Summary
Myoblast fusion is crucial for the formation, growth, maintenance and regeneration of healthy skeletal muscle. Unfortunately, the molecular machinery, cell behaviors, and membrane and cytoskeletal remodeling events that govern fusion and myofiber formation remain poorly understood. Using time-lapse imaging approaches on mouse C2C12 myoblasts, we identify discrete and specific molecular events at myoblast membranes during fusion and myotube formation. These events include rearrangement of cell shape from fibroblast to spindle-like morphologies, changes in lamellipodial and filopodial extensions during different periods of differentiation, and changes in membrane alignment and organization during fusion. We find that actin-cytoskeleton remodeling is crucial for these events: pharmacological inhibition of F-actin polymerization leads to decreased lamellipodial and filopodial extensions and to reduced myoblast fusion. Additionally, shRNA-mediated inhibition of Nap1, a member of the WAVE actin-remodeling complex, results in accumulations of F-actin structures at the plasma membrane that are concomitant with a decrease in myoblast fusion. Our data highlight distinct and essential roles for actin cytoskeleton remodeling during mammalian myoblast fusion, provide a platform for cellular and molecular dissection of the fusion process, and suggest a functional conservation of Nap1-regulated actin-cytoskeleton remodeling during myoblast fusion between mammals and Drosophila.
Keywords: Live imaging, Cell-cell fusion, Myoblast fusion, Muscle, Actin
Introduction
The formation, maintenance and growth of healthy skeletal muscle tissue are dependent upon the fusion and incorporation of mononucleated myoblasts into nascent multinucleate muscle fibers (Chargé and Rudnicki, 2004; Horsley and Pavlath, 2004; Huh et al., 2005; Moss and Leblond, 1970). Diseases and chronic wasting conditions that affect the skeletal musculature are particularly devastating owing to the crucial role of this tissue system in functions such as locomotion, feeding and breathing. Combating skeletal-muscle wasting due to aging, injury, chemotherapy or muscular dystrophies requires an understanding of the molecular mechanisms of myoblast fusion (Chargé and Rudnicki, 2004; Horsley and Pavlath, 2004; Huh et al., 2005; Moss and Leblond, 1970). Despite the importance of this process for muscle development and repair, a clear understanding of the molecular and cellular mechanisms underlying myoblast fusion is lacking.
To date, studies using a variety of experimental approaches have given us a basic framework of the processes that are required for myoblast fusion. On a cellular level, mammalian myoblasts seem to have an initial fibroblast-like morphology, which subsequently develops into an elongated, spindle-like conformation capable of migrating towards other differentiated, elongated myoblasts (Ohtake et al., 2006; Swailes et al., 2006). Following contact, membrane alignment and adhesion occur between partners prior to membrane breakdown and fusion (Horsley and Pavlath, 2004; Ohtake et al., 2006; Peckham, 2008; Swailes et al., 2006; Swailes et al., 2004). In addition, ultrastructural studies of fusing myoblasts from mammalian, insect and avian species have identified fusion interfaces by the presence of lipid-enriched vesicles or electron-dense vesicles close to the membrane surface between myoblasts. These fusion interfaces are often demarcated by large electron-dense plaques at the appositional surface of myoblast membranes (Doberstein et al., 1997; Engel and David, 1985; Kalderon and Gilula, 1979; Rash and Fambrough, 1973). Small fusion pores between lipid bilayers are observed prior to complete membrane breakdown in both mammalian (Engel and David, 1985; Kalderon and Gilula, 1979; Rash and Fambrough, 1973) and insect (Doberstein et al., 1997) myoblasts. The coordination of these subcellular events with the morphological changes observed at the cellular level is not well understood. Likewise, the molecular control and the dynamics of such features, including the time-frame during which the myoblasts change their shape, align membranes, recruit vesicles and fuse, are not known.
Genetic approaches have focused attention on the actin cytoskeleton and its potential roles during myoblast fusion. In Drosophila, a number of genes that are linked to Arp2/3-complex-dependent actin-cytoskeleton remodeling have been shown to be crucial for myoblast fusion (Berger et al., 2008; Kesper et al., 2007; Massarwa et al., 2007; Richardson et al., 2007; Schafer et al., 2007). Moreover, work employing three-dimensional (3D) time-lapse imaging of Drosophila embryos has identified a number of actin-based cellular behaviors during the period of Drosophila myoblast fusion, most notably highlighting a highly dynamic actin structure, termed an actin focus, at the site of myoblast fusion (Richardson et al., 2007). Mutations in several known fusion genes, including myoblast city (mbc), kette and SCAR, lead to enlarged actin foci. These studies indicate that these fusion genes are required for the dissolution of this structure during the fusion process and suggest that removal of this actin structure is crucial for fusion to proceed (Richardson et al., 2008a; Richardson et al., 2007). In addition, several studies proposed that this actin structure at the fusion site is required for vesicle trafficking (Kim et al., 2007) and/or enlarging fusion pores (Berger et al., 2008; Massarwa et al., 2007).
Recent studies have suggested a role for actin remodeling in vertebrate myoblast fusion (Dhawan, 2004; Formigli et al., 2007; Kim et al., 2007; O'Connor et al., 2008). Targeted knockdown of actin regulators Dock180 and Brag2, the mammalian homologs of Drosophila Mbc and Loner, respectively, leads to defects in murine myoblast fusion (Pajcini et al., 2008); correspondingly, skeletal myogenesis is severely reduced in Dock180–/– animals (Laurin et al., 2008). In addition, morpholino-based knockdown of zebrafish Dock1 and Dock5, orthologs of Dock180/Mbc, results in failure of myoblast fusion during fast-twitch myofiber formation (Moore et al., 2007). Together, these studies indicate that remodeling of the actin cytoskeleton has an important role in myoblast fusion and that gene products required for myoblast fusion have a conserved role across species. However, neither the number nor the type of actin-based processes required for mammalian myoblast fusion has been well described. Nor is it known whether the conserved gene products regulate the same cellular, actin-based behaviors in a vertebrate system as are required for Drosophila myoblast fusion.
Identification, recording and study of these actin-based behaviors during myoblast fusion in a developing mammalian embryo unfortunately presents several formidable technical challenges. As a first step to investigate the cellular events that occur during the mammalian fusion process, we report live imaging and identification of actin-based cellular behaviors in differentiated mammalian C2C12 myoblasts before, during and after myoblast fusion and myotube formation. In addition, we found that pharmacological inhibition of actin-cytoskeleton remodeling adversely affected all actin-based behaviors in treated myoblasts during the differentiation and fusion process. However, genetic perturbation of actin remodeling by selective inhibition of one actin regulator, Nck-associated protein 1 (Nap1), a member of the WAVE actin-remodeling complex and the vertebrate ortholog of the Drosophila fusion gene kette, indicates that Nap1-regulated membrane remodeling is crucial for mammalian myoblast fusion. Taken together, these data distinguish distinct roles for the actin cytoskeleton during myoblast fusion. Moreover, these studies suggest a functional conservation of Nap1-regulated actin-cytoskeleton remodeling during myoblast fusion between mammals and Drosophila, and provide an important framework for dissecting the cell biology of myoblast fusion in mammals.
Results
Live imaging of C2C12-myoblast fusion and myotube formation
During differentiation, myoblasts exit the cell cycle and remodel from a fibroblast-like morphology to a spindle-like morphology prior to fusing into myotubes (Swailes et al., 2006). These morphological changes have been previously examined in differentiating H2kb-tsA58 myoblasts using live phase-contrast-based microscopy (Swailes et al., 2006; Wells et al., 1997) and in fixed preparations of C2C12 myoblasts (Ohtake et al., 2006). As an improvement on these previous approaches, we paired fluorescent reporter constructs with live-imaging techniques (see Table 1). Our approach provides several advantages for live imaging, because spectrally distinct reporters can be used to label discrete cellular structures in differentiating and fusing C2C12 myoblasts (see Table 1). Crucially, we found that none of the reporters used in this study interfered with myoblast fusion or myofiber formation.
Table 1.
Reporter constructs used in this study
| Reporter construct | Cell compartment labeled | Reference |
|---|---|---|
| Pleckstrin homology GFP (PH-GFP) | Cell membranes; intensely labels sites of F-actin remodeling | (Tall et al., 2000) |
| Myristoylated RFP (myr-RFP) | Golgi network and internal vesicles | This study |
| Myristoylated Venus (myr-Ven) | Golgi network and internal vesicles | (Rhee et al., 2006) |
| Histone H2B::RFP (H2B-RFP) | Cell nuclei | (Passamaneck et al., 2007) |
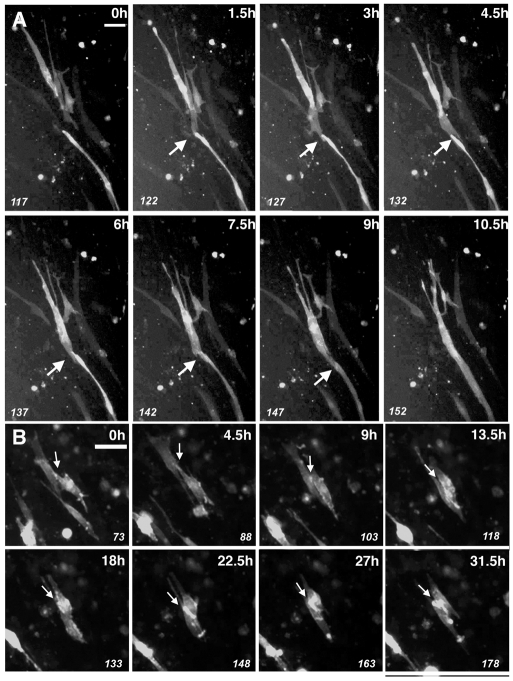
C2C12 myoblasts were transiently transfected with constructs expressing myristoylated red fluorescent protein (myr-RFP). Consistent with published observations, we found that this construct labeled not only the internal membranes and vesicles (Rhee et al., 2006) of transfected myoblasts, but also revealed the shape of transfected myoblasts. Using this marker, we found that C2C12 myoblasts initially have a fibroblast-like conformation during differentiation (Fig. 1A, 25.2 hours).
Fig. 1.
Live time-lapse imaging reveals dynamic C2C12-myoblast behaviors during differentiation and fusion. In all cases, myoblasts were allowed to differentiate for 24 hours and imaged every 18 minutes over a 72-hour period at 37°C. The frame number from the live-imaging movie is given in the lower left. (A) Differentiating myoblasts undergo remodeling of cell morphology during differentiation. Arrowhead indicates myoblast of interest. C2C12 myoblasts were transiently transfected with vectors expressing myr-RFP, which labels internal membranes and cytoplasm (white). Representative panels of live imaging are presented; time in hours post-switch to DM (bottom right) and frame number (bottom left) are given. At 25.2 hours following switch to DM (frame 1), myoblasts exhibit a fibroblast-like morphology that remodels into an elongated spindle-like shape approximately 37.5 hours (frame 41) following the switch to differentiation conditions. Myoblasts remain elongated for several hours prior to fusing with other mononucleated myoblasts at 75.3 hours of differentiation (frame 167; see also supplementary material Fig. S1). (B) C2C12 myoblasts transfected with myr-Ven reporter (white) and fusing with myotubes in culture. Panels shown begin at 53.4 hours following change to DM (supplementary material Movie 1). A mononucleate myoblast (arrowhead) makes contact with a multinucleate myotube (asterisk). Following contact, the myoblast fuses with the myotube (2.4 hours, frame 44) and the nucleus incorporates into the myotube (4.2 hours, frame 50). (C) C2C12 myoblasts were transfected with constructs expressing either myr-Ven (green) or myr-RFP (red) and mixed together prior to differentiation. Panels shown begin at 41.1 hours following medium change to DM (supplementary material Movie 2). Following contact (first detected at 3.6 hours, frame 22) between red (asterisk) and green (arrowhead) myoblasts, rapid mixing is observed between the two different populations (5.4 hours, frame 28), and the two myoblasts fuse to form a myotube (7.2 hours, frame 34) (see also supplementary material Fig. S2). Scale bars: 25 μm (A); 50 μm (B,C).
Within hours of shifting to differentiation medium (DM), myoblast morphology changed to an elongated conformation that is spindle-like in shape (Fig. 1A, 37.5 hours). On average (n=seven events), this morphological remodeling occurred after 31±5 hours of differentiation (range: 23-39 hours following shift to DM). Once committed to this elongated conformation, myoblasts never reverted to the fibroblast-like morphology (n≥50 labeled myoblasts observed). Following this morphological change (see supplementary material Fig. S1 for greater detail), elongated myoblasts were often observed to migrate prior to their fusion with other myoblasts or myotubes (n≥20 labeled myoblasts; Fig. 1A, beginning at 52.2 hours). In all labeled myoblasts examined (n≥20), we never observed fusion events between fibroblast-like myoblasts; all myoblasts participating in fusion events were in the elongated conformation. We found on average that myoblasts fused within 25 ±5 hours of elongation (n=six events; range: 22-31 hours following elongated morphology).
To understand the fusion process in greater detail, we recorded multiple independent fusion events (Fig. 1B; supplementary material Movie 1). In the example given (Fig. 1B), we observed a free myoblast expressing myristoylated Venus (myr-Ven) approach, contact and fuse with another myotube. We found that a fusion event, spanning the time from contact between two fusing cells to the uptake and migration of the myonucleus into the myotube, occurred over an average time interval of 2.5±0.5 hours (n=17). Also, it is important to note that, in this system, we observed myotube-myotube fusion events (supplementary material Movie 1; and data not shown).
To verify that the fusion events we observed in single-fluorophore live imaging were bona fide fusion events, we performed cytoplasmic mixing experiments using spectrally distinct myristoylated-fluorescent-protein reporters. For these experiments, we split C2C12 myoblasts from the same passage into two pools for transfection. One pool of myoblasts was transfected with the same myr-RFP, whereas the other pool was transfected with myr-Ven. By combining these transfected pools and allowing them to differentiate, we imaged cytoplasmic exchange between fusing myoblasts and colocalization of both markers in the newly formed multinucleate myotube (Fig. 1C; supplementary material Movie 2). As reported above in single-fluorophore imaging, we observed a myr-RFP-expressing myoblast approach (0-3.6 hours), contact a myr-Ven-expressing myoblast (3.6 hours) and fuse into a syncytial structure (Fig. 1C; supplementary material Fig. S2, Movie 2). At the level of resolution of our time-lapse movies, we did not observe a concentration of either the Venus or RFP reporter at the site of fusion. Consistent with the previous observations on single-fluorophore and/or unlabeled myoblasts (Fig. 1B; supplementary material Movie 1), fusion between two myoblasts in these binary fluorophore imaging experiments occurs over an average period of 2.5 hours (n=ten fusion events). Our imaging also revealed that fusion between myoblasts was not directionally biased, occurring both in an end-to-end orientation and a perpendicular orientation to each other (Fig. 1C; supplementary material Movie 2; and data not shown).
Live imaging reveals several actin-remodeling-based cellular behaviors during myoblast differentiation and fusion
Actin-cytoskeletal remodeling events have an important role in mammalian myoblast fusion (O'Connor et al., 2008) and have been demonstrated as essential for myoblast fusion in Drosophila (Berger et al., 2008; Kim et al., 2007; Massarwa et al., 2007; Richardson et al., 2007; Schafer et al., 2007). We used live imaging to examine differentiating C2C12 myoblasts for evidence of actin-based behaviors during differentiation and fusion (Fig. 2A,B). At 24 hours after switching to DM, the fibroblast-like C2C12 myoblasts all displayed membrane ruffling and formation of lamellipodia and filopodia, both of which are well-described actin-based behaviors (Kawamura, 2004; Steffen et al., 2006; Steffen et al., 2004) (Fig. 2A). The lamellipodia and filopodia in these myoblasts did not have any readily discernible orientation. At 48 hours of culture in DM, elongated myoblasts extended and retracted filopodia-like membrane protrusions from each tip of the long axis of the myoblast (Fig. 2B). We never observed lamellipodia formation or filopodial extension perpendicular to the main axis of the elongated myoblast (n≥50 elongated myoblasts). Such extensions and retractions are characteristic of extensive actin-cytoskeleton remodeling (Kawamura, 2004; Steffen et al., 2006)
Fig. 2.
Actin-based behaviors observed in differentiating myoblasts. (A,B) Panels shown are representative frames from live imaging of confluent myoblasts in DM at 37°C. Time is given in hours in the top right corner; frame number from live-imaging movies is given in lower left corner. (A) C2C12 myoblasts transfected with myr-Ven constructs (white) display a fibroblast-like morphology initially during differentiation. At this time, C2C12 myoblasts exhibit rapid membrane ruffling and formation of lamellipodia (arrowheads). Panels shown begin at 24 hours of culture in DM. (B) At 48 hours in DM, C2C12 myoblasts transfected with myr-Ven constructs (white) extend large filopodial extensions (arrowhead) that often contact neighboring cells. (C) To indirectly label sites of actin-cytoskeleton remodeling, C2C12 myoblasts were transfected with a pleckstrin-homology-domain variant fused to GFP (PH-GFP), which associates with PtdIns(4,5)P2 phospholipids at sites of actin-cytoskeleton remodeling (Tall et al., 2000). PH-GFP reporter expression (green) labels membranes and colocalizes strongly (arrowheads) with concentrated sites of F-actin, as revealed by phalloidin staining (red). (D) C2C12 myoblasts transfected with PH-GFP (white) exhibited lamellipodia formation initially in DM (*, 24 hours, frame 1), followed by switching to an elongated spindle-like morphology as differentiation proceeds (42 hours, frame 60). Time is indicated in hours following switch to DM. The white arrow indicates the myoblast of interest. (E) The PH-GFP reporter (green) reveals dramatic remodeling of myotube membranes during differentiation. Following transfection, myoblasts were allowed to differentiate for 48 hours and then imaged at 37°C. A total of 10 hours of live imaging is presented. Dynamically active filopodia were detected at the ends of myotubes (arrowhead shown for reference). Asterisk (*) indicates myotube. Scale bars: 20 μm (A-C); 25 μm (D), 50 μm (E).
Direct observation of actin-remodeling events using actin::GFP fusion reporters as well as Lifeact constructs (Riedl et al., 2008) was not possible owing to toxicity of the reporter construct in C2C12 myoblasts (data not shown). To indirectly visualize actin-cytoskeleton remodeling, we transfected myoblasts with plasmids expressing a pleckstrin homology domain fused to GFP (PH-GFP) (Tall et al., 2000). This PLCδ1 PH-domain variant binds preferentially to phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2], which is highly associated with sites of actin-cytoskeleton remodeling (Coppolino et al., 2002; Insall and Weiner, 2001; Rozelle et al., 2000). In agreement with these data, we found that this reporter labels regions of cell membranes containing concentrations of F-actin (Fig. 2C). Therefore, we used this marker to label the membranes of myoblasts as well as the sites of active actin-cytoskeletal remodeling. Using live imaging, we determined that wild-type myoblasts expressing the PH-GFP reporter exhibit the same dramatic remodeling of myoblast morphology as seen with the myr-Ven reporter (Fig. 2D). Importantly, the presence of this construct did not affect the remodeling of unfused C2C12 myoblasts from the fibroblast-like conformation to elongated spindle-like morphology nor subsequent differentiation. Surprisingly, live imaging revealed that membranes of multinucleated myotubes also underwent extensive remodeling (Fig. 2E) with rapid extension and retraction of filopodia, a phenomenon not previously identified in formed myotubes.
Pharmacological inhibition of actin-cytoskeletal remodeling adversely affects myoblast fusion
Our live-imaging data revealed actin-based membrane-remodeling events (e.g. lamellipodia formation, and filopodial formation and extension) that occur during different phases of myoblast differentiation and fusion. These data suggested that actin-cytoskeleton remodeling is required throughout myotube formation, in both unfused myoblasts and myotubes. We next determined the effect of inhibition of actin-cytoskeleton remodeling during the differentiation and fusion of mammalian myoblasts.
For these experiments, differentiating C2C12 myoblasts were treated with either latrunculin B or cytochalasin D, which interfere with F-actin remodeling by binding to actin monomers or actin branches, respectively (Coue et al., 1987; Dhawan, 2004) (Fig. 3A-I). Because contact between myoblasts is essential for their subsequent fusion (Kang et al., 2003; Kang et al., 2004) and both compounds would adversely affect cell migration (Kawamura, 2004), we minimized any potential influence an inhibition of myoblast migration might exert on our fusion analysis by growing myoblasts to complete confluence prior to shifting them to DM containing drugs (supplementary material Fig. S3).
Fig. 3.
Pharmacological inhibition of actin remodeling blocks myoblast fusion. Replicate dishes of confluent myoblasts (see also supplementary material Fig. S3) were incubated in DM containing either 300 nM cytochalasin D (B,E,H) or 100 nM latrunculin B (C,F,I) for 1 (A-C), 2 (D-F) or 3 (G-I) days. Following treatment, cells were fixed, stained with antibodies against sarcomeric Mhc (red) and counterstained with DAPI to visualize nuclei (green). Cytochalasin-D treatment caused severe disruption of cytoskeletal architecture and abnormally shaped myotubes (arrows). Large, myosin-positive, rounded myoblast bodies were also observed (asterisk) as a result of cytochalasin-D treatment. Such morphologies are probably due to loss of strong contacts with the culture dish and not due to increased cell death. Treatment with latrunculin B resulted in abnormally shaped myoblasts, with few fused myotubes present. (J) Fusion index of samples in A-I (see Materials and Methods). (K) Percentage of total nuclei present in each imaging field contained within a myosin-positive structure. The percentage of myosin-positive nuclei increased during the experiment, indicating that drug-treated myoblasts still express markers of differentiated myoblasts. (J,K) Bars indicate standard deviation of fusion indices from the mean. P-values (Student's t-test)<0.0001. Scale bar: 40 μm.
Whereas myoblasts treated with either cytochalasin D (Fig. 3B,E,H) or latrunculin B (Fig. 3C,F,I) adopted elongated morphologies similar to those observed in untreated myoblasts (Fig. 3A,D,G), the fusion index of treated myoblasts was significantly lower than that of untreated myoblasts (Fig. 3J). We found that drug-treated C2C12 myoblasts still expressed sarcomeric myosin heavy chain (Mhc), a marker for both mature myotubes and as-yet-unfused terminally differentiated myoblasts (Pajcini et al., 2008; Yoon et al., 2007), indicating that the decreased fusion of treated myoblasts was not due to an inability of these cells to differentiate (Fig. 3K). We also observed subtle differences between myoblasts treated with either compound: cytochalasin-D-treated myoblasts appeared elongated but thinner compared with untreated control or latrunculin-B-treated myoblasts.
We also examined which actin-based behaviors were altered in both cytochalasin-D- and latrunculin-B-treated myoblasts using live imaging. We found that myoblasts elongated as observed in fixed specimens; however, all actin-based behaviors, such as lamellipodia and filopodia formation and cell migration, were adversely affected as a result of drug treatment during differentiation (data not shown). Taken together, these results suggest that actin-cytoskeleton remodeling is crucial for subsequent fusion and myotube formation.
shRNA-mediated knockdown of Nap1 inhibits myoblast fusion and myotube formation
Given the strong reduction in myoblast fusion after treatment with F-actin-binding compounds, we next sought to determine the role of a specific regulator of actin-cytoskeleton remodeling during myoblast fusion. Recent studies of myoblast fusion in Drosophila have highlighted the important role of the Kette protein for actin-cytoskeletal remodeling during fusion (Richardson et al., 2007; Schröter et al., 2004). These data, together with our data suggesting the importance of actin remodeling during mammalian myoblast fusion, led us to investigate the role of the Kette mammalian homolog Nap1 during fusion. There are two Nap genes in mouse, Nap1 (also known as Nckap1 and Hem2) and Hem1 (Rakeman and Anderson, 2006; Weiner et al., 2006). Hem1 is not expressed in mouse muscle (Hromas et al., 1991). Our reverse transcriptase (RT)-PCR data indicated that Nap1 mRNA is expressed in C2C12 cells both before and after the switch to differentiation conditions (supplementary material Fig. S4) (Kesper et al., 2007).
To test for a role of Kette/Nap1 during mammalian myoblast fusion, we used vectors expressing Nap1-specific short hairpin RNA (shRNA) constructs (Steffen et al., 2004). These constructs have been used to successfully knock down Nap1 function in fibroblasts (Steffen et al., 2004); we therefore used them to reduce Nap1 levels in C2C12 myoblasts. C2C12 myoblast lines were stably transfected either with vectors constitutively expressing Nap1-directed shRNA dimers or control empty vectors. To ensure that any observed effect on fusion was genuine and reproducible, we created multiple clonal cell lines stably transfected with the Nap1-shRNA expression vector. We then evaluated Nap1 knockdown in a subset of these stably transfected cell lines (Fig. 4A-H; supplementary material Fig. S5). Nap1 levels were successfully reduced in C2C12 myoblasts, as verified by western blot analysis (Fig. 4I). We found that Nap1-knockdown cell lines all proliferated at rates indistinguishable from control cells (data not shown) and appeared morphologically identical to control myoblasts prior to differentiating (data not shown). We found that, during incubation in DM, Nap1-knockdown myoblasts failed to fuse normally in comparison to control lines (Fig. 4A-H; supplementary material Fig. S5). Some fusion events between Nap1-knockdown myoblasts were detected; however, large numbers of Nap1-knockdown myoblasts remained unfused (Fig. 4A-H). Correspondingly, the fusion index was significantly lower in Nap1-knockdown lines compared with control myoblasts (Fig. 4J). Importantly, all Nap1-knockdown myoblast lines expressed sarcomeric Mhc following a shift to DM, indicating that these cells were capable of expressing differentiation markers, yet failed to fuse (Fig. 4K). Furthermore, co-culture of Nap1-knockdown myoblasts with wild-type myoblasts did not rescue the fusion defect, because labeled Nap1-knockdown myoblast nuclei failed to incorporate into labeled wild-type myotubes after 5 days of culture (n≥20 myotubes) (supplementary material Fig. S6).
Fig. 4.
shRNA-mediated inhibition of Nap1 blocks mammalian myoblast fusion. Stable cell lines were generated by transfections with constructs constitutively expressing Nap1-directed shRNA oligonucleotides (Nap1 RNAi; B,D,F,H) or empty vectors (control; A,C,E,G). Stable cell lines were allowed to grow to confluence then switched to DM for 1 (A,B), 2 (C,D), 3 (E,F) or 5 (G,H) days. Cells were fixed and immunostained for sarcomeric Mhc (red) and counterstained with DAPI to visualize nuclei (green). For brevity and clarity, one control cell line and one Nap1-knockdown cell line is presented; see supplementary material Fig. S5 for images of other cell lines used in this analysis. Nap1-knockdown cells produced fewer multinucleate myotubes than control cells (arrowheads; D,F,H). Similar to cytochalasin-D treatment, large, myosin-positive, rounded myoblast bodies were also observed (asterisks) in Nap1-knockdown cell lines. Such morphologies are probably due to loss of strong actin contacts with the culture dish and not due to increased cell death (data not shown). Scale bar: 40 μm. (I) Western blot of total cell extracts demonstrates the relative level of Nap1 protein in knockdown cells. (J) Average fusion index in a given image field for the control myoblast line vs Nap1-knockdown myoblast lines. Bars indicate standard deviation of fusion indices from the mean. P-values between all Nap1 cell lines and wild-type control cells (unpaired Student's t-test) are <0.0001. (K) Percentage of total nuclei present in each imaging field that reside within a myosin-positive structure. Bars indicate standard deviation from the mean. The percentage of myosin-positive nuclei remained relatively constant for Nap1-knockdown cells during the experiment, indicating that Nap1-knockdown myoblasts still express differentiation markers.
Nap1 is a member of a conserved complex with Sra-1 (also known as CYFIP), Abi1 and HSPC300 that serves to regulate SCAR/WAVE activity. SCAR/WAVE then regulates Arp2/3-dependent actin polymerization (Ibarra et al., 2005; Machesky and Insall, 1998; Smith and Li, 2004; Vartiainen and Machesky, 2004). Positive regulation of SCAR/WAVE by Kette/Nap1 has been shown to be essential for proper SCAR/WAVE localization and myoblast fusion in Drosophila (Richardson et al., 2007). We therefore next determined whether Nap1 functions through WAVE activity to regulate C2C12-myoblast fusion. We found that Abi1, another member of the WAVE regulatory complex, as well the WAVE1 and WAVE2 isoforms, are expressed in C2C12 myoblasts during differentiation. In Nap1-knockdown myoblasts, Abi1 levels were unaffected, but we did find that levels of both WAVE1 and WAVE2 were strongly reduced (supplementary material Fig. S7). Transient transfection of shRNA constructs, which have been used to successfully knock down WAVE1 (Suetsugu et al., 2003) and WAVE2 (Kawamura, 2004) in other cell types, produced different results: transient knockdown of WAVE2 leads to a reduction in myoblast fusion, whereas WAVE1 is not crucial for C2C12-myoblast fusion (supplementary material Fig. S8). Furthermore, we observe a slight reduction in the levels of Abi1 in transient WAVE2-knockdown myoblasts, which has been described by other groups (Carabeo et al., 2007; Derivery et al., 2008; Ryu et al., 2009). These data suggest that the fusion defect that we observe in Nap1-knockdown myoblasts is due to the action of Nap1 as a component of the WAVE regulatory complex.
In the early stages of differentiation, Nap1-knockdown myoblasts exhibit wild-type morphologies and migration profiles
To pinpoint which actin-based process was affected by Nap1 knockdown, we first addressed whether the failure of Nap1-knockdown myoblasts to fuse was due to a failure of these cells to undergo the same morphological changes observed in differentiating control myoblasts (see Figs 1 and 2). Live imaging of Nap1-knockdown cell lines revealed that these myoblasts displayed a wild-type fibroblast-like morphology with membrane ruffling and lamellipodia formation during the initial period of incubation in differentiation conditions (Fig. 5). Over a period of several hours and comparable to control myoblasts, Nap1-knockdown myoblasts remodeled into an elongated spindle-like morphology, with filopodial extensions that emanated from the `ends' of these myoblasts. As noted above in control cells (Fig. 1A), elongated Nap1-knockdown myoblasts never extended their filopodia perpendicular to the longitudinal axis of the myoblast (n≥40 myoblasts). Consistent with data reported above, we did not observe fusion events in Nap1-knockdown myoblasts during the entire imaging period (>72 hours). We concluded from these data that Nap1-knockdown myoblasts are capable of progressing through the same morphological phases during differentiation as control myoblasts, yet fail to fuse at wild-type rates.
Fig. 5.
Morphological events and migration profiles in Nap1-knockdown myoblasts are identical to wild-type control myoblasts despite fusion block. (A) Nap1-knockdown myoblasts were transfected with constructs expressing PH-GFP (white). Cells were differentiated for 24 hours and then imaged for 72 hours at 37°C. Panels shown are representative frames from live imaging; frame number is given at lower left; arrow indicates the myoblast of interest. PH-GFP-labeled myoblasts exhibited lamellipodia formation during the initial stages of differentiation, followed by adoption of elongated spindle-like morphology as differentiation proceeds. Time is given in hours following shift to DM. Scale bar: 25 μm. (B) A total of 20 control myoblasts (blue) and 20 Nap1-knockdown myoblasts (red) were tracked in live-imaging experiments during 25-48 hours of differentiation. Tracks were plotted and relevant parameters (±standard deviation as indicated) were calculated using Volocity Quantitation software. Nap1-knockdown myoblasts seemed similar to control myoblasts. P-values were calculated from a Student's t-test.
Nap1 is crucial for actin polymerization during cell migration in fibroblasts (Steffen et al., 2004). To ensure that the defect in myoblast fusion observed in Nap1-knockdown myoblasts was not due to impaired or disrupted cellular migration, we tracked the migration of 20 labeled control and 20 labeled Nap1-knockdown myoblasts during 25-48 hours of differentiation using live imaging (Fig. 5B). We chose this interval on the basis of our live-imaging results of control myoblasts, which indicated that myoblasts were highly migratory within a confluent field. We determined that, on average, both control and Nap1-knockdown myoblasts migrated roughly equivalent distances during this interval. Moreover, the average maximum distance per step taken (15.8±0.94 μm vs 18.5±1.66 μm, P≥0.2) as well as the average maximum velocity per step (0.88±0.05 μm/minute vs 1.03±0.09 μm/minute, P≥0.16) were equivalent. Similarly, no discernable difference in the trajectories of Nap1-knockdown or control myoblasts was detected (data not shown). Thus, knockdown of Nap1 does not adversely affect migration in these myoblasts during differentiation, and the lack of fusion observed in these cell lines is not due to a general failure of myoblasts to migrate to their fusing partners.
Impaired fusion of Nap1-knockdown myoblasts is not due to failure to form inter-myoblast contacts or sarcomeric structures
Contact and adhesion of myoblasts with each other is crucial for myoblast fusion (Kang et al., 2003; Kang et al., 2004). These intermyoblast contact sites can be identified by N-cadherin localization during differentiation and fusion (Abramovici and Gee, 2007). To ensure that the reduced fusion index of Nap1-knockdown cells was not due to their inability to form intercellular contacts, both control and Nap1-knockdown cell lines were differentiated for 3 days, fixed and stained with antibodies against N-cadherin (Fig. 6A,B). Nap1-knockdown cell lines were indeed capable of forming contacts, because N-cadherin staining was observed at cell-cell contact points following the shift to differentiation conditions (Fig. 6B). We observed a similar level of N-cadherin staining overall in preparations of Nap1-knockdown myoblasts compared to control myoblasts (data not shown). Furthermore, the reduced fusion of Nap1-knockdown myoblasts is not due to excessive apoptosis, as verified by examining activated caspase-3 in both Nap1-knockdown and control cell lines (data not shown).
Fig. 6.
Nap1-knockdown cell lines are capable of making intercellular contacts and forming sarcomeric structures under differentiation conditions. (A,B) Control (A) and Nap1-knockdown (B) myoblasts were cultured in DM for 72 hours, fixed and immunostained with antibodies against N-cadherin (white). N-cadherin staining (arrows) is observed at contact points between Nap1-knockdown myoblasts, indicating that these cells are capable of forming contacts with neighboring myoblasts. Scale bars: 20 μm. (C-F) Ultrastructural analysis of differentiating control (C,E) and Nap1-knockdown (D,F) myoblasts indicates that both control (C) and Nap1-knockdown (D) myoblasts are capable of forming close membrane junctions (membranes indicated by arrows). (E,F) The appearance of Z bodies (arrows) forming on stress fibers (*) are seen in both control (E) and Nap1-knockdown (F) myoblasts, indicating that Nap1-knockdown cell lines undergo the early ultrastructural hallmarks of myofibrillogenesis.
Ultrastructural examination of Nap1-knockdown myoblasts (Fig. 6D) revealed no obvious differences in membrane structure when compared to control (Fig. 6C) myoblast membranes. Although fusion of Nap1-knockdown myoblasts was significantly impaired, these cells were capable of one or two fusion events to produce sarcomeric myosin-positive multinucleated structures, as our fusion-index data indicated (Fig. 4J). Importantly, these multinucleate Nap1-knockdown structures had several hallmarks of myofibrillogenesis: electron-dense Z bodies were detected along actin stress fibers, and thick filaments were interspersed alongside and in between these densities, a stage known as the premyofibril (Fig. 6F). At the ultrastructural level, myofibrillogenesis in Nap1-knockdown cells was similar to control cells (Fig. 6E), indicating that Nap1-knockdown myoblasts were capable of forming functional myotubes, provided that they were able to fuse into multinucleate structures.
Live imaging of Nap1-knockdown myoblasts reveals abnormal membrane behaviors during myoblast fusion
We next used live imaging to assay the membrane distribution of the PH-GFP reporter in Nap1-knockdown myoblasts. Live imaging revealed a difference in the membrane localization of the PH-GFP reporter between control and Nap1-knockdown myoblasts. We identified large aggregations of the PH-GFP reporter on the surface of Nap1-knockdown myoblasts; these aggregations perdured for a significant period of time (Fig. 7B; supplementary material Movie 3). Noting the prevalence of these structures in Nap1-knockdown myoblasts, we re-examined our live imaging of control myoblasts for similar structures. We found similar PH-GFP reporter aggregations in control myoblasts during an identical imaging period (Fig. 7A; supplementary material Movie 4), but these aggregations were far more transient in control myoblasts. We determined that the average lifetime of these aggregations was 6.5 hours for control myoblasts, as compared with 19.7 hours for Nap1-knockdown myoblast cells (n≥15 of each myoblast type analyzed, P<0.02 in a Student's t-test).
Fig. 7.
Live imaging of Nap1-knockdown myoblasts reveals altered membrane remodeling during differentiation. (A,B) Control (A) or Nap1-knockdown (B) myoblasts were transfected with constructs expressing PH-GFP (white). Cells were cultured in DM for 24 hours and then imaged for 72 hours at 37°C. Time is given in hours following shift to DM; frame number is given on lower left (see supplementary material Movies 3 and 4 for A and B, respectively). Fusion events were detected between control myoblasts. By contrast, in addition to not fusing during the imaging period, Nap1-knockdown myoblasts exhibited large accumulations of PH-GFP at contact points between myoblasts (arrow in B). These structures perdured throughout the imaging period and were highly transient in control myoblasts (arrow in A). For clarity, each panel in B is centered on the myoblast(s) in question. Scale bar: 50 μm.
To determine whether these PH-GFP aggregates were bona fide filament-associated structures or were simply accumulations of GFP-labeled phospholipids, we stained PH-GFP-expressing Nap1-knockdown myoblasts with phalloidin. We found that these aggregates contained F-actin (Fig. 8A). We next addressed whether these structures also contained proteins that have been implicated in myoblast fusion. We immunostained fixed preparations of PH-GFP-transfected Nap1-knockdown myoblasts with antibodies against Dock180, the mammalian homolog of the Drosophila Mbc protein. Mbc is an important component of Drosophila myoblast fusion machinery and was recently identified as a crucial molecule for fusion of macrophages and mammalian myoblasts (Pajcini et al., 2008; Laurin et al., 2008). Antibodies to Dock180 also labeled these membrane PH-GFP aggregates on Nap1-knockdown myoblasts (Fig. 8B). Because Dock180/Mbc immunocolocalizes with regions of concentrated F-actin at Drosophila myoblast-fusion sites (Richardson et al., 2007), it suggests that these regions demarcate a similarly important site for fusion of mammalian myoblasts. On the basis of these data as well as live-imaging data, we conclude that Nap1 activity regulates a key membrane-remodeling event that is essential for myoblast fusion to proceed.
Fig. 8.
PH-GFP reporter aggregates contain F-actin and proteins required for myoblast fusion. (A-C) Projections of 15-μm confocal z-stacks are shown. (A) Phalloidin staining reveals that PH-GFP aggregates (green) observed in Nap1-knockdown myoblasts also contain F-actin (red). These same structures are rarely seen in control myoblasts at the same stage of differentiation (B). (C) Antibodies against Dock180 (red), a regulator of myoblast fusion in both insects and vertebrates, immunocolocalize with PH-GFP aggregates (green) in Nap1-knockdown myoblasts. Scale bars: 20 μm.
Discussion
Myoblast fusion is essential for myotube formation, but the dynamics of this process and the behaviors that are characteristic of differentiating and fusing myoblasts remain poorly understood. In this study, we used live imaging to characterize actin-based behaviors and processes that occur in differentiating myoblasts during fusion and myotube formation. We found that differentiating myoblasts exhibit behaviors such as lamellipodia and filopodia formation, which are highly dependent upon actin-cytoskeleton remodeling prior to fusion. Pharmacological perturbation of actin-cytoskeleton remodeling inhibited myoblast fusion and many of the other behaviors noted in the live-imaging studies. Finally, shRNA-mediated knockdown of Nap1, a key regulator of actin-cytoskeleton remodeling that acts in the WAVE regulatory complex, resulted in a severe reduction of myoblast fusion and of myotube formation. We further determined that membrane remodeling in Nap1-knockdown myoblasts was adversely affected, suggesting that Nap1 regulation of membrane remodeling is a crucial event in the fusion process.
Live imaging of myoblast behaviors and fusion
The application of live-imaging techniques to myogenesis is highly desirable for resolving the temporal and spatial aspects of cellular processes occurring during myoblast fusion. Unfortunately, live imaging of myogenesis in developing mammalian embryos presents several significant technical hurdles, thus necessitating a cultured myoblast-based approach. Live imaging in cultured cell lines has been employed to examine several aspects of myoblast fusion (Pajcini et al., 2008; Straube and Merdes, 2007; Swailes et al., 2006; Wells et al., 1997). The most extensively studied behavior of differentiating and fusing myoblasts is perhaps the remodeling of H2kb-tsA58-myoblast morphology from the fibroblast-like morphology to an elongated spindle conformation. This morphological change is governed by the action of non-muscle myosin 2A, which is thought to increase the surface interaction between myoblasts as they prepare to fuse (Peckham, 2008; Swailes et al., 2006). Our use of live imaging together with fluorescent-protein reporter constructs further refines these data by revealing fine processes, such as small filopodial extensions and membrane remodeling events, that went undetected in live-imaging studies using phase-contrast optics. The previous studies noted above used phase-contrast imaging, necessitating the analysis of small groups of cells in a sparsely seeded environment to allow myoblast visualization. Myoblasts require contact with other myoblasts (Kang et al., 2003; Kang et al., 2004) as well as a minimum culture density for optimal differentiation and fusion (Doherty et al., 2005). By using transient transfection of reporter constructs in our assay, we labeled only a subset of myoblasts in a confluent field, allowing observation of their interactions and behaviors in a denser environment.
Two actin-cytoskeleton-remodeling-based behaviors were noted in our live imaging of myotubes and unfused myoblasts: lamellipodia and filopodia formation in fibroblast-like myoblasts, and filopodia formation and retraction in both elongated spindle-shaped myoblasts and myotubes. Lamellipodia and filopodia formation has been examined in undifferentiated C2C12 myoblasts and is controlled by WAVE2 and N-WASP, respectively (Kawamura, 2004). We were further intrigued by the apparent irreversibility of myoblast morphology during differentiation; as our results indicated, once myoblasts adopted the elongated conformation, we never observed reversion to the fibroblast-like conformation. We did not observe fusion of fibroblast-like myoblasts during our assays. Furthermore, our use of fluorescent reporters to label myoblasts enabled observation of extremely fine filopodial extensions that extended for considerable distances throughout the confluent myoblasts in the dish. In fused myotubes, the most prevalent behavior noted was filopodial extensions from the ends of the myotube. Whereas migration of myotubes has been previously reported (Swailes et al., 2006), in our data we did not detect subsequent migration of the myotube as a result of these filopodial extensions and retractions. Such extensions are highly reminiscent of growth-cone searching behaviors seen in elongating axons (Tornieri et al., 2006) and might serve to attract differentiated myoblasts to each other for fusion, or for attraction of differentiated myoblasts to myotubes for subsequent fusion.
In the fusion events that we recorded by live imaging, we did not detect any orientation preference of fusing partners, or a discernible site of fusion between fusing partners. Live-imaging studies of fusion in H2kb-tsA58 myoblasts have suggested that myoblast fusion occurs in an end-to-end, rather than in a lateral, manner. However, these myoblasts were seeded on an array that constrained their movement along a linear axis (Clark et al., 2002). By contrast, the fusion events that we recorded were unbiased, occurring in multiple orientations in an unrestrained system, with lateral fusion events occurring very readily (Fig. 1; and data not shown). In the two examples that we provide, we observed lateral fusion between a myoblast and a myotube (Fig. 1B) and perpendicular fusion of two myoblasts, followed by perpendicular fusion of the resultant myotube with another multinucleate myotube (Fig. 1C). Furthermore, our live imaging revealed that the myotube diameter changes frequently throughout fusion and myonuclear incorporation in C2C12 cultures (see Fig. 1; supplementary material Movies 1 and 2). Because previous studies were performed with different myoblast lines (Clark et al., 1997; Clark et al., 2002; Swailes et al., 2006; Swailes et al., 2004), it remains to be determined whether these properties of fusing structures are due to peculiarities of each cell line. Final resolution of this matter will probably require live analysis of myoblast fusion in developing animals in vivo.
A role for Nap1 during mammalian myoblast fusion
Nap1 has been previously identified as a crucial component of actin-cytoskeleton remodeling during cellular migration and lamellipodia formation in fibroblasts (Steffen et al., 2004). Furthermore, Nap1–/– mutants exhibit a variety of morphological defects arising from a failure of mesoderm and endoderm to migrate properly, as well as defects in neural-tube closure. Unfortunately, a definitive role for Nap1-regulated actin-cytoskeleton remodeling during mammalian myogenesis is not known, primarily due to the early embryonic lethality of Nap1–/– mice (Rakeman and Anderson, 2006), which precludes analysis of primary myoblasts isolated from these animals.
Our results indicate a crucial role for Nap1 during mammalian myoblast fusion in the C2C12-myoblast model system. As revealed by live imaging, Nap1-knockdown myoblasts are capable of completing the morphological transition from the fibroblast-like to elongated conformation. This result is not altogether surprising, because this remodeling event is regulated by the action of non-muscle myosins (Swailes et al., 2006) as well as the microtubule network (Straube and Merdes, 2007). Interestingly, we observed lamellipodia and filopodia formation in spindle-shaped Nap1-knockdown myoblasts. Although Nap1 is required for the regulation of WAVE activity (Innocenti et al., 2004; Steffen et al., 2004; Stradal et al., 2004) and WAVE2 regulates lamellipodia formation in C2C12 myoblasts (Kawamura, 2004), these lamellipodia formations could also be dependent upon other pathways that regulate actin-cytoskeleton remodeling because lamellipodia formation has been observed in fibroblasts lacking functional WAVE2 and Arp2/3 complexes (Steffen et al., 2006). Our data indicate a role for WAVE2, but not WAVE1, in myoblast fusion (see supplementary material Fig. S8). However, there are three WAVE isoforms in mammals (reviewed by Takenawa and Suetsugu, 2007). Furthermore, studies will be needed to address any specific roles that each of these isoforms might have during the morphological changes that we observe during differentiation, as well as their impact on fusion itself.
In this study, Nap1-knockdown myoblasts underwent the same fibroblast-like-to-polarized morphological transition as wild-type myoblasts. They were able to form myoblast-myoblast contacts, possessed no ultrastructural abnormalities and were capable of forming sarcomeric components. However, the reduction in the fusion index of Nap1-knockdown myoblasts suggests that, at a key point during the fusion process, Nap1 activity is required to complete myoblast fusion. Our live imaging of Nap1-knockdown myoblasts revealed a putative third actin-based behavior in fusing myoblasts as revealed by the localization of the PH-GFP reporter in these cells. These PH-GFP-reporter aggregates seem to be similar to structures previously identified as membrane blebs (Beli et al., 2008; Derivery et al., 2008). Their prevalence suggests that the dynamics of actin-cytoskeleton remodeling at cell membranes is impaired in Nap1-knockdown myoblasts, and that this might be crucial for mammalian myoblast fusion. Recent work describing the actin-based cellular behaviors of fusing Drosophila myoblasts revealed a novel actin structure, a focus of F-actin, at the myoblast-fusion site (Kesper et al., 2007; Richardson et al., 2007). Formation of this structure required contact and adhesion between fusing myoblasts, and the foci dissolved prior to myoblast fusion. Mutations in known fusion genes such as kette, mbc and SCAR/WAVE all lead to enlarged F-actin foci that fail to dissolve (Richardson et al., 2007). It is tempting to equate the aggregations of PH-GFP that we observed in Nap1-knockdown myoblasts with the enlarged F-actin foci identified in kette mutants. As in the Drosophila system, we found that these structures perdure far longer in Nap1-shRNA knockdown cells, seem to localize to contact points between myoblasts, and react with antibodies against Dock180, a known fusion protein in mammalian (Laurin et al., 2008; Pajcini et al., 2008), zebrafish (Moore et al., 2007) and Drosophila (Rushton et al., 1995) myoblasts. These results suggest that these foci are the remnants of a former fusion site that failed to complete the fusion process, and the lack of Nap1 activity results in a failure to disassemble these sites. However, further experiments are needed to confirm that other gene products identified as being essential for mammalian myoblast fusion localize to these aggregates, and whether these aggregates correspond to a site of fusion in mammalian myoblasts.
Despite extensive examination of the data presented in this study, we have failed to identify in our fixed or live-imaging studies the presence of an asymmetric actin wall as has been recently described in differentiating L6 myoblasts (Duan and Gallagher, 2009). One possible reason for this discrepancy could be due to the fact that the actin wall was observed in myoblasts grown on uncoated substrates (Duan and Gallagher, 2009), a culture method that differs widely from protocols used by several other laboratories (Abramovici and Gee, 2007; Holterman et al., 2007; Kafadar et al., 2009; Pajcini et al., 2008; Swailes et al., 2004) (and this study). It remains to be determined whether such a structure is substrate-dependent as a result of the culturing environment used.
Conservation of the insect myogenesis paradigm in vertebrate myogenesis
Our results add Nap1 to the growing list of vertebrate homologs of insect gene products that have a conserved role in myogenesis (Richardson et al., 2008b). For example, knockdown of WASP and Vrp1 in C2C12 myoblasts leads to reduced fusion efficiency, recapitulating the roles of these proteins during insect myoblast fusion (Kim et al., 2007). Dock180 and Brag2, the respective mammalian orthologs of Drosophila Mbc and Loner, are both crucial for both macrophage and myoblast fusion (Pajcini et al., 2008). Kirrel, the zebrafish ortholog of Drosophila Dumbfounded, is required for zebrafish fast-muscle fusion (Srinivas et al., 2007). Additionally, knockdown of Dock1 and Dock5, orthologs of Drosophila Mbc, results in the failure of fast-twitch myoblasts to fuse (Moore et al., 2007). Zebrafish orthologs of Crk, a protein that biochemically interacts with Mbc in Drosophila, also interacts with Dock1 and Dock5, strengthening the conclusion that these proteins are functionally conserved between Drosophila and zebrafish (Moore et al., 2007). Live imaging of mammalian myoblasts that are deficient in each of these gene products will prove indispensable for determining which myoblast cell behaviors and processes are affected by these gene products during the fusion process.
Materials and Methods
Plasmid constructs
Plasmids pCX::PH-GFP and pCX::myr-Ven have been described previously (Rhee et al., 2006; Tall et al., 2000). pCX::myr-RFP was generated by PCR of mRFP1 from pRSET-mRFP1 (Campbell et al., 2002) using an oligo containing a N-myristoylation sequence (5′-CTTGAATTCGCCACCATGGGAAGCAGCAAGAGCAAGCCAAAGGCCTCCTCCGAGGACGTCATCAAGG-3′) and a 3′ oligo (5′-CAAGCTTCGAATTCTTAGGCGCC-3′). The resulting PCR product was cloned into a pCRII-TOPO (Invitrogen) and the insert excised with EcoRI and cloned into pCAGGS (Niwa et al., 1991). Constructs expressing shRNA oligos against WAVE1 (Suetsugu et al., 2003), WAVE2 (Kawamura, 2004) and Nap1 (Steffen et al., 2004) have been described.
Cell culture and drug treatment
C2C12 myoblasts (ATCC) were passaged and proliferated in growth medium [GM; DMEM containing 15% fetal bovine serum supplemented with 1% glutamine, 1% penicillin/streptomycin, 0.1% gentamycin sulfate and 0.5% chick embryo extract (SLI)]. Cells were cultured at 37°C in 5% CO2 in culture dishes coated with 10% Matrigel (BD Biosciences). For differentiation and fusion, cells were rinsed with PBS and cultured in DM (DMEM containing 5% normal horse serum, supplemented with 1% penicillin/streptomycin, 1% glutamine and 0.1% gentamycin sulfate) for the times indicated. For drug treatment, latrunculin B or cytochalasin D (both Calbiochem) was added to DM as indicated. Drugs were replenished every 24 hours to prevent recovery owing to drug titration or hydrolysis.
Transfection and production of stable cell lines
C2C12 myoblasts were transfected using the Lipofectamine/Plus system (Invitrogen). Stable cell lines were generated by transfection with either empty vectors or vectors expressing Nap1-specific shRNA dimers (Steffen et al., 2004) and selected in GM containing 1 μg/ml puromycin. To prevent differences in fusion rates between each population due to variations in cell-passage number, cells from the same passage were used for transfection with control and knockdown vectors, and were passaged at the same time to ensure similar `age' of both control and experimental myoblasts. Clonal cell lines were isolated following 1 week of selection in puromycin, and evaluated for knockdown of Nap1 protein by western blotting.
Immunofluorescence and immunoblotting
Primary antibodies used were: anti-sarcomeric Mhc (MF20, DSHB), anti-Dock180 (Santa Cruz Biotechnology) and anti-N-cadherin (BD Biosciences). For all experiments, myoblasts were grown in 6-cm culture dishes, fixed in 4% paraformaldehyde, permeabilized in PBS + 0.1% Triton X-100 and blocked in PBS containing 1% bovine serum albumin (BSA). Cells were incubated overnight with primary antibodies at 4°C. Secondary detection was performed using Alexa-Fluor-488-conjugated secondary antibodies (Molecular Probes) diluted in PBS-BSA and containing Alexa-Fluor-546-conjugated phalloidin (Molecular Probes) to label the actin cytoskeleton. At the end of immunolabeling, cells were counterstained with DAPI and mounted in Prolong Gold antifade (Molecular Probes). Wide-field immunofluorescent imaging was performed using a Zeiss Axiophot microscope.
Immunoblotting was performed as described (Steffen et al., 2006), with the addition of 1% fish gelatin for membrane blocking. Anti-Nap1 and Anti-Abi1 (Steffen et al., 2004) were gifts of Theresia Stradal (Helmholtz Centre for Infection Research, Braunschweig, Germany); anti-WAVE1 (Millipore), anti-WAVE2 (Millipore) were also used. Anti-α-tubulin (Accurate Chemical) was used as a loading control.
Fusion-index calculation
The fusion index was defined as the percentage of nuclei contained in sarcomeric-Mhc-positive structures (n>three nuclei total), compared to the total number of nuclei contained completely within the given imaging field. For each experimental condition and time point, a minimum of ten fields were randomly imaged at identical magnification from immunofluorescently labeled samples. Average fusion indices and standard error were calculated using Microsoft Excel.
Confocal imaging
Immunostained cells were prepared as described above, mounted on coverslips; supporting tissue-culture plastic sandwiches were excised from tissue culture dishes and attached to glass slides using cyanoacrylate glue. Fluorescent images were acquired using a Zeiss LSM 510 confocal scanning system mounted on an Axiovert 100M microscope with a 63× 1.2 NA C-Apochromat water objective. Pinholes were set to capture optical slices of 1.0 μm. All images were processed using Adobe Photoshop. 3D reconstructions were created using Volocity Visualization (Improvision) software.
Live imaging and migration tracking
For single-fluorophore imaging, C2C12 myoblasts were transfected as described above, and allowed to grow to confluence prior to imaging. For fluorophore-mixing experiments, C2C12 myoblasts from the same passage were split into two separate dishes and transfected (see above) using the indicated reporter constructs. After transfection, the separate cells were trypsinized, mixed together and plated. Once confluent, cells were switched to DM and incubated for 24 hours at 37°C. HEPES was added to a final concentration of 50 mM and cells were immediately imaged on an Olympus inverted microscope equipped with a 37°C chamber. Time-lapse image sequences were acquired using MetaMorph software, and movies were assembled using Volocity Visualization (Improvision). The tracking of myoblasts and calculation of distance traveled, step distance and velocity measurements were performed using Volocity Quantitation software (Improvision).
Electron microscopy
Control and Nap1-knockdown myoblasts were grown on carbon-coated Aclar coverslips and fixed in 2% glutaraldehyde/2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Cells were then washed and post-fixed in 1% OsO4/1% potassium ferrocyanide for 1 hour at room temperature. Following post-fixation, cells were washed and stained en bloc with 2% uranyl acetate (aq) for 1 hour. Cells were washed and dehydrated with an ethanol series before infiltration and embedding in Spurr's resin. Thin (70-90 nm) sections were cut with a diamond knife and stained briefly with 2% uranyl acetate/1% lead citrate. Sections were viewed on an FEI Tecnai Biotwin transmission electron microscope and images were captured with a Gatan 4K×4K digital camera.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/18/3282/DC1
The authors thank Alison North, Shivaprasad Bhuvanendran and Mathieu Marchand of the Rockefeller University BioImaging Center as well as members of the Hadjantonakis laboratory for invaluable advice and technical assistance with live imaging. We also thank Kathryn Anderson, Mario Rebecchi, Theresia Stradal and Mark Tomishima for generously sharing reagents and expertise. Also, we thank David Soffar for technical support, and Geri Kreitzer, Alan Hall, Brian Richardson, Krista Dobi, Shannon Yu and members of the Baylies laboratory for discussion and advice on the manuscript. This work was supported by the Sloan Kettering Institute, NIH grants (GM078318 to M.K.B. and HD 052115 to A.-K.H.) and a MDA Research Development Grant (MDA4153) to S.J.N. Deposited in PMC for release after 12 months.
References
- Abramovici, H. and Gee, S. (2007). Morphological changes and spatial regulation of diacylglycerol kinase-ζ, syntrophins, and Rac1 during myoblast fusion. Cell Motil. Cytoskeleton 64, 549-567. [DOI] [PubMed] [Google Scholar]
- Beli, P., Mascheroni, D., Xu, D. and Innocenti, M. (2008). WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat. Cell Biol. 10, 849-857. [DOI] [PubMed] [Google Scholar]
- Berger, S., Schafer, G., Kesper, D. A., Holz, A., Eriksson, T., Palmer, R. H., Beck, L., Klambt, C., Renkawitz-Pohl, R. and Onel, S. F. (2008). WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J. Cell Sci. 121, 1303-1313. [DOI] [PubMed] [Google Scholar]
- Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A. and Tsien, R. Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabeo, R. A., Dooley, C. A., Grieshaber, S. S. and Hackstadt, T. (2007). Rac interacts with Abi-1 and WAVE2 to promote an Arp2/3-dependent actin recruitment during chlamydial invasion. Cell Microbiol. 9, 2278-2288. [DOI] [PubMed] [Google Scholar]
- Chargé, S. B. and Rudnicki, M. A. (2004). Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209-238. [DOI] [PubMed] [Google Scholar]
- Clark, P., Coles, D. and Peckham, M. (1997). Preferential adhesion to and survival on patterned laminin organizes myogenesis in vitro. Exp. Cell Res. 230, 275-283. [DOI] [PubMed] [Google Scholar]
- Clark, P., Dunn, G. A., Knibbs, A. and Peckham, M. (2002). Alignment of myoblasts on ultrafine gratings inhibits fusion in vitro. Int. J. Biochem. Cell Biol. 34, 816-825. [DOI] [PubMed] [Google Scholar]
- Coppolino, M. G., Dierckman, R., Loijens, J., Collins, R. F., Pouladi, M., Jongstra-Bilen, J., Schreiber, A. D., Trimble, W. S., Anderson, R. and Grinstein, S. (2002). Inhibition of phosphatidylinositol-4-phosphate 5-kinase ialpha impairs localized actin remodeling and suppresses phagocytosis. J. Biol. Chem. 277, 43849-43857. [DOI] [PubMed] [Google Scholar]
- Coue, M., Brenner, S. L., Spector, I. and Korn, E. D. (1987). Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213, 316-318. [DOI] [PubMed] [Google Scholar]
- Derivery, E., Fink, J., Martin, D., Houdusse, A., Piel, M., Stradal, T. E., Louvard, D. and Gautreau, A. (2008). Free Brick1 is a trimeric precursor in the assembly of a functional wave complex. PLoS ONE 3, e2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan, J. (2004). Modulation of acto-myosin contractility in skeletal muscle myoblasts uncouples growth arrest from differentiation. J. Cell Sci. 117, 3735-3748. [DOI] [PubMed] [Google Scholar]
- Doberstein, S. K., Fetter, R. D., Mehta, A. Y. and Goodman, C. S. (1997). Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J. Cell Biol. 136, 1249-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, K. R., Cave, A., Davis, D. B., Delmonte, A. J., Posey, A., Earley, J. U., Hadhazy, M. and McNally, E. M. (2005). Normal myoblast fusion requires myoferlin. Development 132, 5565-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, R. and Gallagher, P. J. (2009). Dependence of myoblast fusion on a cortical actin wall and nonmuscle myosin IIA. Dev. Biol. 325, 374-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel, L. C. and David, J. D. (1985). Temperature-sensitive non-fusing myoblast variant and spontaneous revertant: isolation and characterization. Somat. Cell Mol. Genet. 11, 325-338. [DOI] [PubMed] [Google Scholar]
- Formigli, L., Meacci, E., Sassoli, C., Squecco, R., Nosi, D., Chellini, F., Naro, F., Francini, F. and Zecchi-Orlandini, S. (2007). Cytoskeleton/stretch-activated ion channel interaction regulates myogenic differentiation of skeletal myoblasts. J. Cell. Physiol. 211, 296-306. [DOI] [PubMed] [Google Scholar]
- Holterman, C. E., Le Grand, F., Kuang, S., Seale, P. and Rudnicki, M. A. (2007). Megf10 regulates the progression of the satellite cell myogenic program. J. Cell Biol. 179, 911-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley, V. and Pavlath, G. K. (2004). Forming a multinucleated cell: molecules that regulate myoblast fusion. Cells Tissues Organs 176, 67-78. [DOI] [PubMed] [Google Scholar]
- Hromas, R., Collins, S., Raskind, W., Deaven, L. and Kaushansky, K. (1991). Hem-1, a potential membrane protein, with expression restricted to blood cells. Biochim. Biophys. Acta 1090, 241-244. [DOI] [PubMed] [Google Scholar]
- Huh, M. S., Smid, J. K. and Rudnicki, M. A. (2005). Muscle function and dysfunction in health and disease. Birth Defects Res. C Embryo Today 75, 180-192. [DOI] [PubMed] [Google Scholar]
- Ibarra, N., Pollitt, A. and Insall, R. H. (2005). Regulation of actin assembly by SCAR/WAVE proteins. Biochem. Soc. Trans. 33, 1243-1246. [DOI] [PubMed] [Google Scholar]
- Innocenti, M., Zucconi, A., Disanza, A., Frittoli, E., Areces, L. B., Steffen, A., Stradal, T. E., Di Fiore, P. P., Carlier, M. F. and Scita, G. (2004). Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 6, 319-327. [DOI] [PubMed] [Google Scholar]
- Insall, R. H. and Weiner, O. D. (2001). PIP3, PIP2, and cell movement-similar messages, different meanings? Dev. Cell 1, 743-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafadar, K. A., Yi, L., Ahmad, Y., So, L., Rossi, F. and Pavlath, G. K. (2009). Sca-1 expression is required for efficient remodeling of the extracellular matrix during skeletal muscle regeneration. Dev. Biol. 326, 47-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, N. and Gilula, N. B. (1979). Membrane events involved in myoblast fusion. J. Cell Biol. 81, 411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J. S., Feinleib, J. L., Knox, S., Ketteringham, M. A. and Krauss, R. S. (2003). Promyogenic members of the Ig and cadherin families associate to positively regulate differentiation. Proc. Natl. Acad. Sci. USA 100, 3989-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J. S., Yi, M. J., Zhang, W., Feinleib, J. L., Cole, F. and Krauss, R. S. (2004). Netrins and neogenin promote myotube formation. J. Cell Biol. 167, 493-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura, K. (2004). N-WASP and WAVE2 acting downstream of phosphatidylinositol 3-kinase are required for myogenic cell migration induced by hepatocyte growth factor. J. Biol. Chem. 279, 54862-54871. [DOI] [PubMed] [Google Scholar]
- Kesper, D. A., Stute, C., Buttgereit, D., Kreisköther, N., Vishnu, S., Fischbach, K. F. and Renkawitz-Pohl, R. (2007). Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS). Dev. Dyn. 236, 404-415. [DOI] [PubMed] [Google Scholar]
- Kim, S., Shilagardi, K., Zhang, S., Hong, S. N., Sens, K. L., Bo, J., Gonzalez, G. A. and Chen, E. H. (2007). A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev. Cell 12, 571-586. [DOI] [PubMed] [Google Scholar]
- Laurin, M., Fradet, N., Blangy, A., Hall, A., Vuori, K. and Cote, J. F. (2008). The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc. Natl. Acad. Sci. USA 105, 15446-15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L. M. and Insall, R. H. (1998). Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8, 1347-1356. [DOI] [PubMed] [Google Scholar]
- Massarwa, R., Carmon, S., Shilo, B. and Schejter, E. (2007). WIP/WASp-Based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev. Cell 12, 557-569. [DOI] [PubMed] [Google Scholar]
- Moore, C. A., Parkin, C. A., Bidet, Y. and Ingham, P. W. (2007). A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development 134, 3145-3153. [DOI] [PubMed] [Google Scholar]
- Moss, F. P. and Leblond, C. P. (1970). Nature of dividing nuclei in skeletal muscle of growing rats. J. Cell Biol. 44, 459-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, H., Yamamura, K. and Miyazaki, J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193-199. [DOI] [PubMed] [Google Scholar]
- O'Connor, R., Steeds, C., Wiseman, R. and Pavlath, G. (2008). Phosphocreatine as an energy source for actin cytoskeletal rearrangements during myoblast fusion. J. Physiol. 586, 2841-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake, Y., Tojo, H. and Seiki, M. (2006). Multifunctional roles of MT1-MMP in myofiber formation and morphostatic maintenance of skeletal muscle. J. Cell Sci. 119, 3822-3832. [DOI] [PubMed] [Google Scholar]
- Pajcini, K. V., Pomerantz, J. H., Alkan, O., Doyonnas, R. and Blau, H. M. (2008). Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. J. Cell Biol. 180, 1005-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamaneck, Y. J., Hadjantonakis, A. K. and Di Gregorio, A. (2007). Dynamic and polarized muscle cell behaviors accompany tail morphogenesis in the ascidian Ciona intestinalis. PLoS ONE 2, e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham, M. (2008). Engineering a multi-nucleated myotube, the role of the actin cytoskeleton. J. Microsc. 231, 486-493. [DOI] [PubMed] [Google Scholar]
- Rakeman, A. S. and Anderson, K. V. (2006). Axis specification and morphogenesis in the mouse embryo require Nap1, a regulator of WAVE-mediated actin branching. Development 133, 3075-3083. [DOI] [PubMed] [Google Scholar]
- Rash, J. E. and Fambrough, D. (1973). Ultrastructural and electrophysiological correlates of cell coupling and cytoplasmic fusion during myogenesis in vitro. Dev. Biol. 30, 166-186. [DOI] [PubMed] [Google Scholar]
- Rhee, J. M., Pirity, M. K., Lackan, C. S., Long, J. Z., Kondoh, G., Takeda, J. and Hadjantonakis, A. K. (2006). In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice. Genesis 44, 202-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, B. E., Beckett, K., Nowak, S. J. and Baylies, M. K. (2007). SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development 134, 4357-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, B., Beckett, K. and Baylies, M. (2008a). Visualizing new dimensions in Drosophila myoblast fusion. BioEssays 30, 423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, B. E., Nowak, S. J. and Baylies, M. K. (2008b). Myoblast fusion in fly and vertebrates: new genes, new processes and new perspectives. Traffic 9, 1050-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl, J., Crevenna, A. H., Kessenbrock, K., Yu, J. H., Neukirchen, D., Bista, M., Bradke, F., Jenne, D., Holak, T. A., Werb, Z. et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle, A. L., Machesky, L. M., Yamamoto, M., Driessens, M. H., Insall, R. H., Roth, M. G., Luby-Phelps, K., Marriott, G., Hall, A. and Yin, H. L. (2000). Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10, 311-320. [DOI] [PubMed] [Google Scholar]
- Rushton, E., Drysdale, R., Abmayr, S. M., Michelson, A. M. and Bate, M. (1995). Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development 121, 1979-1988. [DOI] [PubMed] [Google Scholar]
- Ryu, J. R., Echarri, A., Li, R. and Pendergast, A. M. (2009). Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol. Cell. Biol. 29, 1735-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, G., Weber, S., Holz, A., Bogdan, S., Schumacher, S., Muller, A., Renkawitz-Pohl, R. and Onel, S. F. (2007). The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev. Biol. 304, 664-674. [DOI] [PubMed] [Google Scholar]
- Schröter, R. H., Lier, S., Holz, A., Bogdan, S., Klämbt, C., Beck, L. and Renkawitz-Pohl, R. (2004). kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development 131, 4501-4509. [DOI] [PubMed] [Google Scholar]
- Smith, L. G. and Li, R. (2004). Actin polymerization: riding the wave. Curr. Biol. 14, R109-R111. [PubMed] [Google Scholar]
- Srinivas, B. P., Woo, J., Leong, W. Y. and Roy, S. (2007). A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nat. Genet. 39, 781-786. [DOI] [PubMed] [Google Scholar]
- Steffen, A., Rottner, K., Ehinger, J., Innocenti, M., Scita, G., Wehland, J. and Stradal, T. E. (2004). Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23, 749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen, A., Faix, J., Resch, G. P., Linkner, J., Wehland, J., Small, J. V., Rottner, K. and Stradal, T. E. (2006). Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol. Biol. Cell 17, 2581-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal, T. E., Rottner, K., Disanza, A., Confalonieri, S., Innocenti, M. and Scita, G. (2004). Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 14, 303-311. [DOI] [PubMed] [Google Scholar]
- Straube, A. and Merdes, A. (2007). EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Curr. Biol. 17, 1318-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu, S., Yamazaki, D., Kurisu, S. and Takenawa, T. (2003). Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell 5, 595-609. [DOI] [PubMed] [Google Scholar]
- Swailes, N. T., Knight, P. J. and Peckham, M. (2004). Actin filament organization in aligned prefusion myoblasts. J. Anat. 205, 381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swailes, N. T., Colegrave, M., Knight, P. J. and Peckham, M. (2006). Non-muscle myosins 2A and 2B drive changes in cell morphology that occur as myoblasts align and fuse. J. Cell Sci. 119, 3561-3570. [DOI] [PubMed] [Google Scholar]
- Takenawa, T. and Suetsugu, S. (2007). The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37-48. [DOI] [PubMed] [Google Scholar]
- Tall, E. G., Spector, I., Pentyala, S. N., Bitter, I. and Rebecchi, M. J. (2000). Dynamics of phosphatidylinositol 4,5-bisphosphate in actin-rich structures. Curr. Biol. 10, 743-746. [DOI] [PubMed] [Google Scholar]
- Tornieri, K., Welshhans, K., Geddis, M. S. and Rehder, V. (2006). Control of neurite outgrowth and growth cone motility by phosphatidylinositol-3-kinase. Cell Motil. Cytoskeleton 63, 173-192. [DOI] [PubMed] [Google Scholar]
- Vartiainen, M. K. and Machesky, L. M. (2004). The WASP-Arp2/3 pathway: genetic insights. Curr. Opin. Cell Biol. 16, 174-181. [DOI] [PubMed] [Google Scholar]
- Weiner, O. D., Rentel, M. C., Ott, A., Brown, G. E., Jedrychowski, M., Yaffe, M. B., Gygi, S. P., Cantley, L. C., Bourne, H. R. and Kirschner, M. W. (2006). Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 4, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, C., Coles, D., Entwistle, A. and Peckham, M. (1997). Myogenic cells express multiple myosin isoforms. J. Muscle Res. Cell Motil. 18, 501-515. [DOI] [PubMed] [Google Scholar]
- Yoon, S., Molloy, M. J., Wu, M. P., Cowan, D. B. and Gussoni, E. (2007). C6ORF32 is upregulated during muscle cell differentiation and induces the formation of cellular filopodia. Dev. Biol. 301, 70-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.