Abstract
Budbreak in kiwifruit (Actinidia deliciosa) can be poor in locations that have warm winters with insufficient winter chilling. Kiwifruit vines are often treated with the dormancy-breaking chemical hydrogen cyanamide (HC) to increase and synchronize budbreak. This treatment also offers a tool to understand the processes involved in budbreak. A genomics approach is presented here to increase our understanding of budbreak in kiwifruit. Most genes identified following HC application appear to be associated with responses to stress, but a number of genes appear to be associated with the reactivation of growth. Three patterns of gene expression were identified: Profile 1, an HC-induced transient activation; Profile 2, an HC-induced transient activation followed by a growth-related activation; and Profile 3, HC- and growth-repressed. One group of genes that was rapidly up-regulated in response to HC was the glutathione S-transferase (GST) class of genes, which have been associated with stress and signalling. Previous budbreak studies, in three other species, also report up-regulated GST expression. Phylogenetic analysis of these GSTs showed that they clustered into two sub-clades, suggesting a strong correlation between their expression and budbreak across species.
Keywords: Actinidia deliciosa, budbreak, bud dormancy, hydrogen cyanamide, glutathione S-transferase, kiwifruit, microarray
Introduction
Bud dormancy in woody perennials is a complex process that enables plants to survive long periods of adverse conditions, including the extremes of drought, cold, and heat (Faust et al., 1997; Arora et al., 2003). In late summer, declining photoperiods and temperatures cause shoot extension growth to cease and the initiation of apical buds to protect the apical meristem (Li et al., 2003; Heide and Prestrud, 2005). A specific signal (environmental or endogenous) perceived within the bud, induces and maintains these buds in a state of endodormancy (Thomas and Vince-Prue, 1997; Rhode et al., 2002; Espinosa-Ruiz et al., 2004; Böhlenius et al., 2006). In temperate perennial species a period of low temperatures (commonly referred to as winter-chilling), is needed to release buds from endodormancy.
Warm winters in many regions often limit the productivity of temperate fruit crops, including grape, apple, and kiwifruit (Henzell et al., 1991; Erez, 1995; Bound and Jones, 2004). A number of studies have looked at dormancy and dormancy release in grape and apple (Wang et al., 1991; Erez, 1995; Or et al., 2000c, 2002), but work on kiwifruit is limited. In kiwifruit (Actinidia deliciosa), it has been shown that buds enter dormancy in response to shortening daylength (Lionakis and Schwabe, 1984). During winter, kiwifruit buds are likely to be endodormant. Brundell (1976) showed that canes collected in early winter and placed under permissive conditions displayed delayed budbreak relative to canes that had received supplemental chilling or those collected later in the season that had received additional ‘natural’ chilling. This suggests that the ‘dormant-state’ of buds collected in early winter was ‘deeper’ than those collected in late-winter and that the winter-chilling requirement had not been satisfied in the earliest collected material.
During the transition from winter into spring, endormancy is often followed by a period of ecodormancy (Faust et al., 1997), which continues until temperatures rise sufficiently for the resumption of growth. This appears to be the case in kiwifruit as Walton et al. (1991) noted that sap flow in the canes commenced 8 weeks before budbreak and McPherson et al. (1997) reported that bud respiration increased approximately 3–6 weeks before budbreak.
In warmer regions, an application of hydrogen cyanamide (HC) in late winter/early spring is often used to break dormancy in kiwifruit vines and to ensure commercially viable yields are achieved (Linsley-Noakes, 1989; Henzell et al., 1991; Erez, 1995). However, reliance on chemical dormancy-breakers, like HC, is costly and results can be unpredictable, ranging from a limited response to toxicity (Erez, 1995; Richardson et al., 1994). In addition to breaking dormancy in kiwifruit, HC also increases the number of flowers per shoot, reduces the numbers of second-order (side) flowers, and synchronizes flowering (Linsley-Noakes, 1989; Henzell et al., 1991; Walton and Fowke, 1993). The plasticity of the flowering response is possible because flower differentiation in kiwifruit does not commence until the buds begin to swell in spring (Brundell, 1975b). As a result, during winter, the axils of kiwifruit buds contain second-order buds or meristems, of which some of the latter have the potential to differentiate flowers (Walton and Fowke, 1993; Walton et al., 1997, 2001).
An understanding of how the release from dormancy is regulated is essential in order successfully to manipulate flowering of temperate crops like kiwifruit. While many reviews have been published on the physiological aspects of bud dormancy (Saure, 1985; Fuchigami and Nee, 1987; Lang et al., 1987; Faust et al., 1997; Arora et al. 2003), a more detailed understanding of how these processes might be regulated is limited (Horvath et al., 2003). Several recent studies have focused on dormancy release in grapes (Or et al., 2000c, 2002; Pang et al., 2007; Halaly et al., 2008), and have used HC as a tool to modify the breaking of dormancy; however, its mode of action is still unclear. At the metabolic level, there is a significant increase in proline accumulation, which coincides with shoot outgrowth (Walton et al., 1991). There have been suggestions that the putative signalling molecule hydrogen peroxide causes budbreak because of a HC-induced down-regulation of a catalase (Shulman et al., 1986; Perez and Lira, 2005). This is the first step in a cascade that up-regulates several signalling proteins, including transcription factors, protein phosphatases, and protein kinases (Neill et al., 2002). It should be noted that the decreased levels of catalase activity, associated with the increased oxidative stress, were observed only in HC-treated grapevines, suggesting that an alternative mechanism may be involved for the induction of ‘natural’ budbreak. Alternatively, HC may act through a SNF-like protein kinase (GDBRPK) and perceive a stress signal induced by HC application (Or et al., 2000c). More recently, Horvath et al. (2003) suggested that SNF-like protein kinases may function in a more general epigenetic response, with a putative role in the changes in DNA methylation, similar to that observed in the induction and release of dormancy in buds on potato tubers (Law and Suttle, 2003). These reports point to a number of potential modes of HC action in budbreak, but there is still much to be understood. In the work presented here, an assessment of global gene expression was made to identify early transcriptional events following the application of HC, to gain an insight into what triggers budbreak in kiwifruit.
Materials and methods
Plant material and sample collection
Experiments were carried out on kiwifruit [Actinidia deliciosa (A. Chev.) C.F. Liang et A.R. Ferguson ‘Hayward’] vines growing in commercial orchards in Hamilton, New Zealand in 2003, and Kerikeri, New Zealand in 2004 and 2005. Vines were managed using standard orchard practices (Sale and Lyford, 1990). Further information on site, vine management, and sample collection is presented in Table 1.
Table 1.
Summary of information on kiwifruit collection sites in New Zealand, hydrogen cyanamide (HC) application, sample collection and winter temperatures
| 2003 | 2004 | 2005 | |
| Location | Hamilton | Kerikeri | Kerikeri |
| Training systema | T-bar | Pergola | Pergola |
| Date of HC application | 13 August | 25 August | 23 August |
| Rate of HC applicationb | 6%, 600 l ha−1 | 5%, 700 l ha−1 | 5%, 600 l ha−1 |
| Tissue collected | Meristems | Meristems | Buds |
| Number collected/sample | 150 | 50 | 40 |
| Mean winter temperature (°C)c | 9.9 | 11.8 | 12.2 |
| Mean temperature during sampling (°C)d | 8.7 | 7.5 | 11.9 (12.9)e |
Rate as 6% Hi-Cane®, NuFarm, New Zealand, active ingredient hydrogen cyanamide 520 g l−1.
Mean daily temperature from 1 May–31 August.
Mean daily temperature for the period +1 HC through until +6 HC.
Mean daily temperature for the period +1 HC through until +42 HC.
HC was applied in late-winter (Table 1), well before any growth and development would have been observable (Brundell, 1975a). For the microarray analyses in 2003 and 2004, one-year-old canes were collected from a population of 200 vines (either HC-treated and non-treated) at each site on 1, 3, and 6 d after HC application. Upward facing axillary (first-order) buds were selected from canes (Walton et al., 1997), excluding the most distal, to provide a population of buds with similar budbreak characteristics. ‘Meristems’ were removed by making three cuts round the periphery of a bud, and with a fourth cut, flicked off the top of the bud, removing the budscales and most leaf primordia, and parenchyma that surrounds the bud. What remained was the primary bud axis, including the apical meristem, the youngest leaf primordia and any second-order buds and meristems, which together were cut from the cane and snap-frozen in liquid nitrogen.
To validate and extend the expression profiles of genes identified from the microarray experiments, an additional series of samples were collected during in 2005 from vines growing in Kerikeri (Table 1). Samples were collected 1 d before HC application and then 1, 2, 3, 6, 15, 21, 28, and 42 d after application from both treated and non-treated vines. In this series, each sample consisted of entire axillary buds (upward-facing), rather than meristems, as the extended sampling schedule made it impossible to collect meristem samples over the entire time-course.
Determination of budbreak
Budbreak was calculated in terms of the whole vine, as a percentage of those that had broken versus the total number of buds that might have broken (small or malformed buds were ignored). An individual bud was defined as broken once 10 mm of extension growth was observed, with at least some green tissue visible (defined as advanced budbreak by Brundell, 1975b). The expression of the kiwifruit homologue to a Populus cyclin-dependent kinase (AdCDKB1) was also used as a marker of the breaking of dormancy and the resumption of meristematic activity (i.e. cell-division).
Global gene expression analysis
Total RNA was extracted from the kiwifruit meristems and buds following the method of Chang et al. (1993). Non-redundant (NR) contiguous sequences were identified from an Actinidia EST database (Crowhurst et al., 2008) and 45–55mer oligonucleotides, with a constant Tms, were designed for each NR. These oligonucleotides were combined to create a microarray representing 17 472 genes (Crowhurst et al., 2008). Microarray construction, and all labelling and hybridizations followed the methods described by Schaffer et al. (2007). The experimental design for the microarray analysis was direct comparisons of samples collected from HC-treated vines on each day (days 1, 3, and 6) against samples collected from untreated vines on the same day. For the comparisons of the 2003 samples, there were two technical replicates (dye-swaps); for 2004, there were two biological replicates, each with two technical replicates.
The data from each comparison were normalized using global loess normalization, without background correction. Each experiment was then analysed separately using the Linear Models for Microarray Analysis (Limma) package in Bioconductor (www.bioconductor.org), incorporating between gene information. Gene lists were obtained for each comparison. Differential expression was determined using a multiple hypothesis-testing false discovery rate threshold of 0.05 and lists were filtered to remove genes that had less than a 2-fold change in expression. Gene lists from the two years were compared to identify commonality.
Database analysis
Multiple database searches were performed to collect all Arabidopsis thaliana members of the families to which these genes belonged. This was achieved using BLAST programs (TBLASTN and BLASTP) available on the TAIR, MAtD, and TIGR Arabidopsis databases and NCBI Arabidopsis genome database. The nucleotide or translated protein sequences, corresponding to the Actinidia ESTs, were used as the query sequences, Full-length protein sequences were then obtained from The Arabidopsis Information Resource (TAIR) website using AGI ID (www.arabidopsis.org/tools/bulk/sequences/index.jsp).
To identify family members from other plant species, BLAST programs (BLASTP and TBLASTN) against SwissProt and GenBank, respectively, were used. The nearest Arabidopsis protein sequence corresponding to the Actinidia EST was used as the query sequence.
qPCR analysis
Gene specific primers were designed using Primer3 (Rozen et al., 2000) so that the resultant amplicon, preferably, spanned an intron or spanned the stop codon to include a portion of the 3′UTR (see Supplementary Table S1 at JXB online). Quantitative PCR (qPCR) reactions were performed using a rapid-cycle PCR LightCycler (Roche). The total reaction volume of 10 μl, and contained 1× of FastStart SYBR Green Master Mix (Roche), 500 μmol each of the forward and reverse primers, 1 μl of 5-fold diluted cDNA. Each reaction was replicated three times and a negative water control was included in each run. Amplification was carried out with an initial denaturing step at 95 °C for 5 min, then 40 cycles of 95 °C for 5 s, 60 °C for 5 s, and 72 °C for 8 s. The fluorescence signal was measured after each extension step. For each gene, a standard curve was generated using serially diluted cDNA, the qPCR reaction efficiency determined, which was then used during data analysis. A melting curve was assessed to distinguish the expected product from non-specific products. For each primer pair, the expectant size of the PCR products was confirmed by agarose gel electrophoresis. Data were analysed on relative quantification monocolour Lightcycler software 4.0.
Identification of putative Actinidia homologues to known HC-responsive genes
Genes that have been implicated to be HC responsive in other species were used as query sequences (TBLASTN) to identify kiwifruit homologues in the EST database. These candidates were all from grape, namely: catalase (Or et al., 2002; accession number AF236127), SNF-like protein kinase (GDBRPK, Or et al., 2000c; accession number AF178575), two alcohol dehydrogenases (Or et al., 2000a, c; accession numbers AF195866 and AF195867), and pyruvate decarboxylase (Or et al., 2000b, c; accession number AF195868).
Construction of the phylogenetic trees
A multiple alignment analysis was performed with Clustal W (using an opening penalty of 15 and an extension penalty of 0.3) using the AlignX software in Vector NTI 9.0. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 3.1 (Kumar et al., 2004), using minimum evolution phylogeny test and 1000 bootstrap replicates.
Results
Budbreak measurements
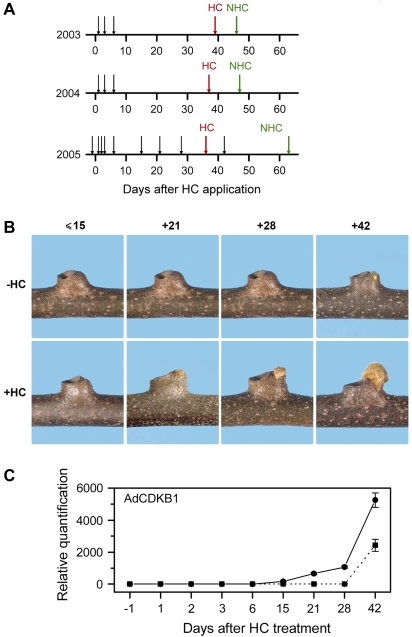
Kiwifruit bud tissue was collected over three years (2003–2005) from two orchards in different locations (Table 1). The timing of budbreak after the application of the bud-breaking chemical HC was consistent between seasons, with 50% budbreak occurring between 36 d and 39 d after application (Fig. 1A), and is similar to that reported by McPherson et al. (2001). However, there was more variation in the timing of budbreak in non-treated vines, with 50% budbreak occurring between 7 d and 27 d later than that on HC-treated vines (Fig. 1A). This spread was most likely due to differences in temperatures between the three seasons (Table 1). During 2005, visible swelling of buds (budswell as defined by Brundell, 1975b) was first observed 21 d after HC application on the treated vines (Fig. 1B), but at the molecular level, using the kiwifruit cell cycle gene AdCDKB1 to determine the start of cell division, transcript accumulation was first detected 15 d after HC treatment (Fig. 1C). In the non-treated plants, no increase of AdCDKB1 was detected at day 28, but by day 42, by which time the buds on these plants had started to swell, AdCDKB1 transcript accumulation could be detected.
Fig. 1.
(A) Summary schematic diagram of microarray (2003 and 2004) and qPCR (2005), experimental designs relative to hydrogen cyanamide (HC) treatment. Each black arrow indicates a day on which a sample was collected; the red and green arrows indicate the dates when HC-treated and non-treated vines achieved 50% budbreak, respectively. (B) Photographic series showing the relative development of buds collected in 2005 from HC-treated and non-treated vines used for qPCR analysis. (C) Quantitative PCR analysis in 2005 of AdCDKB1 expression (a marker for the re-initiation of meristematic activity and growth) for buds from HC-treated (solid line) and non-treated (dashed line) vines.
HC-induced gene expression
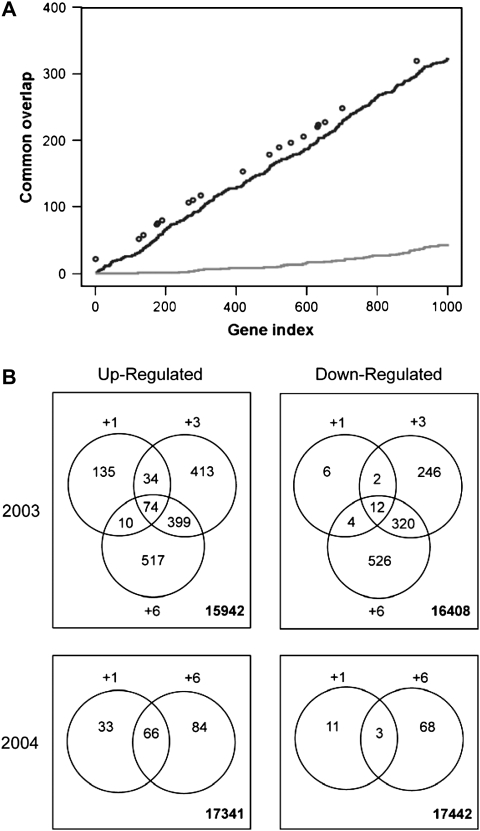
The focus of this study was on early transcriptional events following HC treatment. In both 2003 and 2004, significant changes in transcript accumulation were observed 1 d after HC application (Table 2). Using a FDR threshold of 0.05 and a 2-fold change in expression as significant, in 2003, 1.6% (277) of the genes represented on the array had a significant change in expression 1 d following HC treatment (see Supplementary Table S2 at JXB online). In 2004, that number was 0.66% (113) (see Supplementary Table S3 at JXB online). These numbers increased with time after HC application, and at 6 d after HC application these values were 10.6% and 1.3% of genes, for 2003 (see Supplementary Table S5 at JXB online) and 2004 (see Supplementary Table S6 at JXB online), respectively. (The genes that showed a significant change in expression 3 d after HC application in 2003 are presented in see Supplementary Table S4 at JXB online.) More genes were up-regulated than down-regulated after HC application. Statistically significant global changes in gene expression over the two years yielded not only different lists of genes, but also lists of different lengths. However, comparison of the lists of the top 1000 genes (with the greatest changes in expression, from day 6 in 2003 and 2004), indicated that there was a high degree of commonality between the two lists (Fig. 2A), suggesting that similar molecular events would have occurred in each season.
Table 2.
Numbers of significant gene changes on arrays in response to hydrogen cyanamide (HC) applications to kiwifruit in 2003 and 2004 (in brackets; the numbers of up- and down-regulated genes, respectively)
| Day | 2003 | 2004 | Common |
| 1 | 277 (253, 24) | 113 (99, 14) | 7 (6, 0)a |
| 3 | 1500 (920, 580) | – | – |
| 6 | 1862 (1000, 862) | 221 (150, 71) | 170 (123, 35) |
For lists of genes, see Supplementary Tables S2–S6.
The numbers of genes, up- and down-regulated in both years, respectively.
Fig. 2.
(A) Comparison of the top 1000 (based on adjusted P-values) from day 6 in 2003 and day 6 in 2004 (solid line) comparison of 1000 randomly selected genes (dotted line). The genes selected for further analysis by qPCR are presented by circles above the solid line. (B) Schematic of the 2003 and 2004 microarray experiments, showing the numbers of significantly expressed up-regulated and down-regulated genes on each day and the numbers of genes that were common between individual comparisons.
In 2003, the total number of genes changing consisted of 1582 genes up-regulated and 1116 down-regulated (Fig. 2B; see Supplementary Tables S2, S4, S5 at JXB online). Of the genes up-regulated, approximately 53% of genes identified on day 1 were only identified on that day. The equivalent numbers for days 3 and 6 were 45% and 52%, respectively (Fig. 3B). The same trends were observed with the down-regulated genes. Only a small proportion of all genes identified were up- or down-regulated on all three days (4.7% in 2003 and 6.6% in 2004; Fig. 2B). When comparing between the two years, six genes were up-regulated in both years, within 1 d of HC treatment, and 123 genes within 6 d of HC treatment (Table 2). For down-regulated genes, none were in common in both years at day 1, but 35 were in common at day 6 (Table 2).
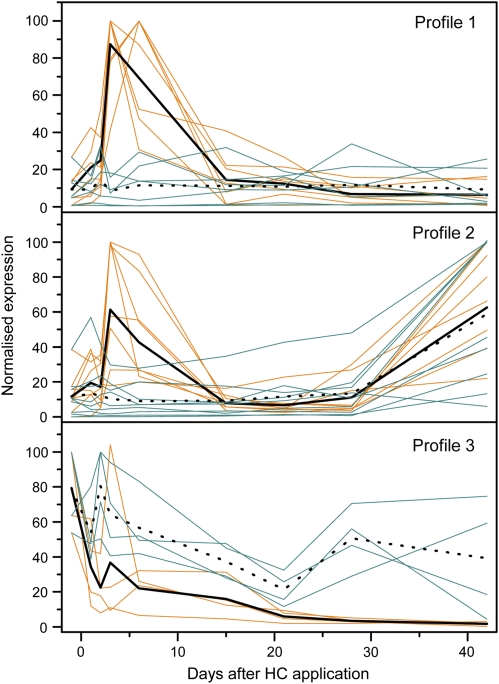
Fig. 3.
Grouping of normalized qPCR expression profiles of those genes selected from the microarray analysis (highest measured expression=100%). Profile 1: genes that were up-regulated in response to hydrogen cyanamide (HC) application, Profile 2: genes that were up-regulated in response to HC application and growth, and Profile 3: genes that were down-regulated in response to HC application and growth. The gold lines are with HC treatment, the blue lines are non-treated, the solid black lines are the mean values for HC treatment and the dotted black lines are the mean values for the non-treated. Expression is relative to that of Actinidia actin (AdActin).
Gene ontologies for each gene on the microarray were identified based on those for Arabidopsis (see Supplementary Table S7 at JXB online). Genes that had a greater than a log2-fold change in expression in the microarray experiments (with an adjusted P value of <0.05) are highlighted.
Expression patterns over budbreak
To investigate further the patterns of gene expression during budbreak in kiwifruit, additional tissue was harvested in 2005, from the day before HC treatment (day –1) until d 42, for both treated and non-treated vines (nine time points in total; Fig. 1B). Fifty-two genes were selected for qPCR verification, based on a combination of their differential expression in the 2003 or 2004 microarrays, the magnitude of change in their expression, and/or their likely function, based on their homology to genes from other species. In addition, five genes homologous to those found to respond to HC treatment in grape (Or et al., 2002) were analysed.
Thirty-five of the 52 genes selected for qPCR analysis (67%), were amplified and showed a single band during qPCR. Of those genes, 29 of the 30 (97%) that were up-regulated on the microarray were also up-regulated in the qPCR analysis. Of the genes selected for qPCR that were down-regulated genes, three of the five (60%) gave similar results. Overall, 32 (91%) of the genes analysed by qPCR gave quantitatively similar results to those obtained through the microarray experiments.
When the extended patterns of expression for all the 32 genes were compared, four distinct profiles were clearly seen. Six genes showed the first profile (Fig. 3, Profile 1; see Supplementary Fig. S1 at JXB online); these genes showed a single early peak in transcript accumulation in buds collected from HC-treated vines, and this occurred before any meristematic activity could be detected (as measured by the AdCDKB1 gene) or external bud growth was observed. Depending on the gene, maximum accumulation occurred either 3 d or 6 d after HC application and returned to baseline levels usually by 15 d after HC application. By contrast, there was no significant change in transcript accumulation in buds collected from the non-treated vines. From these observations, this pattern of transcript accumulation appears to be specific for HC and could be best described as HC-induced transient activation.
The second profile, which included 14 genes (Fig. 3, Profile 2; see Supplementary Fig. S2 at JXB online), showed an early peak in transcript accumulation in response to HC application (as observed with Profile 1 genes), but was followed by a second peak in transcript accumulation which commenced towards the end of the sampling period. In buds collected from non-treated vines, there was only a single peak in transcript accumulation, which appeared to be analogous to the second peak in the buds collected from the HC-treated vines. This profile could be described as an HC-induced transient activation followed by growth-related activation.
Five genes showed the third profile (Fig. 3, Profile 3; see Supplementary Fig. S3 at JXB online), where there was a general reduction in transcript over the sampling period, with the transcript levels in the buds from HC-treated vines declining more rapidly than those from non-treated vines. Again, initial changes in transcript levels were detected before any meristematic activity was detected or external bud growth was observed. The genes that gave this pattern of expression could be described as HC- and growth-repressed. The fourth group (seven genes) included all the remaining genes with expression profiles that did not fit into either Profile 1, 2, or 3 (see Supplementary Fig. S4 at JXB online).
Putative functions for the genes that could be characterised into Profiles 1, 2, or 3 are described in Table 3, based on functions of similar genes from the databases. In all cases the closest Arabidopsis gene was identified (TAIR; www.arabidopsis.org) and the expression patterns of these genes were examined in e-FP browser (bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). From these resources, it was clear that many of the genes had been identified in stress-related studies.
Table 3.
Alphabetical list, by profile, of kiwifruit genes analysed by qPCR using the extended set of samples collected in 2005
| Profile | Top blast hita | Comment |
| 1 | ABC transporter (FG473412; At3g47780.1, e-71) | Likely to be part of the ATH subfamily of the ABC superfamily (Sánchez-Fernández et al., 2001). Members of this subfamily are seemingly disparate but the Arabidopsis gene homologue rapidly accumulates in roots subjected to salt stress. ABC proteins also feature in loss of dormancy arrays in raspberry (Mazzitelli et al., 2007). |
| FAD-binding domain protein (FG470652; At5g44440.1, e-159) | Aligns with both NEC5, a BBE-like protein, and VuCRPD2, a drought-inducible gene. NEC5 exhibits glucose oxidase activity, and is capable of catalysing the oxidation of D-Glu to D-gluconic acid and H2O2 (Carter and Thornburg, 2004). | |
| Galactinol synthase (FG471302; At1g60470.1, e-155) | First and key enzyme in the synthesis of raffinose, the first of a series of soluble galactosyl-sucrose carbohydrates in the raffinose family of oligosaccharides (RFOs), which accumulate in plants when subjected to environmental stresses, including heat, cold, and dehydration (Taji et al., 2002), which are thought to help protect plants from environmental stresses. RFOs also accumulate during seed development and are thought to play a role in desiccation tolerance in seeds (Brenac et al., 1997). Galactinol synthase transcripts were also shown to accumulate in a budbreak and shoot outgrowth microarray experiment in sessile oak (Derory et al., 2006). | |
| In2-1 protein (FG460267; At5g02790.1, 9e-93) | Similar to glutathione S-transferases, the Arabidopsis homologue is up-regulated in response to biotic and abiotic stresses, and during development in leaves and sepals. | |
| SAM:carboxyl methyltransferase (FG446808; At1g68040.1, 5e-20) | Catalyses the conversion of salicylic acid (SA) to methylsalicylate (MSA). MSA benzenoid esters have been proposed to play a role in the SA-mediated plant defence responses (Creelman and Mullet, 1997; Seskar et al., 1998; Shulaev et al., 1997). | |
| 2 | 5′-Adenylylsulphate reductase (FG471388; At4g04610.1, 6e-36) | Key regulatory enzyme in sulphate metabolism in plants. Sulphated metabolites play roles in biotic and abiotic stress tolerance (Kopriva, 2006). Closest Arabidopsis homologue shows high induction under salt stress. Transcripts of this gene were also shown to change during budbreak and shoot outgrowth in oak (Derory et al., 2006). |
| Cinnamyl-alcohol dehydrogenase (FG471467; At4g37980.2, 6e-39) | Catalyses the last step in monolignol synthesis, which is polymerized to form lignin (Kim et al., 2004). Kiwifruit gene most similar to AtCAD7, which is highly expressed in vascular tissues (Kim et al., 2007) and has been shown to be a rapidly inducible defence gene (Kiedrowski et al., 1992). | |
| Curculin-like (mannose-binding) lectin (FG478690; At1g78850.1, 2e-97) | Greatest up-regulation of transcript levels in response to HC. Closest Arabidopsis homologue is up-regulated on arrays to various chemical, biotic, and osmotic stresses. In addition, Horvath et al. (2005) have shown that curculin expression changes during leafy spurge root bud outgrowth. | |
| ERF/AP2 transcription factor (FG479502; At1g53170.1, 6e-33) | Member of subgroup ERFVIIIa/CMVIII-1 (Nakano et al., 2006) which contains a repressor domain (Fujimoto et al., 2000; Ohta et al., 2001). Closest homologue is a negative regulator in the expression of ethylene-, jasmonate-, and ABA-responsive genes (McGrath et al., 2005; Yang et al., 2005). Hypothesized that its expression functions as a negative regulator and modulates the levels of hormonally controlled gene expression, ABA responsive genes which contribute to the initiation of dormancy in Arabidopsis seeds (Garciarrubio et al., 1997), and buds of adult birch (Rinne et al., 1994). | |
| Expressed protein (FG475773; At1g35210.1, 8e-83) | Closest Arabidopsis homologue showed high expression under salt stress. | |
| Expressed protein (FG512494; At2g46150.1, 2e-20) | The closest Arabidopsis homologue has the greatest expression in the salt-stressed root microarray experiments. | |
| Glutathione S-transferase (FG423204; At2g29420.1, 2e-55) | Catalyses the conjugation of the tripeptide gluthathione to a variety of hydrophobic, electrophylic and usually cytotoxic substances and were first identified in plants because of their ability to detoxify herbicides (Marrs, 1996). The most similar Arabidopsis gene is a member of the Tau GST gene family (AtGSTU7), members of which are induced following exposure to biotic and abiotic stresses (Dixon et al., 2002). GSTs were identified in both the raspberry (Mazzitelli et al., 2007) and oak (Derory et al., 2006) arrays. | |
| Glutathione S-transferase (FG523871; At1g78380.1, 5e-92) | As above; catalyse the conjugation of gluthathione to a variety of hydrophobic, electrophylic and usually cytotoxic substances (Marrs, 1996). The most similar Arabidopsis gene is also a member of the Tau GST gene family (AtGSTU17). GSTs were identified in both the raspberry (Mazzitelli et al., 2007) and oak (Derory et al., 2006) arrays. | |
| Magnesium/proton exchanger (FG498083; At2g47600.1, 6e-66) | Tonoplast transporter exchanging protons with Mg2+ and Zn2+ ions and therefore balance the levels of these ions between cytosol and vacuole (Shaul et al., 1999). This is important as excesses or deficiencies in the cytosol can seriously impair cellular function. Highly expressed in vascular tissues (Shaul et al., 1999). In Arabidopsis arrays, the highest expression was osmotic and salt stresses, dry seed and pollen. | |
| Mitogen-activated protein kinase (FG477785; At3g45640.1, 4e-33) | Link external sensors to cellular responses and are known to regulate cell growth and differentiation, the cell cycle, and responses to stress (Jonak et al., 2002). The closest Arabidopsis homologue (AtMPK3) to the kiwifruit gene has been linked with osmotic and oxidative stresses, bacterial elicitor signalling, and ABA signalling (Nakagami et al., 2005). | |
| Myb transcription factor (FG470796; At3g06490.1, 2e-81) | Closest Arabidopsis homologue encodes the BOTRYTIS SUSCEPTIBLE1 (BOS1) (Mengiste et al., 2003), which is up-regulated by applications of ABA, ethylene, jasmonate, and salt stress (Nakagami et al., 2006). | |
| NF-X1-like zinc finger protein (FG510928; At5g05660.1, 7e-09) | In Arabidopsis, these genes appear to have a role in salt and drought stress responses (Lisso et al., 2006). In Arabidopsis arrays, the highest expression was in dry seeds and early during the process of imbibition. | |
| Secretory protein (FG479735; At2g15220.1, 4e-74) | Closest Arabidopsis homologue up-regulated in arrays in response to with osmotic, UVB and genotoxic stresses. | |
| UDP-glycosyltransferase (FG526418; At4g15550.1, 3e-59) | Catalyses the transfer of a glycosyl moiety to an acceptor molecule (Li et al., 2001). In Arabidopsis arrays, the highest expression is the seed, 6 h after imbibition. | |
| 3 | ABC transporter (FG471021; At2g36380.1, e-126) | Nearest Arabidopsis is in the PDR subfamily (Sánchez-Fernández et al., 2001), which are associated with the transport of antifungal agents (van den Brûle and Smart, 2002). |
| CBS domain-containing protein (FG474406l; At2g14520.1, 5e-50) | The precise function of cystathionine-β-synthase (CDS) domains remains to be elucidated. Recent work indicates that CBS domains bind adenosyl domains (Kemp, 2004) and act as sensors of cellular energy status (Scott et al., 2004). The Arabidopsis gene is highest expressed in developing and dry seed. | |
| Expressed protein (FG459066; At2g21180.1, 8e-30) | The closest Arabidopsis homologue accumulates during seed development and declines after imbibition, with germination. | |
| F-box protein (FG402777; At1g68050.1, 7e-94) | Homologous to the FLAVIN BINDING, KELCH REPEAT, F-BOX 1 (FKF1) protein (Imaizumi et al., 2005), which regulates the flowering time gene CONSTANS (CO) (Putterill et al., 1995). A more rapid decline in the kiwifruit FKF1-homologue would result in an increase in CO, which would lead to an increase in flowering. | |
| LEA domain-containing protein (FG474947; At5g06760.1, 1e-24) | Expressed to high levels in the later stages of embryo development and have been associated with desiccation tolerance (Wise, 2003; Tunnacliffe and Wise, 2007), and cold stress (Tunnacliffe and Wise, 2007). LEA proteins came up in both the raspberry bud dormancy release (Mazzitelli et al., 2007) and the oak budburst (Derory et al., 2006) arrays. |
In brackets; Genbank number of kiwifruit sequence, best Arabidopsis protein hit, and expectation value.
When the six genes that showed a significant up-regulation of expression 1 d after HC treatment in both years were analysed by qPCR, it was found that there was two Profile 1 genes and four Profile 2 genes. These were an In2-1 protein (Profile 1), a putative embryo-abundant protein (Profile 1), a glutathione S-transferase (Profile 2), a 5′-adenylylsulphate reductase (Profile 2), a cinnamyl-alcohol dehydrogenase (Profile 2), a UDP-glycosyltransferase (Profile 2), and a magnesium/proton exchanger (Profile 2).
Previous grape expression studies have observed a catalase (VvCat-1) (Or et al., 2002), a SNF-like protein (Or et al., 2000c), an alcohol dehydrogenase (Or et al., 2000a), and a pyruvate decarboxylase as significantly changing in buds that have been treated with HC. More recently, Halaly et al. (2008) observed, in addition, an ascorbate peroxidase, a glutathione S-transferase, a stilbene synthase, a sucrose synthase, and a thioredoxin h that all increased following HC treatment. To establish whether similar mechanisms were occurring in grape and kiwifruit, these genes were investigated further, by array expression analysis and qPCR.
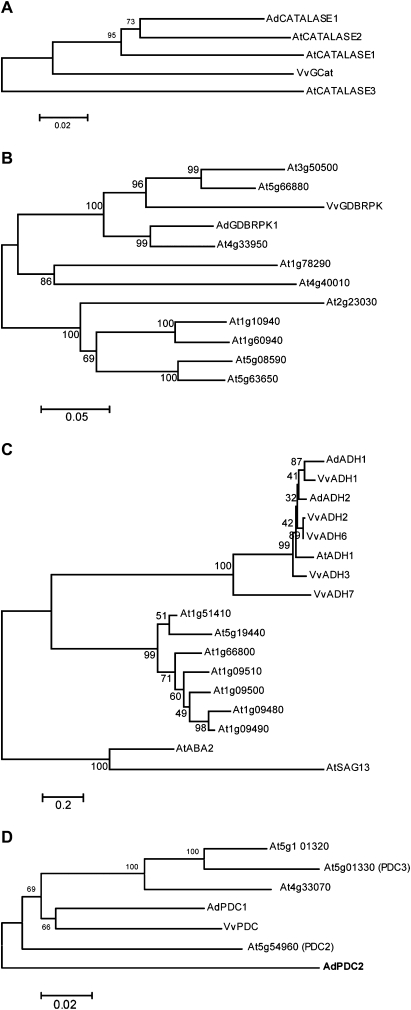
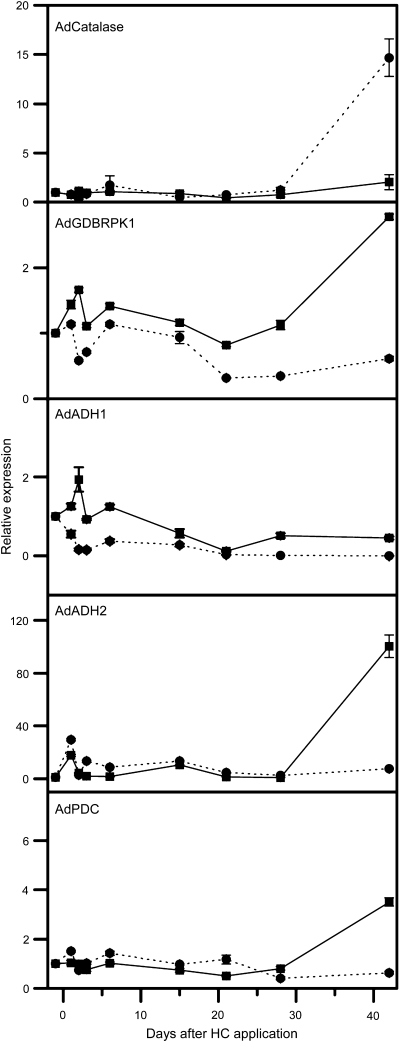
One kiwifruit gene, showing 89.4% identity at the protein level to VvCat-1 (AcCatalase1; AcCat-1), was identified in the EST collection. This gene increased in expression 3 d after HC treatment in the 2003 dataset. When the full-length sequence of this gene was aligned with the VvCat1 and three Arabidopsis catalases (AtCatalase1, AtCatalase2, and AtCatalase3), AcCat-1 found to be most closely related to AtCatalase2 (Fig. 4a). qPCR analysis showed a small increase in AcCat-1 expression late after HC treatment (Fig. 5). One kiwifruit gene (AcGDBRPK1) showed a 78.4% identity to the grape SNF-like protein kinase (GDBRPK; Or et al., 2000c) (Fig. 4b). One microarray oligonucleotide represented that gene and this gene was identified as up-regulated 6 d following HC treatment; qPCR showed a small increase in expression again followed by a larger increase during growth in non-HC buds (Fig. 5). Two kiwifruit alcohol dehydrogenase genes (AcADH1 and AcADH2) clustered in the same clade as VvADH6 (Or et al., 2000a) (Fig. 4c). AcADH2 was represented by an oligonucleotide and was selected as increasing in expression 3 d (2003 only) and 6 d (2003 and 2004) following HC treatment. Analysis of AcAHD2 by qPCR showed an early small increase in expression in both HC- and non-treated buds, followed by a large increase in non-treated buds, while AcADH1 had a Profile 3 pattern of expression (Fig. 5). One kiwifruit pyruvate decarboxylase gene (AcPDC1), clustered very closely to VvPDC1 (Fig. 4d), but had no oligonucleotide represented on the array; qPCR suggested that there was little change in expression during budbreak (Fig. 5). There were two genes showing homology to sucrose synthase on the array that represented the same NR sequence (unique identifiers 324157 and 324394; FG528438); both these genes showed an increase in expression 3 d and 6 d following HC treatment. An oligonucleotide representing an ascorbate peroxidase homologue (unique identifier125576; FG447527) also showed a decrease in expression 6 d after HC induction. The other genes identified by Halaly et al. (2008) were not in the kiwifruit microarray gene lists, i.e. did not show significant changes in expression.
Fig. 4.
Phylogenies of kiwifruit (Ad), grape (Vv) and Arabidopsis (At) genes: (A) catalases, (B) SNF kinase-like, (C) alcohol dehydrogenases, and (D) pyruvate decarboxylases.
Fig. 5.
Expression analysis of kiwifruit homologues to of hydrogen cyanamide (HC)-responsive genes identified in grape using qPCR; solid lines represent buds from HC-treated vines and dashed lines buds from non-treated vines. AdCatalase1 (accession number: FG458399); AdGDBRPK1 (FG521122); AcADH1 (FG466527); AcADH2 (FG525579), and AdPDC (FG475975).
Discussion
The modes of action of dormancy-breaking chemicals, such as HC, have been associated with a sub-lethal stresses, which lead to budbreak (Fuchigami and Nee, 1987). Consistent with this stress hypothesis is the fact that that HC can be phytotoxic on kiwifruit, depending on the concentration and time of application (Linsley-Noakes, 1989; Henzell et al., 1991), which manifests itself as cane burn and/or deformities in the subsequent shoots (Richardson et al., 1994). Sub-lethal stresses have also been used to describe the action of other dormancy-breaking treatments, including high temperatures and sub-lethal freezing temperatures (Fuchigami and Nee, 1987; Halaly et al., 2008). Both these temperatures are effective as short duration treatments, consistent with what is observed with HC treatments as only about 10% of the amount applied is detectable 40 h after application to kiwifruit vines (Alan Cliffe, NuFarm NZ Ltd, personal communication). This swift degradation of HC implies that plants rapidly respond to the chemical, and in fact, apples have been shown respond to HC within 4 h of application (Fuchigami and Nee, 1987).
Of the genes analysed by qPCR, approximately three-quarters could be readily classified into one of the three profiles. Of the six genes showing a Profile 1 expression pattern (Table 3), all are associated with stress events, either by gene function or analysis expression patterns of the closest Arabidopsis homologue. Of the 14 genes showing a Profile 2 expression pattern, 13 can be directly linked to stress events. The one that does not appear to be directly related to stress events is a UDP-glucosyltransferase, the nearest Arabidopsis homologue of which is up-regulated 6 h after seed imbibition. By contrast, all the genes with Profile 3 expression patterns, except the ABC transporter, appear to be associated with developmental processes, as opposed to stress events. Of particular note are the genes that are homologous to Arabidopsis genes involved with the development and maintenance of the dormant state in seeds. There is also a gene homologous to the Arabidopsis gene FKF1 (Flavin binding, Kelch repeat, F-Box 1); that gene regulates CONSTANS (Putterill et al., 1995). In kiwifruit, the homologue could play a role in floral development, a process that is tightly linked to budbreak (Brundell, 1975a; Grant and Ryugo, 1982).
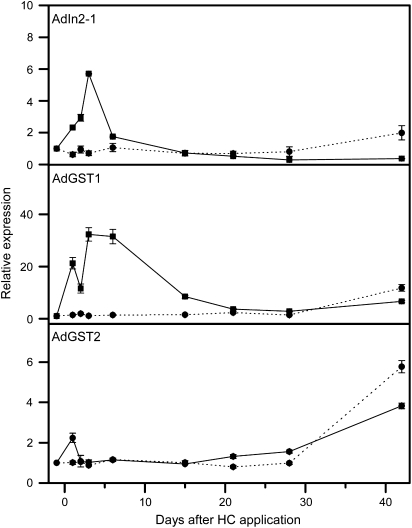
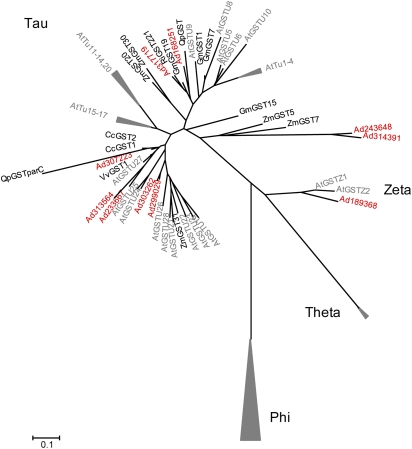
Among the six genes that showed a rapid increase in expression following HC treatment (Profile 1) was a putative glutathione S-transferase (GST) and a GST-like gene (In2-1 protein) (Table 3; Fig. 6). In addition, another GST showed a Profile 2 pattern of expression (Table 3; Fig. 6). Glutathione is a detoxifying agent and has been shown to bind directly to HC in a cell-free system and in germinating mung bean seedlings (Fuchigami and Nee, 1987). Fuchigami and Nee (1987) postulated that glutathione is involved with the breaking of endodormancy in plants. Dormancy-breaking chemicals such as HC, and/or free radicals induced by sub-lethal environmental stresses, bind with free thiol groups on glutathione. A plant's resistance, and therefore response to HC, is related to the ratio of oxidized and reduced glutathione. The amount of rest-breaking agents required to cause a sub-lethal stress increases as the ratios decrease. However, the actual mechanism is unclear. Wang et al. (1991) showed in apples that the ratio of reduced/oxidized glutathione increased after growth induction in apple and that the increase in the amount of the reduced glutathione during chilling was closely associated with the breaking of endodormancy (Siller-Cepeda et al., 1992) and glutathione levels. More recently, the induction of GST has been linked with the conjugation and resultant detoxification of herbicides, the reduction of organic hydroperoxides formed during oxidative stress, and the catabolism of tyrosine (Dixon et al., 2002; Wagner et al., 2002; Moons, 2005). The detoxification role may explain the rapid HC disappearance from treated tissue. Further, when the kiwifruit GSTs are clustered with the grape, oak, and raspberry GSTs (each of which was identified in budbreak experiments), all group within the same two clades, suggesting a common origin (Fig. 7). It is also worth noting that the gene for 5′-adenylylsulphate showed rapid induction after HC-treatment (Profile 2). It is the key regulatory enzyme in sulphate metabolism in plants and sulphur is a key component of glutathione.
Fig. 6.
Expression analysis of AdIn2-1 (accession number; FG470652), AdGST1; (FG423204), and AdGST2 (FG523871) by qPCR; solid lines represent buds from hydrogen cyanamide (HC)-treated vines while dashed lines are buds from non-treated vines.
Fig. 7.
Phylogeny of glutathione S-transferases (GSTs) from Arabidopsis, GSTs from other published budbreak and dormancy microarray experiments [sessile oak (Quercus petraea; Derory et al., 2006), raspberry (Rubus idaeus; Mazzitelli et al., 2007), and grape (Vitis vinifera; Keilin et al., 2007)], and GSTs from kiwifruit identified in the experiments presented in this paper. Focus is given to the Tau class of GSTs, as these are the ones that have been most closely linked to stress events and the breaking of dormancy and shoot outgrowth. The accession numbers for the kiwifruit genes (Adxxxxxx) are as follows: Ad168251(FG423204); Ad189368 (FG456216); Ad233667 (FG478197); Ad243648 (FG460267); Ad299029 (FG510833); Ad303262 (FG496330); Ad307223 (FG519952); Ad313564 (FG523871); Ad317719 (FG501745); and Ad314391 (FG512188).
There have been a number of microarray studies on budbreak in other species. The largest of these was on grape budbreak (Keilin et al., 2007). The other, smaller arrays have looked at the breaking of dormancy and the outgrowth of root buds in leafy spurge (Horvath et al., 2005), raspberry buds during dormancy before budbreak and shoot outgrowth (Mazzitelli et al., 2007), oak buds during budbreak and shoot outgrowth (Derory et al., 2006), and grape (Halaly et al., 2008). Many of the genes identified were associated with shoot and leaf development, rather than the initial reactivation of growth per se, and identifying genes that regulate budbreak remains a challenge.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. List of PCR primers used, product size, and unique identifier for gene.
Supplementary Table S2. Differentially expressed genes identified from a microarray comparison of buds from HC-treated vines compared with those from non-treated vines 1 d after HC treatment in 2003; included are: the log-fold change, the relative expression level, a brief description based on homology to genes in Genbank, and the closest Arabidopsis homologue.
Supplementary Table S3. Differentially expressed genes identified from a microarray comparison of buds from HC-treated vines compared with those from non-treated vines 1 d after HC treatment in 2004; included are: the log-fold change, the relative expression level, a brief description based on homology to genes in Genbank, and the closest Arabidopsis homologue.
Supplementary Table S4. Differentially expressed genes identified from a microarray comparison of buds from HC-treated vines compared with those from non-treated vines 3 d after HC treatment in 2003; included are: the log-fold change, the relative expression level, a brief description based on homology to genes in Genbank, and the closest Arabidopsis homologue.
Supplementary Table S5. Differentially expressed genes identified from a microarray comparison of buds from HC-treated vines compared with those from non-treated vines 6 d after HC treatment in 2003; included are: the log-fold change, the relative expression level, a brief description based on homology to genes in Genbank, and the closest Arabidopsis homologue.
Supplementary Table S6. Differentially expressed genes identified from a microarray comparison of buds from HC-treated vines compared with those from non-treated vines 6 d after HC treatment in 2004; included are: the log-fold change, the relative expression level, a brief description based on homology to genes in Genbank, and the closest Arabidopsis homologue.
Supplementary Table S7. GO analysis of all the genes on the kiwifruit microarray. Genes that had a greater than log2-fold change in expression (with an adjusted P value of <0.05) are highlighted: orange for those up-regulated; blue for those down-regulated.
Supplementary Fig. S1. Expression profiles of genes selected that showed Profile 1 pattern of expression when analysed by qPCR over the extended time-course (day –1 to day 42).
Supplementary Fig. S2. Expression profiles of genes selected that showed Profile 2 pattern of expression when analysed by qPCR over the extended time-course (day –1 to day 42).
Supplementary Fig. S3. Expression profiles of genes selected that showed Profile 3 pattern of expression when analysed by qPCR over the extended time-course (day –1 to day 42).
Supplementary Fig. S4. Expression profiles of genes selected that did not fit into Profiles 1, 2 or 3 when analysed by qPCR over the extended time-course (day –1 to day 42).
Acknowledgments
Thanks to William Laing, Di Barraclough, and John Meekings for their help with the sample collection; to Tim Holmes and Sharlene Cookson with photography and figure drawing, respectively. Thanks to Charles Dwamena and Mirco Montefiori for critically reading this manuscript. This work was funded by the New Zealand Foundation for Research, Science, and Technology contracts C10X0404 and C06X0706.
Glossary
Abbreviations
- qPCR
quantitative PCR
- HC
hydrogen cyanamide
References
- Arora R, Rowland LJ, Tanino K. Induction and release of bud dormancy in woody perennials: a science comes of age. HortScience. 2003;38:911–921. [Google Scholar]
- Böhlenius H, Huang T, Charonnel-Campaa L, Brunner AM, Jansson S, Srauss SH, Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;31:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Bound SA, Jones KM. Hydrogen cyanamide impacts on flowering, crop load, and fruit quality of red ‘Fuji’ apple (Malus domestica) New Zealand Journal of Crop and Horticultural Science. 2004;32:227–234. [Google Scholar]
- Brenac P, Horbowicz M, Downer SM, Dickerman AM, Smith ME, Obsendorf RL. Raffinose accumulation related to desiccation tolerance during maise (Zea mays L.) seed development and maturation. Journal of Plant Physiology. 1997;15:481–488. [Google Scholar]
- Brundell DJ. Flower development of the chinese gooseberry (Actinidia chinensis Planch.). II. Development of the flower bud. New Zealand Journal of Botany. 1975a;13:485–496. [Google Scholar]
- Brundell DJ. Flower development of the Chinese gooseberry (Actinidia chinensis Planch.). II. Development of the flowering shoot. New Zealand Journal of Botany. 1975b;13:473–483. [Google Scholar]
- Brundell DJ. The effect of chilling on the termination of rest and flower bud development of the Chinese gooseberry. Scientia Horticulturae. 1976;4:175–182. [Google Scholar]
- Carter CJ, Thornburg RW. Tobacco Nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiology. 2004;134:460–469. doi: 10.1104/pp.103.027482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:355–381. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, et al. Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics. 2008;9:351. doi: 10.1186/1471-2164-9-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derory J, Léger P, Garcia V, Schaeffer J, Hauser MT, Salin F, Luschnig C, Plomion C, Glössl J, Kremer A. Transcriptome analysis of budburst in sessile oak (Quercus petraea) New Phytologist. 2006;170:723–738. doi: 10.1111/j.1469-8137.2006.01721.x. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-3-reviews3004. reviews 3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez A. Means to compensate for insufficient chilling to improve bloom and leafing. Acta Horticulturae. 1995;395:81–95. [Google Scholar]
- Espinosa-Ruiz A, Saxena S, Schmidt J, Mellerowicz E, Miskolczi P, Bakó L, Bhalerao RP. Differential stage-specific regulation of cyclin-dependent kinases during cambial dormancy in hybrid aspen. The Plant Journal. 2004;38:603–615. doi: 10.1111/j.1365-313X.2004.02070.x. [DOI] [PubMed] [Google Scholar]
- Faust M, Erez A, Rowland LJ, Wang SY, Norman HA. Bud dormancy in perennial fruit trees: physiological basis for dormancy induction, maintenance and release. HortScience. 1997;32:623–629. [Google Scholar]
- Fuchigami LH, Nee C-C. Degree growth stage model and rest-breaking mechanisms in temperate woody perennials. HortScience. 1987;22:836–844. [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. The Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciarrubio A, Legaria JP, Covarrubias AA. Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta. 1997;203:182–187. doi: 10.1007/s004250050180. [DOI] [PubMed] [Google Scholar]
- Grant JA, Ryugo K. Influence of developing shoots on flowering potential of dormant buds of Actinidia chinensis. HortScience. 1982;17:977–978. [Google Scholar]
- Halaly T, Pang X, Batikoff T, Crane O, Keren A, Venkateswari J, Ogrodovitch, Sadka A, Lavee S, Or E. Similar mechanisms might be triggered by alternative external stimuli that induce dormancy release in grape buds. Planta. 2008;228:79–88. doi: 10.1007/s00425-008-0720-6. [DOI] [PubMed] [Google Scholar]
- Heide OM, Prestrud AK. Low temperature, but not photoperiod controls growth cessation and dormancy induction and release in apple and pear. Tree Physiology. 2005;25:109–114. doi: 10.1093/treephys/25.1.109. [DOI] [PubMed] [Google Scholar]
- Henzell RF, Briscoe MR, Gravett I. Improving kiwifruit vine productivity with plant growth regulators. Acta Horticulturae. 1991;297:345–350. [Google Scholar]
- Horvath DP, Anderson JV, Chao WS, Foley ME. Knowing when to grow: signals regulating bud dormancy. Trends in Plant Science. 2003;8:534–540. doi: 10.1016/j.tplants.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Horvath DP, Soto-Suárez M, Chao WS, Jia Y, Anderson JV. Transcriptome analysis of paradormancy release in root buds of leafy spurge (Euphorbia esula) Weed Science. 2005;53:795–801. [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Jonak C, Ökrész L, Bögre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Current Opinion in Plant Biology. 2002;5:415–424. doi: 10.1016/s1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- Kemp BE. Bateman domains and adenosine derivatives form a binding contract. Journal of Clinical Investigation. 2004;133:182–184. doi: 10.1172/JCI20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilin T, Pang X, Venkateswari J, et al. Digital profiling of a grape-bud EST collection leads to new insight into molecular events during grape-bud dormancy release. Plant Science. 2007;173:446–457. [Google Scholar]
- Kiedrowski S, Kawalleck P, Hahlbrook K, Somssich IE, Dagl JL. Rapid activation of a novel plant defence gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO Journal. 1992;11:4677–4684. doi: 10.1002/j.1460-2075.1992.tb05572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-J, Kim M-R, Bedgar DL, Moinuddin SGA, Cardenas CL, Davin LB, Kang C, Lewis NG. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:1455–1460. doi: 10.1073/pnas.0307987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-J, Kim K-W, Cho M-H, Franceschi VR, Davin LB, Lewis NG. Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development; lessons for database annotations? Phytochemistry. 2007;68:1957–1974. doi: 10.1016/j.phytochem.2007.02.032. [DOI] [PubMed] [Google Scholar]
- Kopriva S. Regulation of sulphate assimilation in Arabidopsis and beyond. Annals of Botany. 2006;97:479–495. doi: 10.1093/aob/mcl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lang GA, Early JD, Martin GC, Darnell RL. Endo-, para-, and eco-dormancy: physiological terminology and classification for dormancy research. HortScience. 1987;22:371–377. [Google Scholar]
- Law RD, Suttle JC. Transient decreases in methylation at 5′-CCGG-3’ sequences in potato (Solanum tuberosum L.) meristem DNA during progression of tubers through dormancy precede the resumption of sprout growth. Plant Molecular Biology. 2003;51:437–447. doi: 10.1023/a:1022002304479. [DOI] [PubMed] [Google Scholar]
- Li C, Junttila O, Ernstsen A, Heino P, Palva ET. Photoperiodic control of growth, cold acclimation and dormancy development in silver birch (Betula pendula) ecotypes. Physiologia Plantarum. 2003;117:206–212. [Google Scholar]
- Li Y, Baldauf S, Lim E-K, Bowles DJ. Phylogenetic analysis of the UDP-glycosyltransferase multigene family in Arabidopsis thaliana. Journal of Biological Chemistry. 2001;276:4338–4343. doi: 10.1074/jbc.M007447200. [DOI] [PubMed] [Google Scholar]
- Lionakis SM, Schwabe WW. Bud dormancy in the kiwi fruit, Actinidia chinensis Planch. Annals of Botany. 1984;54:467–484. [Google Scholar]
- Linsley-Noakes GC. Improving flowering of kiwifruit in climatically marginal areas using hydrogen cyanamide. Scientia Horticulturae. 1989;38:247–259. [Google Scholar]
- Lisso J, Altmann T, Mussig C. The AtNFXL1 gene encodes a NF-X1 type zinc finger protein required for growth under salt stress. FEBS Letters. 2006;580:4851–4856. doi: 10.1016/j.febslet.2006.07.079. [DOI] [PubMed] [Google Scholar]
- Marrs KA. The functions and regulation of glutathione s-transferases in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- Mazzitelli L, Hancock RD, Haupt S, et al. Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. Journal of Experimental Botany. 2007;58:1035–1045. doi: 10.1093/jxb/erl266. [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. Repressor- and activator-type ethylene response factors functioning in jasmonate signalling and disease resistance identified via a genome wide screen of Arabidopsis transcription factor gene expression. Plant Physiology. 2005;139:949–959. doi: 10.1104/pp.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson HG, Richardson AC, Snelgar WP, Currie MB. Effects of hydrogen cyanamide on budbreak and flowering in kiwifruit (Actinidia deliciosa ‘Hayward’) New Zealand Journal of Crop and Horticultural Science. 2001;29:277–285. [Google Scholar]
- McPherson HG, Snelgar WP, Manson PJ, Snowball AM. Bud respiration and dormancy of kiwifruit (Actinidia deliciosa) Annals of Botany. 1997;80:411–418. [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R. The BOTRYTIS SUSCEPTIBLE1 gene that encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. The Plant Cell. 2003;15:2551–2565. doi: 10.1105/tpc.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons A. Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs) Vitamins and Hormones. 2005;72:155–202. doi: 10.1016/S0083-6729(05)72005-7. [DOI] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H. Emerging MAP kinase pathways in plant stress signalling. Trends in Plant Science. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Nakagami H, Soukupová H, Schikoraw A, Žárský V, Hirt H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. Journal of Biological Chemistry. 2006;281:38697–38704. doi: 10.1074/jbc.M605293200. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT. Hydrogen peroxide and nitric oxide as signalling molecules in plants. Journal of Experimental Botany. 2002;53:1237–1247. [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. The Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E, Baybik J, Lavee S, Sadka A, Ogrodovitch A. Characterisation of two cDNA clones (accession no AF195866 and AF195867) encoding alcohol dehydrogenase from grape (Vitis vinifera cv. Perlette) developing fruits. Plant Physiology. 2000a;122:619. [Google Scholar]
- Or E, Baybik J, Sadka A, Ogrodovitch A. Fermentative metabolism in grape berries: isolated and characterization of pyruvate decarboxylase cDNA and analysis of its expression throughout berry development. Plant Science. 2000b;156:151–158. doi: 10.1016/s0168-9452(00)00247-8. [DOI] [PubMed] [Google Scholar]
- Or E, Vilozny I, Eyal Y, Ogrodovitch A. The transduction of the signal for grape bud dormancy breaking induced by hydrogen cyanamide may involve the SNF-like protein kinase GDBRPK. Plant Molecular Biology. 2000c;43:483–494. doi: 10.1023/a:1006450516982. [DOI] [PubMed] [Google Scholar]
- Or E, Vilozny I, Fennell A, Eyal Y, Ogrodovitch A. Dormancy in grape buds: Isolation and characterisation of catalase cDNA and analysis of its expression following chemical induction of bud dormancy release. Plant Science. 2002;162:121–130. [Google Scholar]
- Pang X, Halaly T, Crane O, Keilin T, Keren-Keiserman A, Ogrodovitch A, Galbraith D, Or E. Involvement of calcium signalling in dormancy release of grape buds. Journal of Experimental Botany. 2007;58:3249–3262. doi: 10.1093/jxb/erm172. [DOI] [PubMed] [Google Scholar]
- Perez FJ, Lira W. Possible role of catalase in post-dormancy bud break in grapevines. Journal of Plant Physiology. 2005;162:301–308. doi: 10.1016/j.jplph.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene in Arabidopsis promotes flowering and encodes a protein showing similarity to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Rhode A, Prinsen E, De Rycke R, Engler G, van Montagu M, Boerjan W. PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. The Plant Cell. 2002;14:1885–1901. doi: 10.1105/tpc.003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AC, Dawson TE, Hampton RE, Blank RH. The effects of hydrogen cyanamide and mineral oil on kiwifruit performance. Proceedings of the New Zealand Plant Protection Conference. 1994;47:314–319. [Google Scholar]
- Rinne P, Tuominen H, Junttila O. Seasonal changes in bud dormancy in relation to bud morphology water and starch content, and abscisic acid concentration in adult trees of Betula pubescens. Tree Physiology. 1994;14:549–561. doi: 10.1093/treephys/14.6.549. [DOI] [PubMed] [Google Scholar]
- Rozen S, Helen J, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sale PR, Lyford PB. Cultural, management and harvesting practices for kiwifruit in New Zealand. In: Warrington IJ, Weston GC, editors. Kiwifruit: science and management. Wellington, New Zealand: New Zealand Society for Horticultural Science; 1990. pp. 247–296. [Google Scholar]
- Sánchez-Fernández R, Davies TGE, Coleman JOD, Rea PA. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. Journal of Biological Chemistry. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- Saure MC. Dormancy release in deciduous fruit trees. Horticultural Reviews. 1985;7:239–300. [Google Scholar]
- Schaffer RJ, Friel EN, Souleyre EJ, et al. A genomics approach reveals that aroma production in apple is controlled by ethylene predominantly at the final step in each biosynthetic pathway. Plant Physiology. 2007;144:1899–1912. doi: 10.1104/pp.106.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. Journal of Clinical Investigation. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seskar M, Shulaev V, Raskin I. Endogenous methyl salicylate in pathogen-inoculated tobacco plants. Plant Physiology. 1998;116:387–392. [Google Scholar]
- Shaul O, Hilgemann DW, de-Almeida-Engler J, Van Montagu M, Inzé D, Galili G. Cloning and characterisation of a novel Mg2+/H+ exchanger. EMBO Journal. 1999;18:3973–3980. doi: 10.1093/emboj/18.14.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Silverman P, Raskin I. Airborne signalling by methyl salicylate in plant pathogen resistance. Nature. 1997;385:718–721. [Google Scholar]
- Shulman Y, Nir G, Lavee S. Oxidative processes in bud dormancy and the use of hydrogen cyanamide in breaking dormancy. Acta Horticulturae. 1986;179:141–148. [Google Scholar]
- Siller-Cepeda JH, Fuchigami LH, Chen THH. Glutathione content in peach buds in relation to development and release of rest. Plant and Cell Physiology. 1992;33:867–872. [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. The Plant Journal. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in plants. San Diego, USA: Academic Press; 1997. [Google Scholar]
- Tunnacliffe A, Wise MJ. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94:791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- van den Brûle S, Smart CC. The plant PDR family of ABC transporters. Planta. 2002;216:95–106. doi: 10.1007/s00425-002-0889-z. [DOI] [PubMed] [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Molecular Biology. 2002;49:515–532. doi: 10.1023/a:1015557300450. [DOI] [PubMed] [Google Scholar]
- Walton EF, Clark CJ, Boldingh HL. Effect of hydrogen cyanamide on amino acid profiles in kiwifruit buds during budbreak. Plant Physiology. 1991;97:1256–1259. doi: 10.1104/pp.97.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton EF, Fowke PJ. Effect of hydrogen cyanamide on kiwifruit shoot flower number and position. Journal of Horticultural Science. 1993;68:529–534. [Google Scholar]
- Walton EF, Fowke PJ, Weis K, McLeay PL. Shoot axillary bud morphogenesis in kiwifruit (Actinidia deliciosa) Annals of Botany. 1997;80:13–21. [Google Scholar]
- Walton EF, Podivinsky E, Wu R-M. Bimodal patterns of floral gene expression over the two seasons that kiwifruit flowers develop. Physiologia Plantarum. 2001;111:396–404. doi: 10.1034/j.1399-3054.2001.1110318.x. [DOI] [PubMed] [Google Scholar]
- Wang SY, Jiao HJ, Faust M. Changes in metabolic enzyme activities during thidiazuron-induced lateral budbreak of apple. Journal of Horticultural Science. 1991;26:171–173. [Google Scholar]
- Wise MJ. LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics. 2003;4:52. doi: 10.1186/1471-2105-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Molecular Biology. 2005;58:585–596. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.