Abstract
Recent advances in single-molecule detection, nanotechnology and aptameric sensors hold exciting promise for many potential applications. By functionalizing the surface of a quantum dot (QD) with aptamers which can recognize cocaine, and taking advantage of single-molecule detection and fluorescence resonance energy transfer (FRET) between 605QD and Cy5 and Iowa Black RQ, we develop a single-QD-based aptameric sensor that is capable of sensing the presence of cocaine through both signal-off and signal-on modes. In comparison with the established aptameric sensors, this single-QD-based aptameric sensor has the significant advantages of simple sample preparation, high sensitivity and extremely low sample consumption. With the advance in the development of varieties of aptamers for small molecules, nucleic acids, metal ions and proteins, this single-QD-based aptameric sensor might find wide application in forensic analysis, environmental monitoring and clinic diagnostics.
Simple and sensitive detection of cocaine is very important for law enforcement and clinical diagnostics. However, this has remained a critical technology challenge until the discovery of specific aptamers for cocaine.1–9 Aptamers are single-stranded RNA or DNA molecules with ligand-binding ability, which have been developed to detect a variety of molecular targets including small molecules, nucleic acids, proteins and cancer cells.10–12 So far various aptameric sensors based on colorimetric,1–4 electrochemical5 and bulk fluorescence6–9 assay have been developed for cocaine detection. But these reported aptameric sensors either exhibit relatively low detection limits, or involve complicated sample preparation and large sample consumption. Alternative simple and sensitive aptameric sensors are thus highly desirable.
Quantum dots (QDs) are novel semiconductor nanocrystals with the significant advantages of narrow, tunable and highly stable photoluminescence over organic fluorephores.13–18 QDs have been widely used as fluorescent markers in genomic analysis, immunoassay, fluorescence imaging and drug delivery.13–18 Notably, QDs have shown great promise as fluorescence resonance energy transfer (FRET) donors in various biosensors to homogeneously detect nucleic acids, proteins and enzymatic activity.13 The combination of QDs with single-molecule detection19–23 holds exciting promise for many potential applications. A hallmark of single-molecule detection is the unparalleled ability to obtain the distinct fluorescent signal from individual molecules instead of merely their averages in the ensemble measurements.19–23 Recently single-QD-based FRET has been reported for sensitive detection of DNA24, 25 and real-time RNA-peptide interaction.26 We hypothesize that the combination of single-QD-based FRET and specific aptamers might provide a new approach for sensitive cocaine detection. In this paper, by functionalizing the surface of a QD with aptamers 6 which can recognize cocaine, and taking advantage of single-molecule detection19–23 and fluorescence resonance energy transfer (FRET) 24–26 between 605-nm-emitting QD (605QD) and Cy5 and Iowa Black RQ, we developed a single QD-based aptameric sensor that is capable of sensing the presence of cocaine through both signal-off and signal-on modes.
EXPERIMENTAL SECTION
Sample preparation
We designed a biotinylated oligonucleotide (5’-TCA CAG ATG AGT-Biotin-3’), a Cy5-labeled oligonucleotide (5’-GTC TCC CGA GAT-Cy5-3’), a cocaine aptamer (5’-ACT CAT CTG TGATAT CTC GGG AGA CAA GGA AAA TCC TTC AAT GAA GTG GGT CTC CC-3’), a 5’-Cy5-labeled and 3’-biotinylated oligonucleotide (5’-Cy5-TCA CAG ATG AGT-Biotin-3’), and a 3'-Iowa Black RQ-labeled oligonucleotide (5’-GTC TCC CGA GAT- Iowa Black RQ-3’) as models for the detection of cocaine. All oligonucleotides had been purified by high-performance liquid chromatography when purchased from Integrated DNA Technology Inc. (Coralville, IA). For the signaloff single-QD-based aptamer sensor, the aptamer sandwich hybrids were formed by a biotinylated oligonucleotide, a Cy5-labeled oligonucleotide and a aptamer in 100 mM NaCl, 25 mM Tris acetate, 1 mM MgCl2, pH 8.2 from 60 °C to 4 °C. For the signal-on single-QD-based aptamer sensor, the aptamer sandwich hybrids were formed by a 5’-Cy5-labeled and 3’-biotinylated oligonucleotide, a 3'-Iowa Black RQ-labeled oligonucleotide and a aptamer in 100 mM NaCl, 25 mM Tris acetate, 1 mM MgCl2, pH 8.2 from 60 °C to 4 °C. For the detection of cocaine, the aptamer sandwich hybrids were incubated with cocaine (Sigma-Aldrich) at 30 °C, followed by adding streptavidin-functionalized 605QDs (Quantum Dot Corp., Hayward, CA) to capture the aptamer hybrids through the biotin-streptavidin binding. Finally the solution was subjected to single-molecule detection at the QD concentration of 2.5×10−11M.
Experimental Setup for Single-molecule Detection
An argon laser was used as the excitation light source for 605QD. The 488-nm beam was collimated, reflected by a dichroic mirror (Z488RDC, Chroma Technology Corp, Rockingham, VT), and then focused by an oil immersion 100 × /1.30 NA objective lens (Olympus America, Inc., Melville, NY) on the center of a 50-µm ID capillary; the sample was moved through a laser-focused detection volume by the pressure-driven flow from a syringe pump (Harvard Apparatus, Holliston, MA). Photons emitted from 605QD and Cy5 were collected by the same objective, passed through the first dichroic mirror, followed by a 50 µm pinhole (Melles Griot Co, Irvine, CA), and then separated by second dichroic mirror (645DCLP, Chroma Technology Corp, Rockingham, VT). After separation, the signal emitted from Cy5 was filtered by a band-pass filter (D680/30M, Chroma Technology Corp, Rockingham, VT) and detected by an avalanche photodiode (Model SPCM-AQR-13, EG&G Canada, Vaudreuil, PQ, Canada) in the acceptor channel. At the same time, photons emitted from 605QD was filtered by a band-pass filter (D605/20M, Chroma Technology Corp, Rockingham, VT) and detected by an avalanche photodiode in the donor channel. A program written with Labview (National Instruments, Austin, TX) and a digital counter (National Instruments, Austin, TX) were used to perform data acquisition and on-line data analysis. Fluorescence signals from both donor and acceptor channels were integrated in 1-ms interval for a total running time of 100 s for each experiment. In single-molecule detection, a threshold was used to distinguish single-molecule fluorescence signal from random fluctuation in the background. The threshold value was determined by evaluating data from the control sample. In this study, a threshold of 10 photon counts.ms−1 was set for Cy5, and a threshold of 10 photon counts.ms−1 was set for 605QD.
RESULTS AND DISCUSSION
Principle of signal-off single-QD-based aptameric sensor
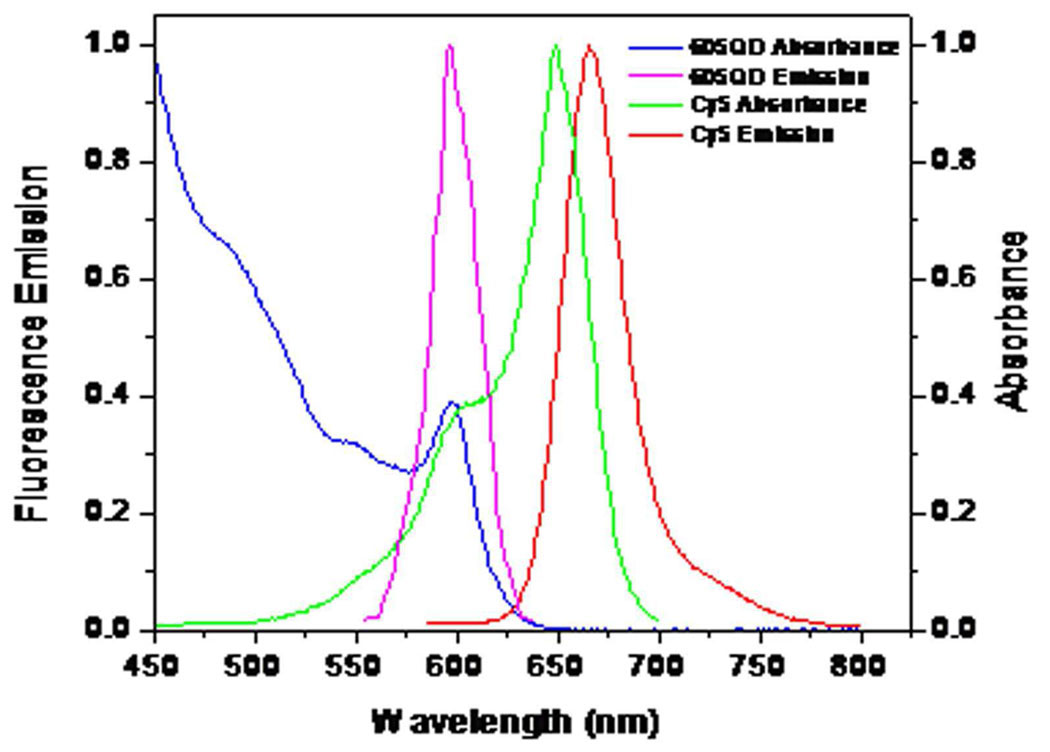
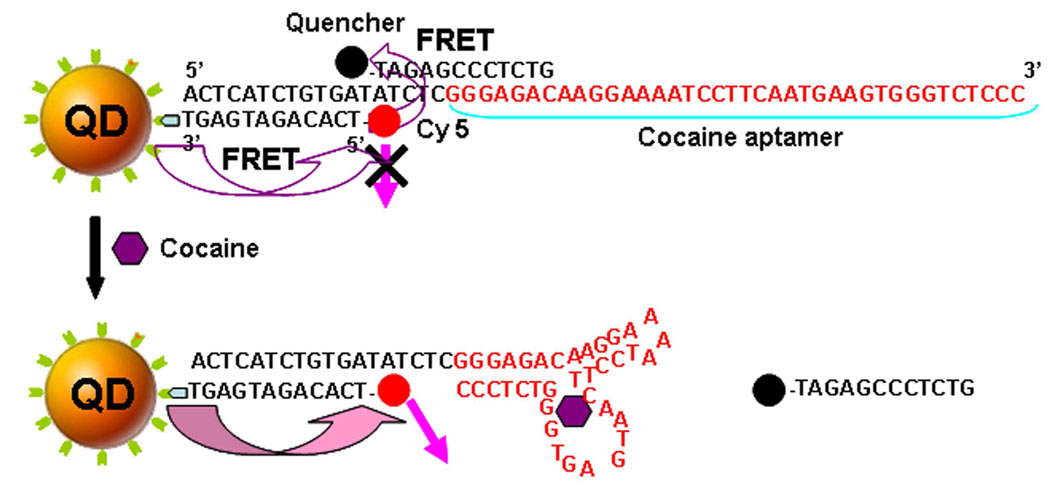
The design of signal-off single-QD-based aptameric sensor is shown in Figure 1. The cocaine aptamer was sandwiched by a biotinylated oligonucleotide and a Cy5-labeled oligonucleotide; this sandwich hybrid was then assembled on the surface of a 605QD through biotin-streptavidin binding to form the 605QD/aptamer/Cy5 complex. In this complex the 605QD served as a FRET donor to transfer energy to Cy5. The selection of 605QD as an energy donor and Cy5 as an acceptor was based on following reasons: (1) There is no cross-talk between the emission spectra of 605QD and that of Cy5; (2) There is no direct excitation of Cy5 at the wavelength of 488 nm (Figure 2); (3) 605QD has a high quantum yield (~0.6); (4) Cy5 has a high extinction coefficient (~250,000 M−1cm−1); The Förster distance (R0) is 69.4 Å for 605QD/Cy5 FRET pair. In addition, a single 605QD can efficiently couple to multiple Cy5-labeled sandwich hybrids; 24–26 therefore 605QD also served as a nanoscaffold to amplify the FRET signals. The nanoscaffold function of 605QD24–26 might created a high local concentration of aptamer, which thermodynamically favoured the formation of more aptamer-cocaine complexes. 27 When this complex was excited by a 488-nm argon laser, Cy5 fluorescence was detected due to FRET between 605QD and Cy5. The presence of cocaine led to the formation of complex structure of a cocaine-aptamer complex,6 which made Cy5-labeled oligonucleotide dissociate from aptamer and 605QD; the subsequent decrease of Cy5 signal due to the absence of FRET between 605QD and Cy5 signified the presence of cocaine (Figure 1).
Figure 1.
Principle of signal-off single QD-based aptameric sensor for cocaine detection. In the absence of cocaine, Cy5 fluorescence was detected due to FRET between 605QD and Cy5. While the presence of cocaine led to the formation of complex structure of a cocaine-aptamer complex, and the subsequent abolishment of FRET between 605QD and Cy5, the decrease of Cy5 signal signified the presence of cocaine.
Figure 2.
The normalized absorption and emission spectra of the 605QD and Cy5. Blue line, absorption spectrum of the 605QD; Magenta line, emission spectrum of the 605QD; Green line, absorption spectrum of Cy5; Red line, emission spectrum of Cy5.
The schematic for the experimental set-up is shown in Figure 3. An argon laser was used as the excitation light source. The 488-nm beam was focused on the center of a 50-µm ID capillary by an oil immersion 100 × /1.30 NA objective; the sample was moved through a laser-focused detection volume at a flow rate of 1.0 µL.min−1 by the pressure-driven flow from a syringe pump. Photons emitted from the 605QD and Cy5 were separated by dichroic mirrors and detected by two avalanche photodiodes (APDs), respectively. The performance of single-QD-based FRET in a microfliudic flow has another significant advantage of improved FRET efficiency, which might even break the FRET limit.28
Figure 3.
Schematic view of the experimental apparatus used for simultaneous detection of the photons emitted from the single 605QD and Cy5. Photons emitted from the 605QD and Cy5 were separated by dichroic mirrors and detected by two avalanche photodiodes (APDs), respectively.
Detection of cocaine with signal-off single-QD-based aptameric sensor
Figure 4A shows the representative trace of fluorescence bursts from 605QD/aptamer/Cy5 complexes; distinct Cy5 bursts were observed with corresponding 605QD bursts, indicating the FRET between 605QD and Cy5. Figure 4B shows the representative trace of fluorescence bursts from 605QD/aptamer complex without Cy5 binding; no Cy5 bursts were observed due to the absence of FRET between 605QD and Cy5. Moreover, in the 605QD/aptamer/Cy5 complexes the Cy5 burst counts increased linearly as a function of increasing Cy5-to-605QD ratio (Figure 4C); even one copy difference of Cy5-labeled oligonucleotide bound to 605QD can be distinguished, suggesting the high sensitivity of this single-QD-based aptameric sensor. In the presence of cocaine, its binding to the aptamer triggered the release of Cy5-labeled oligonucleotide from 605QD and consequently the abolishment of FRET between 605QD and Cy5; as a result, the Cy5 burst counts decreased as a function of increasing cocaine (Figure 4D). The detection limit can reach 0.5 µM. This is at least two orders of magnitude improvements over the conventional nanoparticle-based colorimetric method, where a large amount of nanoparticles and relatively high concentration of cocaine are required to produce observable aggregation or disassembly of nanoparticles.1–2 Compared to established aptamer sensors,1–9 one major advantage of this single-QD-based aptameric sensor lies in its ability to measure extremely low-concentration molecules down to a single particle with near-zero background noise (Figure 4A and B), which significantly improve the detection sensitivity of this single QD-based aptameric sensor.
Figure 4.
Detection of cocaine with signal-off single QD-based aptameric sensor. (A) Representative traces of fluorescence bursts from 605QD/aptamer/Cy5 complexes. The top panel shows the Cy5 fluorescence signals; the bottom panel shows the 605QD fluorescence signals. (B) Representative traces of fluorescence bursts from 605QD/aptamer complexes without Cy5 binding. The top panel shows the Cy5 fluorescence signals; the bottom panel shows the 605QD fluorescence signals. (C) Variance of Cy5 burst counts as a function of increasing Cy5-to-605QD ratio in 605QD/aptamer/Cy5 complexes. (D) Variance of Cy5 burst counts as a function of increasing cocaine concentration. The Cy5-to-605QD ratio is 18:1. Error bars show the standard deviation of three experiments.
Principle of signal-on single-QD-based aptameric sensor
We have further developed a signal-on single-QD-based aptameric sensor for cocaine. As shown in Figure 5, the cocaine aptamer was sandwiched by a 5’-Cy5-labeled and 3’-biotinylated oligonucleotide and a quencher of Iowa Black RQ-labeled oligonucleotide; this sandwich hybrid was then assembled on a 605QD through biotin-streptavidin binding to form 605QD/aptamer/Cy5/Iowa Black RQ complex. Even though FRET could potentially occur between 605QD and Cy5 in this 605QD/apatmer/Cy5/Iowa Black RQ complex when it was excited by a 488-nm argon laser, the Cy5 fluorescence would be quenched by the nearby Iowa Black RQ due to FRET between Cy5 and Iowa Black RQ; therefore, Cy5 would be in the fluorescence ‘OFF’ state. The addition of cocaine induced the release of the Iowa Black RQ-labeled oligonucleotide from the aptamer and the 605QD; subsequently the Cy 5 fluorescence would be activated to ‘ON’ state (Figure 5). In comparison with the signal-off sensor, the signal-on sensor is much more straightforward.
Figure 5.
Principle of signal-on single QD-based aptameric sensor for cocaine detection. In the absence of cocaine, Cy5 was in the fluorescence ‘OFF’ state due to FRET between Cy5 and Iowa Black RQ. While the presence of cocaine led to the formation of complex structure of a cocaine-aptamer complex, and the subsequent abolishment of FRET between Cy5 and Iowa Black RQ, the Cy5 fluorescence was activated to ‘ON’ state.
Detection of cocaine with signal-on single-QD-based aptameric sensor
Figure 6A shows the comparison of Cy5 burst counts from the 605QD/aptamer/Cy5 complex and the 605QD/aptamer/Cy5/Iowa Black RQ complex. Without the quencher of Iowa Black RQ, the Cy5 burst counts increased as a function of the increasing Cy5-to-605QD ratio. In contrast, no change in Cy5 burst counts was observed in the presence of Iowa Black RQ despite the increasing Cy5-to-605QD ratio, suggesting the complete quenching of Cy5 fluorescence by Iowa Black RQ. It was worth noting that Iowa Black RQ not only quenched Cy5, but also quenched 605QD to some extent, which was confirmed by the decrease of 605QD burst counts (Figure 6B). In addition, the selection of Cy5-to-605QD ratio is very important for detection of cocaine with signal-on single-QD-based aptameric sensor, because the Cy5-to-605QD ratio might significantly influence the dynamic range of cocaine detection. To obtain the best dynamic range, the Cy5-to-605QD ratio of 12:1 was used in following experiments.
Figure 6.
Detection of cocaine with signal-on single QD-based aptameric sensor. (A) Variance of Cy5 burst counts as a function of increasing Cy5-to-605QD ratio in 605QD/aptamer/Cy5 complexes (○) and 605QD/apatmer/Cy5/Iowa Black RQ complexes (●). (B) Variance of 605QD burst counts as a function of increasing Cy5-to-605QD ratio in 605QD/aptamer/Cy5 complexes (○) and 605QD/apatmer/Cy5/Iowa Black RQ complexes (●). (C) Variance of Cy5 burst counts as a function of increasing cocaine concentration. The Cy5-to-605QD ratio is 12:1. Error bars show the standard deviation of three experiments.
In the presence of cocaine, its binding to the aptamer triggered the release of Iowa Black RQ-labeled oligonucleotide from the aptamer and 605QD, and consequently the Cy5 fluorescence became detectable; the increase of Cy5 signal signified the presence of cocaine. As shown in Figure 6C, the Cy5 burst counts increased as a function of increasing cocaine concentration; the detection limit of the signal-on single-QD aptameric sensor was of the same order of magnitude as that of the signal-off sensor. It should be noted that the main challenge with an aptameric sensor for cocaine is that the detection sensitivity is limited by their relatively low association constant. 29 The reported best detection limits are 10 µM of cocaine for electrochemical method,5 5.0 µM for an autonomous aptamer-based enzymatic assay,8 10 µM for bulk fluorescence assay,6–7 and 0.5 µM for aptamer-based colorimetric probes.4 The detection limit of single-QD-based aptameric sensor is comparable with those reported aptameric sensors, but does not involve the complicated sample preparation and large sample consumption.
It was worth noting that most analytical labs currently utilized mass-spectral analysis to detect cocaine,30, 31 which involve the use of stable isotopes as internal standards or the available highresolution mass spectra as a reference, long-time sample preparation and application of expensive mass-spectral instruments. In contrast, this single-QD-based aptameric sensor takes advantage of a simple ‘mix and detection’ assay, and has potential to be applied for rapid point-of-care testing and field detection of cocaine. The application of this single-QD-based aptameric sensor to clinical and forensic analysis of cocaine is now underway in our laboratory.
CONCLUSIONS
In conclusion, by functionalizing the surface of a QD with aptamers which can recognize cocaine, and taking advantage of single-molecule detection and bi-FRET between 605QD and Cy5 and Iowa Black RQ, we have developed a single QD-based aptameric sensor that is capable of sensing the presence of cocaine through both a signal-off and signal-on modes. In comparison with established aptameric sensors, this single-QD-based aptameric sensor has the significant advantages of simple sample preparation, high sensitivity and extremely low sample consumption. With the advance in the development of varieties of aptamers for small molecules, nucleic acids, metal ions and proteins, 29, 32 this single-QD-based aptameric sensor might find wide application in forensic analysis, environmental monitoring and clinic diagnostics.
ACKNOWLEDGMENT
This work was supported by NIH (GM08153).
REFERENCES
- 1.Liu J, Lu Y. J. Am. Chem. Soc. 2007;129:8634–8643. doi: 10.1021/ja072075+. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Lu Y. Angew. Chem. Int. Ed. 2006;45:90–94. [Google Scholar]
- 3.Zhang J, Wang L, Pan D, Song S, Boey FYC, Zhang H, Fan C. Small. 2008;4:1196–1200. doi: 10.1002/smll.200800057. [DOI] [PubMed] [Google Scholar]
- 4.Stojanovic MN, Landry DW. J. Am. Chem. Soc. 2002;124:9678–9679. doi: 10.1021/ja0259483. [DOI] [PubMed] [Google Scholar]
- 5.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. J. Am. Chem. Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 6.Stojanovic MN, Parada P, de, Landry DW. J. Am. Chem. Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 7.Stojanovic MN, de Parada P, Landry DW. J. Am. Chem. Soc. 2000;122:11547–11548. doi: 10.1021/ja0022223. [DOI] [PubMed] [Google Scholar]
- 8.Shlyahovsky B, Di L, Weizmann Y, Nowarski R, Kotler M, Willner I. J. Am. Chem. Soc. 2007;12:3814–3815. doi: 10.1021/ja069291n. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Lee JH, Lu Y. Anal. Chem. 2007;79:4120–4125. doi: 10.1021/ac070055k. [DOI] [PubMed] [Google Scholar]
- 10.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 11.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 12.Huang YF, Chang HT, Tan W. Anal. Chem. 2008;80:567–572. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 13.Medintz IL, Uyeda HT, Goldman E, Mattoussi H. Nat. Mater. 2005;4:437–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 14.Gill R, Zayats M, Willner I. Angew. Chem. Int. Ed. 2008;47:7602–7625. doi: 10.1002/anie.200800169. [DOI] [PubMed] [Google Scholar]
- 15.Michalet X, Pinaud FF, Bentolila JL, Tsay AM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan WCW, Nie SM. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 17.Alivisatos AP. Nat. Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 18.Somers RC, Bawendi MG, Nocera DG. Chem. Soc. Rev. 2007;36:579–591. doi: 10.1039/b517613c. [DOI] [PubMed] [Google Scholar]
- 19.Xu XH, Yeung ES. Science. 1997;275:1106–1109. doi: 10.1126/science.275.5303.1106. [DOI] [PubMed] [Google Scholar]
- 20.Hsin TM, Yeung ES. Angew. Chem. Int. Ed. 2007;46:8032–8035. doi: 10.1002/anie.200702348. [DOI] [PubMed] [Google Scholar]
- 21.Sharonov A, Hochstrasser RM. Biochemistry. 2007;46:7963–7972. doi: 10.1021/bi700505h. [DOI] [PubMed] [Google Scholar]
- 22.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 23.Zhang CY, Johnson LW. J. Am. Chem. Soc. 2008;130:3750–3751. doi: 10.1021/ja711493q. [DOI] [PubMed] [Google Scholar]
- 24.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Nat. Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 25.Pons T, Medintz IL, Wang X, English DS, Mattoussi H. J. Am. Chem. Soc. 2006;128:15324–15331. doi: 10.1021/ja0657253. [DOI] [PubMed] [Google Scholar]
- 26.Zhang CY, Johnson LW. Anal. Chem. 2007;79:7775–7781. doi: 10.1021/ac071225w. [DOI] [PubMed] [Google Scholar]
- 27.Lytton-Jean AK, Mirkin CA. J. Am. Chem. Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 28.Zhang CY, Johnson LW. Angew. Chem. Int. Ed. 2007;46:3482–3485. doi: 10.1002/anie.200604861. [DOI] [PubMed] [Google Scholar]
- 29.Hamula CA, Guthrie JW, Zhang H, Li XF, Le XC. Trends Anal. Chem. 2006;7:681–691. [Google Scholar]
- 30.Cone EJ, Hillsgrove M, Darwin WD. Clinic. Chem. 1994;40:1299–1305. [PubMed] [Google Scholar]
- 31.Chinn DM, Crouch DJ, Peat MA, Finkle BS, Jennison TA. J. Anal. Toxicol. 1980;4:37–42. doi: 10.1093/jat/4.1.37. [DOI] [PubMed] [Google Scholar]
- 32.Brody EN, Gold L. Rev. Mol. Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]