Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease with highly variable clinical presentation. Patients suffer from immunological abnormalities that target T cell, B cell and accessory cell functions. B cells are hyperactive in SLE patients. An adaptor protein expressed in B cells called BANK1 (B-cell scaffold protein with ankyrin repeats) was reported in a previous study to be associated with SLE in a European population. The objective of this study is to assess the BANK1 genotype-phenotype association in an independent replication sample. We genotyped 38 single nucleotide polymorphisms (SNPs) in BANK1 on 1892 European-derived SLE patients and 2652 European-derived controls. The strongest associations with SLE and BANK1 were at rs17266594 (corrected p-value=1.97 × 10−5, OR=1.22, 95% C.I.(1.12–1.34)) and rs10516487 (corrected p-value=2.59 × 10−5, OR=1.22, 95% C.I.(1.11–1.34)). Our findings suggest that the association is explained by these two SNPs, confirming previous reports that these polymorphisms contribute to the risk of developing lupus. Analysis of patient subsets enriched for hematological, immunological and renal ACR criteria or the levels of autoantibodies, such as anti-RNP A and anti-SmRNP, uncovers additional BANK1 associations. Our results suggest that BANK1 polymorphisms alter immune system development and function to increase the risk for developing lupus.
Keywords: systemic lupus erythematosus, replication, association, European, BANK1
Introduction
SLE is a prototypic autoimmune disease for which genetic predisposition plays a critical role. Over the past few decades, multiple SLE susceptibility loci have been identified by us and others1. Prior to 2008, confirmed SLE candidate genes included variants in the HLA region, complement component genes, Fc receptors, PDCD1, PTPN22, IRF5, STAT4, and TREX1. Recent genome wide association studies using large numbers of SLE cases and controls have uncovered >10 additional new genes associated with SLE2–5. These genes identified by GWA studies, as well as other candidate genes previously described for SLE, may or may not replicate in the ongoing flurry of genetic research in this area6–8.
Recently, Kozyrev et.al. reported that the nonsynonymous SNP rs10516487 (R61H) and branch point–site SNP rs17266594 in BANK1 are functional disease-associated variants that contribute to the SLE susceptibility in several European populations (Scandinavia, Argentina, Germany, Italy and Spain)9. The BANK1 protein is an adaptor that is predominantly expressed in B cells. The BANK1 gene spans ~284 kilobases (kb) on chromosome 4q24 and consists of 17 exons10–12. This gene encodes a 755 amino acid protein characterized by an ankyrin-repeat-like region and a coiled-coil domain shared with phosphoinositide-3-kinase adapter protein 1 (PIK3AP1, formerly known as B cell adapter protein BCAP) and a protein essential for signal transduction in Drosophila called Dof10–12.
B lymphocyte activation leads to tyrosine phosphorylation of Bank1, resulting in tyrosine phosphorylation of the Type 1 inositol-1, 4, 5-triphosphate (IP(3)R) by the tyrosine kinase Lyn and augmented calcium mobilization10. IP(3)R interacts with Bank1 at exon 2, whereas Lyn associates with the C-terminal domain of Bank110. Bank1 deficient mice show enlarged germinal centers (GC) and enhanced IgM production upon T-dependent (TD) antigen stimulation in vivo, whereas this phenotype is not present in Cd40−/−, Bank1−/− knockout mice12. Bank1 deficient B cells demonstrate enhanced proliferation and survival upon CD40 stimulation through increased Akt activation. Therefore, in mice, Bank1 attenuates CD40-mediated proliferation and survival, thereby inhibiting B cell hyperactivity12.
Using a candidate gene approach in a case-control genetic association study, we independently replicate and confirm that BANK1 is associated with SLE in a European-derived population. The primary association is with a SNP implicated in alternative splicing of BANK1; however, additional SNPs in other parts of the gene may also be associated with lupus in particular subsets of individuals who express disease specific clinical manifestations.
Results
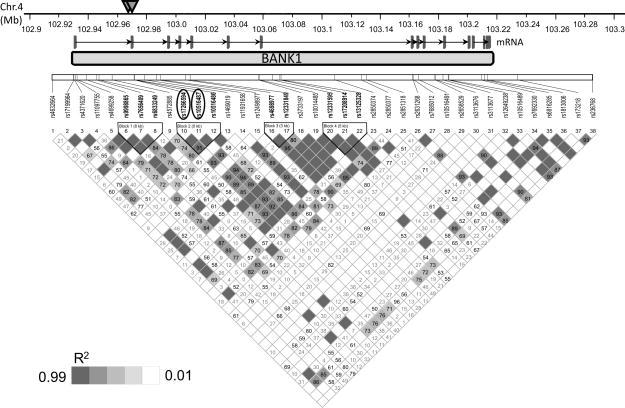
Of the 38 genotyped SNPs spanning the BANK1 gene, including known potentially functional SNPs and SNPs from predicted haplotype blocks (Table 2), we have full genotyping of 35 SNPs on 4544 subjects across this region. Genotypes for three SNPs, rs17266594, rs10516487 and rs4698977, were experimentally determined for 1447 individuals as outlined in the methods below. We were able to impute the genotypes with 97.5% accuracy for the three SNPs in 3097 individuals who we were unable to obtain actual genotyping data. The most significant association was within a 138 kb region of 4q24 (102.929–103.068 Mb). Four SNPs had p-values <10−4 in the combined European-derived group. The two strongest associations were with rs17266594 (p=1.97 × 10−5, OR=1.22 (95% C.I.=1.12–1.34)) and rs10516487 (p=2.59 × 10−5, OR=1.22 (95% C.I.=1.11–1.34)). These two SNPs are in very tight LD (Figure 1) and are consistent with those reported previously9.
Table 2.
Association analysis of 38 SNPs within BANK1 with SLE in a European-derived population.
| Single SNP Association (1892 cases, 2652 controls) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No Correction | Corrected (PCA Covariates used)c | |||||||||
| SNP | BP | Minor Allele | Assoc Allele | Type | Aaf Case | Aaf Ctrl | P-Value | OR (95% C.I.) | P-Value | OR (95% C.I.) |
| rs4632664 | 102911688 | A | G | 5′ UTR | 0.94 | 0.93 | 4.32E-01 | 1.07 (0.9–1.27) | 3.90E-01 | 1.08 (0.91–1.28) |
| rs17199964 | 102926814 | A | G | 5′ UTR | 0.93 | 0.92 | 5.05E-02 | 1.17 (1–1.37) | 1.19E-01 | 1.13 (0.97–1.33) |
| rs4371620 | 102927258 | G | A | 5′ UTR | 0.81 | 0.79 | 1.04E-02 | 1.15 (1.03–1.27) | 1.05E-02 | 1.15 (1.03–1.28) |
| rs11097755 | 102928331 | G | A | 5′ UTR | 0.56 | 0.55 | 2.11E-01 | 1.06 (0.97–1.15) | 1.92E-01 | 1.06 (0.97–1.15) |
| rs4699258 | 102929711 | A | G | 5′ UTR | 0.70 | 0.67 | 3.62E-04 | 1.18 (1.08–1.29) | 2.81E-04 | 1.18 (1.08–1.3) |
| rs9998865 | 102936169 | C | A | Intron | 0.90 | 0.90 | 8.67E-01 | 1.01 (0.88–1.16) | 9.88E-01 | 1 (0.87–1.15) |
| rs7656409 | 102943428 | A | G | Intron | 0.60 | 0.57 | 6.48E-04 | 1.16 (1.06–1.26) | 6.57E-04 | 1.16 (1.07–1.26) |
| rs6833249 | 102945096 | A | G | Intron | 0.90 | 0.90 | 8.42E-01 | 1.01 (0.88–1.17) | 9.57E-01 | 1 (0.87–1.16) |
| rs4572885 | 102954536 | T | A | Intron | 0.63 | 0.60 | 1.09E-02 | 1.12 (1.03–1.22) | 9.32E-03 | 1.12 (1.03–1.22) |
| rs17266594b | 102969945 | C | T | Intron | 0.71 | 0.67 | 2.20E-05 | 1.22 (1.11–1.33) | 1.97E-05 | 1.22 (1.12–1.34) |
| rs10516487b | 102970099 | T | C | Exon | 0.71 | 0.67 | 2.84E-05 | 1.21 (1.11–1.33) | 2.59E-05 | 1.22 (1.11–1.34) |
| rs10516486 | 102970299 | A | G | Exon | 0.64 | 0.61 | 3.54E-04 | 1.17 (1.07–1.28) | 3.89E-04 | 1.17 (1.07–1.28) |
| rs1469019 | 102977483 | A | A | Intron | 0.07 | 0.06 | 3.16E-01 | 1.09 (0.92–1.28) | 2.66E-01 | 1.10 (0.93–1.30) |
| rs11931658 | 102985915 | G | A | Intron | 0.67 | 0.64 | 6.08E-04 | 1.17 (1.07–1.28) | 3.55E-04 | 1.18 (1.08–1.29) |
| rs12498977 | 102988700 | A | A | Intron | 0.06 | 0.06 | 6.64E-01 | 1.04 (0.88–1.23) | 5.27E-01 | 1.05 (0.89–1.25) |
| rs4698977b | 103006446 | T | G | Intron | 0.64 | 0.59 | 4.90E-05 | 1.2 (1.1–1.31) | 2.19E-05 | 1.21 (1.11–1.32) |
| rs12331849 | 103009841 | A | G | Intron | 0.58 | 0.54 | 1.10E-04 | 1.18 (1.09–1.29) | 6.70E-05 | 1.19 (1.09–1.3) |
| rs3733197 | 103058310 | A | G | Exon | 0.66 | 0.63 | 5.46E-03 | 1.13 (1.04–1.24) | 2.96E-03 | 1.14 (1.05–1.25) |
| rs10014485 | 103058902 | G | G | Intron | 0.10 | 0.10 | 8.00E-01 | 1.02 (0.88–1.16) | 8.25E-01 | 1.02 (0.88–1.16) |
| rs12331595 | 103068242 | A | G | Intron | 0.55 | 0.51 | 1.24E-03 | 1.15 (1.06–1.25) | 8.07E-04 | 1.16 (1.06–1.26) |
| rs17208914 | 103073413 | G | G | Intron | 0.47 | 0.44 | 1.81E-02 | 1.11 (1.02–1.20) | 1.58E-02 | 1.11 (1.02–1.20) |
| rs13125328 | 103074133 | C | G | Intron | 0.90 | 0.89 | 2.30E-01 | 1.09 (0.95–1.24) | 2.49E-01 | 1.08 (0.95–1.24) |
| rs2850374 | 103112346 | C | A | Intron | 0.89 | 0.88 | 4.05E-01 | 1.06 (0.93–1.21) | 4.60E-01 | 1.05 (0.92–1.2) |
| rs2850377 | 103131182 | A | G | Intron | 0.59 | 0.56 | 4.99E-02 | 1.09 (1–1.19) | 3.99E-02 | 1.09 (1–1.19) |
| rs2851318 | 103134691 | T | A | Intron | 0.89 | 0.88 | 3.87E-01 | 1.06 (0.93–1.21) | 4.29E-01 | 1.05 (0.92–1.2) |
| rs2631268 | 103167753 | A | A | Intron | 0.29 | 0.27 | 7.27E-02 | 1.09 (0.99–1.19) | 5.26E-02 | 1.10 (1.00–1.20) |
| rs7685012 | 103168014 | G | G | Intron | 0.12 | 0.11 | 7.72E-01 | 1.02 (0.89–1.16) | 6.35E-01 | 1.03 (0.90–1.18) |
| rs10516491 | 103171889 | G | A | Intron | 0.98 | 0.98 | 8.49E-01 | 1.03 (0.77–1.38) | 9.00E-01 | 1.02 (0.76–1.37) |
| rs2658529 | 103183468 | G | G | Intron | 0.12 | 0.10 | 2.86E-02 | 1.16 (1.02–1.33) | 3.02E-02 | 1.16 (1.01–1.33) |
| rs3113676 | 103184066 | A | G | Exon | 0.99 | 0.99 | 2.11E-01 | 1.27 (0.87–1.84) | 2.63E-01 | 1.24 (0.85–1.81) |
| rs3113677 | 103189210 | A | G | Intron | 0.95 | 0.94 | 4.93E-02 | 1.2 (1–1.44) | 7.06E-02 | 1.19 (0.99–1.43) |
| rs12649238 | 103193010 | C | C | Intron | 0.19 | 0.18 | 1.12E-01 | 1.09 (0.98–1.22) | 8.79E-02 | 1.10 (0.99–1.23) |
| rs10516489 | 103193382 | A | A | Intron | 0.59 | 0.60 | 9.69E-01 | 1 (0.92–1.09) | 9.82E-01 | 1 (0.92–1.09) |
| rs7692330 | 103216356 | A | G | 3′ UTR | 0.94 | 0.93 | 1.76E-01 | 1.12 (0.95–1.33) | 2.79E-01 | 1.1 (0.93–1.31) |
| rs6816285 | 103218632 | A | G | 3′ UTR | 0.59 | 0.57 | 7.91E-02 | 1.08 (0.99–1.17) | 7.84E-02 | 1.08 (0.99–1.18) |
| rs1813006 | 103220672 | A | C | 3′ UTR | 0.94 | 0.93 | 1.89E-01 | 1.12 (0.94–1.34) | 3.09E-01 | 1.1 (0.92–1.31) |
| rs173218 | 103242072 | A | A | 3′ UTR | 0.19 | 0.18 | 7.91E-02 | 1.1 (0.99–1.23) | 4.89E-02 | 1.11 (1.00–1.25) |
| rs236768 | 103295006 | G | G | 3′ UTR | 0.19 | 0.18 | 1.47E-01 | 1.09 (0.97–1.2) | 1.14E-01 | 1.09 (0.98–1.22) |
Abbreviations: BP, position; Aaf Case, associated allele frequency in cases; Aaf Ctrl, associated allele frequency in controls; OR, odds ratio; CI, 95% confidence interval; UTR, untranslated region.
Odds Ratio (OR) is shown with 95% confidence interval (CI) and is based on the associated allele
SNPs rs17266594, rs10516487, and rs4698977 are partially imputed as described in the materials and method section.
Covariate correction for European population substructure using PLINK. Additive model log(y)=α+β1*SNP+β2*PC1+β3*PC2+β4*PC3; H0: β1=β2=β3=β4 used.
The significantly associated SNPs (p-value < 0.05) are in bold and italic.
Figure 1.
Genomic organization and LD analyses of BANK1 in the European-derived population. The upper graph summarizes the results from the association analysis within the BANK1 region. The two most significant SNPs are rs17266594 and rs10516487. In genomic structure diagram, rs17266594 is located in Intron 1 and rs10516487 is located in exon2. In the lower graph, it can be seen that the two highly significant SNPs are all found in the region of strong LD (depicted as r2 value).
In total, 15 SNPs were significantly associated with SLE (p<0.05) within three peak areas: one in the intron1/exon2 region as was previously described9, the second in the 5′ untranslated region near rs4699258 and the third in the intron 4 region defined by SNPs rs4698977, rs12331849 and exonic SNP rs3733197 (exon7) nearby. Each of these association peaks represents a distinct haplotype block (Figure 1).
To determine whether each of these peaks of genetic association contributes to SLE development, multivariable logistic regression adjusting for the effect of the other statistically significant BANK1 variant alleles was performed (Table 3). Two SNPs are responsible for the peak association, rs17266594 and rs10516487, and explain the entire effect observed (Table 3), whereas the other associations are a result of weaker linkage with primary SLE associated SNPs. The primary SLE associated SNPs, rs17266594 and rs10516487 are highly correlated (r2 = 96.6%) and are only 154 bp apart and, therefore, pair-wise conditioning of these two SNPs provided zero degrees of freedom for analysis.
Table 3.
Multivariate logistic regression analysis of 18 SLE associated SNPs
| p-value Conditioned on |
|||||||
|---|---|---|---|---|---|---|---|
| SNP | BP | Type | Global | rs17266594 | rs10516487 | rs468977 | rs12331849 |
| rs4371620 | 102927258 | Flanking 5′ UTR | 1.44E-04 | 0.425 | 0.404 | 0.295 | 0.194 |
| rs4699258 | 102929711 | Flanking 5′ UTR | 2.57E-04 | 0.783 | 0.716 | 0.219 | 0.065 |
| rs7656409 | 102943428 | Intron | 1.99E-04 | 0.599 | 0.572 | 0.0522 | 0.171 |
| rs4572885 | 102954536 | Intron | 2.76E-04 | 0.844 | 0.818 | 0.0842 | 0.28 |
| rs17266594 | 102969945 | Intron | 1.81E-05 | * | No variation | 0.0832 | 0.025 |
| rs10516487 | 102970099 | Non-synomyous Coding | 1.44E-05 | No variation | * | 0.101 | 0.03 |
| rs10516486 | 102970299 | Exon | 8.77E-05 | 0.838 | 0.98 | 0.011 | 0.16 |
| rs11931658 | 102985915 | Intron | 2.34E-04 | 0.71 | 0.642 | 0.535 | 0.03 |
| rs4698977 | 103006446 | Intron | 8.44E-05 | 0.244 | 0.235 | * | 0.168 |
| rs12331849 | 103009841 | Intron | 7.11E-05 | 0.204 | 0.198 | 0.57 | * |
| rs3733197 | 103058310 | Exon | 2.15E-04 | 0.649 | 0.592 | 0.657 | 0.791 |
| rs12331595 | 103068242 | Intron | 1.13E-04 | 0.33 | 0.325 | 0.573 | 0.993 |
| rs17208914 | 103073413 | Intron | 8.15E-05 | 0.235 | 0.243 | 0.912 | 0.584 |
| rs2850377 | 103131182 | Intron | 1.22E-04 | 0.358 | 0.313 | 0.543 | 0.597 |
| rs2631268 | 103167753 | Intron | 2.95E-05 | 0.08 | 0.07 | 0.267 | 0.096 |
| rs2658529 | 103183468 | Intron | 2.69E-05 | 0.148 | 0.168 | 0.144 | 0.042 |
| rs3113677 | 103189210 | Intron | 4.10E-05 | 0.133 | 0.113 | 0.07 | 0.056 |
| rs6816285 | 103218632 | Flanking 3′ UTR | 1.69E-04 | 0.505 | 0.472 | 0.617 | 0.393 |
Abbreviation: BP: base pair position; UTR: untranslated region.
Based on the plausible link between B cell hyperactivity and autoantibody production, analyses were performed to assess whether BANK1 polymorphisms were associated with the production of common lupus autoantibodies. Results from a logistic regression analysis evaluating the effect of autoantibody specificities within European-derived subjects as a covariate in the lupus case vs. control association analysis for all 38 SNPs typed are shown in Table 4. Of the 10 lupus specific autoantibodies tested, anti-RNP A (p=0.027, OR=0.77) and anti-SmRNP (p=0.017, OR=0.74) showed the most evidence for increased protective association with rs3733197 when used as covariates in the analysis compared to the borderline association at this SNP (p=0.059, OR=0.80) when no autoantibody covariates were applied. Complete datasets for all 10 autoantibodies and all 38 SNPs are presented in Supplementary Tables 2a–e online.
Table 4.
| Table 4 a). Frequency of BioPlex 2200 Autoantibodies Used in Covariate Analysis | |||||
|---|---|---|---|---|---|
| Autoantibodies | Case (n=341) | Control (n=350) | Chi-square | ||
| + | − | + | − | ||
| dsDNA | 0.33 | 0.67 | 0.01 | 0.99 | 5.28E-28 |
| Chrom | 0.46 | 0.54 | 0.03 | 0.97 | 7.98E-40 |
| Sm | 0.24 | 0.76 | 0 | 1 | 3.88E-22 |
| SmRNP | 0.29 | 0.71 | 0 | 1 | 1.86E-27 |
| RNP 68 | 0.14 | 0.86 | 0 | 1 | 2.22E-13 |
| RNP A | 0.25 | 0.75 | 0.02 | 0.98 | 2.49E-19 |
| Ro 60kD/SSA-60 | 0.36 | 0.64 | 0.01 | 0.99 | 7.98E-40 |
| Ro 52kD/SSA-52 | 0.26 | 0.74 | 0.01 | 0.99 | 1.12E-22 |
| La/SSB | 0.23 | 0.77 | 0.01 | 0.99 | 1.07E-17 |
| Ribo P | 0.16 | 0.84 | 0.01 | 0.99 | 4.31E-13 |
| Scl 70 | 0.12 | 0.88 | 0.02 | 0.98 | 8.33E-08 |
| Cent B | 0.12 | 0.88 | 0.01 | 0.99 | 2.96E-10 |
| Table 4 b) Summary results from a logistic regression analysis in 341 cases and 350 controls of the 38 SNPs in the BANK1 region with autoantibodies as covariates. | ||||||
|---|---|---|---|---|---|---|
| Covariate On |
||||||
| No Covariate | RNP A | SmRNP | ||||
| SNP | P-Value | OR (95% CI) | P-Value | OR (95% CI) | P-Value | OR (95% CI) |
| rs4698977 | 0.07106 | 0.81 (0.65–1.02) | 0.04117 | 0.78 (0.62–0.99) | 0.02248 | 0.75 (0.59–0.96) |
| rs12331849 | 0.1095 | 0.84 (0.67–1.04) | 0.1001 | 0.83 (0.66–1.04) | 0.04019 | 0.78 (0.62–0.99) |
| rs3733197 | 0.05854 | 0.80 (0.64–1.01) | 0.02745 | 0.77 (0.61–0.97) | 0.01714 | 0.74 (0.58–0.95) |
| rs12331595 | 0.0823 | 0.82 (0.66–1.03) | 0.06426 | 0.81 (0.65–1.01) | 0.02507 | 0.77 (0.61–0.97) |
| rs17208914 | 0.1692 | 1.16 (0.94–1.44) | 0.1312 | 1.19 (0.95–1.48) | 0.03366 | 1.28 (1.02–1.61) |
| rs2850377 | 0.09769 | 0.8332 (0.67–1.03) | 0.06088 | 0.81 (0.65–1.01) | 0.02556 | 0.77 (0.61–0.97) |
Chi-square p-value comparing the differences in autoantibody positive frequency in the cases to controls.
Abbreviation: OR, Odds Ratio; 95% CI, 95% Confidence Interval.
The significant increases with covariate in analysis are shown as bold and italicized.
Significant association was observed when evaluating the presence of ACR clinical criteria in European-derived lupus cases. Both BANK1 SNPs shown to be strongly associated prior to stratification showed strongest association in 843 lupus cases who met the immunological criteria, with p-values improving from 10−5 to 10−6 and OR showing a reduction in risk from 0.82 to 0.76. In addition, SNPs in other regions of the BANK1 gene showed associations with immunological disorders (rs13125328, rs2850377, rs2631268, rs12649238, rs173218), renal involvement (rs2631268, rs7685012, rs3113676) and hematological disorders (rs3113677 & rs173218) (Table 5 and Supplementary Tables 3a–e online). Other than the primary SLE associated SNPs, only a few other SNPs showed overlapping subphenotype associations.
Table 5.
Selected results from association analysis of 38 BANK1 SNPs in disease phenotype subsets based on ACR Clinical Criteria
| No subsetting (1892/2652) | Hematological Disorders | Immunological Disorders | Renal Involvement | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | |
| rs4699258 | 3.09E-04 | 0.85 (0.77–0.93) | 6.88E-02 | 0.88 (0.77–1.01) | 5.04E-05 | 0.78 (0.69–0.88) | 8.15E-02 | 0.87 (0.74–1.02) | |
| rs4572885 | 1.01E-02 | 0.89 (0.82–0.97) | 4.40E-01 | 0.95 (0.83–1.08) | 2.30E-03 | 0.84 (0.75–0.94) | 3.66E-01 | 0.93 (0.8–1.08) | |
| rs17266594 | 2.32E-05 | 0.82 (0.75–0.9) | 1.03E-01 | 0.89 (0.78–1.02) | 6.53E-06 | 0.76 (0.67–0.85) | 6.18E-03 | 0.8 (0.68–0.94) | |
| rs10516487 | 2.95E-05 | 0.82 (0.75–0.9) | 1.03E-01 | 0.89 (0.78–1.02) | 6.53E-06 | 0.76 (0.67–0.85) | 6.18E-03 | 0.8 (0.68–0.94) | |
| rs10516486 | 3.84E-04 | 0.86 (0.78–0.93) | 1.71E-01 | 0.91 (0.8–1.04) | 1.91E-04 | 0.8 (0.72–0.9) | 3.13E-02 | 0.85 (0.73–0.99) | |
| rs3733197 | 5.69E-03 | 0.88 (0.81–0.96) | 1.60E-01 | 0.91 (0.8–1.04) | 3.68E-03 | 0.84 (0.75–0.95) | 8.19E-01 | 0.98 (0.85–1.14) | |
| rs13125328 | 2.31E-01 | 0.92 (0.8–1.05) | 2.69E-01 | 0.89 (0.72–1.1) | 8.90E-03 | 0.78 (0.64–0.94) | 4.15E-01 | 0.91 (0.71–1.15) | |
| rs2850377 | 5.11E-02 | 0.92 (0.85–1) | 3.41E-01 | 0.94 (0.83–1.07) | 3.68E-02 | 0.89 (0.79–0.99) | 7.77E-01 | 0.98 (0.85–1.13) | |
| rs2631268 | 7.38E-02 | 1.09 (0.99–1.2) | 7.35E-02 | 1.14 (0.99–1.31) | 3.15E-02 | 1.14 (1.01–1.29) | 1.83E-02 | 1.21 (1.03–1.42) | |
| rs7685012 | 7.69E-01 | 1.02 (0.89–1.16) | 9.73E-01 | 1 (0.82–1.23) | 7.55E-01 | 1.03 (0.86–1.22) | 4.32E-02 | 1.25 (1.01–1.55) | |
| rs3113676 | 2.09E-01 | 0.79 (0.54–1.14) | 9.33E-01 | 0.98 (0.58–1.66) | 6.18E-01 | 0.89 (0.55–1.43) | 1.83E-02 | 0.32 (0.12–0.87) | |
| rs3113677 | 4.80E-02 | 0.83 (0.69–1) | 3.60E-02 | 0.73 (0.54–0.98) | 1.42E-01 | 0.83 (0.65–1.06) | 2.18E-03 | 0.55 (0.37–0.81) | |
| rs12649238 | 1.16E-01 | 1.09 (0.98–1.21) | 1.15E-01 | 1.14 (0.97–1.33) | 4.63E-02 | 1.15 (1–1.32) | 1.53E-01 | 1.14 (0.95–1.37) | |
| rs173218 | 8.20E-02 | 1.1 (0.99–1.22) | 7.73E-02 | 1.15 (0.98–1.35) | 4.89E-02 | 1.15 (1–1.32) | 1.32E-01 | 1.15 (0.96–1.38) | |
Abbreviation: OR, Odds Ratio; 95% CI, 95% Confidence Interval.
The numbers in parenthesis in the headings represent the (# cases meeting the tested criteria/total # controls). The p-values and odds ratios improvements are shown in bold.
Discussion
Our study independently replicates and confirms the strong association of BANK1 variants rs17266594 and rs10516487 with the risk of SLE in the European-derived population. In addition to the previously reported associated SNPs9, rs4699258 (5′UTR), rs7656409 (intron 1), rs4698977 (intron 4) and rs12331849 (intron 4) also suggest a strong association with the susceptibility to SLE in our European-derived subjects. A previous study reported sequencing the proximal promoter regions and exons 1 and 2 in 24 SLE patients and 8 controls9; however, no additional functional SNPs have yet been identified in this region. While there is association at these additional SNPs, our analysis demonstrated that only the SNPs in intron 1 (rs17266594) and exon 2 (rs10516487) drive this association and, therefore, are either the actual causal variations, or are in very tight LD with the yet unidentified causal variant in this region. Clearly, select polymorphisms in the 5′ untranslated region (UTR) of BANK1 demonstrate association (rs4371620 and rs469925); however, the analysis suggests that these variations may not be responsible for the primary genetic association, but they may be exhibiting a secondary association due to LD. It is likely that deep resequencing of portions of BANK1 will be necessary to uncover untyped or novel variants in this region that contribute to associations between BANK1 and SLE.
There appears to be additional diversity in the SLE subphenotype associations compared to those when evaluating the primary SLE phenotype. Clearly, the primary SLE associated SNPs, rs17266594 and rs10516486, also showed strong immunological and renal involvement subphenotype associations. However, several SNPs showed single subphenotype associations, especially with the immunological and renal involvement subphenotypes. It is currently unclear if there is a correlation between the subphenotype associations and the primary SLE associations. More complete clinical data on both lupus cases and controls would be needed to use as interaction terms in logistical regression modeling along with the primary SLE associated SNPs to determine if the primary SLE associated SNPs are markers of the additive effects of the subphenotype associations.
Kozyrev et.al. identified three functional disease-associated variants and elegantly speculated that these variants alter the affinity of BANK1 for IP(3)R. These authors also demonstrated that these SNPs affect the relative splicing efficiency of BANK1 and hypothesized that such splicing differences could lead to B cell hyperactivity or dysregulated B-cell activation9. Our results strongly support the association of the polymorphisms in this region. However, our results do not rule out the possibility of other SNPs in BANK1 also being associated, perhaps through other molecular mechanisms.
Slight changes in BANK1 protein expression or alteration of BANK1 functions, such as altered protein-protein interactions with src-kinases or other signaling molecules, may dramatically impact the autoantibody production and clinical phenotypes associated with SLE development and outcomes. One would predict that decreased BANK1 functions could dampen activating signals that mature B cells receive when signaled through the B cell antigen receptor. Alternatively, altered association with appropriate signaling molecules could lead to aberrant signals that might cause inappropriate B cell development and selection. An exact understanding of the molecular interactions impacted by SLE associated polymorphisms in BANK1 and an understanding of how signals can be attenuated, due to slight differences in expression levels of key B cell signal transduction protein variants, such as BANK1, will be a prerequisite to better understanding how aberrant B cell functions contribute to development and progression of SLE.
Methods
DNA samples
Genomic DNA samples were obtained from 1892 unrelated SLE patients and 2652 controls of European-decent from the Lupus Family Registry and Repository (LFRR) at the Oklahoma Medical Research Foundation (OMRF), the PROFILE Study Group coordinating center organized through the University of Alabama Birmingham, as well as other individual collaborators at OMRF, the Medical University of South Carolina, Feinstein Institute for Medical Research in New York, the United Kingdom and Sweden (Table 1 and Supplemental Table 1s online). All individuals used in this study were confirmed to be independent based upon information provided by the contributors and had IBS sharing proportions <0.5 when evaluating all possible pairwise comparisons at 400 SNPs with minor allele frequencies >0.4. There is no overlap of the 83 European individuals used in this study from our collaborator from Sweden with those used in the Kozyrev study9.
Table 1.
Composition of study group
| Case | Control | Total | |
|---|---|---|---|
| Female | 1708 | 1876 | 3584 |
| Male | 184 | 776 | 960 |
| Total Sample | 1892 | 2652 | 4544 |
All SLE patients met at least 4 of the 11 revised SLE classification criteria of the American College of Rheumatology (ACR)13, 14. DNA was isolated from biological specimens (blood samples, buccal swabs or mouthwash samples) provided from each participant after obtaining the appropriate informed consent as approved by the Institutional Review Boards or ethical committees where the subjects were recruited.
Genotyping
Thirty-eight SNPs spanning the BANK1 gene, including known functional SNPs and SNPs from haplotype blocks were genotyped. Genotypes from 35 of the SNPs were obtained from the complete samples consisting of 1892 European-derived SLE patients and 2652 healthy controls (Table 1 and Table 1s in supplementary data online). The other three SNPs (rs17266594, rs10516487, rs4698977) were genotyped on a subset of cases and controls (891 SLE cases and 556 controls) available (LFRR, James, Merrill, Moser, Gaffney, Gilkeson and those from the Feinstein Institute for Medical Research). For these three SNPs, any missing experimental genotypes or untyped genotypes for the remaining 3097 samples were determined through imputation using European-derived HapMap reference data. To assess the reliability of the imputation, we masked the experimental genotype data from 1447 individuals (~32%) and imputed them with HAPMAP data and then compared them with real genotype data. The imputation predicted correct genotypes 97.5% of the time.
Quality control of genotyping
Genotype data were only used from samples with a call rate greater than 90% of the SNPs screened (98.05% of the samples). The average call rate for all samples was 97.18%. Only genotype data from SNPs with a call frequency greater than 90% in the samples tested and an Illumina GeneTrain score greater than 0.7 (96.74% of all SNPs screened) were used for analysis.
Single SNP analysis
Case-control associations and Hardy-Weinberg Proportions were calculated using PLINK15. Only SNPs with minor allele frequencies (MAF) >0.01 and Hardy Weinberg Proportions in the controls p>0.001 were used for the analysis. The allelic frequencies were calculated for each SNP and case-control associations were analyzed by standard Pearson’s Chi-square test. Principal components were calculated as outlined below and were used as covariates in the association analysis to correct for any residual population substructure. P values of < 0.0013 were considered statistically significant after correcting for multiple testing using the Bonferroni method. Odds ratios (OR) and 95% confidence intervals (CIs) were also calculated for each SNP using logistic regression.
Imputation
Using data for three SNPs (rs17266594, rs10516487 and rs4698977) genotyped on the subset of 2269 individuals and 60 unrelated HAPMAP CEPH parents, we imputed data for these three SNPs for any individuals missing the experimental genotypes as well as the remaining individuals using fastPHASE16. These three SNPs had less than 5% missing data in HAPMAP and the strands were not flipped in HAPMAP release 21 or release 22, making them good candidates for imputation17. To assess the quality of imputation, we checked the MAF of these three SNPs in the successfully experimentally genotyped data (1447 individuals), imputed data (3097 individuals) and overall combined data (4544 study individuals) separately. The MAFs for the three SNPs among the three sets were almost identical. Thus, our final data set had 35 genotyped SNPs and 3 SNPs with both genotype and imputed data.
Linkage Disequilibrium
Both the squared correlation statistic (r2) and Lewontin’s D′ statistic were used as measures of LD strength within the BANK1 region and were calculated using Haploview.
Haplotype Analysis
Haplotype frequencies were estimated using the expectation–maximization algorithm utilized by WHAP18. Haplotype-based association analysis and multivariate logistic regression analysis were used to perform regression-based omnibus haplotype frequency tests and haplotype-specific tests, also implemented in WHAP. Using the two strongest signals in the data, rs17266594 and rs10516487, we performed a pair-wise multivariate logistic regression adjusting for the effects of the other 18 SNPs which were significantly associated. Results are shown in Table 3.
Population Stratification Analysis
All samples used in this study were previously used in a collaborative study where population substructure parameters were defined using two rounds of principle component analysis (PCA) performed on 20,506 SNPs3, 19. Four principal components were initially identified that explained a total of ~60% of the observed genetic variation and allowed identification of outliers from the European cluster. Before outlier removal, the estimated inflation factor (λ) was 1.84. After removal of outliers, the inflation factor was 1.12, indicating that these cleaned data should have a very small population substructure effect on our results. In addition, after trimming of outliers, another round of PCA was performed and three newly calculated PCA values were used as covariates in the association analysis to correct for any residual European population substructure effects. No additional outliers were identified using the new PCA values, which produced a final inflation factor of 1.15.
Logistic Regression Analysis using BioPlex 2200 Normalized Intensity Values as Covariates
The BioPlex 2200 (Bio-Rad, Hercules, CA) is a high throughput automated serological analysis unit that utilizes multiplex bead technology for antibody detection. The BioPlex results are reported on a scale from 0–8. This scale is set relative to calibrator positive and negative control samples provided by the manufacturer. The defined positive cut-off value for each assay is then set to 1.0, with Factor XIII index greater than 0.2 as serum validation control. However, dsDNA is reported in IU/mL with a positive cut-off of 10.0 IU/mL. Ten of 13 autoantibodies commonly associated with lupus (dsDNA, chromatin, ribosomal P, 60kD Ro (SS-A 60), 52kD Ro (SS-A 52), La (SS-B), Sm, Sm/RNP complex, nRNP A, and nRNP 68) were evaluated using BioPlex 2200 in the stored serum from 341 patients and 350 controls of the independent European cohort. Autoantibody levels above the threshold were considered positive and denoted as 1, whereas the negatives samples were denoted as 0 in the dichotomous covariate data set. Each autoantibody was entered individually into the logistic regression model as a covariate. The p-value and odds ratios with 95% confidence interval of the logistic model were calculated using PLINK15.
Association Enhancement in Lupus Disease Phenotypic Subsets Defined by ACR Criteria
To assess the potential role of BANK1 in SLE and disease etiology, cases were stratified based on the presence of the 11 ACR clinical criteria and associations were analyzed comparing the stratified lupus patients to all 2652 unrelated European-derived controls using PLINK15. The ACR clinical criteria information was obtained from the Lupus Family Registry Repository (LFRR) and individual investigators for SLE cases.
Supplementary Material
Acknowledgments
The authors thank the participants, both patients and controls, who graciously agreed to take part in these studies by donating samples to the various collections, including the Lupus Family Registry and Repository (LFRR: http://lupus.omrf.org), PROFILE, BIOLUPUS, Feinstein Institute for Medical Research and many other individual or multicenter collaborator initiated collections. We also thank the recruitment and technical teams at each of the sample procurement sites for their important contributions. We thank the Wake Forest University Health Sciences Center for Public Health Genomics for support of the data analysis efforts by our Wake Forest University collaborators. We thank Dr. Alarcon-Riquelme for her assistance in clearly identifying potential overlapping subjects between our collections to ensure the independence of this study’s observed associations and Dr. Peter Gregersen for providing control samples. Finally, we thank the various funding sources as outlined on the title page for their continued support for the collection of samples and the conduct of this research.
Support: This project was funded by National Institutes of Health RR020143 (JMG and JBH), RR015577 (JMG, JBH, JAJ), NIAID-DAIT-BAA-05-11 (JMG and JAJ), AR053483 (JMG, SKN and JAJ), AI063274 (PMG), AI031584 (JBH, JMG, JAJ), AR052125 (PMG), AR043247 (KLM), Kirkland Scholar awards (JBH and JAJ), AR049084 (SKN, JBH, RPK), AR42460 (JBH), AR12253 (JBH), AR62277 (JBH), AI24717 (JBH), AR48940 (JBH, JAJ), Alliance for Lupus Research (JBH), the US Department of Veterans Affairs (JBH), and OHRS award for project number HR08-037 from the Oklahoma Center for the Advancement of Science & Technology (JMG). Dr. Harley has received consulting fees, speaking fees, and/or director’s fees from Bio-Rad Laboratories, Merck, UCB Inc., ImmunoVision Inc., IVAX Diagnostics and JK Autoimmunity and owns stock or stock options in IVAX Diagnostics.
Footnotes
Supplementary information is available at the Genes & Immunity’s website.
References
- 1.Harley JB, Kelly JA, Kaufman KM. Unraveling the genetics of systemic lupus erythematosus. Springer Semin Immunopathol. 2006 Oct;28(2):119–30. doi: 10.1007/s00281-006-0040-5. [DOI] [PubMed] [Google Scholar]
- 2.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of Systemic Lupus Erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008 Jan 20; doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 3.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008 Feb;40(2):204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008 Aug 1; doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cervino AC, Tsinoremas NF, Hoffman RW. A genome-wide study of lupus: preliminary analysis and data release. Ann N Y Acad Sci. 2007 Sep;1110:131–9. doi: 10.1196/annals.1423.015. [DOI] [PubMed] [Google Scholar]
- 6.Todd JA. Statistical false positive or true disease pathway? Nat Genet. 2006 Jul;38(7):731–3. doi: 10.1038/ng0706-731. [DOI] [PubMed] [Google Scholar]
- 7.Sestak AL, Nath SK, Sawalha AH, Harley JB. Current status of lupus genetics. Arthritis Res Ther. 2007;9(3):210. doi: 10.1186/ar2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forabosco P, Gorman JD, Cleveland C, Kelly JA, Fisher SA, Ortmann WA, et al. Meta-analysis of genome-wide linkage studies of systemic lupus erythematosus. Genes Immun. 2006 Oct;7(7):609–14. doi: 10.1038/sj.gene.6364338. [DOI] [PubMed] [Google Scholar]
- 9.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nature genetics. 2008 Jan 20; doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama K, Su Ih IH, Tezuka T, Yasuda T, Mikoshiba K, Tarakhovsky A, et al. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor. The EMBO journal. 2002 Jan 15;21(1–2):83–92. doi: 10.1093/emboj/21.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battersby A, Csiszar A, Leptin M, Wilson R. Isolation of proteins that interact with the signal transduction molecule Dof and identification of a functional domain conserved between Dof and vertebrate BCAP. Journal of Molecular Biology. 2003 Jun 6;329(3):479–93. doi: 10.1016/s0022-2836(03)00489-3. [DOI] [PubMed] [Google Scholar]
- 12.Aiba Y, Yamazaki T, Okada T, Gotoh K, Sanjo H, Ogata M, et al. BANK negatively regulates Akt activation and subsequent B cell responses. Immunity. 2006 Mar;24(3):259–68. doi: 10.1016/j.immuni.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982 Nov;25(11):1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 14.Hochberg MC, Wallace Dj HBH Anonymous. Dubois‚Äô Lupus Erythematosus. Baltimore: Williams and Wilkins; 1997. The epidemiology of systemic lupus erythematosus; pp. 49–65. [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006 Apr;78(4):629–44. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson CA, Pettersson FH, Barrett JC, Zhuang JJ, Ragoussis J, Cardon LR, et al. Evaluating the effects of imputation on the power, coverage, and cost efficiency of genome-wide SNP platforms. Am J Hum Genet. 2008 Jul;83(1):112–9. doi: 10.1016/j.ajhg.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007 Jan 15;23(2):255–6. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.