Abstract
Autophagy is emerging as a central component of antimicrobial host defense against diverse viral, bacterial, and parasitic infections. In addition to pathogen degradation, autophagy has other functions during infection such as innate and adaptive immune activation. As an important host defense pathway, microbes have also evolved mechanisms to evade, subvert, or exploit autophagy. Additionally, some fungal pathogens harness autophagy within their own cells to promote pathogenesis. This review will highlight our current understanding of autophagy in infection, focusing on the most recent advances in the field, and will discuss the potential implications of these studies in the design of anti-infective therapeutics.
Keywords: autophagy, immunity, infection
Main
Autophagosomes containing intracellular microbes were first observed ultrastructurally in the 1980s, which originally led to the notion that intracellular microbes may be targeted by or subvert the autophagic pathway. However, it was only after the identification of autophagy (ATG) genes in the 1990s that the molecular tools have been available to dissect this pathway and its functions in infection and host immunity. The basic machinery of autophagy and the diverse biological processes in which it is involved are reviewed in the current issue and elsewhere.1, 2 The host cell autophagic machinery can sequester both pathogens that have escaped endocytic or phagocytic vesicles and/ or the pathogen-containing vesicles themselves, as well as microbial proteins and genetic material. The autophagosome containing microbial proteins, nucleic acids, or intact organisms can fuse with other vesicles in the endolysosomal pathway to deliver microbial ligands for adaptive or innate immune activation, or with the lysosome for degradation.
Many aspects of the autophagy pathway during infection may be unique, including the regulatory pathways (pattern recognition receptors (PRRs), pathogen-associated molecular patterns (PAMPs), and downstream signaling pathways), substrates for autophagosome sequestration (i.e., xenophagy of microbes and/or specific cellular contents), and potentially the events in autophagosome formation. For microbes that usurp autophagy-like pathways, it is possible that the cellular components involved are distinct from those of ‘classical’ autophagy and/or that they are modulated by pathogens in a unique manner. Furthermore, autophagy within the pathogen itself is a newly defined virulence strategy for eukaryotic microorganisms. This review will highlight key studies and recent advances in the field of autophagy during infection with bacteria, viruses, protozoa, and fungi and will discuss how the potentially unique aspects of autophagy during infection may be exploited in the treatment of infectious diseases.
Eating for Immunity: Regulatory Signals
Autophagy functions as an innate immune barrier to infection by ridding the cytosol of microbes through lysosomal degradation (discussed below), as well as in other aspects of innate and adaptive immunity (reviewed by Schmid and Munz3 and Levine and Deretic4). Innate sensing and signaling pathways regulate autophagy induction, and an important effector function of autophagosomes may be to deliver ligands for innate and adaptive immune activation. One of the most active areas in autophagy research is the identification of immune signaling pathways that regulate autophagy (see Figure 1).
Figure 1.
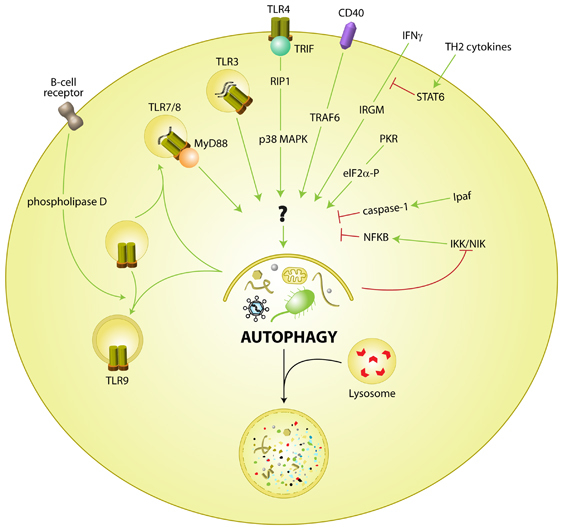
Regulation of autophagy by immune signaling pathways. Examples of positive regulators include TLRs (3, 4, and 7/8), PKR, IFNγ, and CD40. Pathways that negatively regulate autophagy include NF-κB and TH2 cytokines. The mechanisms by which all of these pathways intersect with the autophagy machinery remain unknown
A front line of defense against invading pathogens is the well-conserved recognition of PAMPs by PRRs, including Toll-like receptors (TLRs),5 and these essential innate immune receptors have recently been demonstrated to regulate autophagy (Figure 1). The first study to describe a function of TLRs in autophagy induction found that a component of the Gram-negative bacterial cell wall, lipopolysaccharide (LPS), induces autophagy through its cognate receptor TLR4 in macrophages.6 LPS-induced autophagy requires the TLR4 adaptor TRIF, but not MyD88, and the downstream components RIP1 and p38 mitogen-activated protein kinase (MAPK). Further, LPS stimulation increases the localization of Mycobacterium tuberculosis within autophagosomes suggesting a functional role of LPS signaling in autophagy-mediated pathogen control. Subsequently, Deretic and colleagues7 extended the known repertoire of TLRs and their ligands that induce autophagy to include TLR3/Poly I:C and TLR7/imiquimod and ssRNA, and confirmed TLR4/LPS induction of autophagy. They also demonstrated that TLR7-induced autophagy decreases M. tuberculosis var. bovis Bacille Calmette-Guérin (BCG) survival, and requires the TLR7 adaptor MyD88. An additional recent study suggested a novel function of autophagy in mediating TLR signaling in phagosome maturation.8 Sanjuan et al. found that LPS induces autophagy as well as phagosome maturation (confirming the findings of Xu et al.6 and Delgado et al.7) but, in contrast to Delgado et al., found that autophagy is also stimulated with TLR2 agonists. It is possible that different cell types or agonist preparations may account for these differences, but further studies are needed to more clearly define the full spectrum of PAMPs and PRRs involved in autophagy regulation. Nonetheless, the studies described above convincingly link pathogen recognition, an essential component of innate immunity, with autophagy induction and also underscore the potential importance of autophagy as an innate immune effector in response to PAMPs.
A recent study in Drosophila melanogaster9 further underscores the importance of specific PRRs not only in autophagy induction, but also in mediating autophagic control of bacterial replication in vivo. The PRR molecule PGRP-LE (analogous to Nod-like receptor (NLR) signaling in mammals) was identified as a crucial component of the host antimicrobial autophagic response in Drosophila.9 Yano et al.9 demonstrated that PGRP-LE mutant flies or flies expressing hemocyte-specific RNAi for PGRP-LE or the autophagy gene Atg5 were hypersusceptible to lethal infection with Listeria monocytogenes. Signaling through the IMD and Toll pathways was dispensable for PGRP-LE-mediated autophagy and restriction of bacterial survival in primary hemocytes.9 Additionally, autophagy induction in hemocytes treated with purified tracheal cytotoxin and diaminopimelic-containing peptidoglycan ligands, but not lysine-type peptidoglycan required PGRP-LE.9 Thus, it will be interesting to determine the precise signaling pathways linking PGRP-LE to the autophagy machinery, and the identity of additional PRRs that are involved in autophagy induction in Drosophila.
Beyond the recognition of PAMPs by PRRs, there is a complex interplay between other immune signals and autophagy regulation (Figure 1). For example, NF-κB may negatively regulate TNF-mediated autophagy,10 whereas autophagy may negatively regulate NF-κB, in turn, through selective degradation of the upstream activators IκB kinase11 and NF-κB inducing kinase.12 Additionally, interferon (IFN)γ-mediated autophagy and BCG killing requires the p47 immunity-related GTPase, IRGM1.13 In contrast, TH2 cytokines (e.g., IL-4 and IL-13) negatively regulate IFNγ-mediated autophagic killing of mycobacteria in a STAT6-dependent manner.14 A recent report suggests that B-cell receptor signaling can recruit TLR9 to autophagosomes to promote synergistic signaling through p38 MAPK, and that formation of these compartments requires microtubules and phospholipase D activity.15 Thus, autophagy can be positively and negatively regulated by different immune signaling pathways. Studies in animal models should help to dissect the relative physiological importance of these different immune signaling pathways in the regulation of autophagy during different infectious diseases.
Another emerging theme is that stress-sensing and innate immune sensing pathways may share common components in the induction of autophagy. One of the first immune signaling pathways shown to regulate autophagy was the cytosolic double-stranded viral RNA-sensing kinase PKR, which acts through phosphorylation of eIF2α.16 PKR functions redundantly with the stress-induced kinase, Gcn2, in yeast to mediate starvation-induced autophagy16 and the eIF2α kinase PERK is required for ER stress-induced autophagy.17 The Drosophila homolog, ird1, of the VPS34 regulatory subunit VPS15, an essential component of the autophagy initiation machinery, is required for the IMD immune signaling pathway (similar to TRIF-dependent signaling in mammalian systems), for antimicrobial peptide production (AMP), and for resistance to bacterial infection,18 although it is not yet clear if the autophagy functions of ird1 are responsible for this antimicrobial response or if the IMD or Toll pathways regulate autophagy in Drosophila. Similar to the eIF2α kinase signaling pathway, which functions in both innate immune sensing and general stress responses, Wu et al.18 found that ird1 was required for starvation-induced AMP production.
There are several important unanswered questions with respect to innate immune activation of autophagy. Very little is known about how downstream signaling molecules integrate with the autophagy machinery. The most distal signaling component identified in TLR-mediated autophagy induction is p38 MAPK in response to TLR4 signaling,6 but it is unclear precisely how p38 MAPK results in autophagy induction. Additionally, Xu et al. found that TRIF, but not MyD88, was required for TLR4-mediated autophagy, whereas Delgado et al. demonstrated that TLR7 signaled through MyD88 to induce autophagy. Therefore, it will be important to identify the TRIF- and MyD88-interacting partners that are involved in TLR4- and TLR7-mediated autophagy, respectively. Both TRIF and MyD88 signal through TRAF6, which was also found to mediate CD40-induced autophagy in response to Toxoplasm gondii infection of macrophages, in synergy with TNF-α.19 Thus, it will be interesting to examine whether a common TRAF6 complex is involved in immune activation of autophagy by different receptors and if so, to identify the components of such a complex.
Another important question is whether signaling through additional PRRs, including mammalian cytosolic NLRs, cytosolic DNA and RNA sensors, and/or lectins also regulates autophagy. At least in response to Shigella flexneri infection in macrophages, the NLR Ipaf negatively regulates autophagy through caspase-1 activation, in a Flagellin-independent manner, whereas genetic deletion of the NALP3 adaptor ASC has no effect.20 Yet another question is whether the sites of signaling by immune receptors (pathogen-containing vesicles or cytosolic sensors in contact with pathogens) define targets for sequestration in autophagosomes by recruiting the autophagy machinery to these sites. Also, it remains to be determined if PRR signaling results in the recruitment of any as-of-yet identified molecules that may specify microbes for autophagic sequestration. The answers to these questions will have implications for the development of therapeutics aimed at modulating autophagy to enhance host immune defenses (discussed below).
Eating for Immunity: Effector Functions
Autophagy functions in many aspects of innate and adaptive immunity, including immune activation, survival of infected cells, immune cell homeostasis, degradation of pathogens, and potentially in gut-commensal homeostasis (Figure 2). Many of these functions have been extensively reviewed elsewhere,3, 4 and this section will provide a brief summary of the functions of autophagy in immune activation, infected cell survival, and immune cell homeostasis.
Figure 2.
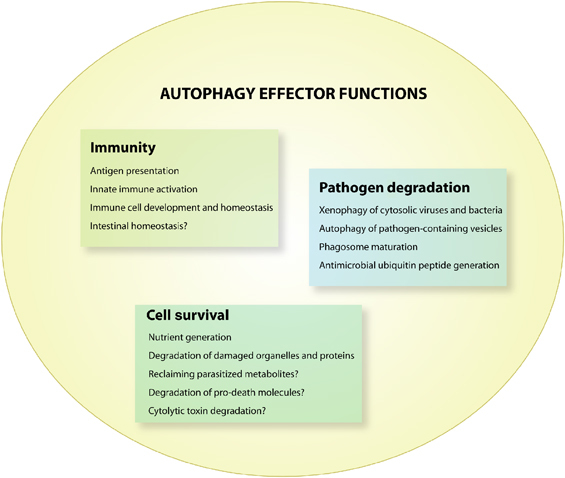
Effector functions of autophagy in infectious diseases
Not only do immune signals regulate autophagy (as discussed above), but autophagy also functions in innate and adaptive immune activation. Autophagy samples certain cytosolic antigens to present on class II MHC molecules for activation of the adaptive immune response.21, 22, 23, 24 Further, specific strategies to target cytosolic antigens to autophagosomes may lead to enhanced vaccine efficacy, as an influenza virus antigen fused to the autophagosomal membrane protein, LC3, elicits higher levels of CD4+ T-cell responses than the antigen alone.24 Autophagy also functions to deliver cytosolic viral replication intermediates to TLR7-containing endosomes to activate type I IFN production.25 In light of the recent study by Delgado et al.,7 which demonstrated that TLR7 itself activates autophagy, it will be interesting to determine whether this represents a positive feedback loop and more broadly, whether IFN-stimulated genes also function to regulate autophagy.
As a stress survival pathway, autophagy may have a protective function during infection by ensuring the survival of infected and/or uninfected cells in infected organisms. This has now been demonstrated in several different types of viral infections. Mouse brains infected with Sindbis virus overexpressing Beclin 126 or with a herpes simplex virus type 1 (HSV-1) mutant strain incapable of inhibiting autophagy27 exhibit decreased neuronal cell death. Additionally, autophagy promotes the survival of B lymphocytes infected with parvovirus B19 in vitro.28 In plants, autophagy gene silencing results in the unrestricted cell death of uninfected cells during the hypersensitive response to tobacco mosaic virus infection.29 In contrast to these prosurvival effects of autophagy during viral infection, there is also evidence that autophagy may contribute to the death of uninfected bystander T lymphocytes during HIV infection.30
The function of autophagy in cell survival during bacterial infections is less well studied. Autophagy may have a function in mediating the death of cells infected with Staphylococcus aureus, as atg5-deleted mouse embryonic fibroblasts (MEFs) showed increased survival following infection with a methicillin-resistant strain of S. aureus.31 However, these effects may be S. aureus strain-specific, as Yoshimori and collegues32 found that methicillin-sensitive S. aureus are targeted and degraded by autophagy in wild-type MEFs, but persist in atg5-deleted MEFs, while resistant strains can escape from autophagosomes. In addition, macrophages undergoing cell death in response to Salmonella infection exhibit increased autophagy, although it is unclear if autophagy is responsible for the observed death.33 Further studies are needed to clarify the function of autophagy in life and death decisions of the cell during bacterial infection.
Autophagy also functions in host immunity by promoting immune cell homeostasis, and potentially contributes to immune tolerance. Peripheral lymphocyte survival and proliferation in response to T-cell receptor (TCR) activation is decreased in atg5−/− bone marrow chimera mice.34 In addition, autophagy is induced by TCR activation in CD4+ T cells, but promotes cell death after growth factor removal.35 Autophagy is also involved in the proper development of specific lineages of B lymphocytes.36 A recently identified function of autophagy in promoting the clearance of apoptotic cells suggests that autophagy may also have a function in preventing inflammation; in atg5−/− embryonic mice, there is a defect in apoptotic corpse clearance which is associated with abnormal inflammation.37
Eating the Enemy: Xenophagy
By sequestering large portions of the cytoplasm in autophagosomes that are destined for lysosomal degradation, autophagy also protects against infection by limiting intracellular microbial survival, a process that has been termed ‘xenophagy’.38 As a recycling pathway to generate nutrients under stress conditions, it is also possible that autophagy targets cytosolic pathogens for degradation not only to rid the cell of the pathogen but also to reclaim metabolites that have been parasitized by microbes – without degrading their own essential components – and thereby promotes host cell survival. Specific mechanisms to selectively sequester microbes remain unidentified, but several characteristics of pathogen-containing autophagosomes suggest that this may occur. For example, bacteria often reside within autophagosomes devoid of other cellular contents, autophagosome membranes are closely opposed to sequestered bacteria, and these vesicles are often larger than typical autophagosomes containing cytoplasmic contents.
The first genetic evidence that autophagy may function as an antimicrobial pathway that restricts intracellular pathogen replication was provided by studies with the neurotropic alphavirus Sindbis virus. Expression of Beclin 1, the mammalian ortholog of yeast Atg6, from a double-subgenomic viral promoter protected mice against fatal Sindbis virus encephalitis and decreased neuronal apoptosis and viral replication in infected mouse brains.26 Studies with another neurotropic virus, the α-herpesvirus, HSV-1, further suggest that the endogenous host autophagy machinery is required for pathogen degradation and protection against CNS viral disease: HSV-1 is incapable of causing neurological disease if a mutation is made in its neurovirulence factor ICP34.5 that cripples its ability to block autophagy (discussed in detail below).27 Xenophagic degradation of HSV-1 virions may contribute to the antiviral effects of autophagy, as ultrastructural and biochemical analyses of neurons infected with a mutant virus deleted of ICP34.5 reveals PKR-dependent degradation of HSV-1 virions in autophagosomes and PKR-dependent autophagic degradation of HSV-1 proteins, respectively.39 The antiviral functions of autophagy are conserved from mammals to plants, since during tobacco mosaic virus infection of plants, the autophagy genes ATG6, ATG3, and ATG7 are required to reduce viral replication and prevent the spread of programmed cell death in uninfected tissues.29 At present, it is unclear if intact virions or viral proteins are specifically targeted for degradation, and if so, what are the identities of putative selectivity determinants.
Bacterial pathogens that invade into the cytoplasm or disrupt phagolysosomal fusion are also targeted by autophagy (Table 1). The first nearly simultaneous findings demonstrating this function of autophagy were with the pathogens, M. tuberculosis52 and Group A Streptococcus.40 Gutierrez et al.52 found that induction of autophagy with physiological (starvation), pharmacological (rapamycin), and immune (IFNγ and LRG-47) stimulation increased maturation of mycobacterial-containing vesicles and localization to autophagosomes, and decreased mycobacterial survival. Nakagawa et al.40 demonstrated that autophagic targeting degrades Group A Streptococci that escape from the endosome and invade intracellularly, as delayed bacterial clearance is observed in atg5-deficient cells. Subsequent studies have demonstrated an important function of autophagy in targeting other intracellular bacterial pathogens including Shigella flexneri, Listeria monocytogenes, Salmonella enterica, Francisella tularensis, and Burkholderia pseudomallei.9, 41, 44, 45, 48, 49 The biological significance of Francisella entry into autophagic compartments is unclear, but a recent study suggests that these compartments may also include MHC class II molecules.46 The recent study by Yano et al.9 of Listeria infection in Drosophila (discussed above) is an important first demonstration that the xenophagic targeting of bacteria can be an integral innate immune effector pathway in vivo. There is an urgent need for similar studies in mammalian hosts.
Table 1.
Interactions between specific microbes and autophagy
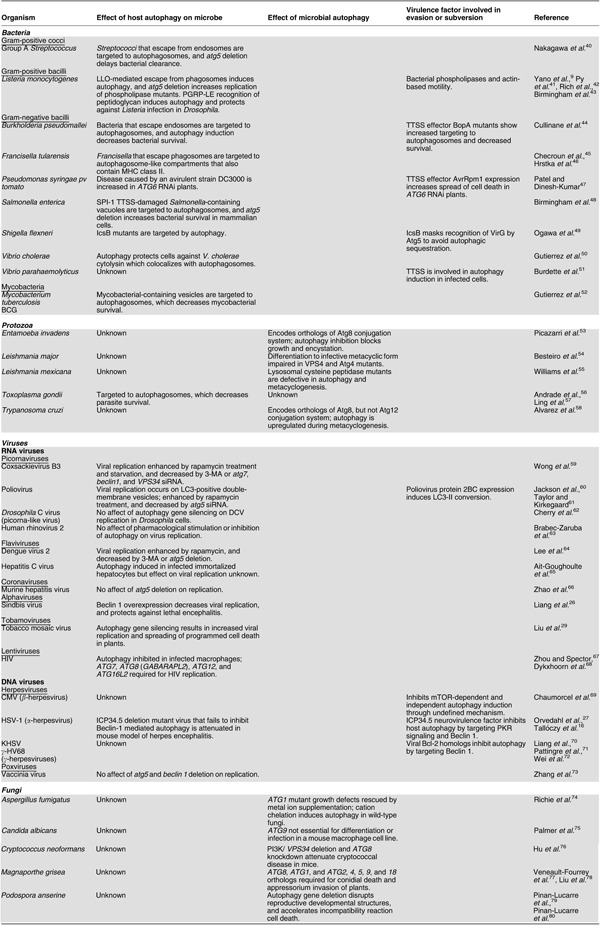
An important question related to bacterial xenophagy is what stimuli lead to autophagy induction. It is possible that disruption of phagosome integrity acts as a signal to induce autophagy to sequester potential invading microorganisms. It is also possible that PRRs in phagosomes or in the cytosol (as demonstrated for PGRP-LE in Drosophila)9 signal for autophagy induction, as discussed above. Additionally, many microbes utilize secretion systems or toxins to escape from phagosomes or invade through endosomes. It is possible that there are mechanisms by which the detection of these toxins or secreted products induces autophagy, or that autophagy is induced as a primary or secondary effect of these bacterial effectors. For example, during infection with Group A Streptococcus, bacteria lacking the toxin Streptolysin O do not escape from the endosome and are not targeted by autophagy;40 it is not yet known whether the escape from the endosome per se or some other effector function of Streptolysin O is essential for autophagy induction. Autophagy induction during Shigella infection requires a type III secretion system (TTSS), presumably during entry into the cytoplasm,20 but a TTSS effector IcsB masks the cell surface protein VirG to block Atg5 binding and potentially blocks recognition by autophagosomes.49 Additionally, the TTSS effector BopA is important for evading autophagic targeting of B. pseudomallei.44 Evidence also exists for induction of autophagy by a TTSS during infection of cells by the food-borne pathogen Vibrio parahaemolyticus, although the precise signals mediating this induction are not yet known.51
Another recent study provides additional evidence for a protective function of autophagy during bacterial infection and in resistance to TTSS effectors; disease caused by avirulent Pseudomonas syringae pv tomato (Pst) DC3000 is increased in Arabidopsis plants transgenic for ATG6 RNAi, and there is increased spread of cell death in ATG6 RNAi transgenic plants when infected with a strain of DC3000 that expresses the TTSS effector AvrRpm1.47 Autophagy induced by microbial toxins may exert a cytoprotective effect, perhaps by enabling the cells to more rapidly clear the toxin. Autophagy protects cells against an exotoxin from Vibrio cholerae; cells treated with V. cholerae cytolysin (VCC) show increased autophagy levels, colocalization of VCC with autophagosomes, and decreased survival in the setting of pharmacological (e.g., 3-MA) or genetic (e.g., atg5 deletion) autophagy inhibition.50 Thus, the antibacterial effects of autophagy may relate both to the degradation of intracellular pathogens as well as to the degradation of toxins that such organisms produce.
In addition to viruses and bacteria, protozoans may be targeted by autophagy. Toxoplasma gondii resides in parasitophorous vacuoles (PV) within macrophages and prevents their fusion with the lysosome. Two nearly simultaneous studies indicated that autophagy could function to overcome this block and restrict T. gondii survival. Andrade et al.56 found that stimulation of the CD40 receptor on macrophages resulted in localization of parasites to GFP-LC3-positive vesicles, vesicle maturation, and decreased intracellular microbial survival. On the basis of ultrastructural studies and chemical autophagy inhibition studies, Ling et al.57 proposed a model in which the PV and parasite membranes are stripped before autophagic sequestration and lysosomal degradation. It will be interesting to explore whether autophagy also protects against other apicomplexan parasites that reside intracellularly.
An Eating Disorder?: Autophagy Defects and Inflammatory Bowel Disease
On the basis of genetic linkage data, defects in host autophagy may contribute to Crohn's disease, an inflammatory bowel disease of the small intestine. In several genome-wide scanning studies performed in large populations of patients in Europe and North America, polymorphisms in two autophagy genes, an autophagy-inducing signaling molecule, IRGM, and a component of the ubiquitin-like protein conjugation system involved in autophagosomal membrane expansion, ATG16L1, are strongly associated with the development of Crohn's disease.81, 82, 83, 84 The pathogenic mechanisms of Crohn's disease are not completely understood, but are postulated to involve a dysregulated immune response to commensal bacteria, altered mucosal barrier function, and/or defects in bacterial clearance. Given the emerging functions of autophagy in immune cell homeostasis, immune cell activation, and bacterial clearance, it seems plausible that defects in autophagy could contribute to one or more of the potential pathogenic mechanisms of Crohn's disease. However, the precise functions of autophagy in degrading translocating bacteria and in immune regulation in the intestine are not yet known. Moreover, it is not yet known whether the polymorphism in Atg16L1 (T300A) alters autophagy function, as this mutation lies in a WD40 repeat domain that is not conserved in yeast Atg16. Further studies are needed to define the precise function of these autophagy-related proteins in host–commensal microbial interactions.
Eating Regulation by the Enemy: Pathogen Evasion and Modulation of Autophagy
As an important antimicrobial pathway, it is not surprising that pathogens have evolved mechanisms to evade or subvert the host autophagy machinery (Table 1). Bacterial pathogens can prevent autophagosome fusion with lysosomes to evade degradation or to utilize nutrients in these vesicles. Viruses from diverse classes inhibit autophagy-inducing signals and autophagy execution genes. Conversely, some RNA viruses usurp autophagosome membranes for virion production.
Many bacteria have been implicated in the evasion or subversion of autophagy; however, until recently, evidence supporting this has been largely limited to colocalization with autophagy markers or chemical manipulation of the autophagy pathway.85, 86 One of the first described examples is Listeria monocytogenes, a bacterium that can evade autophagy after escaping from the phagosome.42 Recent studies indicate that bacterial virulence factors required for modulation of autophagy by L. monocytogenes include the bacterial pore-forming toxin listeriolysin O (LLO) (which induces autophagy) and bacterial phospholipases (which evade autophagy).41, 43 Py et al.41 demonstrated that wild-type Listeria activates autophagy in MEFs, and deletion of atg5 accelerates the kinetics of bacterial growth early during infection. The induction of autophagy, as measured by LC3 lipidation, requires LLO expression, which is also required for bacterial replication in both wild-type and atg5−/− MEFs, whereas in contrast, the replication of phospholipase mutants is restored in atg5−/− MEFs.41 Birmingham et al.43 confirmed LLO expression-dependent autophagy induction and localization of Listeria to GFP-LC3-positive vesicles early during infection in macrophages, and Yano et al.9 confirmed that LLO-mediated entry of Listeria into the cytoplasm is required for autophagy induction in Drosophila. Deletion of the global virulence regulator prfA decreases replication in wild-type MEFs, and LLO expression is insufficient to rescue bacterial growth, whereas atg5 deletion restores Listeria replication, suggesting that additional prfA-regulated virulence factors are required to evade autophagy.43 Additionally, phospholipase mutants colocalize with GFP-LC3 vesicles for a prolonged period, and atg5 deletion restores growth of a plcA mutant.43 Together these studies suggest that LLO-mediated breach of phagosomes induces autophagy, and that additional virulence factors including phospholipases and actin-based motility are important for bacterial escape from cellular autophagy. Furthermore, a recent study suggests that Listeria can replicate slowly in spacious vacuoles that may arise through autophagic targeting of phagosomes that have been damaged by LLO.87 As with studies on the antimicrobial effects of autophagy, animal models are greatly needed to assess the function of bacterial evasion or subversion of autophagy in disease.
As obligate intracellular pathogens, viruses have also evolved mechanisms to evade or subvert host autophagy, and these have also been reviewed elsewhere.85, 88, 89 With respect to autophagy evasion, there is now evidence that all families of herpesviruses possess mechanisms to block host autophagy. As discussed above, HSV-1, an α-herpesvirus, encodes an essential neurovirulence factor, ICP34.5, that antagonizes autophagy. It does this through two mechanisms, including reversal of PKR-mediated eIF2α phosphorylation and direct antagonism of Beclin 1.16, 27 ICP34.5 antagonism of autophagy may be important to restrict xenophagic degradation of virions,39 and is required for fatal HSV-1 encephalitis in mice.27 Recently, cytomegalovirus, a member of the β-herpesvirus family, was shown to inhibit autophagy through an as-of-yet unidentified mechanism.69
γ-herpesviruses, including Kaposi's Sarcoma-associated herpesevirus (KSHV) and murine γ-HV68 encode homologs of cellular Bcl-2 that inhibit autophagy through antagonism of Beclin 1. Although the function of autophagy inhibition in KSHV or γ-HV68 disease remains unclear,70, 71 viral Bcl-2-like proteins seem to have evolved mechanisms to antagonize the autophagy function of Beclin 1 more effectively than their cellular counterparts; viral Bcl-2 proteins have a higher binding affinity than cellular Bcl-2 family members for Beclin 1,90 and escape physiological regulation of binding to Beclin 1 by JNK1-mediated phosphorylation.72 As Beclin 1 is a tumor suppressor gene,91 it will be important to determine whether viral Bcl-2 antagonism of Beclin 1 function contributes to the oncogenic potential of the γ-herpesviruses.
In addition to viral evasion of autophagy, some viruses may have acquired mechanisms to subvert the autophagy pathway for their own benefit. The best-characterized example is poliovirus, a member of the non-enveloped single-stranded RNA picornavirus family. Poliovirus replication occurs on LC3-positive double-membrane vesicles and is decreased by siRNA against components of the autophagic machinery.60 Additionally, expression of poliovirus protein 2BC is sufficient to induce LC3-II conversion, although it is unclear if this is a direct effect of the viral protein on LC3 or perhaps a more indirect effect.61 Autophagy also facilitates replication of another member of the picornavirus family, Coxsackievirus B3; pharmacological inhibition of autophagy with 3-MA or siRNA against atg7, beclin1, or VPS34 decreases viral replication, whereas autophagy stimulation with rapamycin treatment or starvation increases viral replication.59 A recent study suggests that dengue virus 2, a member of the flavivirus family, also utilizes the autophagy machinery for replication.64 A second flavivirus, hepatitis C virus (HCV), induces autophagy in immortalized hepatocytes, but the significance of autophagy in the HCV replication cycle is not yet known.65
The replication of some other viruses seems not to be affected either positively or negatively by autophagy. In contrast to poliovirus and Coxsackievirus B3, studies with Drosophila C virus62 and human rhinovirus 263 suggest that autophagy subversion may not be a universal requirement for picornavirus replication. Further, Atg5 and Beclin 1 are dispensable for the replication of vaccinia virus, a member of the poxvirus family, at least in MEFs and embryonic stem cells, respectively.73 Mouse coronavirus replication does not require Atg5 in low passage MEFs or primary bone marrow-derived macrophages.66 Thus, the relative importance of autophagy and the nature of its function may vary considerably in different types of virus infections.
Of note, increasing evidence suggests cross-talk between the HIV virus and the autophagy pathway. One recent report suggests that HIV can inhibit autophagy in macrophages.67 More complex modulation through viral induction of autophagy and inhibition of autophagosome maturation has been suggested to enhance HIV infection.92 In support of a positive function of autophagy in HIV infection (or at least the protein conjugation systems involved in autophagosomal membrane formation), a recent genome-wide siRNA screen identified several autophagy genes that function in this step of autophagy (e.g., ATG7, ATG8 (GABARAPL2), ATG12, and Atg16L2), as host factors required for HIV replication.68 As noted above, autophagy also mediates cell death of bystander T cells in HIV infection through CXCR4 binding to HIV envelope proteins.30 Thus far, studies performed on HIV infection and autophagy have been performed in vitro and primarily in non-native target cells; therefore, a high research priority will be to assess the functional significance of these diverse interactions in the regulation of HIV replication in primary CD4+ T lymphocytes and macrophages and in the pathogenesis of HIV infection in vivo.
As discussed above, for some viruses, autophagy may be an important antiviral pathway, other viruses may utilize components of the autophagy pathway to foster their own replication, or alternatively, may not intersect with autophagy pathway to any extent. One enigma is that an analysis of the types of viruses that evade, subvert, succumb to, or are not affected by autophagy does not reveal any obvious common characteristics that might provide insight into mechanisms responsible for each of these fates. For example, how do viruses that usurp autophagic machinery avoid the antiviral effects of autophagy? It is possible that viral commandeering of the autophagy machinery may prevent the formation of autophagosomes that would otherwise have antiviral effects. Conversely, it is unclear why some viruses are targeted by autophagy, while others are not. Further clarification of the exact mechanisms by which autophagy recognizes viruses or their components, by which autophagy exerts its antiviral effects (e.g., promoting cellular survival, xenophagic degradation of virions, immune activation, and/or unidentified functions), and by which viruses modulate or evade host autophagy should provide answers to these questions. Moreover, the identification of host factors that function in autophagy specifically in response to viral infection (versus other stress stimuli) may provide insight into the cellular determinants of the antiviral autophagic response.
Eating Within the Enemy: Microbial Autophagy
A largely unexplored area in autophagy and infectious diseases is the function of autophagy that occurs within pathogens that themselves are eukaryotes, such as protozoans, fungi, and helminths. The prosurvival function of autophagy during nutrient stress was originally described in the baker's yeast, S. cerevisaie, and autophagy in the roundworm, C. elegans, is essential for the development, lifespan extension, and stress adaptation.93 These adaptive functions of autophagy in eukaryotic organisms are unlikely to be limited to model organisms studied in the laboratory. Such functions are also likely to be relevant to the pathogenesis of human diseases caused by eukaryotic microbes and indeed, some recent evidence suggests that protozoan and fungal autophagy may function as a virulence pathway during infection.
The molecular machinery of autophagy was originally identified in genetic screens performed in the yeast, S. cerevisiae, and autophagy is also present in disease-causing fungi (see Table 1). The first indication that autophagy was required for fungal disease was in a study of the rice blast fungus Magnaporthe grisea, where MgATG8 is required for conidial death and appresorium invasion during infection.77 Furthermore, M. grisea homologs of ATG1,78 and ATG2, ATG4, ATG5, ATG9 and ATG18 are involved in plant invasion.94 In contrast, autophagy protects against cell death in the filamentous fungus Podospora anserine.79, 80 Autophagy is also important in human fungal pathogens, including in the filamentous Aspergillus fumigatus, where AfATG1 has a function in metal ion homeostasis.74 In contrast, Candida albicans does not require ATG9 for differentiation or infection of a mouse macrophage cell line.75 Of potential medical relevance, a recent study provides strong evidence that autophagy in the dimorphic fungus Cryptococcus neoformans is important for virulence, as Class III PI3K/VPS34 deletion and ATG8 knockdown resulted in decreased replication and lethality in a mouse model of cryptococcosis.76 These findings suggest that specific targeting of fungal, but not mammalian, autophagic machinery could be beneficial in the treatment of human cryptococcal disease.
Protozoan parasites also undergo autophagy, and this response may be important for differentiation during the parasite life cycle and for infection. Autophagy is induced during differentiation of Trypansosoma cruzi,58 although its function in trypanosomatid disease is unknown. Similarly, autophagy is upregulated during encystation in Entamoeba invadens and autophagy inhibition arrests growth and encystation.53 Autophagy is also important for differentiation of Leishmania major54 and Leishmania mexicana.55
To Eat or not to Eat: Therapeutic Considerations
Autophagy is emerging as an attractive therapeutic target for a number of diseases, including infectious diseases.1, 95 As mentioned, pharmacological or immunological activation of autophagy can enhance autophagy of microbes. However, the complex interplay between autophagy and various microbes (evasion, subversion, or susceptibility) raise unique issues in therapeutic intervention of infectious diseases (see Figure 3). For pathogens that are susceptible to autophagic degradation, or for which autophagy promotes cell survival, enhanced induction of autophagy may prove beneficial. Conversely, for pathogens that evade autophagy, enhancement may be sufficient to overcome microbial blockade, although more productive strategies may involve targeting the virulence factors that mediate autophagy evasion. Similarly, for pathogens that subvert autophagy to their own replicative advantage, the identification and inhibition of virulence factors involved in autophagy modulation may also provide benefit and promote antimicrobial effects of autophagy. A more complex situation is presented with protozoan and fungal infections, where host and microbial autophagy may benefit each organism respectively, so that therapeutic intervention in these cases may need to be species-specific.
Figure 3.
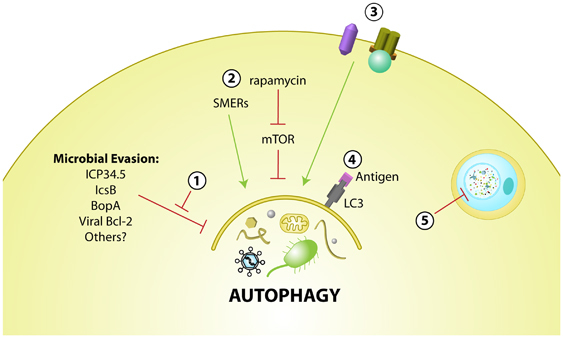
Potential therapeutic targets in modulating autophagy in the treatment or prevention of infectious diseases. (1) Inhibition of microbial virulence factors that evade or subvert host autophagy. (2) Stimulation of autophagy with pharmacological agents (i.e., rapamycin or novel compounds such as SMERs, small molecule enhancers of rapamycin-induced autophagy). (3) Autophagy regulation through activation of innate immunity signaling receptors. (4) Vaccine enhancement by targeting antigens to the autophagy pathway by fusion with the autophagy protein, LC3. (5) Selective inhibition of microbial autophagy in eukaryotic pathogens that utilize their own autophagy as a virulence pathway
Some medically important bacteria and parasites have been shown to be susceptible to autophagy after chemical or immunological induction. Rapamycin, an inhibitor of mTOR and inducer of autophagy, enhances targeting of M. tuberculosis,52 T. gondii,56 and B. pseudomallei44 for autophagolysosomal degradation. Conversely, the replication of some bacteria and viruses is enhanced after rapamycin treatment including Coxiella burnetii,96 Anaplasma phagocytophilum,97 poliovirus,60 and Coxsackievirus B3.59 An additional important consideration is that rapamycin has immunosuppressive effects in vivo, which may have adverse effects in anti-infective treatment, and mTOR inhibition may affect multiple downstream pathways.
The identification of novel compounds that modulate autophagy independently of mTOR may therefore be useful in antimicrobial therapy, as has been demonstrated recently for mycobacterial killing in primary human macrophages.98 Other novel approaches for antimicrobial therapy through autophagy modulation may include immune activation with agonists for autophagy through signaling pathways discussed above7, 52 or enhancement of autophagy to promote microbial killing through autophagic generation of ubiquitin-derived peptides.99 In addition, the efficacy of antigen-based vaccines may be enhanced by targeting such antigens to autophagic compartments by fusion with the autophagy protein, LC3.24
As mentioned, some microbes possess mechanisms to evade autophagy, and others benefit from autophagy induction or utilize their own autophagy pathways to promote virulence. Therapeutic modulation of autophagy in these cases may require inhibition of specific microbial virulence factors; for example IcsB from Shigella,49 BopA from B. pseudomallei,44 or ICP34.5 from HSV-1.16, 27 Further, inhibition of as-of-yet unidentified virulence factors for microbes that subvert autophagy may be beneficial. In addition, combined inhibition of virulence factors with agents that upregulate autophagy may yield synergistic antimicrobial effects. For eukaryotic parasites, strategies to inhibit autophagy specifically within the pathogen may be required to ensure that host autophagy remains functional;100 detailed structural comparisons of pathogen and host autophagy proteins may aid in the design of such targeted therapies.
Conclusion
In the past decade, autophagy has emerged as an important antimicrobial pathway. There is a broad range of immune signals that regulate autophagy, and autophagy has multiple effector functions in immunity and infection, including innate and adaptive immune activation, immune cell homeostasis, and microbial degradation. Despite the immense progress that has been made, our understanding of autophagy in infectious diseases is still quite primitive; most studies have been performed in vitro, which may or may not represent the function of autophagy in vivo, and many questions remain unanswered. An important next step will be to develop animal models for studying the function of autophagy in immunity and infection in vivo and to assess the effects of therapeutic modulation of autophagy on the outcomes of infectious diseases. At a more basic level, we need to better understand how immune signaling pathways intersect with the autophagy pathway, how host molecules identify and target intracellular pathogens to the autophagosome, and how host and microbial factors interact to utilize autophagy to the benefit of the microbe and/or the host. The answers to these questions may not only enhance our general understanding of the molecular mechanisms of autophagy, but may also enable us to translate our knowledge of autophagy as an antimicrobial pathway into new strategies for the treatment of infectious diseases.
Acknowledgements
We apologize to authors whose work could not be included due to space restrictions. We thank Cindy Jozefiak for administrative support, and Angela Diehl for expert medical illustration. The work in the authors’ laboratory was supported by NIH Grants R01 A10151367 (BL) and T32 A1007520 (AO), and an Ellison Medical Foundation Senior Scholars Award in Infectious Diseases (BL).
Glossary
- PRR
pattern recognition receptors
- PAMPs
pathogen associated molecular patterns
- TLRs
Toll-like receptors
- BCG
M. tuberculosis var bovis
- NLRs
Nod-like receptors
- TTSS
Type-III secretion system
- LLO
listeriolysin O
- HSV-1
herpes simplex virus type 1
- LC3
microtubule-associated protein 1 light chain 3
Footnotes
Edited by G Kroemer
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 3.Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 9.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S. NF-κB activation represses tumor necrosis factor-α-induced autophagy. J Biol Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 11.Qing G, Yan P, Xiao G. Hsp90 inhibition results in autophagy-mediated proteasome-independent degradation of IκB kinase (IKK) Cell Res. 2006;16:895–901. doi: 10.1038/sj.cr.7310109. [DOI] [PubMed] [Google Scholar]
- 12.Qing G, Yan P, Qu Z, Liu H, Xiao G. Hsp90 regulates processing of NF-κB2 p100 involving protection of NF-κB-inducing kinase (NIK) from autophagy-mediated degradation. Cell Res. 2007;17:520–530. doi: 10.1038/cr.2007.47. [DOI] [PubMed] [Google Scholar]
- 13.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 14.Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Chaturvedi A, Dorward D, Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity. 2008;28:799–809. doi: 10.1016/j.immuni.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tallóczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc Natl Acad Sci USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Randle KE, Wu LP. ird1 is a Vps15 homologue important for antibacterial immune responses in Drosophila. Cell Microbiol. 2007;9:1073–1085. doi: 10.1111/j.1462-5822.2006.00853.x. [DOI] [PubMed] [Google Scholar]
- 19.Subauste CS, Andrade RM, Wessendarp M. CD40-TRAF6 and autophagy-dependent anti-microbial activity in macrophages. Autophagy. 2007;3:245–248. doi: 10.4161/auto.3717. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33:1250–1259. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 23.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 24.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 26.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G. Protection against fatal Sindbis virus encephalitis by Beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host and Microbe. 2007;1:23–45. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Nakashima A, Tanaka N, Tamai K, Kyuuma M, Ishikawa Y, Sato H. Survival of parvovirus B19-infected cells by cellular autophagy. Virology. 2006;349:254–263. doi: 10.1016/j.virol.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnaith A, Kashkar H, Leggio SA, Addicks K, Kronke M, Krut O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J Biol Chem. 2007;282:2695–2706. doi: 10.1074/jbc.M609784200. [DOI] [PubMed] [Google Scholar]
- 32.Amano A, Nakagawa I, Yoshimori T. Autophagy in innate immunity against intracellular bacteria. J Biochem. 2006;140:161–166. doi: 10.1093/jb/mvj162. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez LD, Pypaert M, Flavell RA, Galan JE. A Salmonella protein causes macrophage cell death by inducing autophagy. J Cell Biol. 2003;163:1123–1131. doi: 10.1083/jcb.200309161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 36.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 37.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 38.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Tallóczy Z, Virgin HWt, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 41.Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- 42.Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- 43.Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–451. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- 44.Cullinane M, Gong L, Li X, Lazar-Adler N, Tra T, Wolvetang E. Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. Autophagy. 2008;4:744–753. doi: 10.4161/auto.6246. [DOI] [PubMed] [Google Scholar]
- 45.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci USA. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hrstka R, Krocova Z, Cerny J, Vojtesek B, Macela A, Stulik J. Francisella tularensis strain LVS resides in MHC II-positive autophagic vacuoles in macrophages. Folia Microbiologica. 2007;52:631–636. doi: 10.1007/BF02932193. [DOI] [PubMed] [Google Scholar]
- 47.Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4:20–27. doi: 10.4161/auto.5056. [DOI] [PubMed] [Google Scholar]
- 48.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 49.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 50.Gutierrez MG, Saka HA, Chinen I, Zoppino FC, Yoshimori T, Bocco JL. Protective role of autophagy against Vibrio cholerae cytolysin, a pore-forming toxin from V cholerae. Proc Natl Acad Sci USA. 2007;104:1829–1834. doi: 10.1073/pnas.0601437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdette D, Yarbrough M, Orvedahl A, Gilpin C, Orth K. Vibrio parahaemolyticus orchestrates a multifacted host cell infection by induction of autophagy, cell rounding and then cell lysis. Proc Natl Acad Sci USA. 2008;105:12497–12502. doi: 10.1073/pnas.0802773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacteriumtuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 53.Picazarri K, Nakada-Tsukui K, Nozaki T. Autophagy during proliferation and encystation in the protozoan parasite Entamoeba invadens. Infect Immun. 2008;76:278–288. doi: 10.1128/IAI.00636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Besteiro S, Williams RA, Morrison LS, Coombs GH, Mottram JC. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J Biol Chem. 2006;281:11384–11396. doi: 10.1074/jbc.M512307200. [DOI] [PubMed] [Google Scholar]
- 55.Williams RA, Tetley L, Mottram JC, Coombs GH. Cysteine peptidases CPA and CPB are vital for autophagy and differentiation in Leishmania mexicana. Mol Microbiol. 2006;61:655–674. doi: 10.1111/j.1365-2958.2006.05274.x. [DOI] [PubMed] [Google Scholar]
- 56.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alvarez VE, Kosec G, Sant′Anna C, Turk V, Cazzulo JJ, Turk B. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J Biol Chem. 2008;283:3454–3464. doi: 10.1074/jbc.M708474200. [DOI] [PubMed] [Google Scholar]
- 59.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM et al. Autophagosome supports Coxsackievirus B3 replication in host cells. J Virol 2008 doi:10.1.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed]
- 60.Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2:e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brabec-Zaruba M, Berka U, Blaas D, Fuchs R. Induction of autophagy does not affect human rhinovirus type 2 production. J Virol. 2007;81:10815–10817. doi: 10.1128/JVI.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Z, Thackray LB, Miller BC, Lynn TM, Becker MM, Ward E. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 67.Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 69.Chaumorcel M, Souquère S, Pierron G, Codogno P, Esclatine A. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy. 2008;4:46–53. doi: 10.4161/auto.5184. [DOI] [PubMed] [Google Scholar]
- 70.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 71.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 72.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang H, Monken CE, Zhang Y, Lenard J, Mizushima N, Lattime EC. Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy. 2006;2:91–95. doi: 10.4161/auto.2.2.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Richie DL, Fuller KK, Fortwendel J, Miley MD, McCarthy JW, Feldmesser M. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot Cell. 2007;6:2437–2447. doi: 10.1128/EC.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palmer GE, Kelly MN, Sturtevant JE. Autophagy in the pathogen Candida albicans. Microbiology. 2007;153:51–58. doi: 10.1099/mic.0.2006/001610-0. [DOI] [PubMed] [Google Scholar]
- 76.Hu G, Hacham M, Waterman SR, Panepinto J, Shin S, Liu X. PI3K signaling of autophagy is required for starvation tolerance and virulence of Cryptococcus neoformans. J Clin Invest. 2008;118:1186–1197. doi: 10.1172/JCI32053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–583. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- 78.Liu XH, Lu JP, Zhang L, Dong B, Min H, Lin FC. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell. 2007;6:997–1005. doi: 10.1128/EC.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pinan-Lucarre B, Balguerie A, Clave C. Accelerated cell death in Podospora autophagy mutants. Eukaryot Cell. 2005;4:1765–1774. doi: 10.1128/EC.4.11.1765-1774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pinan-Lucarre B, Paoletti M, Dementhon K, Coulary-Salin B, Clave C. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol Microbiol. 2003;47:321–333. doi: 10.1046/j.1365-2958.2003.03208.x. [DOI] [PubMed] [Google Scholar]
- 81.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 84.Consortium WTCC Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 87.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 88.Orvedahl A, Levine B. Viral evasion of autophagy. Autophagy. 2007;4:280–285. doi: 10.4161/auto.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Espert L, Codogno P, Biard-Piechaczyk M. Involvement of autophagy in viral infections: antiviral function and subversion by viruses. J Mol Med. 2007;85:811–823. doi: 10.1007/s00109-007-0173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine γ-herpesvirus 68. PLoS Pathog. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 92.He C, Orvedahl A. 2007 keystone symposium on autophagy in health and disease. Autophagy. 2007;3:527–536. doi: 10.4161/auto.4595. [DOI] [PubMed] [Google Scholar]
- 93.Melendez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu XH, Lu JP, Lin FC. Autophagy during conidiation, conidial germination and turgor generation in Magnaporthe grisea. Autophagy. 2007;3:472–473. doi: 10.4161/auto.4339. [DOI] [PubMed] [Google Scholar]
- 95.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 96.Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 97.Niu H, Yamaguchi M, Rikihisa Y. Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell Microbiol. 2008;10:593–605. doi: 10.1111/j.1462-5822.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 98.Floto RA, Sarkar S, Perlstein EO, Kampmann B, Schreiber SL, Rubinsztein DC. Small molecule enhancers of rapamycin-induced TOR inhibition promote autophagy, reduce toxicity in Huntington's disease models and enhance killing of mycobacteria by macrophages. Autophagy. 2007;3:620–622. doi: 10.4161/auto.4898. [DOI] [PubMed] [Google Scholar]
- 99.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci USA. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alvarez VE, Kosec G, Sant Anna C, Turk V, Cazzulo JJ, Turk B. Blocking autophagy to prevent parasite differentiation: a possible new strategy for fighting parasitic infections? Autophagy. 2008;4:361–363. doi: 10.4161/auto.5592. [DOI] [PubMed] [Google Scholar]


