [41]. HS present on tumor cells also contain bioactive sequences that may affect tumor-cell phenotype in relation to cell growth and metastasis [5, 20, 21, 40]. It has been established that growth factor binding to HS which leads to mitogenic activity takes place only when definite structural features are present within the HS chains, such as, sulfation at specific positions within a disaccharide (N, 2-O, 3-O, 6-O) by the enzymes mediating HS synthesis within the Golgi apparatus [41]. On the other hand, it has also been shown that besides the modification that occurs in the Golgi during its synthesis and expression, HS can also be structurally and functionally modulated within the extracellular compartment. The two families of mammalian enzymes currently known to modify HS are the endosulfatases (Hsulf-1 and -2) which remove 6-O sulfation on the HS [20, 21] and HPSE, which cleaves HS into small, biologically active fragments, [4, 7, 8, 9, 36, 42-44]. Recently HPSE has also been shown to promote shedding of cell-surface syndecan-1 and modify tumorigenesis [45, 46].
Elevated levels of HPSE are known to be associated with brain-metastatic melanoma [10-12]. The enzymatic activity of HPSE is characterized by specific intrachain HS cleavage of glycosidic bonds with a hydrolase (but not eliminase) type of action that facilitates the release of several protein modulators of cell function, including migration, adhesion, inflammation, angiogenesis, embryogenesis, and metastatic invasion [2, 4, 7, 47]. When over expressed, HPSE increases tumor cell invasiveness in vitro and in vivo settings [10, 12]; conversely, a downregulation of HPSE by anti-sense or siRNA methodologies decreases tumorigenesis [10-12]. However, at higher concentration HPSE can also inhibit tumorigenesis possibly by extensive remodeling of cell-surface HS that interferes with growth factor binding and signaling leading to subsequent inhibition of biological effects [19, 48, 49].
Extensive evidence suggests that cellular function and phenotype are highly influenced by the composition and size of HS chains on HSPG [4, 5, 23, 40]. A cell can respond to its microenvironment in markedly different ways by dynamically regulating HS structure on its cell-surface as insoluble HS and in the ECM as soluble HS [4, 5, 19, 23, 40]. HSPG and HS chains are present on the surface of all eukaryotic cells, including tumor cells. This is valid also for cells that are important for tumor survival, e.g., endothelial-cell junction surrounding a growing tumor, where HS can participate in the process of angiogenesis. This led us to believe that proangiogenic activity of HPSE could partly be due to generation of bio-active fragments by its enzymatic activity. We found that these fragments are indeed active and probably mediate their effects through melanoma autocrine/paracrine factors.
Highly brain-metastatic B16B15b melanoma cells were chosen as a source of HS since they express large amount of it on the cell-surface [36]. The extent of HS degradation on B16B15b by HPSE was assessed by detecting cell-surface HS on FACS analysis. We were able to remove melanoma (B16B15b) cell-surface HS with HPSE treatment in a dose-dependent manner as indicated by the leftward shift in the profile on the X axis (Fig. 1A). The associated protein core due to shed syndecan-1 by direct action of HPSE [45, 46] and other possible protein contamination were removed by pronase digestion during the isolation process. As expected, HPSE digestion generated HS fragments of about 10 kDa (Fig. 1B).
To directly demonstrate whether the B16B15b cell-surface HS were biologically active, we tested their effects on melanoma and endothelioma biological activity. We reasoned that because VEGF is a HS-binding growth factor and is essential for brain-metastasis formation [28], these fragments would participate in VEGF-mediated activities. Isoforms of VEGF can bind VEGF receptors in the absence of HS, but this interaction is enhanced by cellular or exogenous heparin/HS suggesting that heparin/HS on HSPG regulate the interaction of VEGF to VEGF receptor and subsequent biological activity [50-54]. Interestingly, in our melanoma cell system, the presence of VEGF did not influence the biological activities of the HPSE-degraded melanoma cell-surface HS including migration (Fig. 3) and proliferation (Fig. 4) of melanoma in vitro, and angiogenesis in vivo by Matrigel™ plug assay (Fig. 5). We also investigated effects on melanoma migration by adding FGF2, platelet-derived growth factor (PDGF) and interleukin-8 (IL-8) but no difference in cell migration was observed compared to no growth factor control (data not shown). A possibility could be that VEGF bound to endogenous cell-surface HS and/or because the melanoma cells already have autocrine production of VEGF, we did not observe an added effect with exogenously added growth factor [39]. A recent report by Robinson et al. demonstrated that VEGF requires highly sulfated sites on the HS for binding [55]. According to this study, these sites were exposed by enzymatic action of K5-lyase on HS and it retained significant VEGF165 affinity; in contrast, cleavage of HS by heparinases or HPSE severely reduced VEGF165 binding [55]. This may explain the reason we failed to observe any biological effect with exogenously added VEGF along with HPSE-derived HS in melanoma cell activity. However, melanoma cell-surface HS were able to stimulate melanoma migration (Fig. 3) and angiogenesis (Fig. 5) compared to the controls suggesting that these fragments are tumorigenic although they do not affect melanoma cell proliferation (Fig. 4). The enhanced angiogenesis by these fragments were possibly due to signaling by some other heparin-binding growth factor(s) which could either stimulate tumorigenesis or alternatively, could abolish tumor inhibitory signal. In addition, there were no significant differences observed in tumor weight in all treatment groups, hence the increased vascularity seen with the HPSE-generated cell-surface HS tumors were due to its effects on the tumor microenvironment. These findings are further strengthened by the fact that, following Hep III-mediated digestion into smaller fragments, HPSE-degraded HS lose their proangiogenic properties (Fig. 5).
The HPSE-degraded HS did not have any effects on in vitro b.End3 endothelioma cell migration (Fig. 2), or signaling (data not shown) which could be due to tissue-specific HS structural differences present between the systems [56]. Even though, the same sets of disaccharides are present in most tissues, their relative content varies quantitatively in terms of sulfation or epimerization pattern [56]. Attempts to remove endothelioma cell-surface HS by enzymatic degradation of Hep III or HPSE did not alter response to growth factor in this system (data not shown). This could be due to the fact that removal of cell-surface HS was incomplete and remaining quantities of cell-surface HS were sufficient to arbitrate growth factor-mediated signaling which is known to occur [57]. Nonetheless, the in vivo induction of angiogenesis by the melanoma cell HS fragments could also be due to availability of additional support to the endothelial cells directly or indirectly from the tumor cells.
Remodeling of the ECM and BM is vital for a normal embryonic development, wound healing and tumorigenesis. During tumor progression, this turnover is highly controlled and involves the coordinated action of proteases and endoglycosidases [1, 2]. This process not only contributes to angiogenesis and tumor invasion by altering the integrity of the BM/ECM, but also results in the release of HS-binding molecules such as chemokines and proangiogenic growth factors, initiating numerous downstream signaling cascades. While large families of proteases (matrix metalloproteases, aspartic, cysteine, and serine proteases) mediate the cleavage of protein components of the BM/ECM, cleavage of the HS side chains is performed by a limited set of enzymes, notably HPSE [1, 2]. Characterization of these HPSE-degraded melanoma cell-surface HS would potentially be useful. Devising a method that would isolate individual oligosaccharides is required to test for pro-angiogenic/pro-tumorigenic properties of the fragments. Moreover, the design of novel agents targeted against these HS fragments can be an important addition to developing polysaccharide based anti-tumor therapy in melanoma.
Acknowledgments
We thank Dr. William Henk and Mr. Gregory McCormick for assistance with microscopy, Dr. Jane Reiland for helpful discussions, Dr. Daniel Paulsen for pathology reviews, and Michel T Kearney for statistical analyses (LSU-Baton Rouge). This work was supported by grants from N.I.H. (5R0-1 CA86832 and 1R21 CA 103955 to D.M.).
Abbreviations
- DMEM/F12
Dulbecco's modified Eagle's F-12 medium
- BM
basement membrane
- BSA
bovine serum albumin
- EDTA
ethylenediaminetetraacetic acid
- ECM
extracellular matrix
- FGF2
fibroblast growth factor-2
- Hep III
heparitinase III
- HPSE
heparanase
- HS
heparan sulfate glycosaminoglycan chains
- HSPG
heparan sulfate proteoglycans, P/S, penicillin/streptomycin
- VEGF165
vascular endothelial growth factor isoform 165
- VEGFR
VEGF receptor
References
- 1.Sanderson RD, Yang Y, Kelly T, MacLeod V, Dai Y, Theus A. Enzymatic remodeling of heparan sulfate proteoglycans within the tumor microenvironment: Growth regulation and the prospect of new cancer therapies. J Biol Chem. 2005;96:897–905. doi: 10.1002/jcb.20602. [DOI] [PubMed] [Google Scholar]
- 2.Vreys V, David G. Mammalian heparanase: what is the message? J Cell Mol Med. 2007;11:427–452. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iozzo RV. Heparan sulfate proteoglycans: intricate molecules with intriguing functions. J Clin Invest. 2001;108:165–167. doi: 10.1172/JCI13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elkin M, Ilan M, Ishai-Michaeli R, Friedmann Y, Pappo O, Pecker I, Vlodavsky I. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15(9):1661–1663. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 5.Liu D, Shriver Z, Venkataraman G, El Shabrawi Y, Sasisekharan R. Tumor cell surface heparan sulfate as cryptic promoters or inhibitors of tumor growth and metastasis. Proc Natl Acad Sci (USA) 2002;99:568–573. doi: 10.1073/pnas.012578299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakajima M, Irimura T, Nicolson GL. Heparanases and tumor metastasis. J Cell Biochem. 1988;36:157–167. doi: 10.1002/jcb.240360207. [DOI] [PubMed] [Google Scholar]
- 7.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 9.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, Spector L, Pecker I. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 10.Uno F, Fujiwara T, Takata Y, Ohtani S, Katsuda K, Takaoka M, Ohkawa T, Naomoto Y, Nakajima M, Tanaka N. Antisense-mediated suppression of human heparanase gene expression inhibits pleural dissemination of human cancer cells. Cancer Res. 2001;61:7855–7860. [PubMed] [Google Scholar]
- 11.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- 12.Roy M, Reiland J, Murry BP, Chouljenko V, Kousoulas KG, Marchetti D. Antisense-mediated suppression of heparanase gene inhibits melanoma cell invasion. Neoplasia. 2005;7:253–262. doi: 10.1593/neo.04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedmann Y, Vlodavsky I, Aingorn H, Aviv A, Peretz T, Pecker I, Pappo O. Expression of heparanase in normal, dysplastic, and neoplastic human colonic mucosa and stroma: evidence for Its role in colonic tumorigenesis. Am J Pathol. 2000;157:1167–1175. doi: 10.1016/S0002-9440(10)64632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim AW, Xu X, Hollinger EF, Gattuso P, Godellas CV, Prinz RA. Human heparanase-1 gene expression in pancreatic adenocarcinoma. J Gastrointest Surg. 2002;6:167–172. doi: 10.1016/s1091-255x(01)00087-7. [DOI] [PubMed] [Google Scholar]
- 15.Maxhimer JB, Quiros RM, Stewart R, Dowlatshahi K, Gattuso P, Fan M, Prinz RA, Xu X. Heparanase-1 expression is associated with the metastatic potential of breast cancer. Surgery. 2002;132:326–333. doi: 10.1067/msy.2002.125719. [DOI] [PubMed] [Google Scholar]
- 16.Tang W, Nakamura Y, Tsujimoto M, Sato M, Wang X, Kurozumi K, Nakahara M, Nakao K, Nakamura M, Mori I, Kakudo K. Heparanase: a key enzyme in invasion and metastasis of gastric carcinoma. Mod Pathol. 2002;15:593–598. doi: 10.1038/modpathol.3880571. [DOI] [PubMed] [Google Scholar]
- 17.Kelly T, Miao HQ, Yang Y, Navarro E, Kussie P, Huang Y, MacLeod V, Casciano J, Joseph L, Zhan F, Zangari M, Barlogie B, Shaughnessy J, Sanderson RD. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003;63:8749–8756. [PubMed] [Google Scholar]
- 18.Doweck I, Kaplan-Cohen V, Naroditsky I, Sabo E, Ilan N, Vlodavsky I. Heparanase localization and expression by head and neck cancer: correlation with tumor progression and patient survival. Neoplasia. 2006;8:1055–1061. doi: 10.1593/neo.06577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiland J, Kempf D, Roy M, Denkins Y, Marchetti D. FGF2 binding, signaling, and angiogenesis are modulated by heparanase in metastatic melanoma cells. Neoplasia. 2006;8:596–606. doi: 10.1593/neo.06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narita K, Staub J, Chien J, Meyer K, Bauer M, Friedl A, Ramakrishnan S, Shridhar V. HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res. 2006;66:6025–6032. doi: 10.1158/0008-5472.CAN-05-3582. [DOI] [PubMed] [Google Scholar]
- 21.Dai Y, Yang Y, MacLeod V, Yue X, Rapraeger AC, Shriver Z, Venkataraman G, Sasisekharan R, Sanderson RD. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280:40066–40073. doi: 10.1074/jbc.M508136200. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Rao G, Quiros RM, Kim AW, Miao HQ, Brunn GJ, Platt JL, Gattuso P, Prinz RA. In vivo and in vitro degradation of heparan sulfate (HS) proteoglycans by HPR1 in pancreatic adenocarcinomas: loss of cell surface HS suppresses fibroblast growth factor 2-mediated cell signaling and proliferation. J Biol Chem. 2007;282:2363–2373. doi: 10.1074/jbc.M604218200. [DOI] [PubMed] [Google Scholar]
- 23.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 24.Senger DR, Galli SJ, Dvorak AM, Perruzzi CA, Harvey VS, Dvorak HF. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science. 1983;219:983–985. doi: 10.1126/science.6823562. [DOI] [PubMed] [Google Scholar]
- 25.Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 26.Ranieri G, Patruno R, Ruggieri E, Montemurro S, Valerio P, Ribatti D. Vascular endothelial growth factor (VEGF) as a target of bevacizumab in cancer: from the biology to the clinic. Curr Med Chem. 2006;13:1845–1857. doi: 10.2174/092986706777585059. [DOI] [PubMed] [Google Scholar]
- 27.Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS, Ferrara N. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 28.Yano S, Shinohara H, Herbst RS, Kuniyasu H, Bucana CD, Ellis LM, Davis DW, McConkey DJ, Fidler IJ. Expression of vascular endothelial growth factor is necessary but not sufficient for production and growth of brain metastasis. Cancer Res. 2000;60:4959–4967. [PubMed] [Google Scholar]
- 29.Plouet J, Moro F, Bertagnolli S, Coldeboeuf N, Mazarguil H, Clamens S, Bayard F. Extracellular cleavage of the vascular endothelial growth factor 189-amino acid form by urokinase is required for its mitogenic effect. J Biol Chem. 1997;272:13390–13396. doi: 10.1074/jbc.272.20.13390. [DOI] [PubMed] [Google Scholar]
- 30.Gallagher JT. Heparan sulfate: growth control with a restricted sequence menu. J Clin Invest. 2001;108:357–361. doi: 10.1172/JCI13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchetti D. Specific degradation of subendothelial matrix proteoglycans by brain-metastatic melanoma and brain endothelial cell heparanases. J Cell Physiol. 1997;172:334–342. doi: 10.1002/(SICI)1097-4652(199709)172:3<334::AID-JCP7>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 32.Montesano R, Pepper MS, Mohle-Steinlein U, Risau W, Wagner EF, Orci L. Increased proteolytic activity is responsible for the aberrant morphogenetic behavior of endothelial cells expressing the middle T oncogene. Cell. 1990;62:435–445. doi: 10.1016/0092-8674(90)90009-4. [DOI] [PubMed] [Google Scholar]
- 33.McKenzie E, Young K, Hircock M, Bennett J, Bhaman M, Felix R, Turner P, Stamps A, McMillan D, Saville G, Ng S, Mason S, Snell D, Schofield D, Gong H, Townsend R, Gallagher J, Page M, Parekh R, Stubberfield C. Biochemical characterization of the active heterodimer form of human heparanase (Hpa1) protein expressed in insect cells. Biochem J. 2003;373:423–435. doi: 10.1042/BJ20030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pervin A, Gallo C, Jandik KA, Han XJ, Linhardt RJ. Preparation and structural characterization of large heparin-derived oligosaccharides. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 35.Sinnappah-Kang ND, Kaiser AJ, Blust BE, Mrak RE, Marchetti D. Heparanase, TrkC and p75NTR: their functional involvement in human medulloblastoma cell invasion. Int J Oncol. 2005;27:617–626. [PubMed] [Google Scholar]
- 36.Reiland J, Sanderson RD, Waguespack M, Barker SA, Long R, Carson DD, Marchetti D. Heparanase degrades syndecan-1 and perlecan heparan sulfate: functional implications for tumor cell invasion. J Biol Chem. 2004;279:8047–8055. doi: 10.1074/jbc.M304872200. [DOI] [PubMed] [Google Scholar]
- 37.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 38.Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat Med. 2000;6:389–395. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- 39.Menter DG, Herrmann JL, Marchetti D, Nicolson GL. Involvement of neurotrophins and growth factors in brain metastasis formation. Invasion Metastasis. 1994;14:372–384. [PubMed] [Google Scholar]
- 40.Liu D, Shriver Z, Qi Y, Venkataraman G, Sasisekharan R. Dynamic regulation of tumor growth and metastasis by heparan sulfate glycosaminoglycans. Semin Thromb Hemost. 2002:67–78. doi: 10.1055/s-2002-20565. [DOI] [PubMed] [Google Scholar]
- 41.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 42.Kussie PH, Hulmes JD, Ludwig DL, Patel S, Navarro EC, Seddon AP, Giorgio NA, Bohlen P. Cloning and functional expression of a human heparanase gene. Biochem Biophys Res Comm. 1999;261:183–187. doi: 10.1006/bbrc.1999.0962. [DOI] [PubMed] [Google Scholar]
- 43.Parish CR, Coombe DR, Jakobsen KB, Bennett FA, Underwood PA. Evidence that sulphated polysaccharides inhibit tumour metastasis by blocking tumour-cell-derived heparanases. Int J Cancer. 1987;40:511–518. doi: 10.1002/ijc.2910400414. [DOI] [PubMed] [Google Scholar]
- 44.Dempsey LA, Brunn GJ, Platt JL. Heparanase, a potential regulator of cell-matrix interactions. Trends Biochem Sci. 2000;25:349–351. doi: 10.1016/s0968-0004(00)01619-4. [DOI] [PubMed] [Google Scholar]
- 45.Mahtouk K, Hose D, Raynaud P, Hundemer M, Jourdan M, Jourdan E, Pantesco V, Baudard M, De Vos J, Larroque M, Moehler T, Rossi JF, Reme T, Goldschmidt H, Klein B. Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma. Blood. 2007;109:4914–4923. doi: 10.1182/blood-2006-08-043232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, MacLeod V, Miao HQ, Theus A, Zhan F, Shaughnessy JD, Jr, Sawyer J, Li JP, Zcharia E, Vlodavsky I, Sanderson RD. Heparanase enhances syndecan-1 shedding: A novel mechanism for stimulation of tumor growth and metastasis. J Biol Chem. 2007;282:13326–13333. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- 47.McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Br Journ Pharmacol. 2007;151:1–14. doi: 10.1038/sj.bjp.0707182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy, M., Marchetti, D. Unpublished observations.
- 49.Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Heparanase affects adhesive and tumorigenic potential of human glioma cells. Cancer Res. 2003;63:7733–7741. [PubMed] [Google Scholar]
- 50.Gitay-Goren H, Cohen T, Tessler S, Soker S, Gengrinovitch S, Rockwell P, Klagsbrun M, Levi BZ, Neufeld G. Selective binding of VEGF121 to one of the three vascular endothelial growth factor receptors of vascular endothelial cells. J Biol Chem. 1996;271:5519–5523. doi: 10.1074/jbc.271.10.5519. [DOI] [PubMed] [Google Scholar]
- 51.Terman B, Khandke L, Dougher-Vermazan M, Maglione D, Lassam NJ, Gospodarowicz D, Persico MG, Bohlen P, Eisinger M. VEGF receptor subtypes KDR and FLT1 show different sensitivities to heparin and placenta growth factor. Growth Factors. 1994;11:187–195. doi: 10.3109/08977199409046916. [DOI] [PubMed] [Google Scholar]
- 52.Schlessinger J, Lax I, Lemmon M. Regulation of growth factor activation by proteoglycans: what is the role of the low affinity receptors? Cell. 1995;83:357–360. doi: 10.1016/0092-8674(95)90112-4. [DOI] [PubMed] [Google Scholar]
- 53.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 54.Lake AC, Vassy R, Di Benedetto M, Lavigne D, Le Visage C, Perret GY, Letourneur D. Low molecular weight fucoidan increases VEGF165-induced endothelial cell migration by enhancing VEGF165 binding to VEGFR-2 and NRP1. J Biol Chem. 2006;281:37844–37852. doi: 10.1074/jbc.M600686200. [DOI] [PubMed] [Google Scholar]
- 55.Robinson CJ, Mulloy B, Gallagher JT, Stringer SE. VEGF165-binding sites within heparan sulfate encompass two highly sulfated domains and can be liberated by K5 lyase. J Biol Chem. 2006;281:1731–1740. doi: 10.1074/jbc.M510760200. [DOI] [PubMed] [Google Scholar]
- 56.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krufka A, Guimond S, Rapraeger AC. Two hierarchies of FGF-2 signaling in heparin: mitogenic stimulation and high-affinity binding/receptor transphosphorylation. Biochemistry. 1996;35:11131–11141. doi: 10.1021/bi960125+. [DOI] [PubMed] [Google Scholar]
- 58.Marchetti D, Liu S, Spohn WC, Carson DD. Heparanase and a synthetic peptide of heparan sulfate-interacting protein recognize common sites on cell surface and extracellular matrix heparan sulfate. The Journ Biol Chem. 1997;272(25):15891–15897. doi: 10.1074/jbc.272.25.15891. [DOI] [PubMed] [Google Scholar]