Abstract
The development of multidrug resistance (MDR) to chemotherapy remains a major challenge in the treatment of cancer. Resistance exists against every effective anti-cancer drug and can develop by multiple mechanisms. These mechanisms can act individually or synergistically, leading to multidrug resistance (MDR), in which the cell becomes resistant to a variety of structurally and mechanistically unrelated drugs in addition to the drug initially administered. Although extensive work has been done to characterize MDR mechanisms in vitro, the translation of this knowledge to the clinic has not been successful. Therefore, identifying genes and mechanisms critical to the development of MDR in vivo and establishing a reliable method for analyzing highly homologous genes from small amounts of tissue is fundamental to achieving any significant enhancement in our understanding of multidrug resistance mechanisms and could lead to treatments designed to circumvent it. In this study, we use a previously established database that allows the identification of lead compounds in the early stages of drug discovery that are not ABC transporter substrates. We believe this can serve as a model for appraising the accuracy and sensitivity of current methods used to analyze the expression profiles of ABC transporters. We found two platforms to be superior methods for the analysis of expression profiles of highly homologous gene superfamilies. This study also led to an improved database by revealing previously unidentified substrates for ABCB1, ABCC1 and ABCG2, transporters that contribute to multidrug resistance.
Keywords: Multidrug resistance, ABC transporters, gene expression profiling, diagnosis, personalized medicine, qRT-PCR
Introduction
For many years, multidrug resistance (MDR) has been explained solely as the result of ABCB1 over-expression in a tumor (1–3). This transporter, called P-glycoprotein, was found to be expressed in Chinese hamster ovary cells selected for colchicine resistance. The authors discovered that these cells displayed resistance to a variety of structurally and mechanistically unrelated drugs in addition to colchicine (1). Human P-glycoprotein, the product of the MDR1 or ABCB1 gene, was subsequently shown to confer MDR on drug-sensitive cells (2). More than thirty years later, 14 additional ABC transporters (ABCA2, ABCA3, ABCB1, ABCB4, ABCB5, ABCB11, ABCC1 6, ABCC11 12, and ABCG2) have been associated with drug resistance (4, 5). Of these, ABCB1 (3), ABCC1 (6) and ABCG2 (7) have been the most extensively studied. Yet attempts to translate these transporters into clinical targets have so far been unsuccessful, as evidenced by the failure of trials to modulate ABCB1 expression (5) and the disputed role in vivo of ABC transporters in MDR (5, 8, 9). Among multiple factors, conflicting reports on the role of ABC transporters in the clinic can be explained by the technological limitations of high-throughput gene expression profiling platforms to precisely detect individual genes in a highly homologous gene superfamily such as the ABC transporter superfamily (10). Several recent studies suggest that more than 25 ABC transporters can be involved in chemotherapy-induced resistance (11–16), compounding the challenge of accurate gene expression profiling.
Identifying ABC gene signatures in specific cancers has the potential to improve chemotherapy by offering clinicians the power to predict patient response a priori and avoid administering toxic therapies to patients unlikely to benefit from them. In a previous study, our laboratory utilized qRT-PCR using SYBR Green chemistry to study the expression profile of the 48 human ABC transporters in the NCI-60 panel (16). Correlations were then drawn between these gene expression profiles (16) and the growth inhibitory profiles of 1,429 candidate anticancer drugs tested against the NCI-60 panel (17). This database allowed the identification of lead compounds in the early stages of drug development that are not ABC transporter substrates (16). It also revealed molecules with collateral sensitivity, whose activity is potentiated, rather than antagonized, by ABC transporters (16).
We thus chose to use our previously established database as a model to appraise the accuracy and sensitivity of high-throughput qRT-PCR methods using TaqMan chemistry to analyze the expression profiles of ABC transporters. TaqMan-based qRT-PCR is more sensitive and has the ability to detect genes with a greater specificity than SYBR Green-based qRT-PCR. This is of particular interest for the detection of ABC transporter genes. We found two TaqMan qRT-PCR platforms, based on micro- and nano-fluidic systems, to be superior methods for the analysis of expression profiles of highly homologous gene superfamilies. The wide dynamic range and the sensitivity of TaqMan qRT-PCR in addition to the high-throughput of these platforms make them more applicable to clinical use. Moreover, our study resulted in an improved database by revealing previously unidentified substrates for ABCB1, ABCC1 and ABCG2, transporters that contribute to multidrug resistance.
Materials and Methods
Cell lines
HEK293 cells stably transfected with either empty pcDNA3.1 vector (pcDNA-HEK293) or pcDNA3.1 containing ABCG2 coding arginine 482 (R482-HEK293) or ABCB1-tranfected MDR-19 cells were maintained in Eagle’s MEM (Gibco, Invitrogen), supplemented with 10% FCS, 100 units of penicillin/streptomycin/mL and 2 mg/mL G418 at 37°C in 5% CO2 humidified air (18). 80 μg/mL G418 was added to the cell culture medium for parental pcDNA-HEK293 and MRP1-transfected HEK293 cells (19).
Preparation of total RNA
Total RNA from 59 of the 60 cancer cell lines was prepared and provided by the Developmental Therapeutics Program (DTP) (for details, see http://www.dtp.nci.nih.gov/branches/btb/ivclsp.html). Total RNA for MDA-N was unavailable at DTP. RNA was quantitated using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE). The integrity of the RNA samples was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA) and then stored at −80°C.
Reverse transcription
Synthesis of cDNA from 1 μg total RNA in a 20 μL reaction volume was carried out using the High Capacity cDNA kit with RNAse inhibitor (Applied Biosystems, Foster City, CA) as per the manufacturer’s instructions. The reverse transcription conditions were as follows: 10 minutes at 25°C, 120 minutes at 37°C, 5 seconds at 85°C. Following reverse transcription, cDNA was stored at 4°C.
TaqMan qRT-PCR microfluidic platform (TaqMan Low Density Array (TLDA))
ABC transporter expression levels were measured using custom-made Taqman Low Density Arrays (Applied Biosystems, Foster City, CA). cDNA was mixed with 2X TaqMan Universal PCR Master Mix (Applied Biosystems), loaded on the TLDA card, and run on an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) as per the manufacturer’s instructions.
Correlation of 48 ABC transporter gene expression profiles with three gene expression detection systems
Microarray data for the ABC transporters was obtained from the following site: http://discover.nci.nih.gov/cellminer/datasets.do (17, 20), and the SYBR Green expression profiles were previously reported (16). For the microarray data, in cases where multiple probe sets were reported for the same gene, the Affymetrix probe set yielding the highest average value for all NCI-60 samples was selected. The microarray data was normalized using GC Robust Multi-array Average (GCRMA). SYBR Green values were mean-centered and multiplied by 1 to indicate expression values with reference to the mean expression of each ABC transporter across the 60 cell lines. TLDA values were median-centered as described in Supplemental Figure 1 and Supplemental Table 1. Pearson correlations were calculated from SYBR Green and TLDA normalized data sets expressed in log 2, Supplemental Tables 2 and 3 (note both worksheets).
Cytotoxicity Assay
Sensitivities of cell lines to various chemicals were examined using the Cell-Counting Kit (CCK) technique as detailed previously (21). Cells were plated at a density of 3000 cells/well in 96 well plates containing 100 μl of culture medium. After 24 h incubation at 37 C, drugs were added to wells to a final volume of 200 μl per well and incubated for an additional 72 h. CCK reagent was then added to each well and incubated for 4 h before reading at a wavelength of 450 nm. IC50 values were calculated from dose-response curves obtained from at least three independent experiments.
TaqMan qRT-PCR nanofluidic platform: Linear amplification and sample preparation
1.25 μl of reverse transcribed cDNA from each sample was preamplified with the use of the Applied Biosystems PreAmplification Kit. The samples were preamplified for 14 cycles and then diluted 1:5 with TE Buffer. The BioMark 48.48 Dynamic Array was primed to close the interface valve and to prevent premature mixing. Samples were pipetted into each of the 48 inputs and the TaqMan assays purchased from Applied Biosystems were pipetted into the assay inputs. The array was placed on the NanoFlex IFC controller (Fluidigm, South San Francisco, CA) and loaded. 2.25 μl of the preamplified cDNA was loaded into the Dynamic Array along with 2.5 μl of Applied Biosystems 2x Master Mix and 0.25 μl Fluidigm Loading Agent. Standard 95 C denaturing and 60 C annealing conditions were used for performing the real time PCR experiments on the BioMark instrument.
Results
Comparison of ABC transporter expression profiles derived from oligonucleotide microarrays, SYBR Green qRT-PCR, and the TaqMan qRT-PCR microfluidic platform
We compared the expression profiles of ABC transporters in the NCI-60 panel obtained from three distinct profiling platforms to identify the best technology for transcript analysis. Gene expression profiling of the NCI-60 panel using microarrays was reported previously (17, 20), and the ABC transporter gene expression profiles in this cancer cell line panel using SYBR Green quantitative real-time PCR (qRT-PCR) were also published (16). Here, we assess a high-throughput qRT-PCR microfluidic platform, the TaqMan Low Density Array (TLDA), using TaqMan chemistry (Suppl. Table 4A).
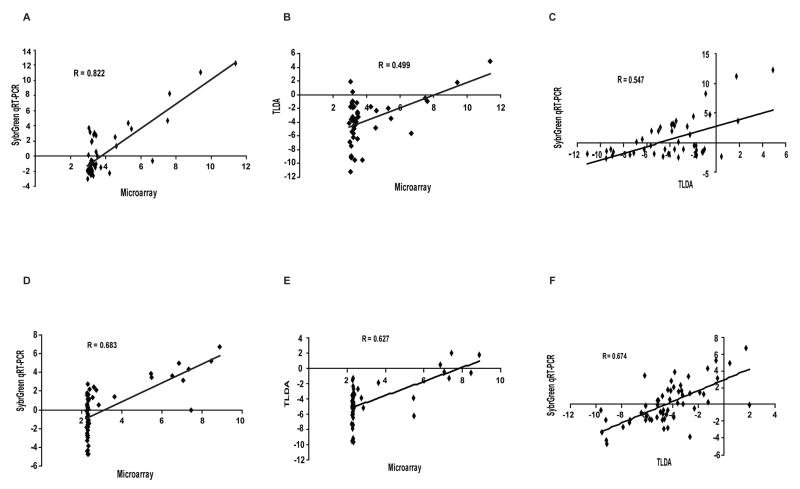
The gene expression profiles for each of the 48 ABC transporters in the NCI-60 cell lines derived from the three different technologies were compared following normalization of the data (Table 1). The data indicate that the latest technology, the TLDA, provides the greatest sensitivity, accuracy, and precision in gene expression profiling (Figure 1). Both qRT-PCR technologies provided a larger dynamic range than microarray (Figure 1A, 1B, 1D and 1E). In addition, the dynamic range for TLDA was found to be larger than that of the SYBR Green method in the analysis of ABCB1 gene expression (Figure 1A and B). When comparing the SYBR Green method directly with TLDA, the expression patterns for ABCB1 (Figure 1C), and to a lesser extent for ABCG2 (Figure 1F) using SYBR Green were found to be less scattered than those obtained using TLDA. Those observations suggest that TLDA provides more sensitivity, yielding a larger dynamic range of measurement, and is the technology best suited for accurately quantitating individual genes in the ABC transporter superfamily in a high-throughput layout.
Table 1.
Comparison of ABC transporter gene expression profiles in the NCI-60 obtained from three technologies*
|
Microarray vs. SYBR Green | ||
|---|---|---|
| r | p | |
| ABCB5 | 0.842 | 6.109E-17 |
| ABCB1 | 0.822 | 1.544E-15 |
| ABCC11 | 0.756 | 4.282E-12 |
| ABCC2 | 0.755 | 5.152E-12 |
| ABCD3 | 0.721 | 1.245E-10 |
| ABCC3 | 0.694 | 1.107E-09 |
| ABCG2 | 0.683 | 2.506E-09 |
| ABCD1 | 0.655 | 1.788E-08 |
| ABCF1 | 0.653 | 2.132E-08 |
| ABCA8 | 0.615 | 2.230E-07 |
| ABCC4 | 0.595 | 6.588E-07 |
| ABCA3 | 0.572 | 2.229E-06 |
| ABCB6 | 0.569 | 2.634E-06 |
| ABCA12 | 0.548 | 6.946E-06 |
| ABCF2 | 0.540 | 1.033E-05 |
| ABCA5 | 0.516 | 2.823E-05 |
| ABCB10 | 0.463 | 2.254E-04 |
| ABCA1 | 0.441 | 4.754E-04 |
| ABCC7 | 0.437 | 5.369E-04 |
| ABCB2 | 0.432 | 6.432E-04 |
| ABCG1 | 0.420 | 9.412E-04 |
| ABCC5 | 0.414 | 0.001 |
| ABCB9 | 0.406 | 0.001 |
| ABCC1 | 0.381 | 0.003 |
| ABCE1 | 0.333 | 0.010 |
| ABCA7 | 0.319 | 0.014 |
| ABCG5 | 0.280 | 0.032 |
| ABCC9 | 0.276 | 0.034 |
| ABCD2 | 0.274 | 0.036 |
| ABCC6 | 0.264 | 0.044 |
| ABCB7 | 0.259 | 0.048 |
| ABCD4 | 0.207 | 0.116 |
| ABCA2 | 0.199 | 0.131 |
| ABCB3 | 0.192 | 0.145 |
| ABCA10 | 0.156 | 0.237 |
| ABCC8 | 0.134 | 0.310 |
| ABCF3 | 0.113 | 0.395 |
| ABCB8 | 0.090 | 0.497 |
| ABCG8 | 0.013 | 0.923 |
| ABCA6 | −0.024 | 0.858 |
| ABCC10 | −0.066 | 0.621 |
| ABCB11 | −0.072 | 0.590 |
| ABCG4 | −0.072 | 0.587 |
| ABCB4 | −0.111 | 0.404 |
| ABCA4 | −0.113 | 0.393 |
| ABCA9 | −0.159 | 0.228 |
|
Microarray vs. TLDA | ||
| r | p | |
|
| ||
| ABCC2 | 0.88 | 1.01E-19 |
| ABCB6 | 0.85 | 7.47E-17 |
| ABCB7 | 0.80 | 8.34E-14 |
| ABCD1 | 0.79 | 1.55E-13 |
| ABCC1 | 0.79 | 2.52E-13 |
| ABCA1 | 0.76 | 3.62E-12 |
| ABCF1 | 0.71 | 5.53E-10 |
| ABCC4 | 0.70 | 1.21E-09 |
| ABCC3 | 0.69 | 1.57E-09 |
| ABCC5 | 0.68 | 5.26E-09 |
| ABCB10 | 0.67 | 9.56E-09 |
| TAP1 | 0.65 | 2.58E-08 |
| ABCG2 | 0.63 | 1.37E-07 |
| ABCD3 | 0.62 | 1.92E-07 |
| ABCA8 | 0.58 | 1.65E-06 |
| ABCF2 | 0.58 | 1.78E-06 |
| TAP2 | 0.57 | 2.63E-06 |
| ABCC10 | 0.56 | 5.53E-06 |
| ABCA3 | 0.50 | 6.28E-05 |
| ABCB1 | 0.50 | 6.70E-05 |
| ABCF3 | 0.47 | 1.84E-04 |
| ABCB9 | 0.47 | 2.02E-04 |
| ABCA7 | 0.42 | 0.001 |
| ABCB5 | 0.41 | 0.001 |
| ABCA4 | 0.40 | 0.002 |
| ABCA5 | 0.40 | 0.002 |
| ABCA2 | 0.38 | 0.003 |
| ABCA12 | 0.38 | 0.003 |
| ABCE1 | 0.36 | 0.006 |
| ABCC11 | 0.33 | 0.011 |
| CFTR | 0.32 | 0.014 |
| ABCC9 | 0.29 | 0.027 |
| ABCC6 | 0.24 | 0.068 |
| ABCG1 | 0.22 | 0.101 |
| ABCD4 | 0.04 | 0.774 |
| ABCD2 | 0.03 | 0.801 |
| ABCA6 | 0.00 | 0.973 |
| ABCA10 | −0.01 | 0.938 |
| ABCB8 | −0.05 | 0.709 |
| ABCB4 | −0.06 | 0.661 |
| ABCG4 | −0.10 | 0.454 |
| ABCA9 | −0.13 | 0.329 |
| ABCG5 | −0.15 | 0.273 |
| ABCB11 | −0.19 | 0.147 |
| ABCG8 | −0.20 | 0.124 |
| ABCC8 | −0.21 | 0.115 |
|
TLDA vs. SYBR Green | ||
| r | p | |
|
| ||
| ABCA3 | 0.88 | 6.21E-20 |
| ABCC2 | 0.85 | 1.84E-17 |
| ABCA8 | 0.78 | 2.23E-13 |
| ABCD1 | 0.72 | 1.48E-10 |
| ABCB9 | 0.70 | 6.17E-10 |
| ABCC3 | 0.69 | 1.26E-09 |
| ABCA5 | 0.68 | 4.37E-09 |
| ABCG2 | 0.67 | 4.79E-09 |
| ABCD3 | 0.60 | 4.92E-07 |
| ABCB1 | 0.55 | 7.39E-06 |
| ABCC6 | 0.54 | 1.14E-05 |
| ABCF1 | 0.52 | 2.32E-05 |
| ABCB6 | 0.50 | 4.85E-05 |
| ABCG1 | 0.49 | 7.88E-05 |
| ABCA12 | 0.47 | 1.53E-04 |
| ABCB10 | 0.47 | 1.72E-04 |
| ABCB4 | 0.46 | 2.75E-04 |
| ABCB5 | 0.43 | 6.14E-04 |
| ABCB2 | 0.43 | 7.71E-04 |
| ABCC9 | 0.42 | 8.82E-04 |
| ABCD4 | 0.42 | 9.15E-04 |
| ABCC4 | 0.41 | 0.001 |
| ABCC11 | 0.41 | 0.001 |
| ABCE1 | 0.40 | 0.002 |
| ABCC5 | 0.40 | 0.002 |
| ABCC1 | 0.40 | 0.002 |
| ABCA7 | 0.39 | 0.003 |
| ABCA1 | 0.38 | 0.003 |
| ABCA13 | 0.34 | 0.008 |
| ABCF2 | 0.31 | 0.015 |
| ABCA4 | 0.31 | 0.016 |
| ABCB7 | 0.31 | 0.016 |
| ABCC7 | 0.31 | 0.017 |
| ABCA2 | 0.24 | 0.064 |
| ABCC12 | 0.18 | 0.164 |
| ABCA10 | 0.18 | 0.167 |
| ABCA6 | 0.16 | 0.224 |
| ABCA9 | 0.14 | 0.296 |
| ABCG4 | 0.12 | 0.353 |
| ABCC8 | 0.10 | 0.460 |
| ABCB3 | 0.08 | 0.531 |
| ABCF3 | 0.07 | 0.575 |
| ABCG8 | −0.06 | 0.637 |
| ABCD2 | −0.07 | 0.622 |
| ABCB11 | −0.09 | 0.509 |
| ABCB8 | −0.09 | 0.500 |
| ABCC10 | −0.09 | 0.479 |
| ABCG5 | −0.22 | 0.096 |
The Pearson correlation (r) was calculated for each comparison, and the p-value (p) was determined using Fisher’s Z-transform.
Figure 1. Correlation of gene expression data from three distinct platforms.
Expression profiles for ABCB1 across all 60 cell lines were compared between: (A) SYBR Green and microarray; (B) TLDA and microarray; (C) TLDA and SYBR Green. Identical comparisons were performed for ABCG2 expression profiles as described in the Supplement: (D) SYBR Green and microarray; (E) TLDA and microarray; (F) TLDA and SYBR Green. The data show that TLDA provides more sensitivity, yielding a larger dynamic range of measurement. The coefficient of correlation is given for each comparison.
Evaluation of a TaqMan qRT-PCR nanofluidic platform
We investigated the use of a TaqMan-based qRT-PCR nanofluidic platform or dynamic array for assessing ABC transporter gene expression profiles in the NCI-60 panel. This platform, the BioMark 48.48 Dynamic Array, requires much smaller quantities of reagent than other approaches, requires fewer pipetting steps, is less labor intensive, and most important, uses nanoscale reaction mixtures (22).
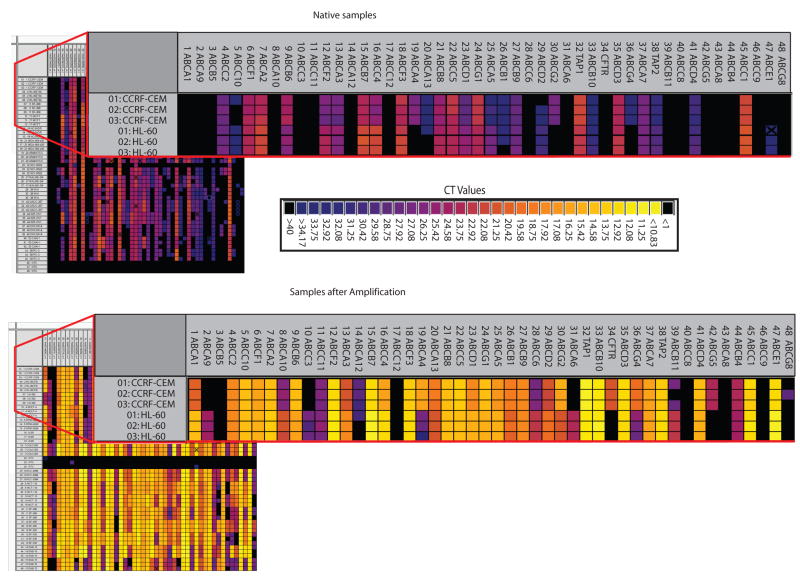
This nanofluidic qRT-PCR platform requires a linear amplification (pre-amplification) of the samples prior to gene expression analysis. A representative group of 15 cancer cells from the NCI-60 panel was analyzed in native state (without the pre-amplification step) and pre-amplified to determine the necessity for linear amplification and reveal any bias related to pre-amplification. Each group was evaluated against the assays for the 48 ABC transporters (Figure 2). Heat maps depicting gene expression across the 48 ABC transporters for each sample were prepared. The native samples showed no or low expression of most of the ABC transporters (Figure 2). Cycle Threshold (CT) values near 28 represent a single copy number, and values greater than 28 are not reproducible using the BioMark platform. Thus, correlations for these 15 samples comparing the gene-expression patterns for the native and pre-amplified samples were extremely low due to the high CT values obtained for the native samples (data not shown). Subsequently, all other samples were pre-amplified using the Applied Biosystems protocol (Suppl. Table 4B).
Figure 2. Comparison of gene expression for the 48 ABC transporters from native and pre-amplified samples using the BioMark 48.48 Dynamic Array.
A representative group of 15 cancer cell lines from the NCI- 60 panel was analyzed, in triplicate, native (without the pre-amplification step) and pre-amplified to determine the necessity for linear amplification. Each group was evaluated against the same 48 assays. CT values and corresponding colors are given in the figure.
To evaluate the reproducibility within a chip, samples were analyzed as triplicates. Intra-card reproducibility was high with minimal standard deviations (Suppl. Table 5). For many of the genes expressed across the 60 cell lines, the coefficient of variability was far less than 3.5%. In addition, the inter-card variability was evaluated using 15 cell lines from the NCI-60. Samples analyzed on 3 different cards also showed high reproducibility (Suppl. Table 6). Here again the variability was less than 3.5% for all samples.
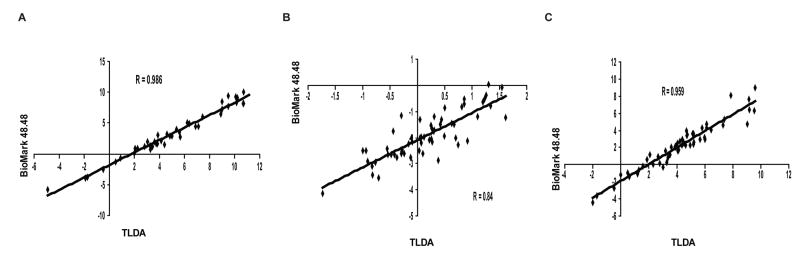
Lastly, we evaluated the ABC gene expression profiles of the NCI-60 panel using the nanofluidic qRT-PCR platform. Pearson correlations were determined for each ABC transporter expression profile across all 60 cell lines found using the TLDA and BioMark platforms. Interestingly, both platforms produced gene profiles which correlated well for this series of genes, as 39 of the 48 ABC transporters showed Pearson correlations greater than .80 (Figure 3 and Suppl. Table 7). To further validate the BioMark 48.48 Dynamic Array, we performed a blinded analysis of 16 cell lines randomly chosen from the NCI-60. The unknown samples were run on a separate day from the original study, and all 16 cell lines were correctly identified from their ABC transporter expression profile, which matched the previous unblinded study (data not shown). The BioMark nanofluidic platform displays very high reproducibility for intra- and inter-card analysis in a high-throughput fashion.
Figure 3. Correlation of NCI-60 gene expression profiles obtained from the TLDA and BioMark 48.48 Dynamic Array.
Expression profiles for ABCB1 (A), -C1 (B) and -G2 (C) across all 60 cell lines were compared between TLDA and BioMark platforms under conditions described in the Methods.
TLDA provides greater predictive power for identifying transporter substrates
A large database of biological information on the NCI-60 panel has been developed that can aid in drug discovery and eliminate compounds that are efflux-transporter substrates (23). We previously reported the expression profiling of the 48 human ABC transporters in the NCI-60 cancer cell line panel using SYBR Green qRT-PCR (16). Correlations were drawn between these gene expression profiles and the growth inhibitory profiles of 1,429 candidate anticancer drugs tested against the NCI-60 panel (17). This resulted in the generation of a database allowing the identification of lead compounds in the early stages of drug development that are not ABC transporter substrates (16). It also revealed molecules with collateral sensitivity, whose activity is potentiated, rather than antagonized, by ABC transporters (16).
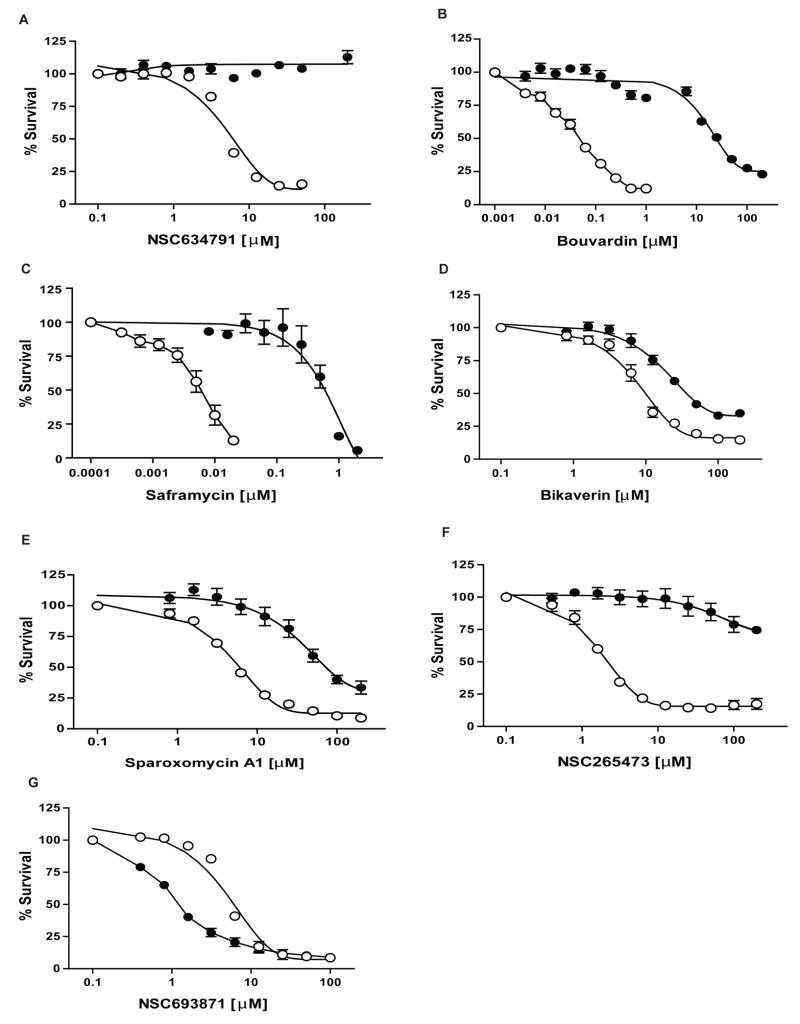
Here we present a refined database improved by correlations with the ABC transporter gene expression profiles obtained by a TLDA microfluidic platform (Suppl. Table 3). We highlight its predictions for the 3 major transporters, ABCB1, ABCC1 and ABCG2 that have been linked to drug resistance. The top 10 substrate predictions for these transporters from the TLDA analysis are given in Table 2 as well as the ranking from the previous database. The new database confirmed several predictions made by our previous study. For example, NSC634791 (Figure 4A) and bouvardin (Figure 4B) are identified as substrates for ABCB1 and bikaverin for ABCG2 (Figure 4D). We also identified additional anticancer agents which display ABC transporter-mediated drug resistance which were poorly ranked in our previous database using SYBR Green qRT-PCR. For instance, we demonstrate that saframycin A (Figure 4C and Suppl. Figure 2A), a quinone antitumor antibiotic, is a potent substrate of ABCC1 while ABCG2-overexpressing cells are resistant to sparoxomycin A1, a pyrimidinylpropanamide antibiotic with antitumor properties (Figure 4E and Suppl. Figure 2B). In addition, besides anticancer agents, the analysis revealed other previously unexplored compounds that are substrates of these ABC transporters, such as NSC265473 (Figure 4F and Suppl. Figure 2C).
Table 2.
Top ten substrate predictions for ABCB1, ABCC1 and ABCG2 transporters
| Transporter | Compound | NSC # | Rank TLDA | Rank qRT-PCR |
|---|---|---|---|---|
| ABCB1 | Phyllanthoside | 328426 | 1 | 4 |
| Bouvardin | 259968 | 2 | 3 | |
| 682066 | 3 | 1 | ||
| 353076 | 4 | 2 | ||
| 645301 | 5 | 17 | ||
| 634791 | 6 | 5 | ||
| Macbecin II | 330500 | 7 | 24 | |
| 676864 | 8 | 15 | ||
| Antibiotic UK 63052 | 630678 | 9 | 10 | |
| Bisantrene | 337766 | 10 | 9 | |
|
| ||||
| ABCC1 | Saframycin A | 325663 | 1 | 46 |
| 652903 | 2 | 107 | ||
| 645033 | 3 | 35 | ||
| (−)-Roehybridine | 626578 | 4 | 19 | |
| 633907 | 5 | 240 | ||
| 6-.alpha.-Senecioyloxychaparrinone | 290494 | 6 | 45 | |
| Neothramycin | 285223 | 7 | 152 | |
| 695636 | 8 | 111 | ||
| 652536 | 9 | 58 | ||
| Streptovaricin A diacetate | 210761 | 10 | 417 | |
|
| ||||
| ABCG2 | Bikaverin | 215139 | 1 | 2 |
| 630684 | 2 | 817 | ||
| 305458 | 3 | 386 | ||
| Sparoxomycin A1 | 251819 | 4 | 914 | |
| 625350 | 5 | 773 | ||
| 5Ph-1, 2-dithiole di-S analog | 641285 | 6 | 22 | |
| 265473 | 7 | 611 | ||
| Fagaronine | 157995 | 8 | 590 | |
| 687496 | 9 | 257 | ||
| TimTec1_000954 | 34445 | 10 | 214 | |
Figure 4. Cytotoxicity assays of compounds predicted with the TLDA expression repository.
(G) Cytotoxicity assays for ABCB1-overexpressing HEK293 (●) and parental HEK293 (○) cells with NSC693871. (A-B) Cytotoxicity assays for ABCB1-overexpressing HEK293 (●) and parental HEK293 (○) cells with NSC634791 and Bouvardin, respectively. (C) Cytotoxicity assays for ABCC1-overexpressing HEK293 (●) and parental HEK293 (○) cells with Saframycin. (D-E-F) Cytotoxicity assays for ABCG2-R482 (WT) overexpressing HEK293 (●) and parental HEK293 (○) cells with Bikaverin, Sparoxomycin A1 and NSC265473, respectively. CCK-8 reagent was used for cytotoxicity assays as described in Materials and Methods. Dose response curves were derived from three independent experiments. In all panels, error bars indicate standard deviation.
The database helps also to reveal molecules with collateral sensitivity (e.g. NSC73306 (24)), even though no evidence has been reported showing a direct interaction of this class of compounds with ABC drug transporters. We show in this manuscript a similar phenomenon with an increased sensitivity of ABCB1-overexpressing cells to NSC693871 (Figure 4G).
Our study demonstrates the utility of such a database for predicting ABC transporter-mediated multidrug resistance in drug discovery. Moreover, the ability to make predictions of substrates for 3 different ABC transporters using mRNA expression data argues strongly that mRNA levels accurately reflect the presence of at least these three functional transporters in the NCI-60 cells.
Discussion
The ATP-binding cassette (ABC) transporter proteins are a large superfamily of membrane proteins comprising 48 members (plus one pseudogene) divided into seven different families based on sequence similarities (4, 5). The nomenclature for human ABC transporter genes is provided at: http://nutrigene.4t.com/humanabc.htm. There is a high sequence homology among all the members, especially those within a particular family. ABC transporters have a wide array of cellular roles including regulation of lipid homeostasis (25) and protection of the organism by effluxing toxins out of the cells (26, 27). Similarly, they are involved in MDR by protecting cancer cells from the toxicity of chemotherapeutic agents. In the last three decades, approximately 25 ABC transporters have been shown to be involved in MDR in in vitro studies; however, numerous studies investigating ABC transporter gene expression in clinical samples have failed to directly link these transporters to drug resistance (5, 28–30). Inappropriately designed studies and poorly chosen cohorts are some of the main reasons for this failure. The limited sensitivity and/or probe specificity of platforms that have been used to assess the expression profiles of highly homologous genes such as ABC transporters is another shortcoming in these studies. The establishment of a standard analytical platform that would allow the precise discrimination of highly homologous genes using a small amount of sample would help to produce a more unified picture of MDR. It could lead to progress not only in understanding the mechanisms governing multidrug resistance but also in the translation of this knowledge to clinical practice, especially in personalized medicine (31–34).
DNA microarrays have been used to explore the relationship between gene expression patterns and drug resistance in cancer cells; however, pinpointing individual genes in gene families possessing high homology represents a major shortcoming of that technology. Although quantitating ABC transporter expression in routine clinical applications is challenging, our previous study indicated that real-time PCR has the ability to discriminate among genes in a complex multigene family, thereby allowing meaningful correlations to be drawn between gene expression and subtle differences in drug sensitivity phenotype (16). We wanted to evaluate the accuracy and sensitivity of TLDA and BioMark 48.48, two currently available high-throughput platforms based on qRT-PCR using TaqMan chemistry to discriminate highly homologous ABC transporter genes. Our previously established database (16) was thus chosen as a model to appraise the accuracy and sensitivity of these micro- and nanofluidic high-throughput TaqMan-based qRT-PCR platforms to analyze the expression profiles of ABC transporters. The NCI-60 panel includes a diverse set of human cancer cell lines derived from nine tissues of origin which have been extensively studied using microarrays, rendering this panel of cancer cell lines ideal for further analysis (20, 23, 35). The present data indicate that the microfluidic TaqMan-based qRT-PCR platform (TLDA) provides the greatest sensitivity, accuracy, and precision for ABC transporter gene expression profiling when compared to SYBR Green-based qRT-PCR. These advantages vis-a-vis current technologies make TLDAs more applicable to clinical use. Merging nanotechnology and biological profiling could enable personalized medicine to advance to the next stage in its development (36). Here we assessed the ability of a nanofluidic TaqMan-based qRT-PCR platform, the BioMark 48.48 Dynamic Array, to precisely detect ABC transporter genes in the NCI-60 panel. Although pre-amplification is a requirement for gene-expression analysis with this platform, it demonstrates reliability and accuracy similar to the TLDA platform.
The National Cancer Institute’s Developmental Therapeutics Program has extensively screened over 100,000 anti-cancer compounds using the NCI-60 cancer cell line panel since 1990 (17, 37). We correlated the gene expression profiles obtained from the microfluidic TaqMan-based qRT-PCR platform (TLDA) and the growth inhibitory profiles of a subset of 1,429 candidate anticancer drugs tested against the panel to establish a database identifying compounds as substrates of one or more ABC transporter(s). Our improved database confirms several predictions made by the previous study, and also highlights formerly unidentified anticancer and yet unexplored compounds that are substrates of ABC transporters. This was demonstrated for three extensively studied ABC transporters: ABCB1, C1 and G2. Also the ability of this database to reveal compounds whose activity is potentiated by ABC transporters was confirmed experimentally with an increased sensitivity of ABCB1 overexpressing cells to NSC693871.
Dramatic advances in gene expression profiling have occurred in the past few years. In this study, two TaqMan qRT-PCR platforms, based on micro- and nano-fluidic systems, TLDA and BioMark 48.48 Dynamic Arrays, were singled out with the potential to be further developed for individualized cancer management. The superiority of these platforms was clearly demonstrated over established technologies in assessing ABC transporter expression profiles. Our investigations led to the refinement of a previously established database with the capability to more precisely identify compounds whose resistance is mediated by ABC transporters as well as ascertain which compounds are responsible for collateral sensitivity. The challenge, now, is to apply these platforms to elucidate the gene signatures for MDR in a well-designed clinical study. That could also lead to progress not only in understanding the mechanisms governing multidrug resistance but also in the translation of this knowledge to clinical practice, especially in personalized medicine.
Supplementary Material
Acknowledgments
We thank Dr. Susan Bates for the ABCB1- and ABCG2-overexpressing HEK293 cells. We thank the DTP for the total RNA from the NCI-60 cell lines and the NSC compounds. We thank Mr. George Leiman for his editorial assistance. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. AMC was supported by the NIGMS Pharmacology Research Associate (PRAT) Program.
References
- 1.Juliano R, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta. 1976;455:152–62. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 2.Ueda K, Cardarelli C, Gottesman MM, Pastan I. Expression of a Full-Length cDNA for the Human ``MDR1'' Gene Confers Resistance to Colchicine, Doxorubicin, and Vinblastine. Proc Natl Acad Sci U S A. 1987;84:3004–8. doi: 10.1073/pnas.84.9.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueda K, Pastan I, Gottesman M. Isolation and sequence of the promoter region of the human multidrug- resistance (P-glycoprotein) gene. J Biol Chem. 1987;262:17432–6. [PubMed] [Google Scholar]
- 4.Gillet JP, Efferth T, Remacle J. Chemotherapy-induced resistance by ATP-binding cassette transporter genes. Biochim Biophys Acta. 2007;1775:237–62. doi: 10.1016/j.bbcan.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–34. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 6.Cole SP, Bhardwaj G, Gerlach JH, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–4. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 7.Doyle LA, Yang W, Abruzzo LV, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A. 1998;95:15665–70. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deeley RG, Westlake C, Cole SP. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev. 2006;86:849–99. doi: 10.1152/physrev.00035.2005. [DOI] [PubMed] [Google Scholar]
- 9.Robey RW, Polgar O, Deeken J, To KW, Bates SE. ABCG2: determining its relevance in clinical drug resistance. Cancer metastasis reviews. 2007;26:39–57. doi: 10.1007/s10555-007-9042-6. [DOI] [PubMed] [Google Scholar]
- 10.Gillet JP, Gottesman MM. Mechanisms of multidrug resistance in cancer. In: Zhou Jun., editor. MultiDrug Resistance in Cancer. Humana Press; Totowa, NJ, USA: 2009. In press. [Google Scholar]
- 11.Annereau JP, Szakacs G, Tucker CJ, et al. Analysis of ATP-binding cassette transporter expression in drug-selected cell lines by a microarray dedicated to multidrug resistance. Molecular pharmacology. 2004;66:1397–405. doi: 10.1124/mol.104.005009. [DOI] [PubMed] [Google Scholar]
- 12.Gillet JP, Efferth T, Steinbach D, et al. Microarray-based detection of multidrug resistance in human tumor cells by expression profiling of ATP-binding cassette transporter genes. Cancer Res. 2004;64:8987–93. doi: 10.1158/0008-5472.CAN-04-1978. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Anderle P, Bussey KJ, et al. Membrane Transporters and Channels: Role of the Transportome in Cancer Chemosensitivity and Chemoresistance. Cancer Res. 2004;64:4294–301. doi: 10.1158/0008-5472.CAN-03-3884. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Peng H, Zhang JT. Expression profiling of ABC transporters in a drug-resistant breast cancer cell line using AmpArray. Molecular pharmacology. 2005;68:430–8. doi: 10.1124/mol.105.011015. [DOI] [PubMed] [Google Scholar]
- 15.Steinbach D, Gillet JP, Sauerbrey A, et al. ABCA3 as a possible cause of drug resistance in childhood acute myeloid leukemia. Clin Cancer Res. 2006;12:4357–63. doi: 10.1158/1078-0432.CCR-05-2587. [DOI] [PubMed] [Google Scholar]
- 16.Szakacs G, Annereau J, Lababidi S, et al. Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell. 2004;6:129–37. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 17.Staunton JE, Slonim DK, Coller HA, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–92. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robey RW, Honjo Y, Morisaki K, et al. Mutations at amino-acid 482 in the ABCG2 gene affect substrate and antagonist specificity. Br J Cancer. 2003;89:1971–8. doi: 10.1038/sj.bjc.6601370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller M, Yong M, Peng XH, Petre B, Arora S, Ambudkar SV. Evidence for the role of glycosylation in accessibility of the extracellular domains of human MRP1 (ABCC1) Biochemistry. 2002;41:10123–32. doi: 10.1021/bi026075s. [DOI] [PubMed] [Google Scholar]
- 20.Scherf U, Ross D, Waltham M, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–44. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 21.Ishiyama M, Tominaga H, Shiga M, Sasamoto K, Ohkura Y, Ueno K. A combined assay of cell viability and in vitro cytotoxicity with a highly water-soluble tetrazolium salt, neutral red and crystal violet. Biol Pharm Bull. 1996;19:1518–20. doi: 10.1248/bpb.19.1518. [DOI] [PubMed] [Google Scholar]
- 22.Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS ONE. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig JA, Szakacs G, Martin SE, et al. Selective toxicity of NSC73306 in MDR1-positive cells as a new strategy to circumvent multidrug resistance in cancer. Cancer Res. 2006;66:4808–15. doi: 10.1158/0008-5472.CAN-05-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi K, Kimura Y, Nagata K, Yamamoto A, Matsuo M, Ueda K. ABC proteins: key molecules for lipid homeostasis. Med Mol Morphol. 2005;38:2–12. doi: 10.1007/s00795-004-0278-8. [DOI] [PubMed] [Google Scholar]
- 26.Fromm MF. Importance of P-glycoprotein at blood-tissue barriers. Trends Pharmacol Sci. 2004;25:423–9. doi: 10.1016/j.tips.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Borst P, Elferink R. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–92. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 28.Clarke R, Leonessa F, Trock B. Multidrug resistance/P-glycoprotein and breast cancer: review and meta-analysis. Seminars in oncology. 2005;32:S9–15. doi: 10.1053/j.seminoncol.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Pakos EE, Ioannidis JP. The association of P-glycoprotein with response to chemotherapy and clinical outcome in patients with osteosarcoma. A meta-analysis Cancer. 2003;98:581–9. doi: 10.1002/cncr.11546. [DOI] [PubMed] [Google Scholar]
- 30.Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert opinion on drug metabolism & toxicology. 2008;4:1–15. doi: 10.1517/17425255.4.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Sandvik AK, Alsberg BK, Norsett KG, Yadetie F, Waldum HL, Laegreid A. Gene expression analysis and clinical diagnosis. Clinica chimica acta; international journal of clinical chemistry. 2006;363:157–64. doi: 10.1016/j.cccn.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 32.Roukos DH, Murray S, Briasoulis E. Molecular genetic tools shape a roadmap towards a more accurate prognostic prediction and personalized management of cancer. Cancer biology & therapy. 2007;6:308–12. doi: 10.4161/cbt.6.3.3994. [DOI] [PubMed] [Google Scholar]
- 33.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 34.Garman KS, Nevins JR, Potti A. Genomic strategies for personalized cancer therapy. Human molecular genetics. 2007;16:R226–32. doi: 10.1093/hmg/ddm184. Spec No. 2. [DOI] [PubMed] [Google Scholar]
- 35.Lee JK, Bussey KJ, Gwadry FG, et al. Comparing cDNA and oligonucleotide array data: concordance of gene expression across platforms for the NCI-60 cancer cells. Genome Biol. 2003;4:R82. doi: 10.1186/gb-2003-4-12-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain KK. Nanomedicine: application of nanobiotechnology in medical practice. Med Princ Pract. 2008;17:89–101. doi: 10.1159/000112961. [DOI] [PubMed] [Google Scholar]
- 37.Weinstein JN, Myers TG, O'Connor PM, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–9. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.