Abstract
The AUXIN RESPONSE FACTORs (ARFs) and the Aux/IAA proteins regulate various auxin responses through auxin perception mediated by the F-box proteins TIR1/AFBs. ARFs are transcription factors that modulate expression of auxin response genes and are negatively regulated by the Aux/IAA proteins. To gain insight into the regulatory mechanisms of Aux/IAA-ARF action at the genome level, the transcriptome regulated downstream of iaa1, a stabilized IAA1 mutant protein, was identified using dexamethasone (DEX)-controlled nuclear translocation of iaa1 during the auxin response. The expression of the iaa1-regulated auxin-responsive genes selected from microarray data was analysed with RNA-gel blot analysis and it was shown that auxin-regulated expression of these genes was significantly inhibited by DEX treatment. While cycloheximide-inducible expression of a majority of these genes was also DEX-suppressible, expression of some genes could not be suppressed by treatment with DEX. Expression analysis in a variety of arf mutant backgrounds suggested that all iaa1-regulated auxin-response genes examined are controlled by ARFs to different extents and that the same ARF protein can regulate the expression of these genes in response to auxin in a positive or a negative manner. However, arf mutations did not affect auxin-mediated down-regulation, indicating that ARFs might not play a critical role in down-regulation. The decrease in auxin-responsive gene expression in arf7 arf19 mutants was more severe than that of tir1/afb quadruple mutants. These results show the diversity and complexity of mechanisms of Aux/IAA-ARF- and auxin-regulated gene expression. These data also provide the opportunity for functional analysis of genes mediating the auxin-response downstream of Aux/IAA-ARFs.
Keywords: ARF, Aux/IAA, auxin, auxin response factor, cycloheximide, dexamethasone, gene expression, IAA1, microarray, TIR/AFB
Introduction
The plant hormone auxin influences almost every aspect of plant growth and development (Davies, 2004). Numerous genetic and biochemical studies have shown that various auxin responses are regulated by two large protein families named the ARF and Aux/IAA proteins (Berleth et al., 2004; Parry and Estelle, 2006). In Arabidopsis, there are 23 ARF proteins. Each ARF contains a conserved DNA-binding domain near their N-terminus, a long middle region, and, in most ARFs, a dimerization domain near the C-terminus (Quint and Gray, 2006). The DNA-binding domain of the ARF proteins binds to the conserved auxin-responsive promoter element and, depending on the structure of the middle region, each ARF functions as a transcriptional activator or repressor. ARF5, ARF6, ARF7, and ARF8 contain a Gln-rich middle region and function as transcriptional activators in transient protoplast transfection assays. By contrast, ARF1, ARF2, ARF3, ARF4, and ARF9 contain a proline- and/or serine-rich region, or lack a Gln-rich region, and act as transcriptional repressors (Ulmasov et al., 1999a; Tiwari et al., 2003).
Different ARFs are involved in various developmental processes ranging from embryogenesis to floral development (Parry and Estelle, 2006). Although many single arf mutants lack obvious phenotypes, which is indicative of the functional overlap among the various family members, some arf single mutants display overt phenotypes. For example, ARF2 is implicated in regulating leaf senescence and floral organ abscission (Ellis et al., 2005; Okushima et al., 2005b). An mnt mutant with dramatically increased seed size and weight was identified as an allele of ARF2 (Schruff et al., 2006). Mutations in ARF2 have also been identified as extragenic suppressors of hookless1 (hls1), suggesting that ARF2 may be regulated downstream of ethylene-regulated HLS1 (Li et al., 2004). ARF3/ETTIN is involved in floral organ and gynoecium development (Sessions and Zambryski, 1995; Sessions et al., 1997). Trans-acting siRNAs mediate regulation of ARF3 gene function during developmental timing and patterning (Fahlgren et al., 2006). ARF5/MONOPTEROS (MP) is involved in vascular strand formation and the initiation of the embryo axis (Hardtke and Berleth, 1998). Loss of function mutations in ARF7/NONPHOTOTROPIC HYPOCOTYL4(NPH4)/MASSUGU1(MSG1)/TRANSPORT INHIBITOR RESPONSE5(TIR5) resulted in impaired hypocotyl phototropism to blue light (Harper et al., 2000). A mutation in ARF8 causes parthenocarpic fruit (Goetz et al., 2006). ARF8 functions as a negative regulator of fruit initiation and is also involved in hypocotyl elongation (Tian et al., 2004; Goetz et al., 2006). MicroRNA (miRNA) 160 targets ARF10, ARF16, and ARF17 and miR167 targets ARF6 and ARF8 (Rhoades et al., 2002). Plants expressing a miRNA-resistant version of ARF17 exhibited various developmental defects, demonstrating the importance of miR160-directed ARF17 regulation (Mallory et al., 2005). Furthermore, arf double mutants exhibited enhanced phenotypes or phenotypes that single mutants did not display, suggesting that there are unique and overlapping functions among related ARF family members. For example, although the arf1 single mutation alone did not confer any phenotype by itself, arf1 mutations enhanced many arf2 pleiotropic phenotypes in arf1 arf2 double mutants, indicating common functions (Ellis et al., 2005). ARF5/MP and ARF7/NPH4 function redundantly during oriented cell differentiation and synergistically in cell expansion (Hardtke et al., 2004). ARF6 and ARF8 function redundantly in flower maturation (Nagpal et al., 2005). While arf6 and arf8 single mutant plants have delayed stamen development and decreased fecundity, arf6 arf8 double mutants have a complete block in flower maturation. Mutations in ARF19 had a little effect on their own, but in arf7 arf19 double mutants, several phenotypes not observed in the single mutants were found, including a decrease in lateral and adventitious root formation, abnormal gravitropism in both hypocotyls and roots, and a decrease in leaf cell expansion (Wilmoth et al., 2005; Okushima et al., 2005b).
The ARF proteins are negatively regulated by the Aux/IAA proteins. In Arabidopsis, there are 29 Aux/IAA proteins that contain four conserved domains (Parry and Estelle, 2006). Domain I is responsible for repression (Tiwari et al., 2004), whereas domains III and IV mediate homo- and hetero-dimerization among the Aux/IAA proteins as well as between Aux/IAA and ARFs (Kim et al., 1997; Ulmasov et al., 1997a, b). The Aux/IAA proteins act as transcriptional repressors by interacting with ARFs through domains III and IV (Tiwari et al., 2001, 2004). Domain II contains a 13-amino acid degron motif that is responsible for the rapid degradation of the Aux/IAA proteins (Worley et al., 2000; Ramos et al., 2001). So far, 13 different loss-of-function aux/iaa mutants have been characterized and none of them have shown obvious developmental defects. Double and triple mutants of the Aux/IAA genes, such as iaa8 iaa9 or iaa5 iaa6 iaa19, within the same clade exhibited wild-type phenotypes, indicating extensive genetic redundancy among Aux/IAA family members (Overvoorde et al., 2005).
Clues as to the biological function of the Aux/IAA genes all came from analysis of gain-of-function mutants. Ten gain-of-function aux/iaa mutants were identified including IAA1/AXR5, IAA3/SHY2, IAA6/SHY1, IAA7/AXR2, IAA12/BDL, IAA14/SLR, IAA17/AXR3, IAA18, IAA19/MSG2, and IAA28, each exhibiting reduced auxin-response in various aspects of development and growth (Berleth et al., 2004; Yang et al., 2004). All of the mutations reside in the 13-amino acid degron motif of domain II. These mutations result in a dramatically increased life span and an increased abundance of the affected protein. This, in turn, leads to inhibition of ARF function, conferring auxin-related phenotypes.
The rapid degradation of the Aux/IAA proteins is proteasome-dependent (Ramos et al., 2001; Gray et al., 2001). Auxin promotes degradation of the Aux/IAA proteins by enhancing the interaction between the ubiquitin-ligase SCFTIR1 complex and domain II of the Aux/IAA proteins (Gray et al., 2001; Zenser et al., 2001). The F-box protein TIR1 was shown to be an auxin receptor that mediates Aux/IAA degradation and transcriptional responses to auxin (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005). Three additional F-box proteins (AFB1, -2, and -3), homologous to TIR1, interact with the Aux/IAA proteins in an auxin-dependent manner (Dharmasiri et al., 2005b). Plants deficient in all four proteins are auxin-insensitive, indicating that the TIR1 and AFB proteins collectively mediate auxin responses throughout plant development. Crystal structures showed that auxin binds to a single surface pocket of TIR1 and on top of auxin, the Aux/IAA peptide occupies the rest of the pocket (Tan et al., 2007). A very recent study showed that TOPLESS (TPL) can interact with IAA12 through the ERF-associated amphiphilic repression motif and can repress transcription in vivo (Szemenyei et al., 2008). Also, direct interaction between TPL and ARF5 is regulated by IAA12, causing a loss-of-function arf5 phenotype. These results demonstrate that TPL is a transcriptional co-repressor for Aux/IAA-ARF-mediated gene regulation during the auxin response.
While there is extensive knowledge on the signal transduction events from auxin perception to gene regulation, little is known about signalling and function of the genes modulated downstream of the Aux/IAA and ARF proteins for mediating the auxin response. We had previously set out to study signalling downstream of the Aux/IAA and ARF proteins using an inducible system that employed the regulatory mechanism of the mammalian glucocorticoid hormone receptor (Park et al., 2002). Transgenic Arabidopsis (Pro35S:iaa1:GR) has been made that expresses the hormone binding domain of glucocorticoid receptor(GR)-fused stabilized-iaa1 proteins harbouring an amino acid change in domain II. Treatment of DEX to this transgenic Arabidopsis evoked dramatic auxin-related phenotypes and repressed auxin induction of various Aux/IAA genes, indicating that the iaa1 protein impaired auxin responses by acting as a negative regulator for the auxin-response pathway. DEX-inducible translocation of the iaa1 protein to the nucleus can induce auxin-related phenotypes that can be linked to downstream signalling of the Aux/IAA proteins at the time of treatment. The effects of DEX-inducible iaa1 on auxin-regulated gene expression are examined here, focusing on early genes and using the Affymetrix full genome array. The transcriptome downstream of iaa1 was identified in an effort to understand the auxin response pathway at the genome level. These microarray data provide valuable resources for understanding specific auxin responses and/or a subset of these responses. Expression of representative auxin-regulated genes affected by iaa1 has also been analysed in terms of induction kinetics using the RNA-gel blot method. The effects of cycloheximide and auxin with or without DEX in wild-type, arf, and tir1/afb mutant backgrounds were examined. The results highlight the complexity and diversity of auxin-regulated gene expression, which cannot be explained by the Aux/IAA-ARF system alone, and it also revealed versatile ARF functions during auxin-regulated gene expression.
Materials and methods
Plant growth and tissue treatment
Arabidopsis thaliana seedlings were grown for 7 d with a 16 h photoperiod on 3MM Whatman filter paper on top of agar plates containing half-strength Murashige–Skoog (MS) media salts with vitamins, 1.5% sucrose, 2.5 mM MES, pH 5.7, and 0.8% phytoagar at 23 °C. The filter paper with the seedlings was then transferred to plates containing half-strength MS with plant hormone or chemicals (20 μM IAA, 10 μM DEX, or 50 μM cycloheximide) without agar and incubated for a given period of time with gentle shaking in the light at 23 °C.
Plasmid construction and Arabidopsis transformation
The promoter region of IAA1 encompassing 1702 bp (from –1705 to –4 bp relative to the AUG initiation codon) was isolated by PCR from genomic DNA of Arabidopsis Col-0, subcloned into pBI101 (Clontech) in place of the CaMV 35S promoter, and transgenic Arabidopsis were made containing this construct (ProIAA1:GUS) by Agrobacterium-mediated transformation (Bechtold et al., 1993). T3 homozygous transformants were made and amplified. All constructs were verified by DNA sequencing prior to plant transformation.
Histochemical GUS assays
Histochemical assays for GUS activity were performed by incubating the treated seedlings in 5-bromo-4-chloro-3-indolyl glucuronide (Duchefa, The Netherlands) at 37 °C for 24 h and removing the chlorophyll from green tissues by incubation in 100% ethanol, as previously described by Jefferson and Wilson (1991).
Confirmation of Arabidopsis ARF T-DNA insertion mutants and tir1/afb mutants
Arabidopsis ARF T-DNA insertion mutants from ABRC were verified by PCR with the primers designed by the T-DNA primer design program available from the Salk Institute Genomic Analysis Laboratory (SIGNAL) (http://signal.salk.edu/) (see Supplementary Table S1 at JXB online). Triple and quadruple tir1/afb mutants were verified by PCR with the primers shown in Supplementary Table S1 at JXB online.
RNA isolation, RT-PCR, and RNA-gel blot analysis
Following treatment, Arabidopsis plants were immediately frozen in liquid nitrogen and stored at –80 °C. Total RNA was isolated from frozen Arabidopsis using the TRI Reagent® (Molecular Research Center, Inc., Cincinnati, OH, USA). Total RNA was separated on 1.2% agarose gels, transferred to nylon membranes, and hybridized with 32P-labelled DNA probes at 68 °C for 3 h using 10 ml of QuickHyb solution (Stratagene, La Jolla, CA, USA) and then washed. The blot was then exposed to X-ray film. For RT-PCR analysis, total RNA was isolated using an RNeasy plant mini kit (Qiagen, Hilden, Germany) and subject to RT-PCR analysis with the Access RT-PCR System (Promega, Madison, WI, USA) according to the manufacturer's instruction. The DNA probes for RNA gel-blot analysis were amplified by RT-PCR, subcloned into the pGEM®-T Easy vector (Promega), and verified by DNA sequencing. RT-PCR conditions, primer sequences, and other DNA vector probes are shown in Supplementary Table S1 at JXB online.
Microarray analysis
For Affymetrix GeneChip analysis, Pro35S:iaa1:GR seedlings (Park et al., 2002) were grown for 7 d on half-strength MS media, treated with 20 μM IAA, 20 μM IAA and 10 μM DEX, or mock-treated for 2 h, as described above in ‘Plant growth and tissue treatment’, and total RNA was isolated with an RNeasy plant mini kit (Qiagen). Five micrograms of RNA were used to make biotin-labelled cRNA products. The Affymetrix Arabidopsis ATH1 genome array GeneChip, which contains >22,500 probe sets representing approximately 20,000 genes, was used. Probe synthesis from total RNA samples, hybridization, detection, and scanning were performed according to standard protocols from Affymetrix, Inc. (Santa Clara, CA, USA). Expression profiles were analysed using the GeneChip Operating Software (Affymetrix, CA, USA). GeneChip Operating Software (Affymetrix) was used to determine the absolute analysis metrics (Detection, Detection P-value) using the scanned probe array data and the different treatment group signals were compared to generate the change, change P-value, and signal log ratio (fold change). The experimental data from the microarray analysis were normalized by global scaling (Statistical Algorithms Reference Guide, Technical Report, Affymetrix, 2001). Differentially regulated genes in response to auxin were selected based on the criteria: | log2(fold-change) |>1 and P-value <0.05 with Welch's t-test (Welch, 1947). The microarray data were analysed using GenPlex™ version 2.6 software (ISTECH, Seoul, Korea). Gene function analysis was performed using the gene ontology mining software, High-Throughput GoMiner (http://discover.nci.nih.gov/gominer/htgm.jsp). Specification of the many gene annotations was also supplemented by further online database searches such as http://www.arabidopsis.org/tools/bulk/go/index.jsp. Microarray data were deposited into ArrayExpress with the accession number E-MEXP-1256 at http://www.ebi.ac.uk/at-miamexpress.
Results
GUS expression of ProIAA1:GUS
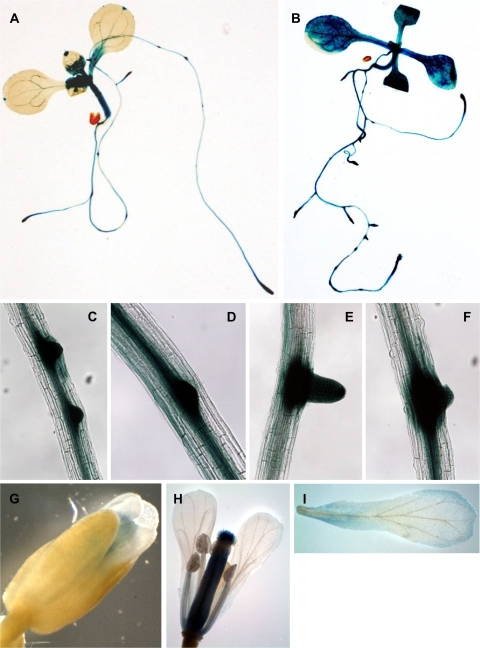
To get an insight into the physiological and functional relevance of iaa1 in auxin-related phenotypes, including gene regulation, it was determined precisely where IAA1 is expressed in Arabidopsis. The 1.7 kbp promoter region of the IAA1 gene was fused to GUS and five independent homozygous transgenic Arabidopsis lines containing the corresponding construct (ProIAA1:GUS) were generated. As shown in representative histochemical analysis of GUS expression (Fig. 1A), strong GUS staining was detected in various tissues including primary roots, lateral roots, petioles, veins, hydathodes, and hypocotyls of 7-d-old seedlings before auxin treatment. The treatment with auxin, indole-3 acetic acid, greatly enhanced the degree of GUS staining throughout the whole seedling (Fig. 1B). Similar expression patterns of GUS staining were observed in 14-d-old plants (data not shown). GUS expression was also observed in the flower of the mature plants (G-I), particularly in the stigma, ovary, filament, and petal. These GUS expression patterns of the IAA1 promoter suggest a role for IAA1 in a variety of tissues during auxin response at early developmental stages.
Fig. 1.
Expression of GUS in ProIAA1:GUS transgenic plants. (A, B) Seven-day-old light-grown seedling without (A) or with 20 μM of IAA (B) for 2 h. (C–F) Emerging (C, D) or emerged (E, F) lateral roots of 7-d-old light grown seedlings without (C, E) or with (D, F) IAA treatment for 2 h. (G, H) Flower of 7-week-old light-grown seedling. (I) Petal of 7-week-old light-grown seedling.
Identification of early auxin-regulated genes affected by iaa1 using microarrays
To identify the transcriptome downstream of iaa1, genome-wide analysis was performed on the effects of the DEX-inducible nuclear localization of iaa1 on auxin-regulated gene expression using the Affymetrix ATH1 Arabidopsis full genome array. A 2 h time point was selected following auxin treatment to investigate the early auxin response. For microarray analysis, Arabidopsis transgenic plants (Pro35S:iaa1:GR) were used that overexpressed the iaa1 mutant protein fused to GR under the CaMV 35S promoter (Park et al., 2002). Pro35S:iaa1:GR seedlings were incubated with mock, IAA, or IAA and DEX. Total RNA was then isolated and the validity of auxin-regulated gene expression was verified by RNA-gel blot analysis using an IAA2 DNA probe (data not shown). This experiment and microarray analysis were performed in triplicate and genes were extracted that satisfied the criteria of both a >2-fold difference on average between auxin- and mock-treated plants and P <0.05 using Welch's t test (Welch, 1947) as a cutoff value (Tables 1, 2). Further, the genes that express the transcripts showing absent calls (A) in all arrays were eliminated. In the case of down-regulated genes, only the genes that express the transcripts showing present calls (P) before auxin treatment in all three hybridizations were included. It has previously been demonstrated that the treatment with DEX for 2 h on wild-type plants and wild-type IAA1:GR plants did not affect expression levels of the IAA genes tested (Park et al., 2002). Also, no phenotypic change in wild-type plants or in wild-type IAA1:GR plants when treated with DEX was observed.
Table 1.
List of genes up-regulated 2 h after treatment with IAA
| Category | At no. | Probe ID | Gene title | IAA |
IAA + Dex |
Impaired in arf mutantse | AuxRE |
|||
| FCa | P-valueb | FCc | P-valued | Af | Bg | |||||
| Auxin-regulated | At1g15580 | 261766_at | IAA5 (Aux2-27) | 92.3 | 0.002 | 21.9 | 0.031 | √ | 1 | 0 |
| At1g29490 | 259785_at | Auxin-responsive protein | 69.2 | 0.017 | 17.9 | 0.260 | √ | 0 | 0 | |
| At2g23170 | 245076_at | GH3.3 | 60.4 | 0.008 | 22.5 | 4.8E-04 | √ | 3 | 2 | |
| At4g37390 | 262099_s_at | GH3.2 (YDK1) | 39.2 | 0.015 | 4.3 | 2.2E-05 | 2 | 3 | ||
| At2g14960 | 266611_at | GH3.1 | 31.5 | 7.2E-05 | 8.9 | 1.1E-03 | √ | 3 | 0 | |
| At3g15540 | 258399_at | IAA19 | 25.7 | 0.007 | 10.9 | 0.024 | √ | 6 | 1 | |
| At4g32280 | 253423_at | IAA29 | 22.0 | 0.039 | 4.4 | 1.2E-03 | √ | 2 | 0 | |
| At4g27260 | 253908_at | GH3.5 | 14.4 | 0.011 | 5.5 | 1.4E-03 | √ | 2 | 1 | |
| At1g52830 | 260152_at | IAA6 | 11.6 | 0.031 | 4.9 | 0.302 | √ | 1 | 0 | |
| At3g62100 | 251246_at | IAA30 | 10.8 | 0.006 | 6.0 | 0.058 | √ | 2 | 0 | |
| At2g18010 | 265806_at | Auxin-responsive protein | 8.6 | 0.043 | 3.5 | 0.247 | √ | 0 | 2 | |
| At4g22620 | 254323_at | Auxin-responsive protein | 8.3 | 0.002 | 2.9 | 0.002 | √ | 1 | 4 | |
| At5g54510 | 248163_at | Dwarf in light 1 (DFL-1, GH3.6) | 8.0 | 0.002 | 4.5 | 0.015 | √ | 2 | 1 | |
| At4g12410 | 254809_at | Auxin-responsive protein | 6.0 | 0.015 | 2.3 | 0.033 | √ | 1 | 0 | |
| At3g23030 | 257766_at | IAA2 | 5.5 | 0.008 | 1.9 | 0.037 | √ | 2 | 1 | |
| At4g36110 | 253103_at | Auxin-responsive protein | 4.5 | 0.033 | 1.8 | 0.091 | √ | 1 | 2 | |
| At1g29500 | 259773_at | Auxin-responsive protein | 4.3 | 0.005 | 1.3 | 0.033 | 4 | 0 | ||
| At2g33310 | 255788_at | IAA13 | 4.1 | 0.022 | 2.4 | 0.038 | √ | 1 | 0 | |
| At5g20820 | 246000_at | Auxin-responsive protein-related | 3.9 | 0.004 | 2.1 | 0.064 | √ | 0 | 0 | |
| At5g43700 | 249109_at | IAA4 (Aux2-11) | 3.5 | 0.003 | 1.9 | 0.010 | √ | 2 | 4 | |
| At4g28640 | 253791_at | IAA11 | 3.3 | 0.003 | 1.5 | 0.012 | √ | 0 | 1 | |
| At4g34770 | 253207_at | Auxin-responsive protein | 3.1 | 0.006 | 1.6 | 0.055 | √ | 0 | 1 | |
| At1g04240 | 263656_at | IAA3, Short hypocotyl 2 (SHY2) | 2.7 | 0.030 | 1.7 | 0.222 | 3 | 4 | ||
| At1g19220 | 256010_at | ARF19 | 2.6 | 0.015 | 1.1 | 0.022 | √ | 1 | 1 | |
| At4g14550 | 245593_at | IAA14, Solitary root (SLR) | 2.6 | 0.020 | 1.4 | 0.012 | √ | 0 | 0 | |
| At3g07390 | 259018_at | Auxin-responsive protein (AIR12) | 2.1 | 0.003 | 1.4 | 0.059 | √ | 0 | 2 | |
| Biotic and abiotic | At4g38410 | 252988_at | Dehydrin | 3.1 | 0.032 | 2.8 | 0.809 | √ | 2 | 0 |
| Development | At3g58190 | 251565_at | LBD29 | 123.7 | 0.015 | 18.7 | 0.039 | √ | 1 | 1 |
| At5g06080 | 250709_at | LBD33 | 14.7 | 6.3E-04 | 8.0 | 0.067 | 3 | 1 | ||
| At2g45420 | 245140_at | LBD18 | 6.3 | 0.007 | 1.7 | 0.009 | √ | 1 | 1 | |
| At2g42430 | 265856_at | LBD16 | 5.6 | 0.003 | 1.6 | 0.025 | √ | 1 | 0 | |
| At3g15370 | 258388_at | EXP12 | 4.3 | 0.023 | 2.6 | 0.021 | 1 | 0 | ||
| At1g73590 | 259845_at | Auxin efflux carrier protein (PIN1) | 3.1 | 0.002 | 2.2 | 0.073 | 0 | 1 | ||
| At2g39700 | 267590_at | EXP4 | 2.4 | 0.013 | 1.8 | 0.295 | √ | 0 | 1 | |
| At3g60550 | 251423_at | Cyclin | 2.2 | 0.026 | 1.4 | 0.081 | 1 | 2 | ||
| Energy | At1g72230 | 259801_at | Plastocyanin-like domain-containing protein | 2.4 | 0.010 | 1.9 | 0.324 | √ | 1 | 1 |
| At1g30760 | 264527_at | FAD-binding domain-containing protein | 2.1 | 0.032 | 1.1 | 0.158 | 1 | 1 | ||
| Hormone responsive | At4g08040 | 255177_at | ACS11 | 36.4 | 1.0E-03 | 7.9 | 0.022 | 1 | 1 | |
| At2g22810 | 266830_at | ACS4 | 17.9 | 0.007 | 5.6 | 0.164 | √ | 1 | 1 | |
| At4g37770 | 253066_at | ACS8 | 5.7 | 0.015 | 2.2 | 0.077 | √ | 3 | 3 | |
| At3g49700 | 252279_at | ACS9 | 5.2 | 0.033 | 2.5 | 0.016 | √ | 0 | 1 | |
| At3g63440 | 251178_at | Cytokinin oxidase (CKX7) | 4.2 | 0.010 | 2.6 | 0.110 | √ | 1 | 0 | |
| At2g41510 | 245108_at | Cytokinin oxidase (CKX1) | 3.7 | 0.018 | 3.0 | 0.494 | √ | 1 | 2 | |
| At1g04310 | 263653_at | Ethylene response sensor 2 (ERS2) | 3.5 | 0.026 | 2.2 | 0.091 | √ | 1 | 0 | |
| At4g11280 | 254926_at | ACS6 | 2.6 | 0.012 | 1.6 | 0.150 | √ | 3 | 1 | |
| Metabolism | At1g74110 | 260376_at | CYP78A10 | 38.4 | 0.002 | 1.9 | 0.004 | 1 | 1 | |
| At1g78970 | 264100_at | Lupeol synthase (LUP1) | 4.8 | 0.010 | 2.6 | 0.139 | 0 | 3 | ||
| At3g26760 | 258253_at | Short-chain dehydrogenase/reductase | 4.7 | 0.008 | 2.8 | 0.134 | √ | 2 | 2 | |
| At5g19530 | 245947_at | Spermidine synthase (ACL5) | 3.9 | 0.009 | 2.4 | 0.031 | 5 | 1 | ||
| At5g18930 | 249951_at | Adenosylmethionine decarboxylase | 3.3 | 0.008 | 2.5 | 0.288 | 1 | 3 | ||
| At2g26710 | 267614_at | CYP72B1 (BAS1) | 3.3 | 0.035 | 1.4 | 0.056 | 1 | 2 | ||
| At1g02660 | 260915_at | Lipase class 3 | 2.8 | 0.021 | 1.4 | 0.015 | √ | 0 | 2 | |
| At3g53450 | 251940_at | Decarboxylase | 2.7 | 0.007 | 2.7 | 0.972 | 2 | 0 | ||
| At5g48175 | 248717_at | Thioglucoside glucohydrolase | 2.7 | 0.013 | 2.3 | 0.252 | √ | 1 | 0 | |
| At1g06080 | 260957_at | Δ-9 desaturase (ADS1) | 2.6 | 0.032 | 3.9 | 0.215 | 1 | 0 | ||
| At2g47130 | 266761_at | Short-chain dehydrogenase/reductase | 2.5 | 0.008 | 1.2 | 0.027 | √ | 1 | 0 | |
| At2g47550 | 245151_at | Pectin esterase | 2.3 | 0.012 | 1.3 | 0.035 | √ | 2 | 1 | |
| At5g64250 | 247283_at | 2-nitropropane dioxygenase | 2.3 | 0.011 | 1.3 | 0.073 | √ | 1 | 0 | |
| At5g54500 | 248162_at | Quinone reductase 1 | 2.1 | 0.012 | 1.5 | 0.090 | √ | 0 | 2 | |
| Protein metaboism | At1g62770 | 262643_at | Invertase/pectin methylesterase inhibitor | 4.2 | 1.9E-05 | 1.5 | 0.002 | √ | 1 | 0 |
| At5g09800 | 250493_at | U-box domain-containing protein | 2.7 | 0.016 | 1.6 | 0.177 | 4 | 1 | ||
| At4g00080 | 255695_at | Invertase/pectin methylesterase inhibitor | 2.3 | 0.003 | 0.7 | 0.057 | √ | 0 | 2 | |
| At4g36880 | 246250_at | Cysteine proteinase | 2.3 | 0.009 | 2.3 | 0.909 | √ | 0 | 1 | |
| Signal transduction | At5g02760 | 251017_at | PP2C family protein | 6.3 | 0.012 | 1.5 | 0.010 | √ | 1 | 2 |
| At5g54490 | 248164_at | Pinoid-binding protein 1 | 5.7 | 0.002 | 2.4 | 0.038 | √ | 0 | 0 | |
| At1g78100 | 260058_at | F-box protein | 4.4 | 0.015 | 1.5 | 0.040 | 3 | 3 | ||
| At3g20830 | 257975_at | Protein kinase | 3.7 | 0.015 | 1.9 | 0.007 | √ | 2 | 4 | |
| At1g51170 | 265144_at | Protein kinase | 3.5 | 0.009 | 2.1 | 0.013 | √ | 2 | 3 | |
| At2g47860 | 266507_at | NPH3 protein | 3.5 | 0.033 | 1.8 | 0.004 | 1 | 4 | ||
| At2g30040 | 266832_at | Protein kinase (MAPKKK14) | 3.5 | 8.3E-04 | 1.4 | 0.084 | √ | 1 | 1 | |
| At2g25790 | 266663_at | LRR protein kinase | 3.0 | 0.015 | 1.9 | 0.015 | 0 | 0 | ||
| At1g77280 | 264479_at | Protein kinase | 2.8 | 0.025 | 1.7 | 0.121 | √ | 1 | 5 | |
| At2g41820 | 260494_at | LRR protein kinase | 2.7 | 0.004 | 1.9 | 0.009 | 1 | 1 | ||
| At5g59010 | 247743_at | Potein kinase-related | 2.5 | 0.029 | 1.5 | 0.033 | 2 | 0 | ||
| At3g13380 | 256981_at | LRR protein kinase | 2.4 | 0.028 | 1.1 | 0.041 | √ | 2 | 1 | |
| At3g50310 | 252212_at | MAPKKK20 | 2.2 | 0.012 | 1.6 | 0.130 | 0 | 1 | ||
| At3g14710 | 258118_at | F-box protein | 2.1 | 0.034 | 1.3 | 0.151 | 0 | 0 | ||
| At5g12940 | 250277_at | LRR protein kinase | 2.1 | 0.023 | 1.3 | 0.083 | 5 | 2 | ||
| At2g26290 | 267372_at | Protein kinase | 2.0 | 0.029 | 1.6 | 0.401 | √ | 1 | 3 | |
| At4g37590 | 253062_at | NPH3 protein | 2.0 | 0.015 | 1.2 | 0.052 | 2 | 0 | ||
| At3g54030 | 251922_at | Protein kinase | 2.0 | 0.008 | 1.6 | 0.103 | 0 | 0 | ||
| Transcription factor | At5g26930 | 246798_at | ZFP | 10.8 | 0.009 | 3.1 | 0.024 | 2 | 0 | |
| At5g67060 | 247023_at | bHLH protein (HEC1) | 5.4 | 8.2E-04 | 1.4 | 3.5E-04 | √ | 1 | 1 | |
| At5g18560 | 249992_at | AP2 domain-containing transcription factor | 4.9 | 1.1E-03 | 2.0 | 0.009 | √ | 1 | 2 | |
| At5g47370 | 248801_at | Homeobox-leucine zipper protein 2 (HAT2) | 4.6 | 0.009 | 2.0 | 0.023 | √ | 1 | 3 | |
| At1g65920 | 261917_at | Regulator of chromosome condensation protein | 4.5 | 0.031 | 1.8 | 0.122 | √ | 1 | 1 | |
| At5g25190 | 246932_at | Ethylene-responsive element-binding protein | 4.5 | 0.018 | 2.6 | 0.172 | √ | 1 | 4 | |
| At1g44830 | 261327_at | AP2 domain-containing transcription factor (TINY) | 3.2 | 0.027 | 1.8 | 0.205 | √ | 1 | 3 | |
| At1g18400 | 261717_at | bHLH protein (BEE1) | 3.1 | 0.029 | 1.3 | 0.066 | 0 | 1 | ||
| At1g28370 | 261470_at | ERF domain protein 11 (ERF11) | 2.9 | 0.038 | 1.9 | 0.295 | √ | 0 | 2 | |
| At5g40590 | 249364_at | DC1 domain-containing protein | 2.6 | 0.030 | 3.5 | 0.095 | √ | 0 | 1 | |
| At2g01430 | 266346_at | Homeobox-leucine zipper protein 17 (HB-17) | 2.6 | 0.022 | 2.5 | 0.908 | 0 | 0 | ||
| At5g15160 | 250155_at | bHLH protein (PRE2) | 2.5 | 0.014 | 1.8 | 0.167 | 0 | 0 | ||
| At4g37890 | 253011_at | C3HC4-type RING finger protein | 2.4 | 0.033 | 1.5 | 3.9E-04 | √ | 1 | 1 | |
| At3g25710 | 257642_at | bHLH protein | 2.4 | 0.042 | 1.9 | 0.227 | 1 | 2 | ||
| At1g73830 | 260070_at | bHLH protein (BEE3) | 2.2 | 0.031 | 1.6 | 0.196 | 0 | 1 | ||
| At4g32880 | 253402_at | Homeobox-leucine zipper protein (HB-8) | 2.2 | 0.033 | 1.8 | 0.056 | 3 | 1 | ||
| At5g58620 | 247795_at | Zinc finger (CCCH-type) | 2.0 | 0.032 | 1.2 | 0.082 | 1 | 4 | ||
| At3g06490 | 258516_at | MYB108 | 2.0 | 0.034 | 1.2 | 0.027 | 0 | 1 | ||
| Transferase | At5g67430 | 246992_at | GCN5-related N-acetyltransferase (GNAT) | 19.8 | 0.043 | 3.9 | 0.050 | √ | 0 | 0 |
| At5g55250 | 248104_at | SAM:carboxyl methyltransferase | 3.8 | 0.001 | 3.1 | 0.205 | 2 | 1 | ||
| At2g39980 | 267337_at | Transferase | 3.7 | 0.030 | 2.0 | 0.019 | √ | 1 | 2 | |
| At5g01210 | 251144_at | Transferase | 2.6 | 0.043 | 1.7 | 0.128 | 0 | 0 | ||
| At1g21980 | 255959_at | AtPIP5K1 | 2.4 | 0.046 | 1.2 | 0.017 | 1 | 0 | ||
| At4g15550 | 245277_at | UDP-glucose:IAA beta-D-glucosyltransferase | 2.3 | 0.024 | 2.0 | 0.584 | √ | 1 | 0 | |
| Transporter | At3g06370 | 258907_at | Sodium proton exchanger (NHX3) | 3.7 | 0.007 | 2.7 | 0.312 | √ | 1 | 1 |
| At1g58340 | 256024_at | MATE efflux protein-related (ZF14) | 3.6 | 0.022 | 2.4 | 0.047 | √ | 2 | 0 | |
| At1g59740 | 262912_at | Proton-dependent oligopeptide transport protein | 3.5 | 0.005 | 2.8 | 0.160 | √ | 0 | 4 | |
| At2g21050 | 264025_at | Amino acid permease | 3.2 | 0.022 | 2.9 | 0.126 | √ | 2 | 0 | |
| At5g27000 | 246802_at | Kinesin 4 (ATK4) | 2.7 | 0.040 | 1.7 | 0.069 | 0 | 1 | ||
| At5g52890 | 248278_at | AT hook motif-containing protein | 2.2 | 0.041 | 0.7 | 0.141 | 1 | 2 | ||
| Unknown | At2g39370 | 266974_at | Expressed protein | 15.2 | 6.E-05 | 4.8 | 1.4E-03 | √ | 3 | 1 |
| At5g62280 | 247474_at | Expressed protein | 9.0 | 0.020 | 1.8 | 0.003 | √ | 3 | 3 | |
| At5g50335 | 248509_at | Expressed protein | 6.8 | 0.045 | 2.3 | 0.130 | √ | 1 | 1 | |
| At2g28690 | 263436_at | Expressed protein | 6.5 | 0.004 | 1.9 | 0.046 | √ | 0 | 2 | |
| At1g64405 | 259735_at | Expressed protein | 6.4 | 2.9E-04 | 1.9 | 0.016 | √ | 2 | 2 | |
| At3g19200 | 257026_at | Hypothetical protein | 5.9 | 0.010 | 4.2 | 0.031 | √ | 0 | 1 | |
| At3g28420 | 257900_at | Expressed protein | 5.5 | 0.003 | 4.1 | 0.361 | 2 | 2 | ||
| At4g35210 | 253180_at | Hypothetical protein | 5.3 | 0.019 | 0.3 | 0.048 | 4 | 3 | ||
| At5g17340 | 250091_at | Expressed protein | 5.2 | 0.007 | 1.6 | 0.022 | √ | 0 | 0 | |
| At4g17350 | 245416_at | Expressed protein | 5.1 | 0.006 | 1.7 | 0.005 | √ | 0 | 1 | |
| At5g52900 | 248282_at | Expressed protein (MXC20_13) | 4.4 | 0.018 | 0.9 | 0.012 | √ | 2 | 0 | |
| At4g37295 | 253047_at | Expressed protein | 3.9 | 0.004 | 1.5 | 0.025 | √ | 0 | 0 | |
| At3g29370 | 256743_at | Expressed protein | 3.8 | 0.017 | 1.9 | 0.006 | √ | 3 | 0 | |
| At3g50340 | 252204_at | Expressed protein | 3.7 | 1.3E-03 | 1.6 | 0.006 | √ | 1 | 0 | |
| At3g59900 | 251436_at | Expressed protein | 3.7 | 0.007 | 2.3 | 0.027 | √ | 3 | 1 | |
| At3g15250 | 257049_at | Expressed protein | 3.6 | 0.002 | 1.7 | 0.049 | 2 | 2 | ||
| At4g35200 | 253179_at | Hypothetical protein | 3.5 | 5.8E-04 | 1.2 | 0.001 | √ | 3 | 4 | |
| At5g57760 | 247878_at | Expressed protein | 3.4 | 0.012 | 1.3 | 0.009 | √ | 1 | 1 | |
| At5g12050 | 250327_at | Expressed protein | 3.3 | 0.027 | 1.2 | 0.019 | 1 | 4 | ||
| At4g22530 | 254318_at | Embryo-abundant protein-related | 3.3 | 0.017 | 1.8 | 0.102 | 1 | 0 | ||
| At4g13195 | 254761_at | Expressed protein | 3.3 | 0.010 | 1.5 | 0.026 | √ | 3 | 3 | |
| At3g55720 | 251751_at | Expressed protein | 3.1 | 0.006 | 0.9 | 0.043 | 1 | 1 | ||
| At1g80240 | 262045_at | Expressed protein (ATGDI1) | 2.8 | 0.012 | 1.3 | 0.099 | √ | 4 | 1 | |
| At1g03820 | 265083_at | Expressed protein | 2.8 | 0.046 | 2.0 | 0.361 | √ | 1 | 1 | |
| At3g03170 | 258878_at | Expressed protein | 2.6 | 0.006 | 1.1 | 0.016 | √ | 1 | 1 | |
| At5g22310 | 249887_at | Expressed protein | 2.6 | 0.016 | 1.7 | 0.014 | 2 | 2 | ||
| At4g09890 | 255028_at | Expressed protein | 2.5 | 0.035 | 1.9 | 0.363 | 2 | 1 | ||
| At3g54000 | 251925_at | Expressed protein | 2.4 | 0.011 | 1.3 | 0.022 | √ | 3 | 1 | |
| At1g23340 | 263042_at | Expressed protein | 2.4 | 0.012 | 1.9 | 0.231 | 0 | 2 | ||
| At5g51670 | 248423_at | Expressed protein | 2.4 | 0.016 | 2.0 | 0.266 | √ | 1 | 0 | |
| At1g29195 | 260841_at | Expressed protein | 2.3 | 0.016 | 1.0 | 0.081 | √ | 2 | 0 | |
| At1g23060 | 264902_at | Expressed protein | 2.2 | 0.029 | 1.4 | 0.039 | 1 | 1 | ||
| At2g40000 | 267357_at | Expressed protein | 2.2 | 0.016 | 1.2 | 0.026 | 1 | 2 | ||
| At5g49170 | 248623_at | Expressed protein | 2.2 | 0.025 | 1.6 | 0.209 | 0 | 2 | ||
| At1g60010 | 263737_at | Expressed protein | 2.1 | 0.010 | 1.3 | 0.015 | √ | 3 | 1 | |
| At3g60520 | 251372_at | Expressed protein | 2.1 | 0.002 | 1.6 | 0.012 | 2 | 0 | ||
| At3g47510 | 252419_at | Expressed protein | 2.0 | 0.007 | 1.7 | 0.251 | √ | 0 | 1 | |
Fold change of gene expresion in auxin-treated 7-d-old iaa1:GR transgenic plants compared to that of mock-treated plants.
P-value calculated from auxin-induced gene expression and mock expression using Welch's t-test.
Fold change of gene expresion in auxin and DEX-treated 7-d-old iaa1:GR transgenic plants compared to that of mock-treated plants.
P-value calculated from auxin-induced gene expression in the presence of DEX and auxin-induced gene expression using Welch's t-test.
Genes showing auxin-induced expression impaired in nph4-1 and/or arf19-1 and/or nph4-1 arf19-1 mutants by microarray analysis (Okushima et al., 2005).
Number of AuxRE, TGTCnC sequences in the 2 kb fragments upstream of the start codon. N, any nucleotide.
Number of AuxRE, GnGACA sequences in the 2 kb fragments upstream of the start codon.
Table 2.
List of genes downregulated 2 h after treatment with IAA
| Category | At no. | Probe ID | Gene title | IAA |
IAA + Dex |
Impaired in arf mutantse | AuxRE |
|||
| FCa | P-valueb | FCc | P-valued | Af | Bg | |||||
| Auxin-regulated | At4g31320 | 253515_at | Small auxin up RNA (SAUR_C) | 0.28 | 0.034 | 1.18 | 0.012 | √ | 1 | 0 |
| At4g17280 | 245412_at | Auxin-responsive protein | 0.45 | 0.013 | 0.73 | 0.036 | 0 | 1 | ||
| Boitic and abiotic | At5g45210 | 248990_at | Disease resistance protein | 0.28 | 5.46E-04 | 0.52 | 0.157 | 0 | 1 | |
| At4g39030 | 252921_at | Salicylic acid induction deficient 1 (SID1) | 0.49 | 0.007 | 0.78 | 0.070 | 1 | 1 | ||
| At1g73620 | 260077_at | Pathogenesis-related protein | 0.49 | 0.039 | 0.56 | 0.637 | 0 | 0 | ||
| Development | At2g20750 | 265443_at | β-expansin (EXPB1) | 0.27 | 0.024 | 0.73 | 0.066 | 3 | 0 | |
| At4g36380 | 246216_at | Rotundifolia3 (ROT3)(CYP90C1) | 0.35 | 0.002 | 0.68 | 0.002 | √ | 1 | 2 | |
| Metabolism | At1g05660 | 263229_s_at | Polygalacturonase | 0.03 | 0.022 | 1.13 | 0.014 | 3 | 0 | |
| At1g64590 | 261956_at | Short-chain dehydrogenase (SDR) | 0.22 | 0.006 | 0.97 | 0.008 | √ | 1 | 1 | |
| At1g67110 | 264470_at | Cytochrome P450 protein (CYP735A2) | 0.27 | 1.80E-04 | 0.62 | 0.068 | 1 | 0 | ||
| At4g13310 | 254767_s_at | Cytochrome P450 protein (CYP71A19) | 0.27 | 0.038 | 0.54 | 0.118 | 0 | 1 | ||
| At5g47990 | 248727_at | Cytochrome P450 protein (CYP705A5) | 0.29 | 0.040 | 0.76 | 0.101 | 3 | 0 | ||
| At2g43880 | 267222_at | Polygalacturonase | 0.34 | 0.010 | 0.82 | 0.038 | 0 | 3 | ||
| At4g21840 | 254385_s_at | Methionine sulfoxide reductase | 0.35 | 0.019 | 0.67 | 0.061 | 0 | 2 | ||
| At1g78090 | 260059_at | Trehalose-6-phosphate phosphatase | 0.39 | 0.017 | 0.85 | 0.029 | 2 | 1 | ||
| At5g42590 | 249203_at | Cytochrome P450 protein (CYP71A16) | 0.39 | 0.050 | 1.35 | 0.060 | 2 | 2 | ||
| At4g39950 | 252827_at | Cytochrome P450 protein (CYP79B2) | 0.41 | 0.003 | 0.62 | 0.041 | √ | 0 | 2 | |
| At4g11290 | 254914_at | Peroxidase | 0.45 | 0.040 | 0.72 | 0.178 | 3 | 0 | ||
| At5g58784 | 247781_at | Dehydrodolichyl diphosphate synthase | 0.48 | 0.047 | 0.55 | 0.359 | 1 | 0 | ||
| At2g23560 | 267123_at | Hydrolase | 0.49 | 0.004 | 0.94 | 0.077 | 0 | 0 | ||
| Protein metabolism | At3g20015 | 256626_at | Aspartyl protease | 0.43 | 0.028 | 0.84 | 0.060 | 0 | 0 | |
| Signal transduction | At5g62920 | 247406_at | ARR6 | 0.22 | 0.017 | 0.71 | 0.038 | 1 | 0 | |
| At3g48100 | 252374_at | ARR5 | 0.24 | 0.039 | 0.57 | 0.080 | 1 | 3 | ||
| At5g24100 | 249768_at | LRR protein kinase | 0.25 | 0.017 | 0.56 | 0.092 | 3 | 1 | ||
| At1g19050 | 259466_at | ARR7 | 0.43 | 0.038 | 0.47 | 0.689 | 3 | 1 | ||
| At1g10470 | 263236_at | ARR4 | 0.49 | 0.011 | 0.74 | 0.038 | 0 | 2 | ||
| Transcription factor | At1g10480 | 263208_at | ZFP5 | 0.21 | 0.007 | 0.96 | 0.005 | 1 | 1 | |
| At3g46130 | 252534_at | MYB111 | 0.31 | 0.042 | 1.09 | 0.080 | 0 | 4 | ||
| At5g25160 | 246933_at | ZFP3 | 0.34 | 0.003 | 0.83 | 7.52E-04 | 1 | 3 | ||
| At1g72200 | 259854_at | Zinc finger (C3HC4-type) protein | 0.50 | 0.013 | 0.95 | 0.020 | √ | 2 | 0 | |
| Transferase | At2g18800 | 266066_at | Xyloglucan:xyloglucosyl transferase | 0.08 | 0.020 | 0.56 | 0.035 | 1 | 0 | |
| At4g12510 | 254820_s_at | Lipid transfer protein (LTP) | 0.15 | 0.008 | 0.54 | 0.010 | √ | 0 | 0 | |
| At5g47980 | 248725_at | Transferase | 0.17 | 0.004 | 0.65 | 0.010 | 1 | 2 | ||
| At1g77530 | 259758_s_at | O-methyltransferase 2 | 0.21 | 0.008 | 0.69 | 0.021 | 0 | 1 | ||
| At5g47950 | 248723_at | Transferase | 0.34 | 1.48E-04 | 0.82 | 0.038 | 0 | 2 | ||
| At1g80050 | 262039_at | Adenine phosphoribosyltransferase 2 (APT2) | 0.39 | 0.004 | 0.66 | 0.016 | √ | 1 | 0 | |
| At1g53680 | 259964_at | Glutathione S-transferase (AtGSTU28) | 0.39 | 0.023 | 0.74 | 0.026 | √ | 0 | 0 | |
| At3g45070 | 252605_s_at | Sulphotransferase | 0.40 | 0.021 | 1.06 | 0.066 | 1 | 0 | ||
| At3g29680 | 257281_s_at | Transferase | 0.40 | 0.007 | 0.75 | 0.076 | 2 | 0 | ||
| At3g08860 | 258983_at | Alanine-glyoxylate aminotransferase | 0.44 | 0.045 | 1.02 | 0.060 | 0 | 0 | ||
| At2g23410 | 267137_at | cis-Prenytransferase | 0.46 | 0.016 | 0.26 | 0.213 | 3 | 1 | ||
| Transporter | At5g47450 | 248790_at | Major intrinsic protein (TIP2;3) | 0.27 | 0.020 | 0.84 | 0.032 | √ | 1 | 2 |
| At5g60660 | 247586_at | PIP2:4 | 0.28 | 0.007 | 0.73 | 0.025 | √ | 0 | 0 | |
| At3g45680 | 252594_at | Oligopeptide transporter | 0.31 | 9.04E-04 | 0.70 | 0.017 | 0 | 2 | ||
| At1g31770 | 246580_at | ABC transporter | 0.35 | 0.007 | 0.76 | 0.002 | 2 | 1 | ||
| At3g23430 | 258293_at | Phosphate transporter (PHO1) | 0.42 | 0.005 | 0.89 | 6.51E-04 | 0 | 0 | ||
| Unknown | At4g26320 | 253957_at | Arabinogalactan-protein (AGP13) | 0.16 | 0.007 | 0.46 | 0.071 | √ | 1 | 1 |
| At5g60520 | 247634_at | Late embryogenesis abundant protein | 0.19 | 0.027 | 0.55 | 0.133 | √ | 0 | 0 | |
| At5g53250 | 248252_at | Arabinogalactan-protein (AGP22) | 0.29 | 0.018 | 0.53 | 0.116 | √ | 2 | 2 | |
| At4g02850 | 255450_at | PhzC/PhzF family protein | 0.33 | 0.019 | 0.61 | 0.065 | 0 | 0 | ||
| At3g21680 | 258178_at | Expressed protein | 0.34 | 0.038 | 1.12 | 0.016 | 1 | 2 | ||
| At4g30460 | 253619_at | Glycine-rich protein | 0.37 | 0.031 | 0.84 | 0.003 | 0 | 1 | ||
| At2g21560 | 263545_at | Expressed protein | 0.37 | 0.019 | 0.88 | 0.029 | 0 | 2 | ||
| At5g28610 | 255945_at | Expressed protein | 0.38 | 0.031 | 0.56 | 0.457 | 1 | 2 | ||
| At3g46880 | 252501_at | Expressed protein | 0.39 | 0.004 | 0.65 | 0.036 | 0 | 2 | ||
| At5g03960 | 250872_at | Calmodulin-binding protein | 0.42 | 0.015 | 1.01 | 0.015 | 1 | 2 | ||
| At5g19970 | 246142_at | Expressed protein | 0.42 | 0.003 | 0.86 | 0.005 | 1 | 0 | ||
| At1g31050 | 265160_at | Expressed protein | 0.44 | 0.035 | 0.89 | 0.055 | 2 | 2 | ||
| At2g09970 | 260453_s_at | Expressed protein | 0.47 | 0.007 | 0.93 | 0.023 | 0 | 1 | ||
Fold change of gene expresion in auxin-treated 7-d-old iaa1:GR transgenic plants compared to that of mock-treated plants.
P-value calculated from auxin-down-regulated gene expression and mock expression using Welch's t-test.
Fold change of gene expresion in auxin and DEX-treated 7-d-old iaa1:GR transgenic plants compared to that of mock-treated plants.
P-value calculated from auxin-down-regulated gene expression in the presence of DEX and auxin-down-regulated gene expression using Welch's t-test.
Genes showing auxin-induced expression impaired in nph4-1 and/or arf19-1 and/or nph4-1 arf19-1 mutants by microarray analysis (Okushima et al., 2005).
Number of AuxRE, TGTCnC sequences in the 2 kb fragments upstream of the start codon.
Number of AuxRE, GnGACA sequences in the 2 kb fragments upstream of the start codon.
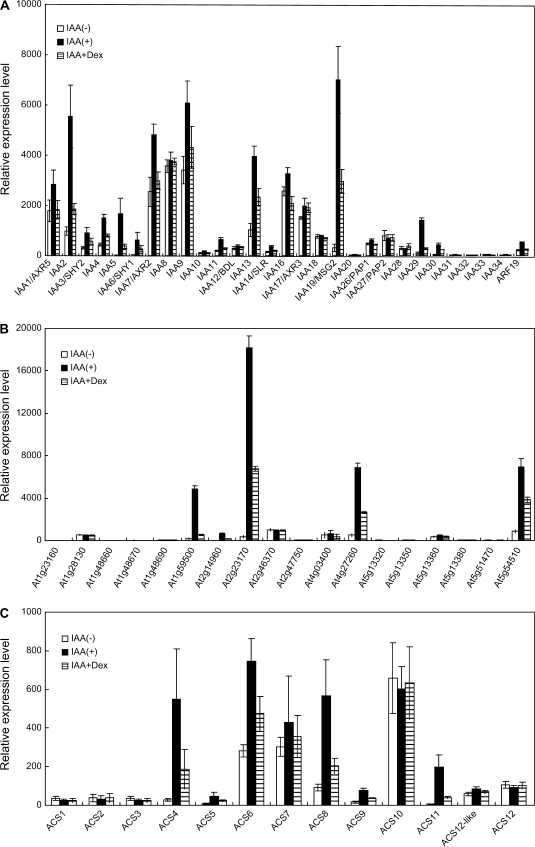
To gain an insight into the function of the auxin-regulated genes displaying both >2-fold differences, on average, between auxin- and mock-treated plants and P <0.05, these genes were classified into 12 functional groups using Gene Ontology (GO) annotation with some manual modifications (Tables 1, 2). It was found that 148 genes were up-regulated and 59 genes were down-regulated more than 2-fold following treatment with auxin and many of these auxin-regulated genes were negatively affected by DEX treatment. Almost all of the Aux/IAA genes, except for a few Aux/IAA members due to late auxin-induced gene expression, were induced by auxin treatment and this auxin-induction was reduced by DEX treatment in triplicate experiments (Table 1; Fig. 2A). These data were similar to the previously reported RNA-gel blot data (Park et al., 2002), confirming validity of our microarray analysis. In addition, a 2 kbp promoter region upstream of the start codon was analysed for the auxin response elements (AuxREs), TGTCnC or GnGACA sequence (Ulmasov et al., 1995, 1997a, b). It was found that most of the auxin-regulated genes affected by iaa1 have the AuxREs in the 2 kbp region (Tables 1, 2).
Fig. 2.
The expression profiles of representative auxin-up-regulated gene families and the effects of DEX on auxin-induced gene expression. (A) Aux/IAA gene family. The data represent the average of the relative expression levels of the samples treated with mock (open bar), treated with auxin (black bar), or treated with auxin in the presence of DEX (shaded bar) from triplicate experiments. Bars indicate SE of the average. (B) GH3 gene family. The legend is the same as (A). (C) ACS gene family. The legend is the same as (A).
Genes encoding a number of auxin-responsive proteins and GH3 are also up-regulated by auxin (Fig. 2B). Development-related genes including lateral organ boundaries domain16 (LBD16), LBD18, LBD29, and LBD33, and PIN1 are up-regulated. ACC synthase (ACS)4, ACS6, ACS8, ACS9, and ACS11 and ethylene response regulator (ERS)2 as well as cytokinin oxidase (CKX)7 are up-regulated (Table 1; Fig. 2C). Many genes involved in signal transduction and transcriptional control are also up-regulated by auxin. A variety of transcription factors are up-regulated, including bHLH proteins, AP2 domain-containing proteins, homeobox leucine-zipper proteins, zinc-finger proteins (ZFPs), MYB108, ethylene response factor (ERF)11, and ethylene-responsive element binding protein (EREBP). A large number of genes encoding metabolic enzymes and unknown proteins are also up-regulated. A number of genes encoding metabolic enzymes and unknown proteins are down-regulated by auxin (Table 2). In particular, the genes encoding various cytochrome P450 proteins are down-regulated. ROTUNDIFOLIA(ROT)3, which catalyses the conversion of teasterone to castasterone in the brassinosteroids (BRs) biosynthetic pathway, is down-regulated, whereas phyB activation-tagged suppressor1-dominant (BAS)1, which inactivates brassinolide by oxidation, is up-regulated by auxin. The type-A response regulator genes such as Arabidopsis response regulator (ARR)4, ARR5, ARR6, ARR7, and Arabidopsis histidine kinase (AHK)4 (the components of the cytokinin two-component signalling system) are co-ordinately down-regulated. Simultaneous treatment of auxin and DEX suppressed the auxin-inducible expression of most of these genes and increased expression of the auxin-down-regulated genes toward their initial levels. Taken together, these results suggest that the iaa1 protein is involved in regulating expression of many early auxin-responsive genes responsible for transcriptional control, signal transduction, and metabolism. It has also been found that auxin-regulated expression of some genes is not affected by the iaa1 protein, indicative of a portion of auxin-regulated genes independent of iaa1 regulation. These include genes encoding dehydrin, CKX1, decarboxylase, cysteine proteinase, homeobox-leucine zipper protein 17 (HB-17), and UDP-glucose:IAA β-D-glucosyltransferase in the case of auxin up-regulation, and pathogenesis-related protein, dehydrodolichyl diphosphate synthase, and ARR7 in the case of auxin down-regulation (Tables 1, 2).
RNA-gel blot analysis of the selected auxin-response genes affected by iaa1
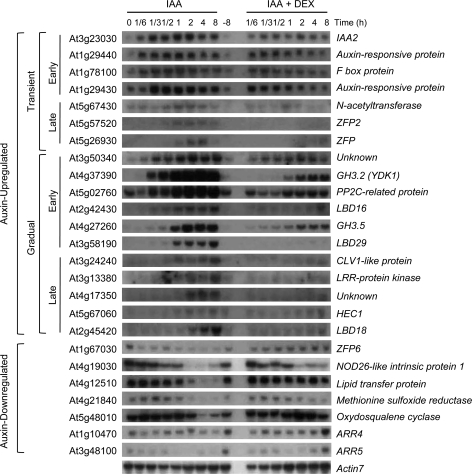
RNA-gel blot analysis was performed with selected auxin-up-regulated gene probes to confirm these microarray results. It should be noted that 5% of the genes listed that changed more than 2-fold in the microarray data are due to change variation and thus a relatively relaxed stringency has been used for the microarray data that set up the RNA-gel blot analyses. To represent the auxin-regulated transcriptome, an attempt was made to pick a range of gene probes that exhibit the highest and lowest auxin-regulated expression as well as genes in between. Also, genes were included with functions related to auxin response, transcription, and development to gain some insights into functional meaning of gene regulation in response to auxin. To study the expression characteristics of the iaa1-regulated auxin-responsive genes, the focus was on the genes showing auxin-regulated expression that were significantly affected by DEX treatment. For the analysis of auxin-up-regulated genes, 18 genes have been chosen in which 15 genes show P <0.05 and P calls in all three hybridizations after auxin treatment, as well as three genes (At3g24240, At5g57520, and At1g29440) that show P calls in all three hybridizations even with P >0.05. Expression of these selected genes was first examined over a time-course in response to auxin and the effects of DEX treatment. Pro35S:iaa1:GR transgenic Arabidopsis was incubated with auxin for varying amounts of time, without auxin for 8 h (–8), or with auxin and DEX, followed by RNA-gel blot analysis (Fig. 3). Quantitative analyses of the RNA-gel blots were performed with Phosphoimager (see Supplementary Figs S1 and S2 at JXB online). The results showed that auxin-induced expression of these genes can be divided into two groups, in which one group shows transient expression patterns and the other group shows a gradually increasing expression pattern without decay of expression until 8 h. These genes are also grouped into early or late genes based on the timing of gene expression in response to auxin. Three genes (At3g24240, At5g57520, and At1g29440) showing P calls in all three hybridizations but with P >0.05, and thus are not listed in the Tables 1 and 2, still exhibited significant auxin-inducible expression patterns. This result indicates the significance of P calls for extracting differentially regulated genes in response to auxin from our microarray data. Expression of all the genes examined in response to auxin was effectively repressed by DEX treatment. Similarly, expression of seven auxin-down-regulated genes was significantly down-regulated at 10 min (the earliest case) or within 2 h. DEX treatment suppressed the auxin-mediated down-regulation of those genes (Fig. 3; see Supplementary Fig. S2 at JXB online). These results show a negative regulation of iaa1 on various early auxin-responsive genes and in part, verify the data obtained by microarray analysis. However, the effect of DEX on auxin-mediated down-regulation of ARR5 was marginal except for the 8 h incubation.
Fig. 3.
Time-course expression analysis of representative auxin-up or down-regulated genes with auxin or auxin and DEX. Pro35S:iaa1:GR seedlings were incubated with auxin (IAA) or auxin and DEX (IAA+DEX) for the indicated time in the light and subject to RNA-gel blot analysis with the given DNA probes. –8 h indicates the samples incubated with mock for 8 h. Representative blots are shown from at least two independent biological replicates. Auxin-induced expression of these genes are divided into two groups, a transient expression group (Transient) and a gradually increasing expression group (Gradual). ‘Early’ or ‘Late’ indicates the genes expressing the transcripts detectable within (Early) or after (Late) 30 min in response to exogenous auxin treatment. The blots were displayed with timing of auxin-responsive gene expression.
Expression of iaa1-regulated auxin-response genes in the presence of cycloheximide
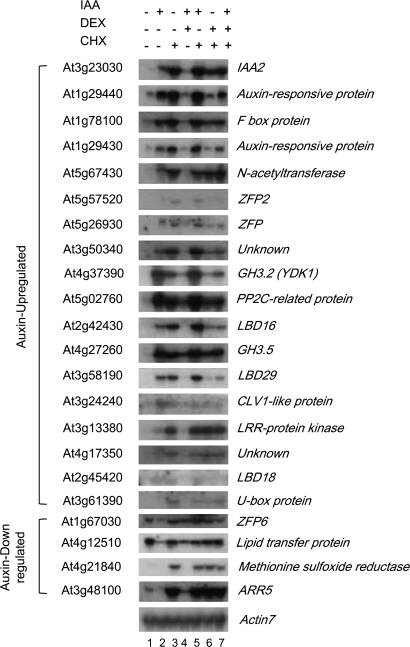
The protein synthesis inhibitor, cycloheximide, has been shown to induce expression of the primary auxin-responsive Aux/IAA genes and pea Aux/IAA genes, PS-IAA4/5 and PS-IAA6 (Abel et al., 1995; Koshiba et al., 1995). According to the current model for auxin action, cycloheximide seems to inhibit the continued supply of labile Aux/IAA repressors being degraded via the proteasome. This results in a release of transcriptional repression that, in turn, allows ARF proteins to modulate auxin-response genes (Quint and Gray, 2006). Translocation of the iaa1 proteins to the nucleus by DEX treatment can suppress auxin- or cycloheximide-inducible Aux/IAA expression (Park et al., 2002). These characteristics were used to gain further insight into how these early auxin-response genes are regulated by iaa1. Pro35S:iaa1:GR transgenic seedlings were incubated with mock treatment, auxin, cycloheximide, auxin and DEX, auxin and cycloheximide, cycloheximide and DEX, or auxin and cycloheximide and DEX for 2 h. The total RNAs were subject to RNA-gel blot analysis using DNA probes against the 18 indicated genes for auxin-up-regulation and the four indicated genes for auxin-down-regulation (Fig. 4). Quantitative analyses of the RNA-gel blots were performed with Phosphoimager (see Supplementary Fig. S3 at JXB online).
Fig. 4.
Expression analysis of representative auxin-response genes in response to cycloheximide, auxin, or DEX treatment. Pro35S:iaa1:GR seedlings were incubated with mock (lane 1), auxin (lane 2), cycloheximide (lane 3), auxin and DEX (lane 4), auxin and cycloheximide (lane 5), cycloheximide and DEX (lane 6), or auxin, DEX and cycloheximide (lane 7) for 2 h in the light, followed by RNA-gel blot analysis with the indicated DNA probes. The transcript levels were determined as described in Fig. 3 legend.
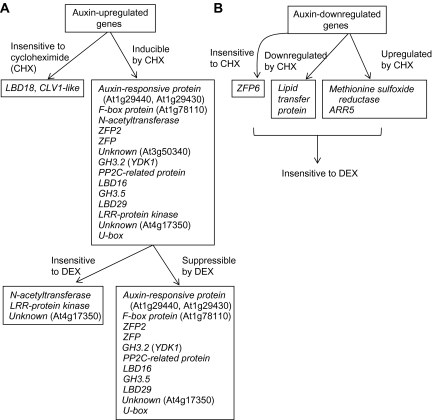
Treatment of cycloheximide to Pro35S:iaa1:GR transgenic seedlings induced most of the up-regulated genes to equal or higher levels than the expression levels of those genes when treated with auxin. As expression of LBD18 and CLV1-like protein was not clear due to the low levels of the transcripts, a longer incubation followed by RT-PCR analysis was performed (see Supplementary Fig. S4 at JXB online). The effect of cycloheximide on the expression of these genes was marginal, indicating that the expression of these genes is not under the direct control of labile repressors or it may be a secondary response as they are late genes (Fig. 3). Treatment of DEX effectively suppressed the expression of most of the iaa1-regulated genes in response to auxin or by cycloheximide. However, cycloheximide-induced expression of some genes encoding proteins such as N-acetyltransferase, LRR-protein kinase, and unknown (At4g17350) could not be suppressed by DEX treatment, even though auxin induction of these genes can be suppressed by DEX. A summary of this result is shown in Fig. 5A. This result indicates that an additional pathway or components might exist, thus effecting the auxin-regulated expression of these genes.
Fig. 5.
Schematic summary of expression patterns of representative auxin-response genes in the presence of cycloheximide and/or DEX. (A) Expression pattern of auxin-up-regulated genes. (B) Expression pattern of auxin-down-regulated genes. The results obtained by the expression analysis in Fig. 4 were schematically summarized.
DEX treatment inhibited auxin-mediated down-regulation of the four genes tested, as expected. Interestingly, genes encoding ZFP6, methionine sulphoxide reductase, and ARR5 were super-induced by cycloheximide treatment alone, indicative of the repression of these genes by labile repressors. Moreover, DEX treatment could not prevent cycloheximide-induced up-regulation of these auxin-down-regulated genes. DEX treatment did not have any effect on either auxin or cycloheximide-induced expression of all four auxin-down-regulated genes. A summary of this result is shown in Fig. 5B. This result suggests that non-Aux/IAA proteins functioning as labile repressors might be involved in the auxin-mediated down-regulation of these genes.
Analysis of iaa1-regulated auxin-response genes in a variety of arf mutants
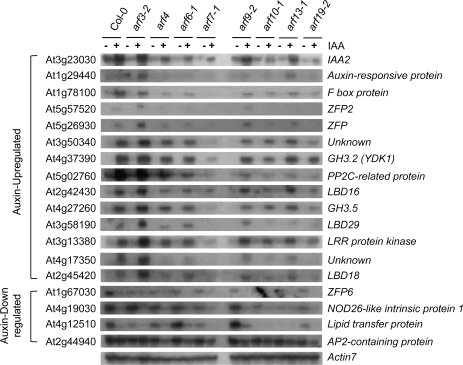
To determine if the auxin response genes identified in the present study are controlled by ARFs, their expression in response to auxin was analysed further in a variety of arf mutant backgrounds including ten single T-DNA insertion mutants and two double mutants previously isolated (Okushima et al., 2005b), and compared with wild-type plants using RNA-gel blot methods (Figs 6, 7). Quantitative analyses of the RNA-gel blots were performed with Phosphoimager (see Supplementary Figs S5 and S6 at JXB online). All single arf mutations affected auxin regulation of iaa1-regulated genes to varying degrees. A summary of these results are shown in Fig. 8. The arf3 mutation resulted in enhanced auxin-induced gene expression, whereas the rest of the arf mutations decreased auxin-induced gene expression (when they demonstrated any effect). Expression of auxin-induced genes is differently affected by arf mutations. For example, arf2-8, arf9-2, arf13-1, or arf14 weakly inhibited auxin-induced expression of most iaa1-regulated genes tested when compared with the other arf mutations. By contrast, arf4, arf6-1, arf10-1, or arf19-2 mutations significantly inhibited auxin-induced expression of a broad range of genes encoding IAA2, auxin-responsive protein, F-box protein, ZFP2, ZFP, unknown (At3g50340), PP2C-related protein, LBD16, LBD29, LRR-protein kinase, unknown (At4g17350), and LBD18, collectively. To gain an insight into the functional relevance of arf mutation effects on iaa1-downstream genes, organ-specific expression patterns of a select set of genes were analysed along with IAA1. Some genes such as LBD16 showed preferential expression patterns in particular organs, while others such as CLV1-like protein showed general expression throughout various tissues (see Supplementary Fig. S7 at JXB online). All the genes examined showed significant expression in the aerial parts as well as in the roots of 14-d-old plants.
Fig. 6.
Expression profiling of auxin-responsive genes regulated by iaa1 in various arf mutant backgrounds. arf mutant seedlings were incubated with mock (–) or auxin (+) for 2 h in the light, followed by RNA-gel blot analysis with the indicated DNA probes. The transcript levels were determined as described in the Fig. 3 legend.
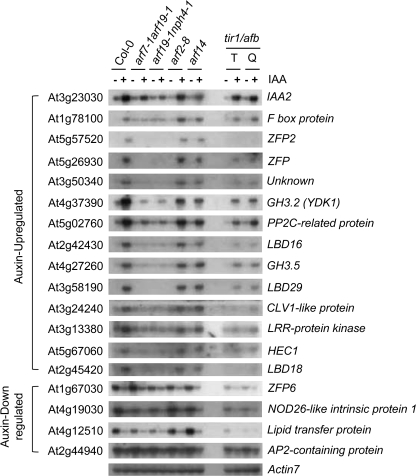
Fig. 7.
Expression profiling of auxin-responsive genes regulated by iaa1 in various arf and tir1/afb mutant backgrounds. arf mutant seedlings were incubated with mock (–) or auxin (+) for 2 h in the light, followed by RNA-gel blot analysis with the indicated DNA probes. The transcript levels were determined as described in the Fig. 3 legend. T and Q indicate tir1 afb2 afb3 triple mutants and tir1-1 afb1 afb2 afb3 quadruple mutants, respectively.
Fig. 8.
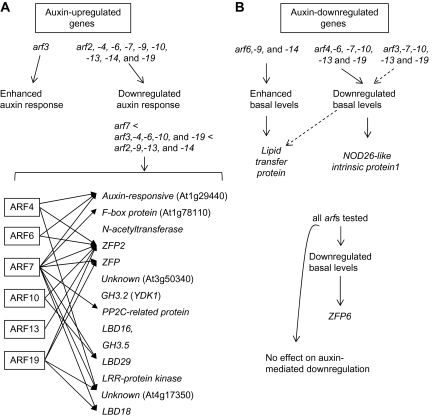
Schematic summary of expression patterns of iaa1-regulated auxin-responsive genes in various arf mutant backgrounds. (A) Expression pattern of auxin-up-regulated genes in arf mutants. (B) Expression pattern of auxin-down-regulated genes in arf mutants. The results obtained by the expression analysis in Figs 6 and 7 were schematically summarized.
The arf7-1 mutation most significantly inhibited the auxin-regulated expression of all 16 auxin-up-regulated genes and the three down-regulated genes tested. The arf7-1 mutation greatly or even completely inhibited the auxin-induced expression of F-box protein, unknown (At3g50340), GH3.2, GH3.5, LBD29, LRR-protein kinase, and LBD18 when compared with the other arf mutations, indicating that ARF7 preferentially regulates the expression of theses genes to auxin more so than the other ARFs do. Auxin-induced expression of the LBD16 and LRR-protein kinase are partially inhibited by the arf7-1 or arf19-2 mutations, but are completely inhibited by double mutations, indicating co-operative action between ARF7 and ARF19 for regulating expression of these two genes. On the other hand, expression of ZFP and ZFP2 to auxin is completely silent in arf7-1 or arf19-2 single mutants, suggesting independent roles for ARF7 and ARF19 in regulating expression of these genes. Auxin-responsive protein (At1g29440), F-box protein, ZFP2, ZFP, PP2C-related protein, LBD29, unknown (At4g17350), and LBD18 are also rendered completely unresponsive to auxin by the arf7-1 mutation. Both arf7-1 arf19-1 mutants and arf19-1 nph4-1 mutants exhibited greatly reduced auxin-induced expression of all auxin-regulated genes tested compared with their corresponding single mutants, except for one auxin-down-regulated gene (At2g44940). Expression of many genes is completely unresponsive to auxin in these two double mutations including unknown (At3g50340), LBD16, GH3.5, CLV1-like protein, LRR-protein kinase, and HECTATE(HEC)1. Expression of various genes is unresponsive to auxin by a single arf mutation as summarized in Fig. 8. Auxin-responsive expression of some genes such as auxin-responsive protein (At1g29440) and ZFP2 are completely suppressed to the basal levels by various arf mutations, indicating redundant role of these ARFs in auxin-regulated expression of these genes.
Interestingly, arf6-1, arf9-2, and arf14 mutations up-regulated the basal levels of the auxin-down-regulated gene encoding lipid transfer protein, while auxin-mediated down-regulation was normal. By contrast, arf4, arf6-1, arf7-1, arf10-1, arf13-1, and arf19-2 mutations resulted in greatly reduced expression of NOD26-like intrinsic protein1 without auxin. arf3-2, arf7-1, arf10-1, arf13-1, and arf19-2 mutations resulted in greatly reduced expression of lipid transfer protein without auxin. However, treatment with auxin decreased their expression levels as in wild-type plants. Similarly, auxin-mediated down-regulation is normal in arf7 arf19 double mutants, whereas basal levels of auxin-down-regulated genes are decreased in the arf7-1 mutant or arf7 arf19 double mutants. The basal levels of ZFP6 were greatly reduced by all arf mutations tested even including arf3. arf7 arf19 double mutations did not have additive effects compared with that of single mutations, suggesting that ARF7 and ARF19 might play an independent role in the regulation of ZFP6 expression level. Normal auxin-mediated down-regulation in arf mutants indicates that ARFs might not play a critical role in mediating gene down-regulation in response to auxin.
Expression of iaa1-regulated auxin-response genes in tir1/afb auxin receptor mutants
TIR1/AFB F-box proteins are auxin receptors mediating auxin responses throughout plant development (Dharmasiri et al., 2005a, b; Kepinski and Leyser, 2005). It was tested whether auxin response of the Aux/IAA-ARF-modulated genes, which had been identified in our microarray analysis, became insensitive by triple or quadruple mutations of these TIR1/AFB genes. It was found that most of these genes exhibited significant levels of auxin responsiveness except for ZFP2 and LBD18 (Fig. 7). Moreover, quadruple tir1-1 afb1 afb2 afb3 mutants had less effect on auxin-regulated gene expression than the arf double mutants, arf7-1 arf19-1 or arf19-1 nph4-1, and was comparable with that of arf7-1 single mutants.
Discussion
Microarray analysis of the effect of iaa1 on early auxin-regulated gene expression
Numerous studies have firmly established the functional importance of the Aux/IAA-ARF proteins during auxin response and their mechanism of action in auxin-mediated gene regulation (Berleth et al., 2004; Woodward and Bartel, 2005; Parry and Estelle, 2006). A stabilized iaa1 had previously been constructed by introducing a point mutation in domain II and expressing it under the CaMV 35S promoter in order to produce auxin-related phenotypes (Park et al., 2002). AXR5 later turned out to be IAA1, whose gain-of-function mutation caused some phenotypes to be comparable with DEX-treated Pro35S:iaa1:GR transgenic plants (Yang et al., 2004). In the present study, the transcriptome downstream of iaa1 was analysed during the auxin response on a genome-wide level by using GR-fused iaa1, which can be induced to translocate to the nucleus following DEX treatment. Putative target genes for iaa1 were determined by identifying the genes that were regulated by auxin and negatively affected by the DEX-inducible translocation of iaa1. The iaa1-regulated auxin-responsive transcriptome was further supported by the analysis showing the presence of AuxREs (TGTCnC or GnGACA) in the promoter regions of most of the genes affected by iaa1 (Tables 1, 2; see Supplementary Table S2 at JXB online). Several exceptions to this rule were noted (see Supplementary Table S2 at JXB online), indicating that other AuxRE variants might exist in the promoter region of auxin-regulated genes.
The constitutive CaMV 35S promoter was used to find any potential auxin-responsive targets regulated downstream of Aux/IAA-ARFs. The putative Aux/IAA-ARF targets were investigated to reveal biological roles in auxin signalling and response. Some of the molecular targets could be due to activation of the mutant iaa1 due to translocation into the nucleus by DEX treatment, as a DEX-triggered appearance of auxin-related phenotypes had been observed by Park et al. (2002). Others might be due to the expression of iaa1 in cells that normally do not express this protein. Expression of GUS of ProIAA1:GUS in a variety of tissues (Fig. 1) as well as constitutive expression of IAA1 in various organs (Abel et al., 1995) suggest that significant portions of the targets identified by microarray could be IAA1-targets, although further molecular characterization is necessary for confirmation of the individual target genes. In addition, cross-regulation and extensive genetic redundancy between Aux/IAA family members (Park et al., 2002; Overvoorde et al., 2005) intrinsically render the identification of the target genes of the individual Aux/IAA proteins complex, especially without any phenotypic changes to be shown in multiple loss-of-function aux/iaa mutants.
The genes regulated by auxin, which have been identified in the present microarray analysis, are generally consistent with previous reports (Tian et al., 2002; Pufky et al., 2003; Goda et al., 2004; Redman et al., 2004; Okushima et al., 2005b; Overvoorde et al., 2005). For example, auxin-regulated expression of a variety of Aux/IAA genes, GH3 genes, and ACS genes has been observed. However, our microarray analysis highlights different aspects of the auxin-responsive transcriptome, as DEX-inducible iaa1 was used to study the auxin response. This approach eliminates permanent effects of constitutive expression or gain-of-function mutations on plant growth and development throughout the whole plant life cycle and allows us to focus on the effect of iaa1 during auxin action. Thus our data have some similarities as well as differences when compared with microarray analysis from gain-of-function aux/iaa mutants. In axr3-1/iaa17-1 seedlings, 108 and 78 genes have been identified as repressed or induced, respectively (Overvoorde et al., 2005). For example, the expression of ARR3, ARR5, ARR6, and ARR7 was suppressed by this mutation. However, the expression profiles of the genes that encode enzymes involved in cytokinin biosynthesis and catabolism were not significantly different from those of the wild type, indicating that a new equilibrium of gene expression, seemingly unrelated to auxin response, is established in the axr3-1/iaa17-1 mutants (Overvoorde et al., 2005). There are significant differences in auxin-regulated gene expression profiles between our present results and these previous reports, and this is possibly because of differences in growth conditions and treatment systems. Similarities as well as significant differences in the negative effects of the axr3-1/iaa17-1 mutation and iaa1 overexpression have also been observed in auxin-regulated gene expression profiles. The genes with reduced auxin-induction shared between axr3-1/iaa17-1 mutants and Pro35S:iaa1:GR are IAA5, IAA6, IAA11, GH3.1, GH3.5, ARF19, LBD16, LBD18, LBD29, EXP4, ACS4, ACS6, ACS8, ERS2, pectin esterase (At2g47550), protein kinase (At1g77280), ERF11, and GNAT (At5g67430). The genes with reduced repression shared are SAUR (At4g31320), TIP2;3 (At5g47450), PIP2;4 (At5g60660), AGP13 (At4g26320), and AGP22 (At5g53250). A large number (94 out of 148 genes) of DEX-suppressible auxin-induced genes has been detected in which their expression is also impaired in nph4-1, arf19-1, and/or nph4-1 arf19-1 mutants (Okushima et al., 2005b) (Table 1). Such shared expression patterns might contribute to the common phenotypes observed among these mutant plants (Leyser et al., 1996; Park et al., 2002; Okushima et al., 2005b). Likewise, unshared expression patterns could explain part of their distinct phenotypes or varying degree of phenotypes.
Recent results provide evidence for the involvement of some genes identified by our microarray analysis, particularly transcription factors, in auxin response and plant development. HEC1 (At5g67060) and other HEC genes (HEC2 and HEC3) encoding basic helix-loop-helix (bHLH) proteins were shown to be involved in auxin-mediated control of gynoecium patterning (Gremski et al., 2007). HEC1 was also found to be up-regulated by auxin (Overvoorde et al., 2005). The AP2 domain-containing transcription factor (At5g18560) named PUCHI has been shown to be required for morphogenesis in the early lateral root primordium of Arabidopsis (Hirota et al., 2007). Overexpression of HAT2 (At5g47370), encoding a homeobox-leucine zipper protein, caused the opposite effects on the shoot and root tissues in regulating auxin-mediated morphogenesis (Sawa et al., 2002). Two (At1g18400 and At1g73830) of the three brassinosteroid early response genes, BR ENHANCED EXPRESSION1, -2, and -3 (BEE1, BEE2, and BEE3), encoding bHLH proteins, are up-regulated by auxin in our microarray analysis. These genes were found to be functionally redundant positive regulators of BR signalling and also were shown to be up-regulated by other hormones including auxin (Friedrichsen et al., 2002), indicating auxin-mediated positive regulation of BR signalling. PACLOBUTRAZOLE RESISTANCE2 (PRE2) (At5g15160), homologous to PRE1 that has been proposed to play a regulatory role in gibberellin-dependent plant development (Lee et al., 2006), is up-regulated, indicating crosstalk between auxin and gibberellin signalling. Auxin-inducible MYB77 has been shown to modulate auxin signal transduction via an interaction between MYB77 and ARFs (Shin et al., 2007). MYB108 was identified in the present study and might be a possible MYB transcription factor involved in auxin signalling. Genes related to lateral organ development, LBD16, LBD18, LBD29, and LBD33, are up-regulated. Previously, LBD16, LBD17, LBD18 or LBD29 have been reported to be up-regulated by auxin (Redman et al., 2004; Okushima et al., 2005b; Overvoorde et al., 2005). A recent study showed that lateral root formation of Arabidopsis is regulated by ARF7 and ARF19 via direct activation of LBD16 and LBD29 (Okushima et al., 2007). The expression of all these genes in response to auxin was suppressed by iaa1, suggesting that these putative transcription factors are likely to be modulated downstream of the Aux/IAA-ARF proteins during plant development. Up-regulation of PIN1, encoding an auxin efflux carrier, by auxin (Table 1) has also been reported (Heisler et al., 2005). The rest of the genes that encode transcription factors and signal transduction components need to be investigated further for their potential functional roles in the auxin response.
Four ACS genes encoding the key enzymes in ethylene biosynthesis are up-regulated by auxin, consistent with a recent report (Stepanova et al., 2007). Up-regulation of ACS4, ACS6, and ACS8 by auxin has previously been reported (Tian et al., 2002; Goda et al., 2004). We and another group (Okushima et al., 2005b) found that ERS2, encoding a second subfamily of the ethylene receptors, is also up-regulated. Up-regulation of these ACS genes and ERS2 might contribute to an auxin-mediated response to ethylene effects. Up-regulation of CKX1 and CKX7, responsible for the degradation of cytokinins, and down-regulation of various cytokinin-inducible A-type response regulator genes (ARR4, ARR5, ARR6, and ARR7) and a cytokinin receptor gene (AHK4) have been observed. In addition to CKX1 and CKX7, CKX6 is up-regulated by auxin (Overvoorde et al., 2005). Down-regulation of CKX4 has also been reported (Goda et al., 2004). Thus, auxin plays a complex role in the regulation of cytokinin signal transduction and cytokinin action. It was found that ROT3, involved in the biosynthesis of BRs, is down-regulated, whereas BAS1 that inactivates BRs is up-regulated by auxin. This is consistent with previous observations (Goda et al., 2004) and indicative of an antagonistic regulation of auxin in BR action. The expression of all these hormone-related genes in response to auxin was suppressed by iaa1. These results provide additional evidence for crosstalk between auxin and other plant hormones, ethylene, cytokinin, and brassinosteroids, by auxin-mediated transcriptional response via the Aux/IAA-ARF system.
It is clear that these microarray data will be valuable resources for investigations towards understanding specific auxin responses or a subset of auxin responses as well as hormonal crosstalk. Loss-of-function studies on single or multiple mutants of those identified target genes will also be important, as many auxin responses are constitutively affected by mutations in Aux/IAAs or ARFs (Okushima et al., 2005b; Overvoorde et al., 2005), or can be affected in an inducible way (Park et al., 2002).
Verification of microarray data by RNA-gel blot analysis
RNA-gel blot analysis was used to confirm our microarray data and to analyse the expression characteristics under various conditions so as to further our understanding of auxin-regulated gene expression. Transient expression patterns have been observed from a group of these genes in response to auxin (Fig. 3), indicating a negative feedback loop on their expression which is reminiscent of the response of A-type ARR genes to cytokinin (D'Agostino et al., 2000). Expression of all 18 auxin-up-regulated genes tested from the iaa1-regulated transcriptome was suppressed by DEX treatment. Similarly, auxin-mediated down-regulation was also suppressed by DEX. These results suggest that iaa1 might have promiscuous interactions with a variety of ARFs, resulting in inhibiting ARF functions. Auxin-induced expression of LBD16, LBD18, and LBD29 are effectively suppressed by DEX. LBD16 and LBD29 have been shown to be involved in lateral root formation (Okushima et al., 2007). DEX treatment greatly reduced lateral root formation in Pro35S:iaa1:GR plants (Park et al., 2002). Thus simultaneous suppression of those three LBD genes to auxin might contribute to a reduction in lateral root numbers. DEX treatment effectively suppressed auxin-inducible expression of two GH3 genes, typical marker genes for early auxin response, demonstrating the validity of the microarray data.
Expression analysis of iaa1-regulated genes in the presence of cycloheximide indicates the complexity and diversity of auxin-regulated gene expression
Cycloheximide-induced expression of many of the auxin-up-regulated genes examined were suppressed by DEX, consistent with the current model in which labile Aux/IAA repressors mediate transcriptional repression for the ARF proteins that regulate auxin-response genes. Interestingly, cycloheximide-induced expression of some of the iaa1-regulated genes, N-acetyltransferase, LRR-protein kinase, and unknown (At4g17350) was not suppressible by DEX (Figs 4, 5). It has been reported that optimized pairs of interacting Aux/IAA-ARF proteins generate developmental specificity (Knox et al., 2003; Weijers et al., 2005: Muto et al., 2007). It is thus possible that there might be specificity in the protein–protein interactions between Aux/IAA and ARF, and iaa1 might not be able to interact with ARFs that are involved in this gene regulation. This result might also indicate the existence of an additional pathway for auxin-regulated gene expression in addition to the Aux/IAA-ARF pathway. One possibility is that some auxin-responsive genes downstream of Aux/IAA-ARF proteins might require a labile co-repressor for their regulation. If this is true, then iaa1 alone may not suppress cycloheximide-induced gene expression because of the lack of this hypothetical corepressor. The recent identification of the TPL corepressor function in Aux/IAA-ARF-mediated gene regulation during the auxin response (Szemenyei et al., 2008) might support this hypothesis.
ARR5, methionine sulphoxide reductase, and ZFP6, which are auxin-down-regulated, were super-induced by cycloheximide treatment alone, indicative of the repression of these genes by labile repressors. However, super-induction of these genes by cycloheximide could not be repressed by DEX, suggesting involvement of non-Aux/IAA proteins as the labile repressors in auxin-regulated down-regulation of this gene expression. These results show the complexity and diversity of auxin-regulated gene expression that cannot be explained by the Aux/IAA-ARF system alone.
Expression analysis of the iaa1-regulated genes in arf mutants suggests iaa1-regulated transcriptome as ARF targets and versatile ARF functions in auxin-regulated gene expression
The representative iaa1-target genes were auxin-responsive and auxin induction of these genes was repressible by DEX-triggered nuclear translocation of iaa1, suggesting that these genes are regulated by Aux/IAA-ARFs. This proposal is further supported by the analysis showing the presence of AuxREs in the promoter regions of theses iaa1-target genes where ARFs directly bind (Table 1; see Supplementary Table S2 at JXB online). To confirm that these genes are regulated by ARFs in the auxin response, the effect of auxin on gene regulation at the early time of 2 h was examined in various arf mutants. Expression analysis of the iaa1-regulated genes in a large set of arf mutants showed that expression of all the genes tested in response to auxin was affected by any of the arf mutations (with varying impacts depending on the genes and arf mutations), indicating that each of these ARFs has a role in modulating the expression of auxin-regulated genes in plants. Mutation in ARF3 caused a super-induction of gene expression to the auxin treatment compared with that of wild-type Col-0, but the rest of the arf mutations, arf2, -4, -6, -7, -9, -10, -13, -14, and -19, reduced auxin-induced gene expression, suggesting that ARF3 might function as a negative component and the other ARFs tested might function as positive components in regulating auxin-responsive gene expression. Mutations in ARFs greatly overexpressed or reduced the basal levels of lipid transfer protein and NOD26-like intrinsic protein1, respectively, which are both auxin-down-regulated. Expression analysis of these genes in arf mutants indicates that ARF9 and ARF14 function as negative components, while ARF3, -4, -7, -10, -13, and -19 as positive components. Moreover, ARF6 might act as a negative component for lipid transfer protein and as a positive component for NOD26-like intrinsic protein1 in maintaining their basal expression levels. Therefore, ARFs could act either as a transcriptional activator or as a repressor depending on the nature of expression of the auxin-responsive genes. These results suggest that although several steps of positive and/or negative regulation may be involved at any point, ARFs could have versatile roles in auxin-regulated gene expression. This analysis was based on the differences in steady-state RNA levels being monitored. Thus further molecular studies are needed to draw conclusions about the roles of ARFs for the regulation of these genes in response to auxin.
ARF1 to ARF9 mRNA are each present in roots, rosette leaves, cauline leaves and flowers (Ulmasov et al., 1999b). In ProARF1:GUS and ProARF2:GUS lines, GUS staining appeared throughout 8-d-old seedlings and in rosette leaves, as well as in the sepals and carpels of young flower buds (Ellis et al., 2005). GUS expression of ProARF2:GUS was also detected in the vascular tissues and the initiation sites of lateral roots (Okushima et al., 2005a). ETTIN (ARF3) is expressed in floral organs (Sessions et al., 1997). Low levels of MP (ARF5) were detected in all major organs in RNA-gel blot analyses (Hardtke and Berleth, 1998). GUS expression of both ProARF6:GUS and ProARF8:GUS was detected in flowers at multiple stages (Nagpal et al., 2005). ProARF7:GUS was expressed in petioles and blades of cotyledons and leaves, and in roots (Wilmoth et al., 2005). The root expression was strongest in the vasculature and in more mature parts of the root, and also appeared in root meristems. ProARF19:GUS was expressed in vasculature of cotyledons and leaves, stems and roots. Both GUS fusion genes were also expressed in flower organs. Another study has shown that the expression patterns of these two GUS fusion genes are distinct, with partial overlap (Okushima et al., 2005b). Strong GUS expression was observed in the hypocotyls and petioles of ProARF7:GUS seedlings, whereas ProARF19:GUS expression was restricted to the vascular tissue in the aerial parts. In root tissue, ProARF19:GUS is strongly expressed throughout the meristematic region, root cap, root hair, and the sites of newly forming lateral toots (Okushima et al., 2005b; Li et al., 2006). ProARF7:GUS expression in the primary root is restricted to the vascular tissues. The GUS expression of various ARF-GUS fusions detected in the early stages of seedlings, as well as throughout various tissues, and overlapping expression of iaa1-regulated genes (see Supplementary Fig. S1 at JXB online) and ARFs further strengthen the argument that various ARFs might be involved in regulating representative iaa1-target genes.
Mutations in the ARF genes did not affect auxin-mediated down-regulation per se. However, those mutations had significant effects on the basal levels of the down-regulated genes, indicating that these ARFs are involved in maintaining basal levels as positive components or negative components depending on the ARFs and/or target genes, but not in the process of down-regulation itself.
Mutations in ARF2, -9, -13, and -14 had weak effects on auxin-induced gene expression, whereas mutations in ARF3, -4, -6, -7, -10, and -19 had strong effects, suggesting that some ARFs play a major role in auxin-induced gene expression, but others may play an auxiliary or redundant role at the young seedling stage. In particular, arf7 mutation greatly reduced auxin-inducible expression of a majority of genes examined (Fig. 6). Auxin-responsive protein (At1g29440), F-box protein, ZFP2, ZFP, PP2C-related protein, LBD29, unknown (At4g17350), and LBD18 were completely unresponsive to auxin by arf7 mutation, suggesting that these genes might be the primary targets for ARF7. Double mutations in ARF7 and ARF19 dramatically reduced auxin-induced expression of all 14 genes examined. The obvious growth phenotype of arf3 (Sessions et al., 1997), arf7 (Harper et al., 2000), or arf7 arf19 (Okushima et al., 2005b) mutants correlates with the degree of inhibition of auxin-induced gene expression. Our results showing impaired auxin-induced gene expression in arf7, arf19, and arf7 arf19 mutants generally agree with the global gene expression profiling of these mutants. In the case of arf19, RNA-gel blot analysis of a selected set of genes in the present study showed more significant inhibition of auxin-induced gene expression compared with global expression profiling. Noticeable difference in RNA-gel blot data might be due to the sensitive auxin-induced expression of the chosen genes or might be due to different growth conditions and tissue treatment methods. Auxin-induction of 11 out of the 14 auxin-induced genes used for RNA-gel blot analysis in the present study was found to be impaired in nph4-1, arf19-1, and/or nph4-1 arf19-1 mutants based on microarray data (Okushima et al., 2005b). Expression analysis of auxin-induced genes in arf7 arf19 mutants using RNA-gel blot analysis in the present study thus confirms the previous microarray data. Three genes (At5g57520, At1g78100, and At4g37390) are absent from the list of genes with reduced auxin induction, and this might be due to the fold-change cut-off used to make the gene list in their microarray analysis.
No obvious growth-phenotype for arf4, -6, -10, or -19 single mutants had previously been reported (Okushima et al., 2005b), although these mutations significantly impaired auxin-induced gene expression in the present study. arf6 had delayed stamen development and decreased fecundity (Nagpal et al., 2005). arf19 showed an altered phototropic response and a mild but significant resistance to exogenous auxin in the roots (Okushima et al., 2005b). Thus these ARFs may function in specific aspects of the auxin response.
Potential mechanisms for an ARF to act as a positive or a negative component in auxin-regulated gene expression
Our expression analysis in the arf mutant backgrounds suggests that ARFs have a positive or a negative function (or both) in auxin-regulated gene expression depending on the target gene. Previous studies with transient gene expression assays in protoplasts showed that, depending on the structure of the middle region, each ARF functions as a transcriptional activator or a repressor (Ulmasov et al., 1999a; Tiwari et al., 2003). While the molecular function of ARFs could be revealed in that protoplast reporter gene system, it is still poorly understood how the ARFs regulate their target gene expression in plants. For example, the arf3 mutation caused enhanced auxin-responsive expression of auxin-induced genes, while expression of auxin-down-regulated genes, ZFP6 and lipid transfer protein, was reduced by this mutation (Fig. 5), suggesting dual functions for ARF3 in its target gene regulation. Another example is the observation that the arf6 mutation up-regulated lipid transfer protein, but down-regulated NOD26-like intrinsic protein1 in their basal expression levels. It is not without precedent that transcription factors have dual functions as activators or repressors. A variety of studies on transcription factors of mammalian cells such as hormone receptors, Pit-1, and Oct-1 transcription factors have demonstrated that, although some factors are pure activators or repressors, many others can both activate and repress transcription in a manner that is dependent on the particular situation (Latchman, 2001). For example, Pit-1 can act as either an activator or a repressor of cell-type specific genes depending on the nature of its binding sites. This distinction depends on a two-base-pair hormone promoter site. The allosteric effect on Pit-1, in combination with other DNA binding factors, results in the recruitment of a corepressor N-CoR for active repression of the growth hormone gene in the lactotrope, while it activates growth hormone gene expression in the other cell type, the somatotrope (Scully et al., 2000). It is therefore possible that interaction between ARFs and coactivators or corepressors depending on the sequence variation on the promoters may determine whether the same ARFs activates or represses transcription of a given target gene.
arf7 arf19 mutants exhibit additionally reduced auxin-induction relative to tir1/afb quadruple mutants
The arf7 arf19 double mutants exhibited stronger auxin-related phenotypes than those of arf7 and arf19 single mutants (Okushima et al., 2005b). Consistently, global expression profiling of the arf7 arf19 double mutants displayed severely impaired auxin-induced gene expression. Likewise, it was found that all of the iaa1-regulated genes tested via RNA-gel blot analyses had severely inhibited auxin-induced gene expression. tir1 afb2 afb3 triple and tir1-1 afb1 afb2 afb3 quadruple mutants of auxin receptor genes had reduced auxin-induced gene expression, but significantly less than that of the arf7 arf19 double mutants. The quadruple mutant showed an extremely strong phenotype in plant growth and development from embryos to adults (Dharmasiri et al., 2005b). Even so, the degree of impairment to auxin-induced expression of Aux/IAA-ARF-regulated genes was much milder than that of the arf7 arf19 double mutants, and was comparable to that of arf7 or arf19 single mutants. This may be because two other additional TIR1-like proteins, AFB4 and AFB5, might play significant roles during auxin signalling mediated by the Aux/IAA-ARF proteins. This could also be due to some of the receptor mutants utilized not being complete nulls (Dharmasiri et al., 2005b). The other explanation is that the lack of TIR1/AFBs or ARF7 and ARF19 might result in very distinct patterns of gene expression being established and the effect of auxin might then be superimposed onto these patterns.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Quantification of time-course expression of representative auxin-up-regulated genes selected from microarray analysis with auxin or with auxin and DEX.
Supplementary Fig. S2. Quantification of time-course expression of representative auxin-down-regulated genes selected from microarray analysis with auxin or with auxin and DEX.
Supplementary Fig. S3. Quantification of expression analysis of representative auxin-response genes selected from microarray analysis in response to cycloheximide, auxin, and DEX treatment.
Supplementary Fig. S4. Expression analysis of LBD18 and CLV1-like protein in response to cycloheximide, auxin, or DEX treatment.
Supplementary Fig. S5. Quantification of expression profiling of auxin-responsive putative target genes regulated by iaa1 in various arf mutant backgrounds.
Supplementary Fig. S6. Quantification of expression profiling of auxin-responsive putative target genes regulated by iaa1 in various arf and tir1/afb mutant backgrounds.
Supplementary Fig. S7. Organ-specific expression profiles of auxin-responsive genes regulated by iaa1.
Supplementary Table S1. Oligonucleotides and PCR conditions used for RT-PCR and genotyping and DNA probes for RNA-gel blot analysis.
Supplementary Table S2. List of genes examined for AuxREs beyond 2 kbp promoter region or whole promoter region.
Acknowledgments
We are grateful to Dr Mark Estelle for triple and quadruple tir1/afb mutants and to ABRC for arf T-DNA insertion mutants. We thank Jin-Young Park for her initial help in microarray analysis. This work was supported by grants from the Korea Science and Engineering Foundation (KOSEF) (R01-2005-000-10943-0), from the Plant Diversity Research Center of 21st Century Frontier Research Program (PF06302-01), from the Agricultural Plant Stress Research Center (R11-2001-092-04001-0) funded by the Korean Government (MOST), from the World Class University project of the Ministry of Science and Technology of Korea (R31-2009-000-20025-0) to J Kim. This work was also supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (KRF-2006-353-C00046).
References
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. Journal of Molecular Biology. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Academie Science (Paris) 1993;316:1194–1199. [Google Scholar]
- Berleth T, Krogan NT, Scarpella E. Auxin signals-turning genes on and turning cells around. Current Opinion in Plant Biology. 2004;7:553–563. doi: 10.1016/j.pbi.2004.07.016. [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Seruère J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiology. 2000;124:1706–1717. doi: 10.1104/pp.124.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ. A1. The plant hormones: their nature, occurrence, and functions. In: Davies PJ, editor. Plant hormones biosynthesis, signal transduction, and action! Dordrecht. The Netherlands: Kluwer Academic Publishers; 2004. pp. 5–6. [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005a;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Developmental Cell. 2005b;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132:4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Current Biology. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiology. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM. AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. The Plant Cell. 2006;18:1873–1886. doi: 10.1105/tpc.105.037192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development. 2007;134:3593–3601. doi: 10.1242/dev.011510. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO Journal. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. The Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Current Biology. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Hirota A, Kato T, Fukaki H, Aida M, Tasaka M. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. The Plant Cell. 2007;19:2156–2168. doi: 10.1105/tpc.107.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Wilson KJ. The GUS gene fusion system. Plant Molecular Biology Manual. 1991;B14:1–33. [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. Protein–protein interactions among the Aux/IAA proteins. Proceedings of the National Academy of Sciences, USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O. AXR3 and SHY2 interact to regulate root hair development. Development. 2003;130:5769–5777. doi: 10.1242/dev.00659. [DOI] [PubMed] [Google Scholar]
- Koshiba T, Ballas N, Wong LM, Theologis A. Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum) Journal of Molecular Biology. 1995;253:396–413. doi: 10.1006/jmbi.1995.0562. [DOI] [PubMed] [Google Scholar]
- Latchman DS. Transcription factors: bound to activate or repress. Trends in Biochemical Sciences. 2001;26:211–213. doi: 10.1016/s0968-0004(01)01812-6. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee S, Yang KY, Kim YM, Park SY, Kim SY, Soh MS. Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant and Cell Physiology. 2006;47:591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dhamasiri S, Estelle M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. The Plant Journal. 1996;10:403–413. doi: 10.1046/j.1365-313x.1996.10030403.x. [DOI] [PubMed] [Google Scholar]
- Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR. Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Developmental Cell. 2004;7:193–204. doi: 10.1016/j.devcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Li J, Dai X, Zhao Y. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiology. 2006;140:899–908. doi: 10.1104/pp.105.070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. The Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT. Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiology. 2007;144:187–196. doi: 10.1104/pp.107.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Mitina I, Quach HL, Theologis A. AUXIN RESPONSE FACTOR2 (ARF2): a pleitropic developmental regulator. The Plant Journal. 2005a;43:29–46. doi: 10.1111/j.1365-313X.2005.02426.x. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. The Plant Cell. 2005b;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. The Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. The Plant Cell. 2005;17:3282–3300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Kim HJ, Kim J. Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. The Plant Journal. 2002;32:669–683. doi: 10.1046/j.1365-313x.2002.01459.x. [DOI] [PubMed] [Google Scholar]
- Parry G, Estelle M. Auxin receptors: a new role for F-box proteins. Current Opinion in Cell Biology. 2006;18:152–156. doi: 10.1016/j.ceb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Pufky J, Qiu Y, Rao MV, Hurban P, Jones AM. The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Functional Integrative Genomics. 2003;3:135–143. doi: 10.1007/s10142-003-0093-7. [DOI] [PubMed] [Google Scholar]
- Quint M, Gray WM. Auxin signaling. Current Opinion in Plant Biology. 2006;9:448–453. doi: 10.1016/j.pbi.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. The Plant Cell. 2001;13:2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD. Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. The Plant Journal. 2004;38:545–561. doi: 10.1111/j.1365-313X.2004.02061.x. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. The Plant Journal. 2002;32:1011–1022. doi: 10.1046/j.1365-313x.2002.01488.x. [DOI] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carrière C, Rose DW, Hooshmand F, Aggarwal AK, Rosenfeld MG. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- Sessions RA, Zambryski PC. Arabidopsis gynoecium structure in the wild and in ettin mutants. Development. 1995;121:1519–1532. doi: 10.1242/dev.121.5.1519. [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. The Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM. Multilevel interactions between ethylene and auxin in Arabidopsis roots. The Plant Cell. 2007;19:2169–2185. doi: 10.1105/tpc.107.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tian CE, Muto H, Higuchi K, Matamura T, Tatematsu K, Koshiba T, Yamamoto KT. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. The Plant Journal. 2004;40:333–343. doi: 10.1111/j.1365-313X.2004.02220.x. [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. The Plant Cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. The roles of auxin response factor domains in auxin-responsive transcription. The Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. The Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. The Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. The Plant Cell. 1995;10:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997a;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences, USA. 1999a;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. The Plant Journal. 1999b;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell. 1997b;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO Journal. 2005;24:1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch BL. The generalization of ‘student's’ problem when several different population variances are involved. Biometrica. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. The Plant Journal. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Annals of Botany. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. Degradation of Aux/IAA proteins is essential for normal auxin signalling. The Plant Journal. 2000;21:553–562. doi: 10.1046/j.1365-313x.2000.00703.x. [DOI] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M. The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. The Plant Journal. 2004;40:772–782. doi: 10.1111/j.1365-313X.2004.02254.x. [DOI] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J. Auxin modulates the degradation rate of Aux/IAA proteins. Proceedings of the National Academy of Sciences, USA. 2001;98:11795–11800. doi: 10.1073/pnas.211312798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.