Abstract
Human deoxycytidine kinase (dCK) is responsible for the phosphorylation of a number of clinically important nucleoside analogue prodrugs in addition to its natural substrates, 2′-deoxycytidine, 2′-deoxyguanosine, and 2′-deoxyadenosine. To improve the low catalytic activity and tailor the substrate specificity of dCK, we have constructed libraries of mutant enzymes and tested them for thymidine kinase (tk) activity. Random mutagenesis was employed to probe for amino acid positions with an impact on substrate specificity throughout the entire enzyme structure, identifying positions Arg104 and Asp133 in the active site as key residues for substrate specificity. Kinetic analysis indicates that Arg104Gln/Asp133Gly creates a “generalist” kinase with broader specificity and elevated turnover for natural and prodrug substrates. In contrast, the substitutions of Arg104Met/Asp133Thr, obtained via site-saturation mutagenesis, yielded a mutant with reversed substrate specificity, elevating the specific constant for thymidine phosphorylation by over 1000-fold while eliminating activity for dC, dA, and dG under physiological conditions. The results illuminate the key contributions of these two amino acid positions to enzyme function by demonstrating their ability to moderate substrate specificity.
Human deoxycytidine kinase (dCK;1 EC 2.7.1.74) catalyzes the phosphorylation of 2′-deoxycytidine (dC), 2′-deoxyadenosine (dA) and 2′-deoxyguanosine (dG) to their corresponding monophosphates using nucleoside triphosphates as phosphoryl donors. This reaction is the first step of the deoxyribonucleoside salvage pathway, an alternative to de novo nucleotide biosynthesis, which, in combination with deoxyribonucleoside mono- and diphosphate kinases, provides triphosphate anabolites for DNA replication and repair (1).
In addition to recycling natural 2′-deoxyribonucleosides, dCK catalyzes the initial, often rate-determining phosphorylation of several chemotherapeutic nucleoside analogue (NA) prodrugs such as gemcitabine (2′,2′-difluorodeoxycytidine), AraC (1-β-D-arabinosylcytosine), and clofarabine [2-chloro-9-(2-deoxy-2-fluoro-β-D-arabinofuranosyl)-9H-purin-6-amine], as well as antiviral prodrugs including ddC (2′,3′-dideoxycytidine), 3TC (2′-deoxy-3′-thiacytidine), and FTC (5-fluoro-2′-deoxy-3′-thiacytidine) whose pharmacological activity depends on their triphosphate form (2–7). Given the critical role of dCK in phosphorylating 2′-deoxyribonucleosides and NA, the enzyme has undergone extensive structural and functional characterization in recent years.
Multiple sequence alignments and crystallographic studies have identified dCK as a member of the type-1 deoxynucleoside kinase (dNK) subfamily (8). Type-1 dNKs show a wide range of distinct substrate profiles including the purine-specific human deoxyguanosine kinase (dGK) and the pyrimidine-specific human thymidine kinase-2 (TK2), as well as dNK from Drosophila melanogaster (DmdNK), which is a highly efficient phosphoryl transfer catalyst for all natural 2′-deoxyribonucleosides. Such functional versatility suggests that these enzymes make an excellent framework for engineering novel substrate specificity, a thought that is further supported by their striking structural similarities. Recent crystallography studies of dCK and other family members show highly similar orientation of the protein backbone and substrate binding in the different kinases (9–13). The fact that pyrimidine, purine, and nucleoside analogue substrates bind in superimposable conformation in the active site has helped in identifying a number of enzyme–substrate interactions, yet a detailed analysis of their contributions in individual enzymes is complicated by the low sequence identity among family members (typically 30–50%). As a consequence, it has inspired a series of random mutagenesis studies on DmdNK. Early studies included mutagenesis over the entire length of the fruit fly enzyme and led to the successful identification of multiple residues throughout the kinase structure with impact on substrate specificity. However, the limited sample set and the mutation’s distance to the active site complicated a conclusive analysis (14). Follow-up experiments focused on positions Val84, Met88, and Ala110 in the DmdNK active site (Figure 1C,) substituting these residues to Ala, Arg, and Asp, respectively (15). These rational changes were inspired by the residues found in the same position in human dGK and converted the native broad-specificity enzyme into a purine-specific kinase. Similar efforts in dCK were reported in the context of the first crystal structure, demonstrating the importance of the corresponding three active site residues in dCK, Ala100, Arg104, and Asp133, in regard to substrate specificity (11). Separately, rational design studies of the enzyme’s lid region (16) and deletion of a distinct surface loop (17) have been reported but had no significant impact on the phosphoryl acceptor specificity of the enzyme.
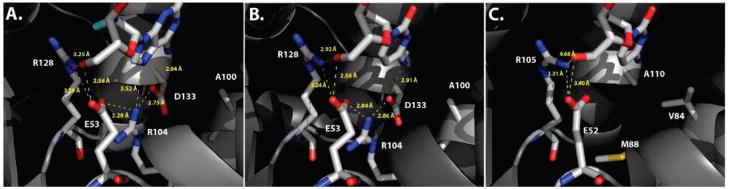
Figure 1.
Active site comparison of human dCK and DmdNK with pyrimidine and purine nucleoside substrates. Detailed view of the active sites of dCK in the presence of the 2′-deoxyadenosine analogue clofarabine (A; PDB code: 2A7Q (13) and dC (B; PDB code: 1P6O (11), as well as DmdNK with dC (C; PDB code: 1J90 (9). In all three enzymes, the nucleoside is bound in the phosphoryl-acceptor site. Hydrogen-bonding interactions between the substrate’s 5′-hydroxyl group, Arg128 (Arg105 in DmdNK) and Glu53 (Glu52) are highly conserved. The latter functions as a general base in the phosphoryl transfer reaction. In dCK, both purine and pyrimidine nucleosides form an elaborate network of hydrogen bonds between Glu53 and the exocyclic amino group on the substrate nucleobase, involving Arg104 and Asp133. The same residues in DmdNK, Met88 and Ala110, create a significantly larger binding pocket with no specific contacts, presumably allowing for the broader substrate specificity in the fruitfly enzyme. Position Ala100 (Val84 in DmdNK) flanks Asp133.
Enhancing the activity and tailoring the substrate specificity in dCK are desirable features for the creation of orthogonal NA kinases with potential future therapeutical applications. We have therefore initiated a more systematic exploration of the impact of active site mutations on the catalytic performance of dCK using rational design and combinatorial protein engineering. Our experiments confirm the critical role of amino acid positions 104 and 133 in regard to substrate specificity. Selected mutations in these two positions created “generalist” catalysts with broad specificity for the natural 2′-deoxyribonucleosides. The simplicity of converting dCK into a generalist dNK supports the notion that the human enzyme is a close relative of the last common ancestor dNK (18, 19). Furthermore, the generalist serves as an excellent template for subsequent enzyme engineering, best demonstrated by the complete reverse of dCK’s substrate specificity to an exclusive thymidine kinase in one of the characterized enzymes. Besides enhancing the phosphorylation of the four natural substrates, alternate amino acid substitutions also introduce novel catalytic activity toward thymidine analogues such as 3′-azido-thymidine (AZT) and 2′,3′-dideoxythymidine (ddT), a first step toward the creation of orthogonal NA kinases.
EXPERIMENTAL PROCEDURES
Materials
All reagents were purchased from Fisher (Pittsburgh, PA) and Sigma & Aldrich (St. Louis, MO). Enzymes were purchased from New England Biolabs (Beverly, MA) unless indicated otherwise. DNA samples were purified using the QIAquick and QIAprep purification kits (Qiagen, Valencia, CA) according to the manufacturer’s protocols. Primers were purchased from Integrated DNA Technologies (Coralville, IA). All DNA manipulations were performed in Escherichia coli DH5α-E (Invitrogen, Carlsbad, CA) using standard methodologies.
Gene Isolation and Cloning of hdck
The gene for human dCK (hdck: NCBI access # P27707) was isolated with gene-specific primers G-1 and G-2 (Table S-1, Supporting Information) from a human thymus cDNA library (Clontech, Palo Alto, CA). Prior to subcloning of hdck, an internal NdeI restriction site was removed by introducing a silent mutation in Thr98 (ACA to ACG) via primer overlap extension (20), using the mutagenic primers G-3 and G-4. The corrected PCR product was then digested with NdeI and SpeI and ligated into pDIM-PGX (21) for in vivo complementation and pET-14b (Novagen, Madison, WI) for protein overexpression. All constructs were confirmed by DNA sequencing.
Random Mutagenesis of hdck
Random mutagenesis of hdck was performed by error-prone PCR using the Gene-Morph II system (Stratagene, La Jolla, CA). Following the manufacturer’s protocol, the mutation frequency was controlled via the template concentration in the PCR. Three libraries using 1 pg, 10 pg and 100 pg of pDIM-hdck in combination with the primer pair G-1/G-2 were generated in 50 μL reaction volume. The amplification products were digested with NdeI and SpeI and ligated back into linearized pDIM vector, followed by transformation into electrocompetent E. coli KY895 cells (22). The cells were plated on LB-agar plates containing ampicillin (100 μg/mL) and incubated at 37 °C overnight. Colonies were harvested by rinsing the agar plates twice with 10 mL of 2YT medium (20), supplemented with glucose and glycerol to a final concentration of 2% and 15% respectively. After flash freezing in liquid nitrogen, library aliquots were stored at −80 °C. The mutation frequency in the three libraries was determined by DNA sequence analysis of twenty randomly picked colonies per library.
Site-Saturation Mutagenesis of hdck
Libraries with all nineteen amino acids in positions 104, 133, as well as 104 and 133 of hdck were created by primer overlap PCR amplification, using primers Arg104NNS_f/r and Asp133NNS_f/r (Table S-1). The degenerate codons (N = 1:1:1:1 mixture of dA, dG, dC, and T; S = 1:1 mixture of dG and dC) were introduced during automated primer DNA synthesis, using hand-mixing deoxynucleotide phosphoramidites to minimize library biases. The resulting PCR products were subcloned into the pDIM vector via the NdeI and SpeI restriction sites as described above, and the gene sequences were confirmed by DNA sequencing. The libraries were transformed into E. coli KY895, plated on LB-agar plates containing ampicillin (100 μg/mL) and incubated overnight at 37 °C. The resulting colonies were harvested and stored as outlined above.
In Vivo Complementation. Functional Selection for Kinase Activity
An aliquot of the frozen pDIM-hdck libraries was thawed, and 20 μL cell suspension was used to inoculate 50 mL of LB medium containing ampicillin (100 μg/mL) and incubated at 37 °C. When the culture reached an OD600 of ~0.2, a 1 mL aliquot was diluted with 9 mL of LB media and 1 mL of that solution (~2 × 106 cfu) was plated on selection plates (2% bacto-peptone; 0.5% NaCl; 1.5% noble agar; 0.2% glucose; 50 μg/mL carbenicillin; 60 μg/mL 5-fluoro-2′-deoxyuridine;2μg/mLT;12.5μg/mLuridine)(23,24). For selection experiments with the random mutagenesis libraries, the plates were incubated at ambient temperature for 36 h while site-saturation libraries were tested at 37 °C for 16 h. Colonies that appeared over that time period were picked and restreaked on fresh selection plates to confirm the tk activity, followed by DNA sequence analysis of active mutants.
Site-Directed Mutagenesis of hdck
Site-specific amino acid substitutions in hdck were introduced via primer overlap extension mutagenesis. The corresponding primer pairs (M-133f/r, M-104f/r, M-100f/r) are listed in Table S-1. The resulting PCR products were subcloned into the pDIM and pET vectors via the NdeI and SpeI restriction sites as described above, and the correct gene sequences were confirmed by DNA sequencing.
Protein Overexpression and Purification
For in vitro characterization, WT dCK and mutant kinases were overexpressed as fusion proteins with a N-terminal poly His tag. Individual kinase genes, subcloned in pET-14b, were transformed into E. coli strain BL21(DE3)pLysS (Novagen, Madison, WI). Cell cultures were grown in 200 mL of 2YT media containing ampicillin (100 μg/mL) and chloramphenicol (70 μg/mL) to an OD600 ~ 0.5 at 37 °C, and protein expression was induced with 0.1 mM IPTG for 4 h at 30 °C. The cells were centrifuged (4000g, 4 °C, 20 min) and the cell pellet resuspended in 12 mL of buffer A (50 mM Tris-HCl, pH 8; 0.3 M NaCl; 10 mM imidazole) and mixed with 60 μL of protease inhibitor cocktail (Sigma), 6 μL benzonase (Novagen) and 60 μL of lysozyme (20 mg/mL, Sigma). After 15 min incubation on the orbital shaker at 4 °C, cells were lyzed by sonication and the suspension centrifuged (10000g, 4 °C, 20 min).
The clear supernatant was mixed with 1 mL of Ni-NTA agarose resin (Qiagen), pre-equilibrated in buffer A, and incubated for 90 min at 4 °C. The resin was loaded on a Prep-column (BioRad, Carlsbad, CA) and washed with 10 column volumes (CV) of buffer A. Unspecific bound protein was then eluted with 4 CV of buffer A containing 20 mM imidazole and 2 CV of buffer A containing 50 mM imidazole, followed by protein elution in 3 CV of buffer A containing 250 mM imidazole. The combined product fractions were concentrated in an Amicon Ultra-4 ultrafiltration unit (Millipore, Bedford MA; MWCO: 10 kDa; 5000g at 4 °C) and buffer-exchanged into storage buffer (50 mM Tris-HCl, pH 8; 0.2 M NaCl, 5 mM MgCl2, 2 mM DTT), yielding protein of >95% purity as determined by SDS–PAGE. For stability reasons, the NaCl concentration in the storage buffer for mutant dCKs was increased to 0.5 M. Aliquots were stored at −80 °C after flash freezing in liquid nitrogen. The protein concentration was determined by A280 measurements (ε = 5.6 × 104 M−1 cm−1) (25).
Steady-State Kinetic Assay
The kinase activity of recombinant enzymes was determined using a spectrophotometric coupled-enzyme assay (26, 27). Briefly, 2′-deoxyribonucleosides and NAs at 1 to 5000 μM were prepared in reaction buffer containing 50 mM Tris-HCl (pH 7.5), 0.1 M KCl, 5 mM MgCl2, 1 mM DTT, 1 mM ATP, 0.21 mM phospho-enolpyruvate, 0.18 mM NADH, and 2 units/mL pyruvate kinase and 2 units/mL lactate dehydrogenase (Roche Biochemicals, Indianapolis, IN). Assays were performed at 30 or 37 °C (as indicated), measuring the absorbance change at 340 nm in the presence of 0.3–4.3 μg of enzyme per reaction. The enzyme amount was adjusted to limit NADH turnover to 10% over the time of the experiment. All experiments were performed in triplicate, and kinetic data was determined by nonlinear regression analysis using the Michaelis–Menten equation in Origin7 (OriginLab, Northampton, MA).
Circular Dichroism Spectroscopy
All CD experiments were performed on a Jasco J-810 spectropolarimeter equipped with a Peltier unit for temperature control. Samples were prepared in 50 mM potassium phosphate buffer (pH 7.4) with 0.5 M KF, 5 mM MgCl2 and 2 mM DTT. Spectra were collected at 20 °C, in a 0.1 cm path length quartz cuvette. Protein concentrations ranged from 2.3 to 3.5 μM as determined by the absorbance at 280 nm. All spectra were corrected for background buffer contribution.
Far UV spectra were collected using a scan rate of 20 nm/min, bandwidth of 2 nm and a response time of 2 s. Each spectrum represents the average of 5 accumulation scans. Thermal denaturation was monitored from 10 to 85 °C by the change in ellipticity at 222 nm with a temperature gradient of 0.85 °C/min. The data from each sample is the average of two independent experiments. The protein denaturation was irreversible for all constructs.
RESULTS
Thymidine Kinase Activity by Rational Design
Two rational design mutants of dCK with tk activity were prepared based on previous reports in the literature. Guided by the consensus sequence from multiple sequence alignments and crystal structure information, Lavie and co-workers mutated three positions in the active site, changing Ala100Val, Arg104Met, and Asp133Ala in dCK to expand the enzyme’s substrate specificity (Figure 1) (11). The substitutions in positions 104 and 133 are critical for T binding as the Arg side chain will clash with the methyl group in thymine while Asp is unable to form hydrogen-bonding interactions. In the absence of detailed kinetic data for the rational dCK mutants, we constructed the double mutant Arg104Met/Asp133Ala (rTK2) (Table 1) and triple mutant Ala100Val/Arg104Met/Asp133Ala (rTK3) by site-directed mutagenesis and determined the catalytic properties of these two enzymes with natural 2′-deoxyribonucleosides as substrates.
Table 1.
Summary of Amino Acid Substitutions in dCK and Effects on Protein Stabilitya
| amino acid residue |
||||||||
|---|---|---|---|---|---|---|---|---|
| mutant | 47 | 100 | 104 | 133 | 163 | 202 | 242 | TM (°C) |
| wild-type | D | A | R | D | N | L | F | 59.5 |
| rTK2 | M | A | 54.5 | |||||
| rTK3 | V | M | A | 51.0 | ||||
| epTK16 | Q | N | H | nd | ||||
| epTK16A | Q | N | 45.5 | |||||
| epTK6 | E | Q | G | I | L | 47.5 | ||
| epTK6A | Q | G | 44.2 | |||||
| ssTK1 | M | S | 54.5 | |||||
| ssTK1A | V | M | S | 46.1 | ||||
| ssTK2 | M | T | 51.5 | |||||
| ssTK2A | V | M | T | 48.4 | ||||
| ssTK3 | M | N | 55.6 | |||||
Single-letter abbreviations for all amino acids are used to indicate substitutions in the protein sequence. The temperature of unfolding (TM) was determined by heat denaturation in the CD spectrophotometer. All measurements were performed in duplicate and analyzed by fitting the curve to a two-state model. The TM data have a standard error of ±0.5 °C (nd = not determined).
Overall, the amino acid substitutions in rTK2 and rTK3 result in a significant enhanced pyrimidine activity with a simultaneous drop in purine specificity (Table 2). Most noticeable, the specificity constants (kobs/KM) for T increased 263-fold (rTK2) and 1105-fold (rTK3), respectively. The gains are largely due to lower KM values with an additional 5- to 10-fold increase in T turnover rates in both mutants. Besides establishing catalytic activity for T in dCK, the mutations also significantly enhanced the enzymes’ catalytic performance with dC. Although the binding constant for dC is slightly higher in the mutants compared to WT enzyme, the rate of turnover increases 10- to 20-fold, improving the specificity constant by up to an order of magnitude.
Table 2.
Rational Design: Kinetic Properties of dCK Variants with Natural 2′-Deoxyribonucleosidesa
| substrate | enzyme | KM (μM) | kobs (s−1) | kobs/KM (M−1 s−1) × 103 |
|---|---|---|---|---|
| dC | wild-type | 1.0 ± 0.2 (1) | 0.064 ± 0.002 (1) | 63 (1) |
| rTK2 | 2.0 ± 0.4 (0.5) | 0.58 ± 0.03 (9) | 287 (4.6) | |
| rTK3 | 1.54 ± 0.17 (0.7) | 1.2 ± 0.03 (19) | 777 (12) | |
| T | wild-type | 3485 ± 482 (1) | 0.38 ± 0.03 (1) | 0.095 (1) |
| rTK2 | 74 ± 11 (47) | 1.88 ± 0.09 (5) | 25 (263) | |
| rTK3 | 31.4 ± 7.4 (112) | 3.29 ± 0.29 (9) | 105 (1105) | |
| dA | wild-type | 81 ± 6 (1) | 3.10 ± 0.06 (1) | 38 (1) |
| rTK2 | 412 ± 68 (0.2) | 2.71 ± 0.19 (0.9) | 7 (0.2) | |
| rTK3 | 598 ± 95 (0.1) | 3.40 ± 0.27 (1.1) | 6 (0.16) | |
| dG | wild-type | 154 ± 10 (1) | 3.26 ± 0.07 (1) | 21 (1) |
| rTK2 | 1280 ± 63 (0.1) | 1.23 ± 0.04 (0.4) | 0.96 (0.05) | |
| rTK3 | 1364 ± 399 (0.1) | 0.31 ± 0.06 (0.1) | 0.23 (0.01) |
The experiments were performed in triplicate at 37 °C, and kinetic parameters were determined by nonlinear fit to the Michaelis–Menten equation. Relative specificity changes compared to WT enzyme are shown in parentheses.
As intended by Sabini et al. (11), the amino acid substitutions create a pyrimidine-specific human dNK by lowering the substrate specificity for purine nucleosides. In the case of dA, the steady-state kinetics indicates that mutagenesis leaves the turnover rates largely unaffected but causes an almost 5- to 7.5-fold reduction of the binding affinity from a moderate KM of 81 μM for the wild type enzyme to 412 μM (rTK2) and 598 μM (rTK3), respectively. For dG, the effects of the mutagenesis are more dramatic, showing a similar decline in the KM values as seen for dA in addition to a 10-fold drop in the kobs values. Overall, the catalytic performance of rTK2 and rTK3 with dG, compared to WT dCK, decreases 20-fold and 100-fold, respectively.
In parallel with the kinetic characterization of the dCK mutants, we evaluated the effects of mutagenesis on the overall protein structure by CD spectroscopy. Spectral scans in the far UV region did not detect any significant secondary structure changes at ambient temperatures (data not shown). Nevertheless, denaturation experiments showed a decline in the temperature of unfolding (TM) by 5 and 9 °C for the double and triple mutants, respectively (Table 1).
Random Mutagenesis and Functional Selection
Random mutagenesis was performed by PCR amplification of the 747-bp dck with low-fidelity polymerases (GeneMorphII system, Stratagene, TX). Ligation into pDIM generated a library of 4 × 105 members. We targeted a mutation frequency of ~2–3 amino acid changes per gene by adjusting the template concentration in the PCR reaction. DNA sequence analysis of 23 randomly picked library members (~18,000 bases) showed 2.67 ± 0.4 nucleotide substitution per gene, translating into 2.1 ± 0.2 amino acid changes on the protein level. Given these colony counts, our library covers approximately 30% of the theoretical diversity (~1.2 million) (28).
Functional selection for tk activity among random mutagenesis library members was carried out in E. coli KY895 (22) and yielded two unique dCK mutants. For the genetic complementation experiment, approximately 2 million library members were plated on selection plates and incubated at either 37 °C or ambient temperature. The lower temperature reduces the selection pressure for kinase activity as it slows bacterial growth, facilitating proper folding of mutant proteins and better accommodating enzymes with compromised overall stability. While no colonies were found at 37 °C, 14 colonies grew at room temperature. Among the successful candidates, two sets of sequences with distinct mutations were identified. The first mutant (epTK6) carried five amino acid substitutions (Asp47Glu/Arg104Gln/Asp133Gly/Asn163Ile/Phe242Leu). The second mutant (epTK16) encoded three amino acid changes (Arg104Gln/Asp133Asn/Leu202His). To further evaluate the catalytic performance of the two functional dCK mutants, we subcloned their corresponding genes into pET-14b and overexpressed the protein product in E. coli BL21 as described in Experimental Procedures.
In Vitro Characterization of Selected dCK Mutants from Random Mutagenesis
The overexpression of epTK6 and epTK16 yielded soluble protein at WT dCK levels for epTK6 but resulted in protein aggregates in the case of epTK16. Inspection of the three mutations in epTK16 by multiple sequence alignment against other type-I dNKs and the dCK crystal structure suggested that mutagenesis of the highly conserved Leu202 could at least in part be responsible for inclusion body formation. To stabilize epTK16, we reversed Leu202His by site-directed mutagenesis, creating the double mutant epTK16A (Arg104Gln/Asp133Asn). The new construct also complemented the E. coli auxotroph and, more importantly, produced soluble protein in good yields upon overexpression in the pET-system at 30 °C. Subsequent analysis of epTK16A by CD showed no significant deviation from the WT dCK spectra at room temperature, yet TM measurements indicated that the two remaining mutations still cause a dramatic decrease in protein thermostability by 14°C (Table 1). We reengineered epTK6 in a similar fashion, removing all but Arg104Gln/Asp133Gly to create epTK6A. In contrast to epTK16A, the removal of the three distant substitutions in epTK6 seems to reduce protein stability compared to the parental sequence. While the five mutations in epTK6 cause a 12 °C decline in protein stability, the CD analysis of epTK6A showed a reduction of its TM by 15 °C, suggesting that the three distant mutations actually compensate in part for the protein’s destabilization by the mutations in the active site. The conservative nature of the substitutions and their distance to the active site make it difficult to further rationalize these effects.
The kinetic properties of epTK6, as well as the two redesigned mutants epTK6A and epTK16A, were determined for all four natural dNs (Table 3). Similar to the rational design mutants, the specificity constants for dC and T increase significantly. For dC, all three mutants show 2- to 6-fold increases in kobs/KM. In the case of epTK6 and epTK6A, the apparent binding constants are unchanged and gains are largely due to higher turnover numbers. In contrast, epTK16A shows an almost 10-fold higher KM value, possibly caused by unfavorable steric and electrostatic interactions of the Asp133Asn mutation with the exocyclic amine on the substrate. The decline in apparent binding affinity is compensated by a 20-fold increase in kobs, resulting in an overall 2-fold improvement in catalytic performance. For T, all three candidates show increases in their specificity constants by more than 2 orders of magnitude, a change that is mostly due to 100-fold lower KM values. The similarity of the kinetic parameters in all three enzymes suggests that the observed improvements in T phosphorylation are not resulting from newly created beneficial interactions but more likely arise from eliminating unfavorable steric (Arg104) and electrostatic interactions (Asp133).
Table 3.
Random Mutagenesis: Kinetic Properties of dCK Variants with Natural 2′-Deoxyribonucleosidesa
| substrate | enzyme | KM (μM) | kobs (s−1) | kobs/KM (M−1s−1) × 103 |
|---|---|---|---|---|
| dC | wild-type | 1.0 ± 0.2 (1) | 0.064 ± 0.002 (1) | 63 (1) |
| epTK16A | 8.6 ± 1.5 (0.1) | 1.30 ± 0.06 (20) | 152 (2.4) | |
| epTK6 | 1.41 ± 0.36 (0.7) | 0.21 ± 0.01 (3.3) | 150 (2.4) | |
| epTK6A | 1.04 ± 0.12 (1) | 0.37 ± 0.01 (5.8) | 356 (5.7) | |
| T | wild-type | 3485 ± 482 (1) | 0.38 ± 0.03 (1) | 0.095 (1) |
| epTK16A | 36 ± 7 (97) | 1.51 ± 0.08 (4) | 42 (442) | |
| epTK6 | 25 ± 5 (139) | 0.68 ± 0.04 (1.8) | 27 (284) | |
| epTK6A | 25.7 ± 5.5 (134) | 1.01 ± 0.05 (2.7) | 39 (410) | |
| dA | wild-type | 81 ± 6 (1) | 3.10 ± 0.06 (1) | 38 (1) |
| epTK16A | 532 ± 99 (0.2) | 2.00 ± 0.15 (0.6) | 4 (0.1) | |
| epTK6 | 91 ± 21 (0.9) | 1.08 ± 0.07 (0.3) | 12 (0.3) | |
| epTK6A | 231 ± 25 (0.4) | 1.75 ± 0.07 (0.6) | 8 (0.2) | |
| dG | wild-type | 155 ± 10 (1) | 3.26 ± 0.07 (1) | 21 (1) |
| epTK16A | 240 ±23 (0.6) | 1.92 ± 0.06 (0.6) | 8 (0.4) | |
| epTK6 | 79 ± 10 (2) | 1.18 ± 0.05 (0.4) | 15 (0.7) | |
| epTK6A | 81 ± 11 (2) | 1.50 ± 0.07 (0.5) | 19 (0.9) |
The experiments were performed in triplicate at 30 °C, and kinetic parameters were determined by nonlinear fit to the Michaelis–Menten equation. Relative specificity changes compared to WT enzyme are shown in parentheses.
While rational design and random mutagenesis appear to affect pyrimidine substrate specificity similarly, the trends in the kinetic parameters for purines are noticeably different. In the case of dA, the lower catalytic activity is not caused by higher Michaelis–Menten constants alone as seen for the rational design mutants but results equally from a reduction in the kobs and KM values. For dG, all three random mutagenesis enzymes show WT-like catalytic performance, generally experiencing less than 2-fold changes in the kinetic parameters. These results are in sharp contrast to the 20- to 100-fold lower kobs/KM values measured for rTK2 and rTK3.
In summary, we have used random mutagenesis to search for alternative solutions to introducing tk activity in dCK. The tk selection in a large library with multiple random amino acid substitutions shows a clear preference for library members with mutations in position 104 and 133, suggesting that these two residues are major determinants for T specificity in dCK. Nevertheless, our results also reveal some limits to the chosen experimental design. The application of random mutagenesis did not yield either one of the rational design variants rTK2 or rTK3. The result can be explained by the fact that an Arg104Gln change is possible by a single nucleotide substitution (CGA to CAA) while Arg104Met (CGA to ATG) requires simultaneous nucleotide changes in all three codon positions (29). Confronted with the possibility that alternative amino acid substitutions in these two residues could produce enzymes with tk activity, we applied site-saturation mutagenesis to completely randomize both positions.
Site-Saturation Libraries
We generated three site-saturation libraries covering amino acid positions 104 and 133 individually, as well as the double mutant 104 and 133. These experiments were motivated by our findings that rational design and random mutagenesis produced two distinct solutions for tk activity in dCK.
The individual site-saturation libraries were prepared with the help of degenerate NNS primers which offers close to natural distributions for all amino acids while minimizing the occurrence of stop codons (30). The sizes for the Arg104NNS and Asp133NNS single-site libraries (theoretical library size = 20) were 7000 cfu and 1500 cfu, respectively. The Arg104NNS/Asp133NNS library consisted of 1.5 × 105 members, providing almost 400-fold coverage of the maximum diversity (theoretical library size = 400). To evaluate the codon distribution in both positions in the naive libraries, DNA sequences from 48 random samples of the Arg104NNS/Asp133NNS library were analyzed (Table S-2, Supporting Information). In position 104, the nucleotide distribution is relatively well balanced. We found 17 of the 20 natural amino acids in our small sample set. Consistent with the degeneracy of the genetic code, the most frequently found amino acids were Ser and Arg, closely followed by Leu, Pro, Ala, Val, and Gly. Furthermore, multiple codons for individual amino acids could be identified. The dA in the third nucleotide position maybe the result of contamination during oligonucleotide synthesis. The nucleotide distribution at Asp133 is less ideal, showing clear biases in all three codon positions. Despite the skewed distribution, resulting in the overrepresentation of Pro and Leu, our sample set still contained 14 different amino acids, suggesting that all natural amino acids are represented in the current libraries.
The three libraries were subsequently tested for members with tk activity, using the previously described E. coli auxotroph. For both single-site mutagenesis libraries, the genetic complementation experiments did not yield any colonies. As both libraries are comprehensive and were sampled at 10-fold redundancy, we concluded that individual amino acid substitutions in either location are insufficient to establish tk activity for in vivo complementation. In contrast, the double mutant library yielded 16 cfu when incubated at 37 °C. DNA sequence analysis shows that Met in position 104 dominates among the functional variants (13 out of 16) while Gln is a less frequent alternative (3/16). Position 133 tolerates a broader range of amino acid substitutions, showing Thr (7/16), Ser (4/16), and Asn (3/16) as the most common replacements while Ala and Ile were each found once. In a separate experiment, we tested the same library for growth complementation at 30 °C. Interestingly, the lower incubation temperature yielded fewer colonies, forming only 5 cfu. DNA sequence analysis revealed that about half the functional genes carried the previously observed Arg104Met (2/5) substitution while the rest encoded for Arg104Asn (3/5). The reason for the absence of Arg104Gln at lower temperature is unclear. In position 133, these mutants carried the previously identified Asp133Ala, Thr or Ser substitutions. The diversity of amino acid substitutions at position 133 seems unaffected by the change in growth temperature.
Focusing on the functional mutants from the 37 °C selection experiment, we identified and characterized three new amino acid combinations, Arg104Met/Asp133Ser (ssTK1), Arg104Met/Asp133Thr (ssTK2), and Arg104Met/Asp133Asn (ssTK3). As these mutants were among the most frequently selected library members, we hypothesized that such dominance reflects superior function. We therefore overexpressed all three mutants in the pET system and purified them to homogeneity. The three proteins show equal or higher temperatures of unfolding than the proteins from rational and random mutagenesis with transition midpoints at 52 °C (ssTK2) and 55 °C (ssTK1 and ssTK3), respectively. As for previous dCK mutants, no significant changes in the far UV CD spectra were observed.
The kinetic data for these three new mutants reveal distinct substrate profiles, showing a conspicuous shift toward general pyrimidine kinase activity (Table 4). The substitution of Asp133Ser in ssTK1 increases the catalytic turnover for T and dC by 6- and 26-fold over WT while showing a substantially lower KM value for T (19 μM, 183-fold) and only a moderate increase in the binding affinity for dC (4.6 μM, 4.5-fold). The same mutations result in a 5- to 8-fold increase of the apparent binding constants for purines while leaving their turnover rates largely unaffected (<2-fold). A similar trend can be observed for ssTK2 where the Asp133Thr substitution further enhances the tk activity by reducing the KM for T to 5.8 μM. Combined with a 4-fold increase in turnover, ssTK2’s specificity constant for T surpasses even the rational design triple mutant rTK3 by 2.5-fold, reaching a 2700-fold improvement in kobs/KM. Interestingly, the methyl side chain in Thr133 seems to interfere with dC binding, increasing its KM from 1 μM in WT to 21 μM in ssTK2. The reduced apparent binding constant is in part compensated by a 45-fold increase in kobs, still doubling the specificity constant for dC in respect to WT dCK. Finally, ssTK3 represents a hybrid of rational and random mutagenesis, carrying the consensus substitution Arg104Met of the former in combination with the unconventional Asp133Asn mutation found in the latter. The significantly higher stability of ssTK3 in comparison to epTK16A suggests that Arg104Gln is responsible for the destabilization of the mutant enzymes identified in the random approach. Otherwise, the mutant enzyme shows similar rate changes for pyrimidine and purine substrates as the other two ssTK constructs.
Table 4.
Site-Saturation Mutagenesis: Kinetic Properties of dCK Variants with Natural 2′-Deoxyribonucleosidesa
| substrate | enzyme | KM (μM) | kobs (s−1) | kobs/KM (M−1 s−1) × 103 |
|---|---|---|---|---|
| dC | wild-type | 1.0 ± 0.2 (1) | 0.064 ± 0.002 (1) | 63 (1) |
| ssTK1 | 4.55 ± 0.6 (0.2) | 1.70 ± 0.06 (26) | 373 (5.9) | |
| ssTK1A | 5.25 ± 0.45 (0.2) | 2.10 ± 0.06 (33) | 400 (6.3) | |
| ssTK2 | 20.9 ± 1.5 (0.05) | 2.86 ± 0.07 (45) | 137 (2.1) | |
| ssTK2A | 71 ± 8 (0.01) | 3.54 ± 0.11 (55) | 50 (0.8) | |
| ssTK3 | 11.8 ± 1.5 (0.08) | 2.23 ± 0.10 (35) | 189 (3) | |
| T | wild-type | 3480 ± 482 (1) | 0.38 ± 0.03 (1) | 0.095 (1) |
| ssTK1 | 18.9 ± 2.3 (183) | 2.33 ± 0.08 (6) | 123 (1294) | |
| ssTK1A | 5.58 ± 0.58 (625) | 1.91 ± 0.05 (5) | 342 (3600) | |
| ssTK2 | 5.8 ± 0.6 (603) | 1.49 ± 0.04 (4) | 258 (2715) | |
| ssTK2A | 3.88 ± 0.22 (902) | 1.57 ± 0.02 (4) | 406 (4273) | |
| ssTK3 | 25.6 ± 3.6 (134) | 2.27 ± 0.10 (6) | 88 (926) | |
| dA | wild-type | 81 ± 6 (1) | 3.10 ± 0.06 (1) | 38 (1) |
| ssTK1 | 398 ± 27 (0.2) | 4.85 ± 0.13 (1.6) | 12 (0.3) | |
| ssTK1A | 843 ± 80 (0.1) | 3.37 ± 0.10 (1.1) | 4 (0.1) | |
| ssTK2 | 691 ± 128 (0.1) | 4.14 ± 0.35 (1.3) | 6 (0.16) | |
| ssTK2A | 1437 ± 179 (0.05) | 2.67 ± 0.13 (0.9) | 2 (0.05) | |
| ssTK3 | 452 ± 88 (0.2) | 3.47 ± 0.30 (1.1) | 8 (0.2) | |
| dG | wild-type | 155 ± 10 (1) | 3.26 ± 0.07 (1) | 21 (1) |
| ssTK1 | 1174 ± 109 (0.13) | 4.38 ± 0.23 (1.3) | 4 (0.2) | |
| ssTK1A | 739 ± 61 (0.21) | 1.72 ± 0.04 (0.5) | 2 (0.1) | |
| ssTK2 | 1203 ± 179 (0.13) | 2.5 ± 0.18 (0.8) | 2 (0.1) | |
| ssTK2A | 1164 ± 107 (0.13) | 2.29 ± 0.08 (0.7) | 2 (0.1) | |
| ssTK3 | 324 ± 31 (0.5) | 3.59 ± 0.12 (1.1) | 11 (0.5) |
The experiments were performed in triplicate at 37 °C, and kinetic parameters were determined by nonlinear fit to the Michaelis–Menten equation. Relative specificity changes compared to WT enzyme are shown in parentheses.
In a follow-up experiment, we introduced Ala100Val as a third mutation in ssTK1 and ssTK2 via site-directed mutagenesis, creating ssTK1A and ssTK2A (Table 4). The introduction of Ala100Val did reduce the overall thermostability of ssTK2 and ssTK1 by 3 and 8 °C, respectively, similar to the drop observed for rTK3. At the same time, the third substitution seemed to magnify the trend in substrate specificity change observed for the double mutants. The additional amino acid change raised kobs and KM for dC in both enzymes to the extent of leaving their specificity constants largely unaffected (<2-fold). The most dramatic change in the kinetic parameters is a 3-fold raise in the apparent binding constant in ssTK2A. We speculate that the more bulky valine side chain increases the steric rigidity of the adjacent Thr133, orienting its side chain in such a way that steric clash with the exocyclic amino group of dC prevents effective substrate binding. In contrast, the reduction in side chain flexibility seems to benefit T as an enzyme substrate as Ala100Val results in even lower KM values. At 3.88 μM, ssTK2A shows a reduction in KM by almost 3 orders of magnitude, the lowest apparent binding constant for T of any dCK mutants characterized so far. The effect can be rationalized by a more favorable orientation of the hydroxyl group of Thr for hydrogen bonding with the carbonyl in the 4-position on thymine. Overall, the three mutations in ssTK2A raise the kobs/KM for T to 4.06 × 105 M−1 s−1, more than 4000-fold above WT dCK. For purine substrates, the additional substitution in position 100 lowers the specificity constants for dA about 3-fold while leaving dG phosphorylation largely unaffected. The decline in activity for dA can mostly be attributed to increased KM values. Interestingly, the Ala100Val substitution in rTK3 resulted in exactly the opposite effect, reducing the kobs/KM for dG but not significantly altering the kinetic parameters for dA.
Nucleoside Analogue Activity of dCK Mutants
Beyond our primary study of the effects of active site mutations on the substrate specificity of natural dNs, we evaluated WT dCK and six of the dCK mutants for their activities on five pyrimidine nucleoside analogues with distinct sugar modifications; ddT, AZT, 3TC, AraC, and gemcitabine (Table 5). Although none of the dCK mutants were selected for those particular substrates, our active site changes likely extend beyond nucleobase recognition and affect binding interactions with the sugar moiety of the substrate as well.
Table 5.
Substrate Specificities of Wild-Type and Mutant dCKsa
| substrate | ddT | AZT | 3TC | AraC | gemcitabine |
|---|---|---|---|---|---|
| wild-type | 6.5 (1) | 1.0 (1) | 88 (1) | 984 (1) | 1648 (1) |
| rTK3 | 64 (10) | 43 (43) | 149 (1.7) | 3308 (3.4) | 4530 (2.7) |
| epTK6 | 206 (32) | 136 (136) | 815 (9.3) | 4400 (4.5) | 6460 (3.9) |
| ssTK1 | 28 (4.3) | 8.5 (8.5) | 545 (6.2) | 3339 (3.4) | 5624 (3.4) |
| ssTK1A | 186 (29) | 107 (107) | 270 (3.1) | 3177 (3.2) | 6009 (3.6) |
| ssTK2 | 108 (17) | 45 (45) | 114 (1.3) | 1387 (1.4) | 4267 (2.6) |
| ssTK2A | 135(21) | 113 (113) | 26 (0.3) | 599 (0.6) | 2262 (1.3) |
Activities (μM/min per mg enzyme) were determined by spectrophotometric assay at a constant substrate concentration of 500 μM (standard error: ±10%). Relative specificity changes compared to WT enzyme are shown in parentheses.
The kinetic data for AraC and gemcitabine show moderate improvements in activity among the engineered kinases. We attribute the observed 2- to 4-fold activity increases for both NAs, which are already good substrates for the WT dCK, mostly to changes in position 104. Substitution of Arg104 might be beneficial to directly or indirectly (by repositioning the neighboring Glu53 and Arg128) reduce unfavorable steric and electrostatic interactions with the additional substituent at the 2′-position of the ribose (11). The same rationale could also explain the up to 10-fold activity enhancement observed for 3TC (10). The dramatic improvement in activity for NAs with thymine nucleobases is consistent with the newly acquired T specificity in the dCK mutants. While WT dCK shows only residual activity for either analogue, the six mutant enzymes show up to 30-fold higher activity for ddT and a 100-fold increase for AZT turnover. Assuming that the enzymes operate under Vmax conditions at 500 μM substrate, it is interesting to note that the activity changes for the NAs do not correlate with the changes measured for the natural counterparts T and dC. In epTK6, dC activity increases 3.3-fold while 3TC turnover raises 9.3-fold. In contrast, ssTK2A has 55-fold higher activity for dC but a 3-fold decline in 3TC activity. Similarly, epTK6 shows 1.8-fold higher turnover for T and 136-fold activity increase for AZT while ssTK1 shows similar moderate rate increases for T and AZT of 6- and 8.5-fold, respectively. The absence of a clear correlation between data from the native substrates and the NAs supports the notion that the amino acid substitutions in position 104 and 133 extend beyond the nucleobase binding portion of the active site.
Finally, a comparison of the kinetic properties of dCK mutants based on the engineering strategy reveals some interesting differences in the enzyme’s response. To introduce tk activity, rational design based on multiple sequence alignments correctly identifies residues Arg104 and Asp133 in human dCK as the critical amino acids for the desired changes in substrate specificity. However, faster and more specific mutants could be isolated from site-saturation mutagenesis libraries, targeting these same two positions, in combination with genetic selection in the tk-deficient E. coli KY895. On the other hand, random mutagenesis led to the identification of a generalist, a mutant with substantial activity for all four natural dNs as well as NAs. EpTK6 consistently shows the highest activity for all tested NA substrates.
DISCUSSION
Initial work by Lavie and co-workers suggested that the introduction of tk activity in dCK was relatively straightforward. The authors hypothesized that the WT enzyme’s inability to phosphorylate T was related to steric clashes and adverse electrostatics of the thymine nucleobase in the active site. Supporting their hypothesis, the authors identified amino acid positions 100, 104, and 133 as responsible for the unfavorable interactions. Inspired by the broad substrate specificity of DmdNK, they mutated the three residues in dCK to the corresponding amino acids in the fruit fly kinase improved the dCK activity for T. As our subsequent kinetic study of these mutants shows, the 1000-fold enhanced catalytic activity is largely due to a lower KM value. These findings are consistent with the model of an enlargement of the active site binding pocket. Nevertheless, our experiments also reveal a drop in catalytic activity for purine nucleosides associated with the expansion of the active site which seems at first counterintuitive. The moderate decline in activity for dA can be rationalized by the removal of a hydrogen-bonding interaction between the nucleoside’s exocyclic amino group and the side chain carboxylates on Asp133. The 10-fold drop in the specificity constant for dA can be accounted for by the higher Michaelis–Menten constants, a trend also seen with dC which, according to the crystal structure, benefits from the same noncovalent interaction. In contrast, the reduced specificity constant for dG by an additional order of magnitude is more difficult to explain. Given the similar trends in the kinetics of dA and dC, we expected the size difference between pyrimidines and purines to be of lesser importance to catalysis. This led us to predict that an expanded active site and favorable enzyme–substrate interactions, which greatly improves tk activity, should be beneficial for dG phosphorylation as well. However, our steady-state kinetics does not support such a hypothesis. The rationale for these inconsistencies is unclear. Potential steric conflicts, arising from dG’s exocyclic amino group, are ruled out based on the structural data for clofarabine (13). The dCK/clofarabine structure shows a sufficiently large binding pocket for substituents in the 2-position of the purine base. Furthermore, multiple sequence alignments indicate a high degree of conservation in that region of the active site among all members of the type-I dNK family, providing no indication of special accommodation for dG in dedicated purine kinases such as human dGK. A review of our current dG binding model, based on crystallographic information for dA and its derivatives, which places the heterocycle in anticonformation relative to the sugar moiety, might be necessary. Alternative binding modes to consider might include the syn-orientation of the guanine base in the bound state, as well as the relocation of protein side chains and backbone elements. The data analysis would also greatly benefit from crystal structure information of dG bound in the phosphoryl acceptor site of dCK or other type-I dNK family members.
Another interesting test of the functional relevance of positions 100, 104, and 133 in dCK is the comparison of our kinetic data with previously published results for the reverse experiment in DmdNK. Introducing Val84Ala, Met88Arg, and Ala110Asp in DmdNK which corresponds to positions 100, 104, and 133 in dCK, Knecht and co-workers observed a substantial decline in T and dC kinase activity while purine substrates were largely unaffected and, in the case of dG, even slightly improved (15). Overall, these findings suggest that the current model captures the critical interaction of the enzyme with pyrimidine deoxyribonucleosides, providing a relatively accurate predictive framework, but is less reliable to forecast effects on purine substrates. The data confirm that residues 104 and 133 are critical for substrate specificity while alterations in position 100 generally enhance the trend in specificity seen in the double mutants.
Naturally, the results from the rational design study did raise the question whether (a) Arg104Met and Asp133Ala are the only two mutations leading to tk activity in dCK and (b) there are alternative amino acid substitutions in these two positions or elsewhere in the protein that could accomplish the same change in substrate specificity. We addressed these questions via two complementary techniques: whole-gene random mutagenesis and site-saturation mutagenesis at positions 104 and 133 of human dCK.
In searching for mutations throughout the entire dCK sequence that allowed for T phosphorylation in the E. coli auxotroph, our random mutagenesis library yielded two unique variants with three and five amino acid substitutions, respectively. Besides amino acid changes in positions distant to the active site, both variants showed mutations in positions 104 and 133. Amino acid Arg104 was changed to Gln in both mutants while Asp133 was replaced by either Asn or Gly. In subsequent experiments, the reversion of the distant mutations to WT showed little change in catalytic performance, clearly linking the gains in tk activity to the two active site residues.
Interestingly, the kinetic analysis of these two variants obtained by random mutagenesis shows very distinct substrate specificity profiles compared to the rational design constructs. Even though both groups of enzymes were selected via in vivo genetic complementation, the mutants from the error-prone PCR experiment behave less like pyrimidine kinases but instead show a more balanced substrate profile than rTK2 and rTK3. The Arg104Gln/Asp133Gly mutations in epTK6A improve tk activity 400-fold, mostly by lowering the KM value for T, but cause less than 6-fold change in the catalytic parameters for dC, dA and dG. With only two amino acid changes, we have been able to create a human dNK with respectable turnover rates for all four natural dNs. The ease of such transformation is intriguing in light of the recently put forward idea of enzyme evolution via a generalist as intermediate state of functional divergence (31). Along the same lines, evidence that changes in position 104 and 133 are not limited to enzymes with broader substrate specificity was found in ssTK2A whose substrate profile indicates a complete inversion, turning the dCK into a thymidine kinase with no significant catalytic activity for the other three natural 2′-deoxynucleosides (see below).
Given the perceived importance of positions 104 and 133 in dCK with regard to nucleobase specificity, we chose site-saturation mutagenesis of those two residues to overcome the shortcomings of random mutagenesis and obtain a complete set of functional amino acid combinations. The results from this study have helped to elucidate the functional role of these two residues. Position 104 tolerates very limited diversity, accommodating only Met and Gln at 37 °C, as well as Asn at 30 °C selection conditions. In the case of Met104, the loss of hydrogen-bonding interactions with Glu53, whose role as a general base involves deprotonating the 5′-hydroxyl group of the substrate, can account for the moderate 5 °C decline of TM in Arg104Met. Nevertheless, the crystal structure of DmdNK suggests that the new residue fits well into a hydrophobic region near Phe126 below the substrate binding pocket. The consequences of the Arg104Gln substitution are less certain. A comparison of epTK16A and ssTK3 which share Asp133Asn but carry either Arg104Met or Gln links the observed decline in protein stability to the Gln residue. We speculate that Gln104 assumes a similar downward orientation as Met, possibly interacting with Gln108 on the adjacent helical turn. Although accommodating Gln104’s polar side chain is less favorable, the strikingly similar trend in the kinetic parameters for epTK16A and ssTK3 argues against a direct involvement of the amino acid in that position in substrate binding or catalysis. As such, our kinetic data are consistent with the proposed orientation of the side chain, points away from the active site. The high degree of conservation at this position despite no direct role in substrate binding might reflect the residue’s structural role and its contribution to overall protein stability. Furthermore, the identification of these specific mutations could be skewed by the auxotrophic selection system for T phosphorylation.
In position 133, the substitution of Asp with small hydrophobic residues such as Ala or Gly seems a suitable compromise for substrate binding and catalysis with all four natural dNs. The amino acid change eliminates unfavorable electrostatic interactions for binding of T while only slightly compromising the Michaelis–Menten constants for dC and dA that benefit from an additional hydrogen bonding interaction in the WT enzyme. In the case of dG, all mutations except Asp133Gly appear detrimental to enzyme activity. An argument for steric constraints seems unsupported by the kinetic data and the structural data for dCK with bound clofarabine as discussed above. In the absence of direct structural evidence, we hypothesize that the introduction of Gly133 can result in higher backbone flexibility, which could aid conformational changes in the enzyme along the reaction coordinate.
Site-saturation mutagenesis in combination with selection for tk activity also identified a second category of functional substitutions in position 133: small polar amino acids such as Ser, Thr, and Asn. In contrast to the rational design mutants with pyrimidine kinase specificity and the random mutagenesis variants with broad dNK specificity, the introduction of Ser and Thr resulted in a complete inversion of dCK’s substrate specificity. Best demonstrated by ssTK2, Thr133 raises the apparent binding affinity for dC and the purine substrates which we attribute to unfavorable interactions between the amino acid side chain and the nucleobase moiety of the substrate. At the same time, the hydroxyl side chain can establish hydrogen-bonding interactions with the thymine base, explaining its dramatically lower KM value for T. The introduction of Ala100Val in ssTK2A further amplifies the trend, resulting in a dCK variant that, at physiological substrate concentrations in the low micromolar range, functions primarily as a thymidine kinase.
In summary, our study demonstrates that positions 104 and 133 are responsible for the nucleobase specificity of dCK. Substitutions in position 133 can directly influence substrate specificity via steric and electrostatic effects, allowing for tailoring of dCK’s native substrate profile to a “generalist” kinase and to a thymidine kinase. In contrast, position 104 seems to play only an indirect role in substrate specificity by shaping the substrate-binding pocket of the mutant enzymes. Even though the two residues in this study are part of the active site region near the phosphoryl acceptor’s nucleobase, our experiments with NAs confirm that the effects of mutagenesis are not locally restricted. The five antiviral and cancer prodrugs in our tests (Table 5) all carry native thymine or cytosine moieties, yet have undergone various degrees of modifications in the ribose portion. The results of our activity measurements clearly show in some cases substantial rate enhancements for the phosphorylation of these NAs. Furthermore, the previously discussed “generalist” behavior of error-prone PCR mutant epTK6 extends to these substrates. The Arg104Gln/Asp133Gly mutations yield the most active kinase for all five NAs.
Our experiments demonstrate the simplicity by which the substrate specificity of dCK, and presumably other members of the type-I dNK family, can be manipulated. These results are encouraging in light of various hypotheses about the evolutionary trajectory of dNKs. Nevertheless, a major limitation in the current study is the inability to select or screen for nucleoside kinase activity other than T. Our experiments rely on the genetic complementation of the tk-deficient E. coli strain KY895 for library analysis, inevitably biasing the results toward tk activity. The absence of screening and selection protocols that allow direct testing for dC, dA and dG activity is a major problem and severely restricts progress in the field. Future work would greatly benefit from the development of novel screening techniques, not only allowing for more comprehensive tests on the enzyme’s residues responsible for nucleobase specificity but also offering the opportunity to expand the active site probing to residues responsible for recognizing and discriminating the sugar moiety of the substrate. As many nucleoside analogue prodrugs carry key modifications in the ribose portion, such a protocol would be invaluable to directly identifying dNKs with higher NA activity and even generate orthogonal NA kinases.
Supplementary Material
Footnotes
The authors would like to acknowledge financial support in part by NIH Grant GM69958 and by a grant to the Emory Center for AIDS Research (AI050409) from the NIH and by institutional funding from the Emory University Health Science Center.
Abbreviations: dCK, deoxycytidine kinase; dNK, deoxynucleoside kinase; tk, thymidine kinase; ATP, adenosine triphosphate; dN, 2′-deoxyribonucleoside; dC, 2′-deoxycytidine; T, thymidine; dA, 2′-deoxyadenosine; dG, 2′-deoxyguanosine; NA, nucleoside analogue; WT, wild type.
SUPPORTING INFORMATION AVAILABLE
Oligonucleotide sequences used as primers for the site-directed mutagenesis (Table S-1) and statistical data on the codon distribution of the site-saturation mutagenesis experiments (Table S-2). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Arner ES, Eriksson S. Mammalian deoxyribonucleoside kinases. Pharmacol Ther. 1995;67:155–186. doi: 10.1016/0163-7258(95)00015-9. [DOI] [PubMed] [Google Scholar]
- 2.Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues and nucleobases in cancer treatment. Lancet Oncol. 2002;3:415–424. doi: 10.1016/s1470-2045(02)00788-x. [DOI] [PubMed] [Google Scholar]
- 3.Johansson M, Karlsson A. Differences in kinetic properties of pure recombinant human and mouse deoxycytidine kinase. Biochem Pharmacol. 1995;50:163–168. doi: 10.1016/0006-2952(95)00129-n. [DOI] [PubMed] [Google Scholar]
- 4.Kewn S, Veal GJ, Hoggard PG, Barry MG, Back DJ. Lamivudine (3TC) phosphorylation and drug interactions in vitro. Biochem Pharmacol. 1997;54:589–595. doi: 10.1016/s0006-2952(97)00189-5. [DOI] [PubMed] [Google Scholar]
- 5.Kierdaszuk B, Krawiec K, Kazimierczuk Z, Jacobsson U, Johansson NG, Munch-Petersen B, Eriksson S, Shugar D. Substrate/inhibitor specificities of human deoxycytidine kinase (dCK) and thymidine kinases (TK1 and TK2) Adv Exp Med Biol. 1998;431:623–627. doi: 10.1007/978-1-4615-5381-6_120. [DOI] [PubMed] [Google Scholar]
- 6.Lotfi K, Mansson E, Spasokoukotskaja T, Pettersson B, Liliemark J, Peterson C, Eriksson S, Albertioni F. Biochemical pharmacology and resistance to 2-chloro-2′-arabino-fluoro-2′-deoxyadenosine, a novel analogue of cladribine in human leukemic cells. Clin Cancer Res. 1999;5:2438–2444. [PubMed] [Google Scholar]
- 7.Shewach DS, Liotta DC, Schinazi RF. Affinity of the antiviral enantiomers of oxathiolane cytosine nucleosides for human 2′-deoxycytidine kinase. Biochem Pharmacol. 1993;45:1540–1543. doi: 10.1016/0006-2952(93)90058-5. [DOI] [PubMed] [Google Scholar]
- 8.Eriksson S, Munch-Petersen B, Johansson K, Eklund H. Structure and function of cellular deoxyribonucleoside kinases. Cell Mol Life Sci. 2002;59:1327–1346. doi: 10.1007/s00018-002-8511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson K, Ramaswamy S, Ljungcrantz C, Knecht W, Piskur J, Munch-Petersen B, Eriksson S, Eklund H. Structural basis for substrate specificities of cellular deoxyribonucleoside kinases. Nat Struct Biol. 2001;8:616–620. doi: 10.1038/89661. [DOI] [PubMed] [Google Scholar]
- 10.Sabini E, Hazra S, Konrad M, Burley SK, Lavie A. Structural basis for activation of the therapeutic L-nucleoside analogs 3TC and troxacitabine by human deoxycytidine kinase. Nucleic Acids Res. 2007;35:186–192. doi: 10.1093/nar/gkl1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat Struct Biol. 2003;10:513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 12.Soriano EV, Clark VC, Ealick SE. Structures of human deoxycytidine kinase product complexes. Acta Crystallogr. 2007;63:1201–1207. doi: 10.1107/S0907444907048834. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Secrist JA, 3rd, Ealick SE. The structure of human deoxycytidine kinase in complex with clofarabine reveals key interactions for prodrug activation. Acta Crystallogr. 2006;62:133–139. doi: 10.1107/S0907444905034293. [DOI] [PubMed] [Google Scholar]
- 14.Knecht W, Munch-Petersen B, Piskur J. Identification of residues involved in the specificity and regulation of the highly efficient multisubstrate deoxyribonucleoside kinase from Drosophila melanogaster. J Mol Biol. 2000;301:827–837. doi: 10.1006/jmbi.2000.3990. [DOI] [PubMed] [Google Scholar]
- 15.Knecht W, Sandrini MP, Johansson K, Eklund H, Munch-Petersen B, Piskur J. A few amino acid substitutions can convert deoxyribonucleoside kinase specificity from pyrimidines to purines. EMBO J. 2002;21:1873–1880. doi: 10.1093/emboj/21.7.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Usova EV, Eriksson S. Identification of residues involved in the substrate specificity of human and murine dCK. Biochem Pharmacol. 2002;64:1559–1567. doi: 10.1016/s0006-2952(02)01389-8. [DOI] [PubMed] [Google Scholar]
- 17.Godsey MH, Ort S, Sabini E, Konrad M, Lavie A. Structural basis for the preference of UTP over ATP in human deoxycytidine kinase: illuminating the role of main-chain reorganization. Biochemistry. 2006;45:452–461. doi: 10.1021/bi0518646. [DOI] [PubMed] [Google Scholar]
- 18.Gentry GA. Viral thymidine kinases and their relatives. Pharmacol Ther. 1992;54:319–355. doi: 10.1016/0163-7258(92)90006-l. [DOI] [PubMed] [Google Scholar]
- 19.Piskur J, Sandrini MP, Knecht W, Munch-Petersen B. Animal deoxyribonucleoside kinases: ‘forward’ and ‘retrograde’ evolution of their substrate specificity. FEBS Lett. 2004;560:3–6. doi: 10.1016/S0014-5793(04)00081-X. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 21.Lutz S, Ostermeier M, Benkovic SJ. Rapid generation of incremental truncation libraries for protein engineering using alpha-phosphothioate nucleotides. Nucleic Acids Res. 2001;29:E16. doi: 10.1093/nar/29.4.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Igarashi K, Hiraga S, Yura T. A deoxythymidine kinase deficient mutant of Escherichia coli. II Mapping and transduction studies with phage phi 80. Genetics. 1967;57:643–654. doi: 10.1093/genetics/57.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munir KM, French DC, Dube DK, Loeb LA. Herpes thymidine kinase mutants with altered catalytic efficiencies obtained by random sequence selection. Protein Eng. 1994;7:83–89. doi: 10.1093/protein/7.1.83. [DOI] [PubMed] [Google Scholar]
- 24.Munir KM, French DC, Loeb LA. Thymidine kinase mutants obtained by random sequence selection. Proc Natl Acad Sci USA. 1993;90:4012–4016. doi: 10.1073/pnas.90.9.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mani RS, Usova EV, Eriksson S, Cass CE. Hydrodynamic and spectroscopic studies of substrate binding to human recombinant deoxycytidine kinase. Nucleosides Nucleotides Nucleic Acids. 2003;22:175–192. doi: 10.1081/NCN-120019513. [DOI] [PubMed] [Google Scholar]
- 26.Munch-Petersen B, Knecht W, Lenz C, Sondergaard L, Piskur J. Functional expression of a multisubstrate deoxyribonucleoside kinase from Drosophila melanogaster and its C-terminal deletion mutants. J Biol Chem. 2000;275:6673–6679. doi: 10.1074/jbc.275.9.6673. [DOI] [PubMed] [Google Scholar]
- 27.Schelling P, Folkers G, Scapozza L. A spectrophotometric assay for quantitative determination of kcat of herpes simplex virus type 1 thymidine kinase substrates. Anal Biochem. 2001;295:82–87. doi: 10.1006/abio.2001.5191. [DOI] [PubMed] [Google Scholar]
- 28.Bosley AD, Ostermeier M. Mathematical expressions useful in the construction, description and evaluation of protein libraries. Biomol Eng. 2005;22:57–61. doi: 10.1016/j.bioeng.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Hayes RJ, Bentzien J, Ary ML, Hwang MY, Jacinto JM, Vielmetter J, Kundu A, Dahiyat BI. Combining computational and experimental screening for rapid optimization of protein properties. Proc Natl Acad Sci USA. 2002;99:15926–15931. doi: 10.1073/pnas.212627499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaBean TH, Kauffman SA. Design of synthetic gene libraries encoding random sequence proteins with desired ensemble characteristics. Protein Sci. 1993;2:1249–1254. doi: 10.1002/pro.5560020807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aharoni A, Gaidukov L, Khersonsky O, Mc QGS, Roodveldt C, Tawfik DS. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.