Abstract
Above-optimal temperatures reduce yield in tomato largely because of the high heat stress (HS) sensitivity of the developing pollen grains. The high temperature response, especially at this most HS-sensitive stage of the plant, is poorly understood. To obtain an overview of molecular mechanisms underlying the HS response (HSR) of microspores, a detailed transcriptomic analysis of heat-stressed maturing tomato microspores was carried out using a combination of Affymetrix Tomato Genome Array and cDNA-amplified fragment length polymorphism (AFLP) techniques. The results were corroborated by reverse transcription-PCR (RT-PCR) and immunoblot analyses. The data obtained reveal the involvement of specific members of the small heat shock protein (HSP) gene family, HSP70 and HSP90, in addition to the HS transcription factors A2 (HSFA2) and HSFA3, as well as factors other than the classical HS-responsive genes. The results also indicate HS regulation of reactive oxygen species (ROS) scavengers, sugars, plant hormones, and regulatory genes that were previously implicated in other types of stress. The use of cDNA-AFLP enabled the detection of genes representing pollen-specific functions that are missing from the tomato Affymetrix chip, such as those involved in vesicle-mediated transport and a pollen-specific, calcium-dependent protein kinase (CDPK2). For several genes, including LeHSFA2, LeHSP17.4-CII, as well as homologues of LeHSP90 and AtVAMP725, higher basal expression levels were detected in microspores of cv. Hazera 3042 (a heat-tolerant cultivar) compared with microspores of cv. Hazera 3017 (a heat-sensitive cultivar), marking these genes as candidates for taking part in microspore thermotolerance. This work provides a comprehensive analysis of the molecular events underlying the HSR of maturing microspores of a crop plant, tomato.
Keywords: cDNA-AFLP, gene expression, heat stress response, microarray, microspore maturation, tomato
Introduction
Most crop plants are exposed to heat stress (HS) during some stage of their life cycle. HS, defined as the temperatures above normal optimum, is expected to become a more frequent and acute problem in the coming years (Sato et al., 2000). Exposure to HS reduces yield and decreases the quality of many crops, including vegetable crops (Kinet and Peet, 1997; Wien, 1997; Boote et al., 2005). Peet et al. (1998) demonstrated in tomato that at daily mean temperatures of 29 °C (32/26 °C day/night), fruit number, fruit weight per plant, and seed number per fruit were markedly decreased compared with at 25 °C. Plants also encounter high temperature damage during spring and autumn when grown in the warmer regions of the world. During these seasons, short waves of high temperatures may be detrimental. Impaired pollen development under high temperature conditions has been implicated in reduced yields across a large number of crop systems (Saini and Aspinall, 1982; Peet et al., 1998; Sato et al., 2000, 2002; Aloni et al., 2001; Porch and Jahn, 2001; Stone, 2001; Prasad et al., 2006; Jain et al., 2007). In tomato, developing pollen grains are highly HS sensitive (Pressman et al., 2002, 2006; Firon et al., 2006). Heat-tolerant tomato cultivars (exhibiting higher yield under HS) produced larger numbers of high-quality pollen grains under HS compared with all tested heat-sensitive cultivars (Firon et al., 2006). Despite the anticipation that HS is becoming a more frequent problem and despite the accumulated data on the high HS sensitivity of developing pollen grains, data on the molecular mechanisms involved in pollen HS response (HSR) and thermotolerance are very limited (Pressman et al., 2007).
The various stages of pollen development and functioning, including the release of mature pollen grains from the anther, are HS sensitive (Pressman et al., 2007). This work focuses on microgametogenesis—the microspore maturation phase. During this phase, symplastic discontinuity requires that the individual microspores be programmed with appropriate signals, or at least be activated for major functions, including the two mitotic divisions, intracellular vacuolar biogenesis, and major metabolic changes such as starch biosynthesis (Gorman and McCormick, 1997, and references therein; Pressman et al., 2007).
Thermotolerance is an essential component of the acclimation response of different organisms and it is generally divided into acquired thermotolerance (i.e. the ability to acquire tolerance to otherwise lethal HS) and basal thermotolerance (i.e. the inherent ability to survive temperatures above the optimal growth temperatures) (Suzuki et al., 2008, and references therein). The capacity for acquired thermotolerance may be achieved by elevating expression levels of ‘protective’ genes prior to HS exposures (Larkindale and Vierling, 2008). Plants have evolved a variety of responses to elevated temperatures that minimize damage and ensure protection of cellular homeostasis. Heat shock transcription factors (HSFs) play an important role in both basal and acquired thermotolerance, regulating heat shock proteins (HSPs) (Kotak et al., 2007). Emerging evidence shows that HS is accompanied by some degree of oxidative stress, and that a cross-talk exists between heat and oxidative stress signalling. Increased protection from HS-mediated oxidative stress might be a component of the acquired thermotolerance trait as the mRNAs and activities of reactive oxygen species (ROS) scavengers such as ascorbate peroxidase (APX) increase under HS conditions and are controlled by HSFs (Suzuki and Mittler, 2006). In recent years, several other signalling pathways involving plant hormones, such as ethylene, were proposed to play an important role in plant thermotolerance (Larkindale et al., 2005a; Kotak et al., 2007; Suzuki et al., 2008). Recently, the transcriptional coactivator multiprotein bridging factor 1c (MBF1c) has been shown to function upstream of salicylic acid (SA) and ethylene and regulate basal thermotolerance in Arabidopsis (Suzuki et al., 2005, 2008). These studies implied that MBF1c is not required for the expression of HSFA2, APX1, and HSP genes during HS, thus raising the possibility for the existence of two separate HS-regulatory pathways. Other responses to HS involve compatible solute production, thought to stabilize proteins and membrane bilayer structure (Sung et al., 2003) and include additional factors such as Ca2+ and Ca2+-dependent signalling (Kotak et al., 2007, and references therein). Despite the wealth of recently accumulated data on HSR and thermotolerance mechanisms in vegetative parts of various plants systems, to our knowledge such data are not available with regard to developing pollen, in general, and in tomato pollen in particular (Pressman et al., 2007).
In this work, a genome-level approach was undertaken, using heat-sensitive and heat-tolerant genotypes of tomato (Hazera 3017 and Hazera 3042 cultivars, respectively) in order to identify HS-responsive genes in maturing tomato microspores. A combination of Affymetrix Tomato Genome Array and cDNA-amplified fragment length polymorphism (AFLP) techniques was used to obtain an overview of molecular mechanisms that participate in the HSR of microspores. This approach was aimed at the identification of genes that represent the classical HSR and thermotolerance mechanisms, as well as novel factors, including pollen-specific genes that represent pollen-specific components, that may be recruited for coping with HS. The results indicate HS regulation of specific HSF genes, HSP genes, ROS scavengers and genes that control the levels of sugars, as well as homologues of Arabidopsis thaliana pollen-specific CDPK and vesicle trafficking machinery gene family members. A specific role for ethylene in the HSR of microspores is suggested in view of the high HS-induced elevation in a number of ethylene-related genes, including 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, several ethylene-responsive factors, and the transcriptional co-activator MBF1. For several genes, differences in basal expression levels between microspores of the heat-tolerant and heat-sensitive cultivars were detected and their putative functions are discussed.
Materials and methods
Plant material and growth conditions
Tomato plants (Solanum lycopersicum L.) of two cultivars, Hazera 3017 (heat sensitive) and Hazera 3042 (heat tolerant; Hazera Genetics, Israel), were grown in two temperature-controlled greenhouses at the Volcani Center, Bet Dagan, Israel, with day/night temperatures of 26/22±2 °C, day length of 13–14 h, and under natural illumination conditions. In one of the greenhouses, after the development of the second truss, plants bearing 2–3 inflorescences were exposed to short-term HS conditions (43–45 °C for 2 h). During the heat treatment, to avoid drought stress and wilting, plants were watered every 60 min. In both greenhouses, plants produced flowers and fruits continuously for the next 3 months. To obtain enough biological material for the molecular analyses detailed below, pollen grains were collected during two summer seasons (2005 and 2006).
Pollen quality determination and preparation of microgametophytes
Heat stress was applied to flower buds at 7, 6, 5, and 3 d before anthesis (A), corresponding to microspore developmental stages: A-7 and A-6, post-miotic microspore stages; A-5, vacuolated microspore stage; and A-3, early binucleate stage (Pressman et al., 2002). To determine pollen quality, microspores were allowed to mature and pollen grains were collected at anthesis from the heat-stressed and control plants. Total number, number of viable, non-viable, and germinating pollen grains was determined as described by Pressman et al. (1998).
For RNA and protein extractions, flower buds at A-7, A-5, and A-3 were sampled from heat-stressed (immediately following the treatment) and control plants, and microspores were separated from the anther tissues, as described by Pressman et al. (2002). At least 100 flower buds were used for each sample preparation. These samples were collected during the two seasons and each such sample served as a biological replicate. The isolated pollen grains were plunged into liquid nitrogen and kept at –70 °C until use.
RNA isolation and labelling for microarray hybridization experiments
Microspores were ground to a fine powder using liquid nitrogen and sea sand (Merck, Darmstadt, Germany) and total RNA was extracted using the Tri reagent (Sigma-Aldrich, Israel). Array hybridizations were performed using two biological replicates of RNA samples extracted from microspores of the two cultivars that were exposed to either control or HS conditions at three developmental stages, A-7, A-5, and A-3. Affymetrix GeneChip® Tomato Genome Array, designed specifically to monitor gene expression in tomato, was used. All procedures for probe preparation, hybridization, washing, staining, and scanning of the GeneChip® Tomato Arrays, as well as data collection, were performed at the Microarray Core Facility, Department of Biological Services, The Weizmann Institute of Science, Rehovot, Israel. A 10 μg aliquot of total RNA was used as starting material and cRNA was prepared using the Affymetrix GeneChip Exp 3’ One-Cycle kit according to the relevant Affymetrix GeneChip® Expression Analysis Technical Manual (No. 701021 Rev. 5).
Array hybridization and statistical analysis
The cRNA was fragmented before hybridization and hybridized to the probe array for 16 h at 45 °C. Independent hybridizations were performed for each developmental stage, cultivar, and treatment sample (detailed above): a total of 24 hybridizations. Immediately after hybridization, the probe array underwent an automated washing and staining protocol on the fluidics station FS450. The probe array was scanned on a GC7000 scanner. Initially, probe signal summarization, normalization, and background subtraction were performed using robust multichip analysis (RMA; Irizarry et al., 2003) in the ‘affy’ package with default parameters. The statistical test for differentially expressed genes was performed using the linear models for microarray (LIMMA) package (Smyth, 2004), which allows a better variance estimation by calculating the moderated t-statistic using empirical Bayesian techniques.
Twenty-four samples were hybridized to separate arrays. For statistical analysis, these hybridizations were arranged into 12 groups, which included two biological replicates each. The 12 groups are: heat-sensitive cultivar (Hazera 3017) after HS treatment in all three developmental stages (3017_HS_A-7, 3017_HS_A-5, and 3017_HS_A-3) and their respective controls, namely the heat-sensitive cultivar exposed to control conditions (C) in all stages (3017_C_A-7, 3017_C_A-5, and 3017_C_A-3), and the corresponding heat-tolerant cultivar (Hazera 3042) after HS treatment in all three stages (3042_HS_A-7, 3042_HS_A-5, and 3042_HS_A-3), and their respective controls, namely the heat-tolerant cultivar exposed to control conditions in all stages (3042_C_A-7, 3042_C_A-5, and 3042_C_A-3). This arrangement, called factorial design, enables detection of (i) the effect of HS treatment (sample size n=12 for each group: stress or control); (ii) the effect of cultivar (sample size as in previous effect n=12); and (iii) the effect of developmental stages (sample size n=8 for each stage), as well as all possible interactions among the three effects, i.e. synergism or antagonism.
To control the level of false discoveries that result due to multiple comparisons, the approach of Benjamini and Hochberg (1995) was applied, to generate adjusted P-values (q-values). The putative function of each gene was predicted by the Affymetrix annotation, the Institute for Genomic Research (TIGR) definition, and the National Center for Biotechnology Information (NCBI) database.
Preparation of poly(A)+ RNA and cDNA synthesis for cDNA-AFLP experiments
AFLP-based transcript profiling (cDNA-AFLP) was carried out as described by Breyne et al. (2003) with some modifications, starting with 75 μg of total RNA. Poly(A)+ RNA was prepared using a Dynabeads® mRNA Purification Kit (Dynal, Oslo, Norway) according to the manufacturer's instructions. First-strand cDNA was synthesized using the SuperScript™ First-strand Synthesis System for reverse transcription-PCR (RT-PCR; Invitrogen, Carlsbad, CA, USA) and oligo(dT)12–18 (Promega, Madison, WI, USA). Second-strand synthesis was performed in second-strand synthesis buffer (20 mM TRIS-HCl, pH 7.5, 100 mM KCl, 10 mM NH4SO4) by adding 5 mM MgCl2, 0.15 mM β-NAD+, 0.5 mg ml−1 bovine serum albumin (BSA), 0.2 mM dNTPs, 10 μl of template single-stranded cDNA, 6 U of DNA polymerase I, 0.25 U of RNase-H, and 1.2 U of Escherichia coli DNA ligase (New England BioLabs, Ipswich, MA, USA). The reaction was incubated at 16 °C for 2 h, after which 6 U of T4 DNA polymerase (New England BioLabs) were added and the reaction was incubated for an additional 30 min at 16 °C. The reaction was terminated by adding 10 mM EDTA. Glycogen (1 mg ml−1) was added and the resulting double-stranded cDNA was phenol extracted by adding phenol:chloroform:isoamyl alcohol (25:24:1) and using Phase Lock Gel (Eppendorf, Hamburg, Germany).
Template preparation, AFLP reactions, and PAGE analysis of products
Templates for AFLP were prepared by performing cDNA digestion and adaptor ligation in the same reaction mixture as follows: 500 ng of cDNA were added to a reaction mixture which included 50 mM NaCl, 1 mg ml−1 BSA, E. coli DNA ligase buffer [3 mM TRIS-HCl, pH 8, 0.4 mM MgCl2, 100 μM dithiothreitol (DTT), 2.6 μM NAD+, 5 μg ml−1 BSA], EcoRI, and MseI (2.5 U each), EcoRI and MseI adaptors (Invitrogen™, Life Technologies; see the sequences in Supplementary Table S1 available at JXB online) and 1 U of T4 DNA ligase (New England BioLabs). The reaction was incubated at 37 °C for 2 h in a final volume of 10 μl. The resultant fragments were subjected to pre-amplification as described in the AFLP Analysis System I and Starter Primer Kit (Invitrogen™, Life Technologies) instruction manual using pre-selective amplification primers (Supplementary Table S1 at JXB online), except for the use of 1 U of Ex Taq DNA polymerase (TAKARA, Otsu, Shiga, Japan). The obtained amplification mixture was diluted 50-fold and 3 μl was used for selective amplification. End labelling of one of the selective primers (listed in Supplementary Table S1) and selective amplifications were carried out essentially as described in the AFLP Analysis System I and Starter Primer Kit (Invitrogen™, Life Technologies) instruction manual. The EcoRI primer was end labelled with 2 μCi of [γ-33P]ATP per reaction using T4 polynucleotide kinase (New England BioLabs). Selective amplification was performed using 0.5 pmol of labelled primer, EcoRI, or MseI, and 0.5 pmol of MseI or EcoRI selective primer (Supplementary Table S1), respectively, 0.1 mM dNTPs, and 1 U of Ex Taq polymerase (TAKARA) in a 20 μl reaction volume. Amplification was done using a touchdown program for 11 cycles (the first cycle being 94 °C for 60 s, 65 °C for 60 s, and 72 °C for 90 s). The annealing temperature was then lowered by 1 °C each cycle, during 11 cycles, followed by 23additional regular program cycles (94 °C for 30 s, 56 °C for 30 s, and 72 °C for 60 s). Amplifications were performed in an ATC401 thermal cycler (CLP, San Diego, CA, USA).
Isolation and sequencing of transcript-derived fragments
Electrophoresis was performed in an Amersham Biosciences SQ3 Sequencer Apparatus (Amersham, CA, USA). Reactions were run on 30×40 cm gels using 6% acrylamide. Dried gels were exposed to X-ray film (Kodak Biomax MR-1 Film) at –80 °C for 3 d. The cDNA expression profiles were determined by PCR-selective amplification using 29 primer combinations. The AFLP fragments ranged in length from 50 bp to ∼800 bp. For each primer combination, 60 bands were observed, with a total of 4520 fragments being screened. Bands corresponding to differentially expressed transcripts were eluted from the gel. AFLP fragments were recovered by PCR under the same conditions that were used for pre-amplification. Resultant amplified fragments were cloned into pGEM-T easy (Promega), transformed into E. coli DH5-α (BioLab Ltd, Israel) and individual clones were sequenced (Danyel Biotech, Rehovot, Israel). Usually 3–5 clones were sequenced from each transformation. Nucleotide sequences and translated sequences were compared with nucleotide and translated sequences of GenBank databases and sequences of expressed sequence tag (EST) databases, including the tomato database of The Institute of Genomic Research (TIGR), by using the TBLASTX sequence alignment program (Altschul et al., 1990).
Validation of the microarray and cDNA-AFLP results using semi-quantitative RT-PCR analyses
All RNA samples were cleaned of DNA contamination using RQ1 RNase-free DNase (Promega). cDNA was synthesized using 1 μg of total RNA and Superscript II (Invitrogen) reverse transcriptase, according to the manufacturer's instructions. First-strand cDNA (1 μl) was used for semi-quantitative RT-PCR that was performed in three biological replicates. Transcript accumulation of genes under investigation was evaluated using primers designed according to the respective microarray probe and AFLP fragment sequences (Hy-labs, Rehovot, Israel) in a final volume of 20 μl, using 0.05 U of Ex Taq (TAKARA) and 0.1 mM dNTPs in an ATC401 thermal cycler (CLP, San Diego, CA, USA). For each reaction, a set of a different number of cycles, ranging between 23 and 35, was tested to choose those corresponding to the exponential phase. Each cycle consisted of 30 s denaturation at 94 °C, 30 s annealing at the Tm, and 30 s extension at 72 °C; cycles were followed by a final 5 min extension at 72 °C. Expression levels are compared with the expression of the 18S gene (accession X51576). Primer sequences are listed in Supplementary Table S2 at JXB online.
Protein electrophoresis and immunoblotting
Microspores and pollen grains were ground to a fine powder as described above and total proteins were extracted in SDS sample buffer [60 mM TRIS-HCl pH 8.0, 60 mM DTT, 2.0% (w/v) SDS, 15% (w/v) sucrose, 5 mM 6-amino-n-caproic acid, 1.0 mM phenylmethylsulfonyl fluoride (PMSF)]. The protein concentration was determined as described in Larkindale et al. (2005a) using a Coomassie Brilliant Blue dye-binding assay with BSA as a standard. Proteins (2 μg) were resolved by SDS–PAGE using 12% (w/v) polyacrylamide gels. For western analysis, proteins were blotted onto nitrocellulose and processed for detection using peroxidase-conjugated secondary antibodies (Sigma-Aldrich, Israel) and a chemiluminescence detection kit (EZ-ECL, Biological Industries Co., Beit Haemek, Israel), and were visualized on film (Super RX X-ray film, Fujiilm, Japan). Antiserum against class I and class II Arabidopsis 17.6 small HSPs (sHSPs; a gift from Professor E Vierling, University of Arizona, Tucson, AZ, USA) and against Chenopodium mitochondrial sHSP (a gift from Professor A Gau, University of Hannover, Hannover, Germany) was used at a dilution of 1:1000 (v/v), and antiserum against spinach chloroplast sHSP (a gift from Professor A Perl, The Volcani Center, ARO, Israel) was used at a dilution of 1:3500 (v/v).
Results and Discussion
Effect of heat stress on tomato pollen characteristics
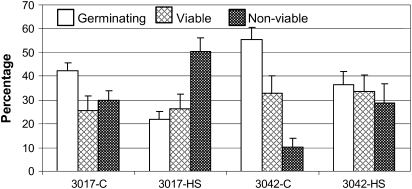
Exposure of tomato plants to season-long chronic high temperature conditions (day/night temperatures of 32/26 °C) has been previously shown to reduce yield by greatly affecting male gametophyte development (Firon et al., 2006). In the present study, in order to identify early HS-responsive genes in maturing tomato microspores, short-term HS conditions (43–45 °C for 2 h) were used. These threshold conditions were selected to minimize massive death of pollen. The results of pollen quality, recorded in at least five biological replicates, are summarized in Fig. 1. The total number of pollen grains in both cultivars under control and HS conditions was similar and ranged between 38 and 50×103 (data not shown). The short-term HS treatment reduced pollen germinability and increased the percentage of non-viable pollen grains in the two cultivars tested (Fig. 1). The differences between the cultivars with respect to the effect of HS on pollen germination was small: an ∼48% and 33% decrease in the number of germinating pollen grains in the heat-sensitive (cv. Hazera 3017) and heat-tolerant (cv. Hazera 3042) cultivars, respectively (Fig. 1). This is in contrast to the effect of chronic HS conditions where heat-tolerant cultivars produced about three times as many viable and about 10 times as many germinating pollen grains as the heat-sensitive cultivars (Firon et al., 2006).
Fig. 1.
Effect of heat stress on tomato pollen quality. Tomato plants of cv. Hazera 3017 (heat sensitive) and cv. Hazera 3042 (heat tolerant) were exposed to a short-term HS (2 h at 43–45 °C). Pollen grains were collected from the heat-stressed (HS) plants of both cultivars (3017-HS, 3042-HS) and from control (C) plants (3017-C, 3042-C) 7, 6, 5, and 3 d after stress application. Mean values ±SE of the percentage germinating pollen grains, percentage viable pollen grains, and percentage non-viable pollen grains are presented. The mean values were calculated from the combined results of all tested developmental stages, derived from at least five biological replicates. The total number of pollen grains in both cultivars under control and HS conditions was similar and ranged between 38 and 50×103.
Transcriptome profiling of maturing microspores’ response to HS using Affymetrix Tomato Genome Array
Using a microarray platform consisting of ∼10 000 tomato probe sets, the following effects were compared: (i) HS versus control; (ii) heat-tolerant cultivar versus heat-sensitive cultivar; and (iii) the gene expression differences among three microspore developmental stages, as well as the synergism and antagonism among all effects. It was found that only genes expressed in heat-stressed microspores (n=12) compared with those expressed in control microspores (n=12) were statistically significant. Results yielded a total of 104 genes that exhibited significant (P <0.05) up-regulation (>2-fold) following the HS treatment. No genes exhibited reduced expression levels in heat-stressed compared with non-treated (control) microspores (Table 1 and Supplementary Table S3 at JXB online). This may be due to the limited number of genes on the microarray and lack of genes that are expressed in pollen tissues. The complete data set of the HS-dependent up-regulated expression is presented in Supplementary Table S3 at JXB online. To avoid false discoveries, multiple test correction (BH multiple comparison correction; Benjamini and Hochberg, 1995) was used, which reduced the number of the HS-responsive genes to 30. These 30 heat-regulated genes are presented in Table 1. Table 1 also contains 19 genes that were selected from the original list of 104 HS-up-regulated genes for their potential involvement in HS signalling based upon recent reports (Kotak et al., 2007) and genes selected due to high expression of their Arabidopsis orthologues in pollen grains (obtained by the Genevestigator Gene Atlas tool; Zimmermann et al., 2004). To characterize further heat-regulated genes, microarray probe region sequences were used to query available databases. In Supplementary Table S3 and Table 1, for each Affy ID number, either the NCBI accession number, GenBank homologue, or Arabidopsis genome initiative number of the best hit from TBLASTX search is indicated, using a significance threshold of 1.00E-4. Of the genes included in Table 1, the Arabidopsis orthologues of seven genes (AT3G22370, AT2G46140, AT4G12040, AT3G27330, AT2G18440, AT1G24530, and AT5G63100) were found to be highly expressed in Arabidopsis pollen grains, using the Genevestigator Gene Atlas tool (Zimmermann et al., 2004; Table 1). In addition to specific members of the classical HS-responsive genes, a significant proportion of the genes presented in Table 1 are those that were only recently implicated to contribute to HSR and/or HS signalling in other plant systems (Kotak et al., 2007). The data presented deal with the effect of HS on the transcriptome of developing microspores (HS-sensitive phase) in tomato, adding information to existing transcriptomic studies that deal with non-stressed mature pollen grains of Arabidopsis (Becker et al., 2003; Honys and Twell, 2003; Pressman et al., 2007).
Table 1.
Summary of genes with significant (P <0.05) differential expression (>2.0-fold) between heat-stressed and control maturing microspores
| Affy ID | Accession/AGI no.* | E-value† | Gene description‡ | Arabidopsis orthologue§ | Fold¶ |
| Stress response | |||||
| HSPs | |||||
| LesAffx.69957.1.S1_atBH | AT1G52560 | 8E-50 | 26.5 kDa class I small heat shock protein-like (AtHSP26.5-P-CI); A. thaliana | 158 | |
| Les.269.1.S1_atBH | U66300 | 1E-132 | Chloroplast heat shock protein (LeHSP21); S. lycopersicum | AT4G27670 | 155 |
| Les.3739.1.S1_atBH | AB026983 | 5E-116 | Endoplasmic reticulum-located small heat shock protein (LeERHSP21.5); S. lycopersicum | AT4G10250 | 154 |
| LesAffx.10596.1.S1_atBH | AT5G59720 | 1E-63 | 18.2kDa heat shock protein (AtHSP18.2); A. thaliana | 141 | |
| Les.3677.1.S1_atBH | AY128100 | 1E-134 | Small heat shock protein (vis1); S. lycopersicum | AT4G27670 | 132 |
| Les.4004.1.S1_a_atBH | AF123256 | 2E-66 | 17.8 kDa class I small heat shock protein (LeHSP17.8-CI); S. lycopersicum | AT1G07400 | 93 |
| Les.4150.1.S1_atBH | AB017134 | 2E-131 | Mitochondrial small heat shock protein (LeMTHSP23.8); S. lycopersicum | AT4G25200 | 82 |
| Les.3578.1.S1_atBH | AF090115 | 2E-99 | Cytosolic class II small heat shock protein HCT2 (LeHSP17.4-CII); S. lycopersicum | AT5G12020 | 75 |
| LesAffx.70264.1.S1_atBH | AB333795 | 6E-49 | Peroxisomal small heat shock protein (GmPHS); Glycine max | AT5G37670 | 64 |
| Les.3550.1.S1_atBH | AF096251 | 7E-7 | Ethylene-responsive heat shock protein cognate 70 (LeHSC70/ER21); S. lycopersicum | 83 | |
| LesAffx.56637.1.S1_atBH | AT3G09700 | 2E-29 | DNAJ heat shock N-terminal domain-containing protein; A. thaliana | 70 | |
| LesAffx.34336.1.S1_atBH | AT2G32120 | 1E-112 | HSP70T-2; A. thaliana | 36 | |
| LesAffx.10807.1.S1_at | X13301 | 3E-29 | Heat shock protein hsp70 (PhHSP70); Petunia×hybrida | AT5G02490 | 7 |
| Les.3134.1.S1_at | AF123259 | 7E-60 | Heat shock protein 90 (LeHSP90); S. lycopersicum | 5 | |
| Oxidative stress | |||||
| Les.5622.1.S1_atBH | AT3G16050 | 1E-143 | Pyridoxal phosphate synthase protein (AtPDX1); A. thaliana | 64 | |
| LesAffx.3918.1.S1_atBH | DQ096286 | 4E-108 | Cytosolic ascorbate peroxidase 3 (SlAPX3); S. lycopersicum | AT3G09640 | 53 |
| Les.1132.1.A1_atBH | AT1G17870 | 2E-43 | Ethylene-dependent gravitropism-deficient and yellow-green-like (ATEGY3); A. thaliana | 32 | |
| Les.4709.1.S1_at | AT2G27680 | 0 | Aldo/keto reductase family protein; A. thaliana | 6 | |
| Les.4223.1.S1_at | AY034148 | 0 | Alternative oxidase 1a (LeAOX1a); S. lycopersicum | AT3G22370p | 5 |
| Les.1724.1.S1_at | AB087837 | 9E-65 | Glutathione S-transferase (PsGST); Pisum sativum | AT5G02790 | 4 |
| Les.1724.2.S1_at | AI776156 | 1E-12 | Glutathione S-transferase (PsGST); Pisum sativum | AT5G02790 | 4 |
| Others | |||||
| LesAffx.47187.1.S1_atBH | AT3G03270 | 4E-55 | Universal stress protein (USP) family protein/early nodulin ENOD18 family protein; A. thaliana | 28 | |
| Hormone metabolism and response | |||||
| Ethylene | |||||
| Les.3642.1.S1_at | U17972 | 0 | 1-Aminocyclopropane-1-carboxylate synthase 3/ACC synthase 3 (LeACS3); S. lycopersicum | AT4G37770 | 6 |
| Les.3766.1.S1_at | U77719 | 1E-108 | Ethylene-responsive late embryogenesis-like protein (ER5); S. lycopersicum | AT2G46140p | 4 |
| Abscisic acid | |||||
| Les.4807.1.S1_atBH | AT5G13200 | 1E-111 | ABA-responsive protein-related/GRAM domain-containing protein; A. thaliana | 25 | |
| Jasmonic acid | |||||
| Les.368.1.S1_atBH | DQ359730 | 6E-34 | Jasmonate resistance 1-like protein (NaJAR6); Nicotiana attenuate | AT2G46370 | 43 |
| Defence | |||||
| LesAffx.66384.1.S1_atBH | AT1G12060 | 2E-20 | BCL-2-associated athanogene 5 (AtBAG5); A. thaliana | 56 | |
| LesAffx.58019.1.S1_atBH | AT5G20740 | 1E-29 | Invertase/pectin methylesterase inhibitor family protein; A. thaliana | 44 | |
| LesAffx.1881.1.S1_atBH | AT4G36010 | 6E-73 | Pathogenesis-related thaumatin family protein; A. thaliana | 19 | |
| Carbohydrate metabolism | |||||
| Les.3522.1.S1_at | AF071786 | Sucrose-phosphate synthase; S. lycopersicum | AT5G20280 | 7 | |
| Les.3069.1.S1_at | AT2G22900 | 4E-22 | Galactosyl transferase GMA12/MNN10 family protein; A. thaliana | 5 | |
| Les.3696.1.S1_at | AF311943 | 0 | Galactinol synthase 1 (LeGolS-1); S. lycopersicum | AT2G47180 | 4 |
| LesAffx.10299.1.A1_at | DQ104196 | 2E-77 | Sorbitol transporter (ST1); Nicotiana langsdorffii×Nicotiana sanderae | AT3G18830 | 4 |
| Transcription regulation | |||||
| LesAffx.24696.1.S1_atBH | X67601 | 2E-119 | Heat stress transcription factor 30 (LpHSF30/HSFA2); L. peruvianum | AT2G26150 | 200 |
| Les.3551.1.S1_atBH | EU240881 | 5E-113 | Ethylene-responsive transcriptional coactivator multiprotein bridging factor ER24 (LeMBF1); S. lycopersicum | AT3G24500 | 156 |
| Les.3985.1.S1_atBH | AF500011 | 3E-179 | Dehydration responsive-element binding protein (LeDREB1); S. lycopersicum | AT5G05410 | 14 |
| LesAffx.3163.1.S1_at | AT4G12040P | 5E-48 | Zinc finger (AN1-like) family protein; A. thaliana | 10 | |
| Les.2322.1.A1_at | AY044235 | 2E-103 | Jasmonate and ethylene responsive factor 1 (LeJERF1); S. lycopersicum | AT1G53910 | 9 |
| Les.5292.1.S1_at | AT3G27330P | 5E-03 | Zinc finger (C3HC4-type RING finger) family protein; A. thaliana | 8 | |
| Les.2876.2.S1_at | AF208544 | 3E-79 | Heat stress transcription factor A3 (LpHSFA3); L. peruvianum | AT5G03720 | 4 |
| Unclassified | |||||
| LesAffx.15004.1.S1_atBH | AW737975 | cDNA, clone: FC25AB05, HTC in fruit | 380 | ||
| Les.3726.1.S1_atBH | AF204783 | 4E-112 | Ripening-regulated protein DDTFR8; S. lycopersicum | 130 | |
| Les.3822.1.S1_atBH | AF204795 | 1E-33 | Ripening-regulated protein DDTFR17; S. lycopersicum | 55 | |
| Les.1718.1.A1_atBH | AT2G18440P | 6E-03 | Gene with unstable transcript 15 (GUT15); A. thaliana | 50 | |
| Les.195.1.S1_atBH | BM411019 | Transcribed locus | 46 | ||
| Les.5053.1.S1_atBH | BT013184 | Clone 134261F, mRNA sequence | 43 | ||
| Les.1910.1.S1_at | AT2G18440P | 6E-03 | Gene with unstable transcript 15 (GUT15); A. thaliana | 18 | |
| LesAffx.66316.1.S1_at | AT1G24530P | 8E-41 | Transducin family protein/WD-40 repeat family protein; A. thaliana | 10 | |
| Les.4752.1.S1_at | BT013346 | Clone 135282R, mRNA sequence | AT5G63100P | 9 | |
Results are presented as the fold difference between the mean value derived from 12 replicates of heat-stressed versus the mean value derived from 12 replicates of control microspores. Genes found to be statistically significant (P <0.05) are presented in this table.
Either the tomato NCBI accession number, GenBank homologue, or Arabidopsis genome initiative number of best hit from TBLASTX search, using a significance threshold of 1E-4 are given for each Affy ID number.
E-value indicating the significance level of homology to the corresponding gene.
Gene function predicted by the Affymetrix annotation, TIGR definition, and NCBI database.
The Arabidopsis genome initiative number is given for Arabidopsis gene orthologues.
The RMA-normalized ratio of gene expression in heat-stressed (n=12) versus control (n=12) microspores.
To ensure no more than 5% false discovery rate, the BH approach for multiple comparison correction (Benjamini and Hochberg, 1995) was used. Genes that were less than the adjusted P-value of 5% are marked by ‘BH’, the rest (unmarked) are statistically significant (P <0.05, not corrected).
Gene sequences found to be highly expressed in Arabidopsis pollen grains under control conditions (Zimmermann et al., 2004).
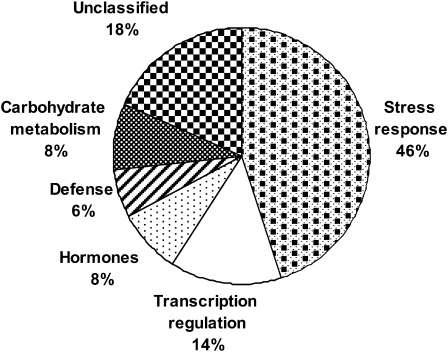
Functional classification of the 49 HS-regulated genes (Table 1) indicates that the largest group (46%) consists of genes that were previously shown to be involved in stress response in other plant systems (Sung et al., 2003; Larkindale et al., 2005a; Kotak et al., 2007), including HSP genes and genes involved in oxidative stress response (Fig. 2). The second largest group of nine genes (18%) includes unclassified genes. The majority of the genes in the other functional groups (including defence- and hormone-related genes as well as genes involved in transcription regulation and in carbohydrate metabolism) were also stress- or defence-related genes. The same categories and similar proportions were obtained for functional classification of the 104 genes that are included in Supplementary Table S3 (Supplementary Fig. S1 at JXB online).
Fig. 2.
Graphical representation of the percentage of genes belonging to a given functional group for the 49 heat-regulated genes that are presented in Table 1.
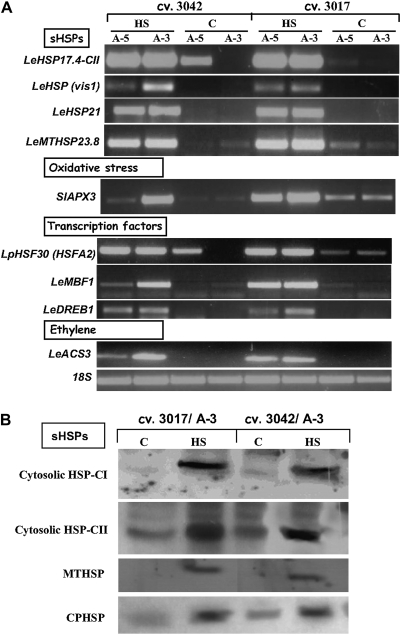
The validity of expression differences detected by the microarray was examined at both the RNA and protein levels. Validation at the RNA level was done by semi-quantitative RT-PCR and results for nine selected genes are presented (Fig. 3A). These genes represent four groups of notable differentially regulated genes that seem to be involved in the HSR of maturing microspores: genes encoding HSPs, genes encoding ROS-scavenging enzymes, genes involved in hormone metabolism or hormonal response, and regulatory genes. Validation at the protein level was done by immunoblot analysis, and the results for four selected genes are presented (Fig. 3B). These genes represent members of the sHSP family, and include mitochondrial, chloroplast, and two cytosolic members. In all cases tested, semi-quantitative RT-PCR and immunoblot analyses confirmed the differences detected by the microarray analyses (Table 1, Supplementry Table S3 at JXB online, Figs 3, 4), namely HS-induced expression of the respective genes, supporting the reliability and accuracy of the microarray profiles and data analysis.
Fig. 3.
Validation of the microarray analysis results by semi-quantitative RT-PCR (A) and immunoblot (B) analyses. Expression levels were tested using either RNA (A) or protein (B), extracted from maturing microspores (5 d and/or 3 d before anthesis; A-5 and/or A-3, respectively) of at least 100 flowers, derived from plants of cv. Hazera 3017 and cv. Hazera 3042, which had been either kept under control conditions or exposed to HS (2 h at 43–45 °C). (A) Expression values were determined by semi-quantitative RT-PCR using gene-specific primers and at least three biological replicates. Several dilutions of template cDNA were tested in order to ensure that gene amplification was in the linear range, and expression levels are compared with expression of the 18S gene (accession X51576). (B) Expression was determined by immunoblot analysis, using 2 μg of total protein, extracted from maturing microspores at A-3, as detailed above. The blot was probed with polyclonal antibodies raised against class I and class II Arabidopsis 17.6 sHSPs (cytosolic HSP-CI and cytosolic HSP-CII, respectively; a gift from Professor E Vierling, University of Arizona, Tucson, AZ, USA), Chenopodium mitochondrial sHSP (MTHSP; a gift from Professor A Gau, University of Hannover, Hannover, Germany), and spinach chloroplast sHSP (CPHSP; a gift from Professor A Perl, The Volcani Center, ARO, Israel). The presented blot represents the results of three biological replicates. Gene names are abbreviated according to the nomenclature used by the NCBI, and the corresponding accession or AGI numbers as well as the Affy ID numbers appear in Table 1.
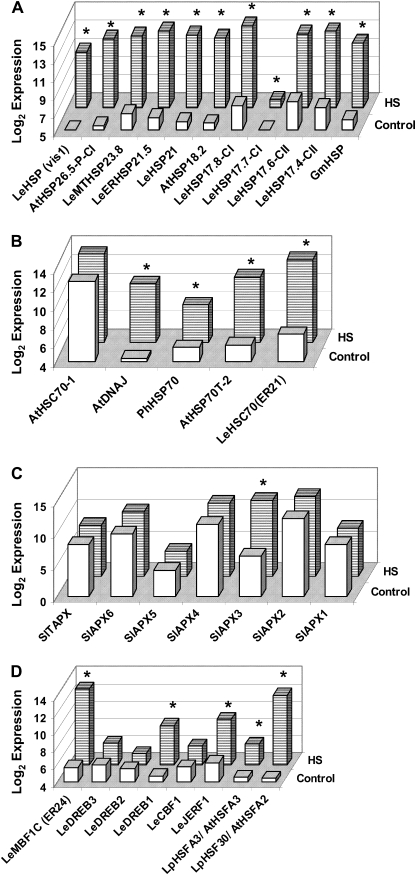
Fig. 4.
Effect of heat stress on the expression profiles of small heat shock protein (sHSP; A), HSP70 (B), and ascorbate peroxidase (APX; C) family members and of genes involved in transcription regulation (D) in maturing microspores. Expression values presented here correspond to the normalized, log base 2, mean expression values of heat-stressed (12 replicates) and control (12 replicates) microspores. Genes whose expression levels differ significantly (P <0.05) between HS and control conditions [based on the procedure described in the section ‘Array hybridization and statistical analysis’, using the LIMMA package (Smyth, 2004)] are marked by an asterisk. More details regarding the plant material and the experimental procedure are given in the legend to Fig. 3 and in Materials and methods. Only the genes represented on the Tomato Affymetrix array were analysed. Gene names are abbreviated according to the nomenclature used by the NCBI, and the corresponding accession or AGI numbers as well as Affy ID numbers appear in Table 1 and in Supplementary Table S3 at JXB online. CI, class I; CII, class II; MT, mitochondrial; LeER, L. esculentum endoplasmic reticulum. HSC-70-1 represents the BT013586 sequence. The gene sequences of the ascorbate peroxidase (APX) family members present on the microarray were annotated according to Najami et al. (2008) as follows: BM4119001 (SlAPX1), BG626096 (SlAPX2), BM410706 (SlAPX3), BG630221 (SlAPX5), AI775047 and BI208755 were annotated as SlAPX4-like and SlAPX6-like genes because of 89% and 84% identity to the DQ131130 and DQ029334 sequences, respectively, and AF413573 as SlTAPX. Affy ID numbers for all the genes that do not appear in either Table 1 or Supplementary Table S3 are as follows: AtHSC70-1, Les.4819.1.S1_at; SlAPX1, Les.247.3.S1_at; SlAPX2, Les.247.1.S1_a_at; SlAPX4, LesAffx.14867.1.S1_at; SlAPX5, Les.1230.1.A1_at; SlAPX6, LesAffx.25588.1.S1_s_at; SlTAPX, Les.2999.1.S1_at; LeCBF1, Les.124.1.S1_at; LeDREB2, Les.3984.1.S1_at; LeDREB3, Les.3379.1.S1_at.
In order to provide a better insight into the nature of genes that respond to HS, following are the expression results, extracted from the microarray data, of notable differentially regulated genes (Fig. 4).
Expression profiles of HSPs
The accumulation of HSPs under the control of HSFs was suggested to play a central role in the HSR in plants and other organisms (Kotak et al., 2007 and references therein). HSPs are assumed to aid other proteins to maintain or regain their native conformation by stabilizing partially unfolded states (Low et al., 2000, and references therein). The increased expression of 11 members of the sHSP family, including two types of cytosolic sHSP genes (class I and class II) as well as mitochondrial, endoplasmic reticulum, and chloroplast members, following exposure of plants to a short-term high temperature treatment (2 h at 43–45 °C; Table 1 and Supplementary Table S3 at JXB online, Fig. 4A) points to their involvement in the HSR of the different cellular compartments of maturing pollen grains. In tobacco, Volkov et al. (2005) demonstrated that sHSP genes are differentially regulated during pollen development and following HS, with three genes of class CI (sHSP-6, -2B, and -3C) exhibiting high HS-induced up-regulation. Current models propose that sHSPs act as ATP-independent chaperones by binding to aggregating proteins and maintaining them accessible for refolding by HSP70 (and co-chaperones) and, under some circumstances, by HSP100/ClpB proteins (Larkindale et al., 2005b). In accord with this model, expression of four members of the HSP70 gene family has been found to be significantly elevated in maturing microspores upon exposure of the plants to HS conditions (Table 1, and Supplementary Table S3 at JXB online, Fig. 4B). These include an ethylene-responsive member (HSC-70; ER21) and an HSC70-2 member. HSC-1-70 is found, however, to be constitutively expressed under both control and HS conditions. A dual role was suggested for HSP70 in plants: a protective role in thermotolerance and a regulatory effect on HSF activity and hence the autoregulation of the heat shock response (Low et al., 2000).
Considerable progress has been made with regard to the function of HSP100/ClpB and sHSPs during stress. HSP100/ClpB proteins are members of the AAA+ family of ATPases and are involved in resolubilizing protein aggregates (Kotak et al., 2007, and references therein). A cytosolic member of this family was found to be essential for tolerance to high temperature in plants (Hong and Vierling, 2001). Genetic analysis in Arabidopsis indicates that HSP101 interacts with the sHSP chaperone system to resolubilize protein aggregates after HS (Lee et al., 2005). Recently, it has been demonstrated by us that the transcript levels of HSP101 increased in maturing pollen grains of tomato (cultivars Hazera 3017 and Hazera 3042) in response to HS (Pressman et al., 2007). Furthermore, HS-induced HSP101 transcript levels were several fold higher in maturing pollen of the heat-tolerant cultivar (cv. Hazera 3042) than in the heat-sensitive cv. Hazera 3017, pointing to its potential involvement in pollen thermotolerance. Because the Affymetrix GeneChip® Tomato Genome Array does not contain a HSP101 probe set, quantitative RT-PCR analysis was used to follow this gene expression, where the HSP101 primers were based upon the available EST database information (TIGR) (Pressman et al., 2007). In addition, LeHSP90 was found to be up-regulated by HS in microspores, exhibiting >4-fold elevated expression (Table 1). The accumulated data thus point to the involvement of specific members of the sHSP genes, HSP70, HSP101 and HSP90 families in the HSR of maturing tomato microspores.
Heat stress factors
Previous studies have shown that HSFs promote thermotolerance by regulating HSPs (Kotak et al., 2007). Plants possess multiple HSF-coding genes, with at least 17 members in tomato (Koskull-Döring et al., 2007). Plant HSFs comprise three conserved evolutionary classes, A, B, and C, which are distinguished from each other mainly by their oligomerization domains (Kotak et al., 2007). In tomato, HSFA1, HSFA2, and HSFB1 form a regulatory network that controls the expression of HS-responsive genes (Mishra et al., 2002; Schramm et al., 2006; Koskull-Döring et al., 2007). HSFA1 is constitutively expressed and regulates the HS-induced expression of HSFA2 and HSFB1 as well as synthesis of HSP genes including sHSPs and HSP101 (Mishra et al., 2002). HSFA1 was therefore defined as a ‘master regulator’ of the HSR in tomato (Mishra et al., 2002), whereas HSFA2 (HSF30; Scharf et al., 1990, 1993; Baniwal et al., 2004) is the major HSF gene in thermotolerant cells. The data presented in the present study show that both HSFA2 and HSFA3 are highly HS up-regulated in tomato maturing microspores (Table 1, Figs 3A, 4D). The presented data of HS-induced expression of HSFA2, sHSP genes, HSP70, and HSP101 in microspores are in agreement with accumulating data indicating that in the course of a HSR, the ongoing accumulation of HSFA2 and other HS-inducible proteins (mainly sHSPs classes CI and CII and HSP70) results in a unique storage form of the transcription factor in cytoplasmic multichaperone complexes composed of the 40 nm HS granules (HSGs; Baniwal et al., 2004). Dissociation of HSG complexes with liberation of HSFA2 needs cooperation with the ATP-dependent HSP70 and/or HSP101 chaperone machineries (Baniwal et al., 2004). A role for HSP90, in addition to HSP17-CII and HSP70, in the restoration of HSFs was also suggested (Baniwal et al., 2004).
Arabidopsis HSFA3 was recently shown to be regulated by DREB2A, a transcription factor involved in regulation of dehydration-responsive genes (Sakuma et al., 2006). These findings suggest HSF-mediated cross-talk between HS and other abiotic stress signalling. Notably, the high HS up-regulation in microspores of another member of the DREB gene family, LeDREB1, has been detected by both microarray and semi-quantitative RT-PCR analyses (Figs 3, 4D).
ROS-scavenging genes
High constitutive expression levels of the following five APX genes were detected in maturing microspores: two chloroplastic (thylakoid) members (TAPX and APX6-like), a peroxisomal member (APX4-like), and two cytosolic members (APX2 and APX1) (Fig. 4). High HS up-regulation of SlAPX3 (Table 1 and Figs 3, 4C) is in accordance with recent data indicating that HS is accompanied by some oxidative stress, and that there is a cross-talk between heat and oxidative stress signalling. Increased protection from HS-mediated oxidative stress was previously shown to be a component of the acquired thermotolerance trait in that the mRNAs and activities of ROS scavengers, such as APX, increased under HS conditions and were controlled by HSFs (Suzuki and Mittler, 2006). Also, overexpression of HSF constructs in Arabidopsis resulted in a moderate increase in basal thermotolerance that was also associated with protection from oxidative bleaching of seedlings (Lee et al., 1995; Prandl et al., 1998). These findings suggest that increased ability either to prevent or to repair heat-induced oxidative damage is an aspect of thermotolerance. This suggestion is in accord with the HS-induced expression of APX genes (references cited in Panchuk et al., 2002; Volkov et al., 2006) and the identification of novel heat-tolerant, HSF-dependent expression of an APX isoform in Arabidopsis (Panchuk et al., 2002). ROS may also function as signal transduction molecules and cause HSF activation (Volkov et al., 2006); hence, APX may play a role in regulating H2O2 signalling. Also, as recently demonstrated, ROS may contribute to pollen tube growth (Potocký et al., 2007). The data thus point to a delicate balance between the harmful and beneficial effects of ROS in heat-stressed microspores and suggest the need for developmental regulation. Indeed, using semi-quantitative RT-PCR analysis, lower SlAPX3 levels were detected in A-3 compared with A-5 microspores of cv. 3042 (Fig. 3A).
Ethylene and MBF1 may be involved in the HSR of microspores
The microarray analysis indicates HS induction of several ethylene-responsive genes in maturing pollen grains, including ER5, ER21, and LeJERF1 (Table 1, and Supplementary Table S3 at JXB online) as well as ER24 (Figs 3, 4D). The latter has been previously isolated from tomato fruit (Zegzouti et al., 1999) and found to be a homologue of MBF1, which is involved in transcriptional activation (Suzuki et al., 2005, 2008). HS-regulated expression of the gene coding for ACC synthase, the enzyme involved in ethylene biosynthesis, was also detected in maturing tomato microspores (Table 1, Fig. 3A). Evidence for the involvement of ethylene in plant thermotolerance was amply demonstrated. For instance, pre-treatment of a cool-season grass (Agrostis stolonifera var. palustris) with ACC, prior to exposure of plants to HS, increased heat tolerance (Larkindale and Huang, 2004). Similarly, ACC, when added exogenously, protected Arabidopsis against heat-induced oxidative damage (Larkindale and Knight, 2002). Larkindale et al. (2005a) showed that the Arabidopsis ethylene signalling mutants, ein2 and etr1, are defective in basal thermotolerance. Yet, these mutants were found to accumulate wild-type levels of HSP101 and small HSP genes in response to HS, indicating the involvement of an additional pathway in thermotolerance, separate from the HSF-regulated pathway. Recent data suggest that a member of the MBF1 gene family in Arabidopsis, MBF1c, may act as a regulator of thermotolerance upstream of SA and ethylene (Suzuki et al., 2008). These authors suggest that MBF1c expression enhances the tolerance of transgenic plants to heat by perturbing or partially activating the ethylene response signal transduction pathway (Suzuki et al., 2005). MBF1c was not required for the expression of HSFA2 and different HSP genes (Suzuki et al., 2008). To our knowledge, the involvement of ethylene and/or the regulatory gene MBF1 in pollen HSR and thermotolerance has not been studied previously and there are no relevant data available (Pressman et al., 2007).
Involvement of carbohydrate metabolism and transport in HSR and thermotolerance of microspores
The HS-up-regulated expression of sucrose phosphate synthase, a key enzyme in the sucrose synthesis pathway, detected in the present study, is in accord with the suggested role for sucrose as an osmoprotectant, protecting cell membranes and proteins during exposure to stress conditions (Kaplan et al., 2004; Guy et al., 2008). During microspore maturation, sucrose was found to accumulate in pollen of several members of the Solanaceae family, including tomato (Firon et al., 2006; and unpublished data). Speranza et al. (2007) suggested that high sucrose and cytoplasmic carbohydrate content in mature pollen grains confer longer viability. Pacini (2006) stressed the importance of pollen carbohydrates in regulating the water balance with the surrounding environment and maintaining their viability during dehydration. A similar mechanism may thus be used by the maturing pollen grains to cope with other stresses, such as extreme temperatures. Busch et al. (2005) suggested that HS-induced production of specific sugars, such as galactinol, by galactinol synthases, and raffinose, is involved in the Arabidopsis HSR. The galactinol synthase gene, GolS1, was found to be under regulation of HSF (Busch et al., 2005). The induced expression of galactinol synthase in maturing microspores (LeGolS-1; Table 1) is in agreement with a role for galactinol and/or raffinose in the HSR of maturing tomato microspores. In addition, HS-induced elevation in the expression of a homologue of a sorbitol transporter (ST1; Table 1) may point to the participation of yet another potential compatible solute, the sugar alcohol sorbitol, in pollen HSR. Alternatively, as shown in Arabidopsis for the sugar alcohol permease homologue AtPLT5, the potential tomato sorbitol transporter may function as a non-specific sugar uptake transporter (Reinders et al., 2005).
Isolation and identification of pollen heat stress-responsive genes by cDNA-AFLP analysis
As cDNA-AFLP allows the identification of tissue-specific and low abundance sequences with no prior knowledge of gene sequences, this approach was adopted for the identification of pollen-expressed genes that are not represented by the Affymetrix GeneChip® Tomato Genome Array. Comparison of transcription profiles obtained from heat-stressed and non-stressed (control) microspores allowed identification of transcript-derived fragments (TDFs) from mRNAs that were either up- or down-regulated following the HS treatment (Supplementary Fig. S2 at JXB online). A total of 94 TDFs were extracted from the polyacrylamide gels, reamplified with the pre-selective primers, and subcloned for sequencing. The sequence obtained for each TDF was compared with the GenBank non-redundant public sequence database using the TBLASTX program (Altschul et al., 1997). Among these, 29 (31%) showed close matches [TBLASTX expectation values (E) of <10−4] to database entries with assigned identities (Table 2). The sequences of most fragments showed no homology with known sequences. They may represent previously uncharacterized genes, or the AFLP fragments may have been too short to reveal significant homology. Future characterization of these sequences could lead to the identification of novel pathways involved in mechanisms that cope with HS. Supplemenatry Table S4 at JXB online includes the data for the unknown proteins and all the unidentified sequences. Eight of 94 TDFs (TDF3.1, TDF3.2, TDF32.1, TDF4.2, TDF27.2, TDF42.1, TDF12.1, and TDF32.2; Table 2 and Supplementary Table S4) were found to be represented by the Affymetrix GeneChip® Tomato Genome Array, with two TDFs (TDF3.1 and TDF3.2, similar to AtHSP81-1 and LeHSP90, respectively) exhibiting significant HS-induced expression in the Affymetrix tomato array analyses. Functional classification of the 29 annotated TDFs shows that 12 (42%) genes are involved in stress response, including HSPs and genes associated with oxidative stress response (Table 2). One gene (3%) is involved in cell wall modification. Signal transduction and metabolism and transport-related sequences comprise 17%, including five genes each. Transcription regulation sequences comprise 21% and include six genes.
Table 2.
Summary of HS-responsive transcript-derived fragments (TDFs) of maturing tomato pollen grains and their homologies to sequences in the databases
| TDF* | Primer combination† | Length (bp)‡ | HS response§ | Exp. pattern¶ | Gene description** | Accesion/AGI no.†† | E-value‡‡ |
| Stress response | |||||||
| HSPs | |||||||
| TDF7 | E-ACA/M-CAA | 320 | + | A | Heat shock protein 81-1 (AtHSP81-1); A. thaliana | AT5G52640p | 3E-48 |
| TDF3.1¶¶ | E-ACA/M-CAT | 390 | + | A | Heat shock protein 81-1 (AtHSP81-1); A. thaliana | AT5G52640p | 5E-44 |
| TDF3.2¶¶ | E-ACA/M-CTT | 299 | + | A | Heat shock protein 90 (LeHSP90); S. Lycopersicum | AF123259 | 3E-13 |
| Oxidative stress | |||||||
| TDF17.1 | E-ACT/M-CTT | 268 | + | A | Glutathione S-transferase T4 (LeGSTT4); S. lycopersicum | AY007561 | 2E-8 |
| TDF6.1 | E-ACT/M-CTT | 523 | – | B | Adenylyl-sulphate reductase; S. lycopersicum | AY568717 | 3E-92 |
| TDF6.2 | E-ACT/M-CTT | 447 | – | B | Adenylyl-sulphate reductase; S. lycopersicum | AY568718 | 8E-68 |
| ER stress response | |||||||
| TDF3.3 | E-ACT/M-CTG | 616 | + | A | Cell division cycle protein 48, putative (AtCDC48); A. thaliana | AT3G53230 | 5E-109 |
| Vesicle fusion | |||||||
| TDF18.1 | E-ACT/M-CTG | 262 | + | C | Vesicle-associated membrane protein 725 (AtVAMP725); A. thaliana | AT2G32670p | 2E-27 |
| TDF17.2 | E-ACA/M-CTT | 298 | + | C | Vesicle-associated membrane protein 726 (AtVAMP726); A. thaliana | AT1G04760p | 8E-24 |
| Ubiquitination | |||||||
| TDF17.3 | E-ACT/M-CTG | 283 | + | D | Kelch repeat-containing F-box family protein; A. thaliana | AT1G15670 | 4E-23 |
| TDF4.1 | E-ACA/M-CTT | 483 | + | A | Protein kinase family protein; A. thaliana | AT5G51270 | 4E-13 |
| TDF32.1¶¶ | E-ACT/M-CTT | 173 | + | C | Ubiquitin-associated (UBA)/TS-N domain-containing protein; A. thaliana | AT1G04850 | 3E-13 |
| Defence | |||||||
| Cell wall modification | |||||||
| TDF10.1 | E-ACT/M-CTG | 334 | – | B | Tomato LAT56 gene for protein P56; S. lycopersicum | X15500 | 3E-57 |
| Metabolism and transport | |||||||
| TDF4.2¶¶ | E-ACT/M-CTG | 565 | + | A | UDP-D-glucuronate 4-epimerase 5 (AtGAE5); A. thaliana | AT4G12250 | 1E-77 |
| TDF27.1 | E-ACA/M-CAG | 143 | + | A | Glucosyl transferase; N. tabacum | AB000623 | 5E-4 |
| TDF27.2¶¶ | E-ACT/M-CTG | 179 | – | B | GL2-expression mudulator (AtGEM); A. thaliana | AT2G22475 | 7E-14 |
| TDF28 | E-ACA/M-CAA | 135 | + | A | Mitochondrial NAD+-dependent malic enzyme (malate dehydrogenase) precursor; S. tuberosum | Z23023 | 4E-5 |
| TDF17.4 | E-ACT/M-CTT | 287 | + | A | FH protein-interacting protein 2/voltage-gated potassium channel (AtFIP2); A. thaliana | AT5G55000 | 4E-13 |
| Signal transduction | |||||||
| TDF31 | E-ACT/M-CAA | 146 | + | A | Calcium and calmodulin-dependent protein kinase 2 (AtCDPK2); A. thaliana | AT1G35670p | 1E-8 |
| TDF26 | E-ACT/M-CAC | 202 | + | A | Calcium-dependent protein kinase 2 (NtCDPK2); N. tabacum | AJ344154 | 1E-8 |
| TDF4.3 | E-ACT/M-CTG | 633 | + | A | Protein kinase; A. thaliana | AT1G68690 | 7E-13 |
| TDF42.1¶¶ | E-ACT/M-CTT | 116 | – | E | AKIN gamma; M. truncatula | AY247269 | 8.6E-9 |
| TDF12.1* | E-ACT/M-CAC | 323 | + | A | 14–3–3 protein (LeTFT9); S. lycopersicum | X98865 | 7E-50 |
| Transcription regulation | |||||||
| TDF14.2 | E-ACA/M-CTC | 301 | + | A | RNA-binding protein RZ-1; N. sylvestris | D83696 | 1E-5 |
| TDF43 | E-ACT/M-CAG | 122 | + | A | DNA-binding protein phosphatase 2C (NtDBP1); N. tabacum | AF520810 | 3E-4 |
| TDF4.4 | E-ACA/M-CTT | 480 | + | A | Argonaute1 (AtAGO1); A. thaliana | AT1G48410 | 8E-76 |
| TDF4.5 | E-ACT/M-CTT | 570 | + | A | Zinc finger protein, putative; G. hirsutum | AY919286 | 6E-27 |
| TDF1 | E-ACT/M-CTG | 811 | + | C | Zinc finger (GATA type) family protein; A. thaliana | AT5G25830 | 8E-46 |
| TDF11.1 | E-ACA/M-CTA | 286 | + | A | TITAN8 (TTN8); ATP binding; A. thaliana | AT3G54670 | 4E-17 |
TDF number, representing the relative location on the acrylamide gel. Fragments in the same location representing different treatment groups or primer combinations are marked.
Primer combination used for the fragment amplification: E indicates EcoRI, M indicates MseI.
Length of the TDF sequence.
Induction (+) or repression (–) of expression by heat stress, detected in at least one of the tested stages of microspore development and one of the cultivars.
Gene expression (Exp.) pattern. Expression patterns were divided into five categories, A–E. A represents HS-induced expression in either or both developmental stages tested, in both cultivars. B represents HS-down-regulated expression in both cultivars in microspores at either the A-3 or A-5 developmental stage. C represents constitutive expression in microspores of cv. 3017 and HS-induced expression in maturing microspores of cv. 3042 at either or both developmental stages. D represents constitutive expression in cv. 3042 and HS-induced expression in microspores of cv. 3017 at either or both developmental stages. E represents constitutive expression in microspores of cv. 3017 and HS-down-regulation in microspores of cv. 3042 at A-3.
Gene description according to NCBI, TAIR, or TIGR database annotation.
Either tomato NCBI accession number (No.), GenBank homologue, or Arabidopsis genome initiative number (AGI) of best hit from TBLASTX search, using a significance threshold of 1.00E-4 are given for each TDF.
E-value indicating the significance level of homology to the corresponding gene.
TDF sequences found on the Affymetrix GeneChip® Tomato Genome Array. All TDF sequences were checked using the NETAFFX program of the Affymetrix service site (http://www.affymetrix.com/analysis/index.affx) for identifying TDFs which are presented on the Affymetrix chip.
Gene sequences found to be highly expressed in Arabidopsis pollen grains (Zimmermann et al., 2004).
Five main expression patterns could be distinguished, designated A, B, C, D, and E. In the first group (Table 2, A), 19 TDFs (TDF7, TDF3.1, TDF3.2, TDF17.1, TDF3.3, TDF4.1, TDF4.2, TDF27.1, TDF28, TDF17.4, TDF31, TDF26, TDF12.1, TDF14.2, TDF43, TDF4.3, TDF4.4, TDF4.5, and TDF11.1) exhibited HS-induced expression in either one or both developmental stages tested, in both cultivars. The second group included four TDFs (TDF6.1, TDF6.2, TDF10.1, and TDF27.2) that exhibited HS-down-regulated expression in both cultivars in microspores at either the A-3 or A-5 developmental stage, respectively (Table 2, B). The remaining seven TDFs, assigned into groups C, D, and E, exhibited expression patterns that differed between the cultivars: four TDFs (TDF18.1, TDF17.2, TDF32.1, and TDF1) were constitutively expressed in microspores of cv. 3017 and HS induced in maturing microspores of cv. 3042 at either or both developmental stages (Table 2, C). One TDF (TDF17.3) was found to be constitutively expressed in cv. Hazera 3042 and HS induced in microspores of cv. Hazera 3017 at either or both developmental stages (Table 2, D). TDF42.1 exhibited a constitutive expression pattern in microspores of cv. 3017 and HS down-regulation in microspores of cv. 3042 at A-3 (Table 2, E).
Validation of differential expression revealed by cDNA-AFLP
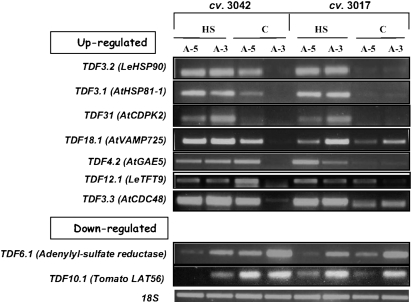
After identification by cDNA-AFLP, expression patterns of six out of the nine selected TDFs tested by RT-PCR analyses (TDF3.2, TDF3.1, TDF31, TDF4.2, TDF3.3, and TDF6.1; Fig. 5) matched the expression categories assigned to these TDFs by the cDNA-AFLP results (A, A, A, A, A, and B, respectively; Table 2). TDF18.1, corresponding to AtVAMP725, belongs to group C with respect to the expression pattern of the cDNA-AFLP results, demonstrating constitutive expression in microspores of cv. 3017 and HS-induced expression in maturing microspores of cv. 3042 at either or both developmental stages (Table 2). The RT-PCR results showed that it was HS induced in both cultivars at A-3 and in cv. Hazera 3042 also at A-5 (Fig. 5). TDF10.1, corresponding to LAT56, exhibiting similarity to pectate lyase and suggested to play a role in pectin degradation during pollen tube growth (Wing et al., 1990), belongs to group B with respect to the expression pattern of the cDNA-AFLP results, demonstrating HS-down-regulated expression in microspores of both cultivars at A-3 (Table 2). The RT-PCR results showed its constitutive expression in microspores of cv. 3017 and HS down-regulation in microspores of cv. Hazera 3042, at both developmental stages (Fig. 5). In the case of TDF12.1, a homologue of a 14–3–3 protein family member, the cDNA-AFLP results indicated that it belongs to group A, exhibiting HS-induced expression in either or both developmental stages, in both cultivars (Table 2). The RT-PCR results showed that it was HS induced in cv. Hazera 3017 at both microspore developmental stages and in cv. Hazera 3042 at A-3, but was down-regulated in cv. Hazera 3042 at A-5 (Fig. 5). The differences detected between the cDNA-AFLP and semi-quantitative RT-PCR analyses might be due to the presence, at least in some cases, of more than one fragment of the same size in the polyacryamide gel and isolation of a non-relevant fragment.
Fig. 5.
RT-PCR analysis of the steady-state transcript levels of selected TDFs. Expression levels were tested using RNA extracted from maturing microspores (5 and 3 d before anthesis; A-5 and A-3, respectively) of at least 100 flowers, derived from plants of cv. 3017 and cv. 3042, which had been either kept under control conditions (C) or exposed to HS (2 h at 43–45 °C). Expression values were determined by semi-quantitative RT-PCR using TDF-specific primers and at least three biological replicates. Several dilutions of template cDNA were tested in order to ensure that gene amplification was in the linear range, and expression levels are compared with the expression of the 18S gene (accession X51576).
The cDNA-AFLP results supported the microarray results by adding genes to the functional groups and gene families identified by the Affymetrix GeneChip® Tomato Genome array analyses, including A. thaliana homologues of pollen-specific HSP80/90 family members (Zimmermann et al., 2004) and genes involved in oxidative stress scavenging, such as glutathione S-transferase (GST; Table 2). Plant GSTs are encoded by a large and diverse gene family and are involved in multiple functions, including detoxification of herbicides, the reduction of organic hydroperoxides, as well as regulatory functions involved in the oxidative stress response (Dixon et al., 2002; Kilili et al., 2004). No members of the small HSP family were detected. This is due to the lack of EcoRI and MseI restriction sites in all small HSP gene sequences tested here.
In addition, the cDNA-AFLP results enabled identification of pollen-expressed HS-regulated genes that are not represented by the microarray and may represent novel mechanisms involved in the HSR of maturing pollen grains. Indeed, several identified homologues of A. thaliana pollen-specific genes point to involvement of Ca2+-dependent signalling and vesicle trafficking in pollen HSR as detailed below. In addition, identified by the cDNA-AFLP analysis and found to be HS up-regulated, is a homologue of Arabidopsis CDC48 (Table 2, Fig. 5). Arabidopsis CDC48, a conserved homohexameric AAA-ATPase chaperone, was suggested to be directly involved in cell division and expansion, as well as in pollen germination and tube elongation (Park et al., 2008, and references therein). Also, in Caenorhabditis elegans, CDC48 is required for relocation of misfolded proteins across the endoplasmic reticulum membrane and subsequent ubiquitin-dependent degradation by the 26S proteasome in the cytosol (Mouysset et al., 2006). CDC48 may have a similar function(s) in heat-stressed microspores, namely the clearance of denatured proteins.
Ca2+-dependent signalling and vesicle trafficking may have a role in maturing pollen HSR
Four gene sequences (four TDFs), homologues of two different gene families, vesicle-associated membrane protein (VAMPs) and calcium-dependent protein kinase 2 (CDPK2), respectively, were found to be HS up-regulated by the cDNA-AFLP analysis (Table 2, Fig. 5). The Arabidopsis VAMP725 and CDPK2 homologues were found to be expressed in a pollen-specific manner (Zimmermann et al., 2004), suggesting their involvement in pollen grain-specific functions and pointing to the recruitment of pollen-specific mechanisms for coping with stress conditions. The vesicle trafficking machinery is highly conserved among eukaryotes. Its major components include lipids and integral membrane proteins, such as VAMPs. The trafficking process is regulated by proteins that assist the vesicle budding, trafficking, and fusion with target membranes. VAMPs constitute the major component of a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex, which functions in facilitating the fusion between the vesicle and the target membranes (Leshem et al., 2006, and references therein). Vesicle trafficking has been traditionally viewed as a housekeeping process, but recent findings in plant, yeast, and animal cells show that it can also play a role in stress responses (Leshem et al., 2006, and references therein). Recently, three novel SNARE genes were isolated in rice, OsNPSN11, OsNPSN12, and OsNPSN13 (Bao et al., 2008). Transgenic tobacco plants overexpressing 35S:OsNPSN11 showed more tolerance to H2O2 and higher sensitivity to NaCl and mannitol than non-transgenic tobacco, suggesting that OsNPSN genes may be involved in different aspects of the signal transduction in plant responses to abiotic stresses (Bao et al., 2008). In addition, the Arabidopsis hit1-1 (heat-intolerant) mutant, found to be inhibited by both heat and water stress, is defective in a homologue of yeast Vps53p which is involved in vesicle trafficking (Lee et al., 2006). A highly efficient vesicular transport system is present in maturing pollen grains and is used during pollen tube elongation (Hepler et al., 2001). Pollen germination and tube growth require directional transport of vesicles containing cell wall materials and is driven by a dynamic actin system that is regulated by a tip-localized Ca2+ gradient (Krichevsky et al., 2006). One of the largest and most differentiated group of calcium sensors are protein kinases, among them CDPKs, which were identified only in plants and protists (Klimecka and Muszyńska, 2007). Recently, Yoon et al. (2006) have identified two pollen-expressed calcium-dependent protein kinase isoforms in Petunia; one (CDPK1) appears to be a key regulator of growth polarity and may participate in maintaining Ca2+ homeostasis, whereas the second (CDPK2) appears to be more generally involved in pollen tube extension. Taken together, presumably upon exposure to HS, maturing microspores may recruit available components of the pollen tube elongation machinery and signalling network and use them for other purposes, namely in the response to HS conditions. If this is the case, other important questions remain to be addressed, such as what cargo do the vesicles carry under stress conditions and whether there is specificity in the triggering stimulus that may affect the cargo loaded onto this secretory machinery.
Differences in gene expression between cultivars
Although no significant differences in gene expression between the cultivars were detected by the Tomato Affymetrix Genome Array hybridizations, higher expression levels of HSFA2 and LeHSP17.4-CII genes were detected by semi-quantitative RT-PCR analyses in non-stressed (‘control’) microspores of cv. Hazera 3042 (the heat-tolerant cultivar) versus microspores of cv. Hazera 3017 (the heat-sensitive cultivar) (Fig. 3A). These results may point to a potential benefit for microspores that exhibit higher basal expression levels of ‘protective’ genes, such as HSP genes, prior to exposure of plants to HS. In line with this hypothesis is the higher expression levels of HSP101 in non-stressed microspores of cv. 3042 compared with microspores of cv. 3017, as has been shown recently (Pressman et al., 2007). Indeed, elevated thermotolerance by constitutive expression of HSP101 was demonstrated (Queitsch et al., 2000; Katiyar-Agrawal et al., 2003). It should be noted that, at the protein level, results showing higher basal expression levels of sHSP-CII in cv. Hazera 3042 microspores compared with cv. 3017 microspores are less pronounced. This may be due to the interaction of the antibodies used with another sHSP family member (Fig. 3B). Notably, in cv. 3042 SlAPX3 exhibited HS up-regulation in A-3 but not in A-5 microspores, pointing to its developmental regulation and to potential differences between the cultivars with respect to H2O2 balance during microspore maturation. Using cDNA-AFLP, differences between the cultivars were observed, with six of the TDFs (TDF3.2, TDF3.1 TDF18.1, TDF4.2, TDF12.1, and TDF3.3) exhibiting higher expression levels in microspores derived from flowers at A-5 of cv. 3042, kept under control conditions, compared with microspores of cv. 3017 (Fig. 5). These TDFs included the Arabidopsis VAMP725 and CDC48 homologues as well as LeHSP90, indicating developmental regulation in the HSR of these genes and pointing to their potential involvement in thermotolerance. Additional studies are necessary, however, including functional analyses, to verify their contribution.
Conclusion
The data presented here provide genome-wide expression profiles of maturing tomato microspores following their exposure to a short-term HS treatment. A combination of Affymetrix Tomato Genome Array and cDNA-AFLP techniques was successfully used to identify HS-regulated genes representing the classical HSR and thermotolerance mechanisms, as well as novel factors, including pollen-specific genes, representing pollen-specific components, that may be recruited for coping with HS. The results indicate HS regulation of HSF and HSP family members, ROS scavengers, genes that control the levels of specific carbohydrates, ethylene-related genes, as well as a pollen-specific CDPK and pollen vesicle trafficking machinery. For several of these genes, higher basal expression levels were detected in non-stressed (‘control’) microspores of a heat-tolerant cultivar compared with microspores of a heat-sensitive cultivar, marking such genes as candidates for participating in microspore thermotolerance. More complete understanding of the molecular mechanisms that contribute to pollen thermotolerance requires, however, additional data, including the functional analyses of a large part of the above-specified genes, under both short-term HS and longer durations of moderate HS conditions. Such studies are underway by means of constructing transgenic tomato plants with modulated expression of part of the respective genes using a pollen-specific promoter.
Supplementary data
Supplementary data can be found at JXB online.
Table S1. List of adaptors and primers used for cDNA-AFLP application.
Table S2. List of primer sequences used for semi-quantitative RT-PCR analysis for validation of the microarray and cDNA-AFLP results.
Table S3. Complete list of genes with significant (P <0.05) differential expression (>2.0-fold) between heat-stressed and control maturing microspores.
Table S4. Summary of HS-responsive transcript-derived fragments (TDFs) of maturing tomato pollen grains representing unknown proteins and unidentified sequences.
Fig. S1. Graphical representation of the percentage of genes belonging to a given functional group for the 104 heat-regulated genes that are presented in Table S3.
Fig. S2. Autoradiography of a representative cDNA-AFLP polyacrylamide gel.
Acknowledgments
This work was supported by the US–Israel Binational Agricultural Research and Development Fund (grant no. IS-3738-05 R) and by the India Israel Cooperation Fund (grant no. 3-2299), and the Israel Ministry of Science. The authors thank Dr Ron Ecker, Hazera Genetics, Israel, for providing us with the tomato heat-tolerant and heat-sensitive cultivars. The authors acknowledge the assistance of Dr S Horn-Saban, Head of the Microarray Facility at The Weizmann Institute of Science, Rehovot, Israel. The authors thank Professor E Vierling (University of Arizona, Tucson, AZ, USA) for the cytosolic sHSP antibodies, Professor A Gau (University of Hannover, Hannover, Germany) for the mitochondrial sHSP antibodies, and Professor A Perl (The Volcani Center, ARO, Israel) for the chloroplast sHSP antibodies.
References
- Aloni B, Peet M, Pharr M, Karni L. The effect of high temperature and high atmospheric CO2 on carbohydrate changes in bell pepper (Capsicum annuum) pollen in relation to its germination. Physiologia Plantarum. 2001;112:505–512. doi: 10.1034/j.1399-3054.2001.1120407.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bao YM, Wang JF, Huang J, Zhang HS. Cloning and characterization of three genes encoding Qb-SNARE proteins in rice. Molecular Genetics and Genomics. 2008;279:291–301. doi: 10.1007/s00438-007-0313-2. [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Bharti K, Chan KY, et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. Journal of Biosciences. 2004;29:471–487. doi: 10.1007/BF02712120. [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiology. 2003;133:713–725. doi: 10.1104/pp.103.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- Boote KJ, Allen LH, Prasad PVV, Baker JT, Gesch RW, Snyder AM, Pan D Thomas JMG. Elevated temperature and CO2 impacts on pollination, reproductive growth, and yield of several globally important crops. Journal of Agricultural Meteorology of Japan. 2005;60:469–474. [Google Scholar]
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inze D, Zabeau M. Quantitative cDNA-AFLP analysis for genome-wide expression studies. Molecular Genetics and Genomics. 2003;269:173–179. doi: 10.1007/s00438-003-0830-6. [DOI] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schoeffl F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. The Plant Journal. 2005;41:1–14. doi: 10.1111/j.1365-313X.2004.02272.x. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Adrian Lapthorn A, Edwards R. Plant glutathione transferases. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-3-reviews3004. reviews 3004.1–3004.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firon N, Peet MM, Pharr DM, Zamski E, Rosenfeld K, Althan L, Pressman E. Pollen grains of heat tolerant tomato cultivars retain higher carbohydrate concentration under heat stress conditions. Scientia Horticulturae. 2006;109:212–217. [Google Scholar]
- Gorman SW, McCormick S. Male sterility in tomato. Critical Reviews in Plant Science. 1997;16:31–53. [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. Metabolomics of temperature stress. Physiologia Plantarum. 2008;132:220–235. doi: 10.1111/j.1399-3054.2007.00999.x. [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY. Polarized cell growth in higher plants. Annual Review of Cell and Developmental Biology. 2001;17:159–187. doi: 10.1146/annurev.cellbio.17.1.159. [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E. Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. The Plant Journal. 2001;27:25–35. doi: 10.1046/j.1365-313x.2001.01066.x. [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiology. 2003;132:640–652. doi: 10.1104/pp.103.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jain M, Prasad PV, Boote KJ, Hartwell AL, Jr, Chourey PS. Effects of season-long high temperature growth conditions on sugar-to-starch metabolism in developing microspores of grain sorghum (Sorghum bicolor L. Moench) Planta. 2007;227:67–79. doi: 10.1007/s00425-007-0595-y. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiology. 2004;136:4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agrawal S, Agarwal M, Grover A. Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Molecular Biology. 2003;51:677–686. doi: 10.1023/a:1022561926676. [DOI] [PubMed] [Google Scholar]
- Kilili KG, Atanassova N, Vardanyan A, Clatot N, Al-Sabarna K, Kanellopoulos PN, Makris AM, Kampranis SC. Differential roles of Tau class glutathione S-transferases in oxidative stress. Journal of Biological Chemistry. 2004;279:24540–24551. doi: 10.1074/jbc.M309882200. [DOI] [PubMed] [Google Scholar]
- Kinet JM, Peet MM. The physiology of vegetable crops. In: Wien HC, editor. Tomato. Wallingford UK: CAB International; 1997. pp. 207–258. [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. Complexity of the heat stress response in plants. Current Opinion in Plant Biology. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Koskull-Döring P, Scharf KD, Nover L. The diversity of plant heat stress transcription factors. Trends in Plant Science. 2007;12:452–457. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Klimecka M, Muszyńska G. Structure and functions of plant calcium-dependent protein kinases. Acta Biochimica Polonica. 2007;54:219–233. [PubMed] [Google Scholar]
- Krichevsky A, Kozlovsky SV, Tian GW, Chen MH, Zaltsman A, Citovsky V. How pollen tubes grow. Developmental Biology. 2006;303:405–420. doi: 10.1016/j.ydbio.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Larkindale J, Hall JD, Knight MR, Vierling E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiology. 2005a;138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Huang B. Thermotolerance and antioxidant systems in Agrostis stolonifera: involvement of salicylic acid, abscisic acid, calcium, hydrogen peroxide, and ethylene. Journal of Plant Physiology. 2004;161:405–413. doi: 10.1078/0176-1617-01239. [DOI] [PubMed] [Google Scholar]
- Larkindale J, Knight MR. Protection against heat-stress induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiology. 2002;128:682–695. doi: 10.1104/pp.010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Mishkind M, Vierling E. Plant responses to high temperature. In: Jenks MA, Hasegawa PM, editors. Plant abiotic stress. Oxford: Blackwell Publishing; 2005b. pp. 100–144. [Google Scholar]
- Larkindale J, Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiology. 2008;146:748–761. doi: 10.1104/pp.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CF, Pu HY, Wang LC, Sayler RJ, Yeh CH, Wu SJ. Mutation in a homolog of yeast Vps53p accounts for the heat and osmotic hypersensitive phenotypes in Arabidopsis hit1-1 mutant. Planta. 2006;224:330–338. doi: 10.1007/s00425-005-0216-6. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hübel A, Schöffl F. Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. The Plant Journal. 1995;8:603–612. doi: 10.1046/j.1365-313x.1995.8040603.x. [DOI] [PubMed] [Google Scholar]
- Lee U, Wie C, Escobar M, Williams B, Hong SW, Vierling E. Genetic analysis reveals domain interactions of Arabidopsis Hsp100/ClpB and cooperation with the small heat shock protein chaperone system. The Plant Cell. 2005;17:559–571. doi: 10.1105/tpc.104.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y, Melamed-Book N, Cagnac O, Ronen G, Nishri Y, Solomon M, Cohen G, Levine A. Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proceedings of the National Academy of Sciences, USA. 2006;103:18008–18013. doi: 10.1073/pnas.0604421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low D, Brandl K, Nover L, Forreiter C. Cytosolic heat-stress proteins Hsp17.7 class I and Hsp17.3 class II of tomato act as molecular chaperones in vivo. Planta. 2000;211:575–582. doi: 10.1007/s004250000315. [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes and Development. 2002;16:1555–1567. doi: 10.1101/gad.228802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouysset J, Kähler C, Hoppe T. A conserved role of Caenorhabditis elegans CDC-48 in ER-associated protein degradation. Journal of Structural Biology. 2006;156:41–49. doi: 10.1016/j.jsb.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Najami N, Janda T, Barriah W, Kayam G, Tal M, Guy M, Volokita M. Ascorbate peroxidase gene family in tomato: its identification and characterization. Molecular Genetics and Genomics. 2008;279:171–182. doi: 10.1007/s00438-007-0305-2. [DOI] [PubMed] [Google Scholar]
- Pacini E, Guarnieri M, Nepi M. Pollen carbohydrates and water content during development, presentation, and dispersal: a short review. Protoplasma. 2006;228:73–77. doi: 10.1007/s00709-006-0169-z. [DOI] [PubMed] [Google Scholar]
- Panchuk, Volkov RA, Schoffl F. Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiology. 2002;129:838–853. doi: 10.1104/pp.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Rancour DM, Bednarek SY. In planta analysis of the cell cycle-dependent localization of AtCDC48A and its critical roles in cell division, expansion, and differentiation. Plant Physiology. 2008;148:246–258. doi: 10.1104/pp.108.121897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet MM, Sato S, Gardner RG. Comparing heat stress effects on male-fertile and male-sterile tomatoes. Plant, Cell and Environment. 1998;21:225–231. [Google Scholar]
- Porch TG, Jahn M. Effects of high-temperature stress on microsporogenesis in heat-sensitive and heat-tolerant genotypes of Phaseolus vulgaris. Plant, Cell and Environment. 2001;24:723–731. [Google Scholar]
- Potocký M, Jones MA, Bezvoda R, Smirnoff N, Zárský V. Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytologist. 2007;174:742–751. doi: 10.1111/j.1469-8137.2007.02042.x. [DOI] [PubMed] [Google Scholar]
- Prändl R, Hinderhofer K, Eggers-Schumacher G, Schöffl F. HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Molecular and General Genetics. 1998;258:269–278. doi: 10.1007/s004380050731. [DOI] [PubMed] [Google Scholar]
- Prasad PVV, Boote KJ, LH Adverse high temperature effects on pollen viability, seed-set, seed yield and harvest index of grain sorghum (Sorghum bicolor (L.) Moench) are more severe at elevated carbon dioxide. Agricultural and Forest Meteorology. 2006;139:237–251. [Google Scholar]
- Pressman E, Harel D, Zamski E, Shaked R, Althan L, Rosenfeld K, Firon N. The effect of high temperatures on the expression and activity of sucrose cleaving enzymes during tomato (Lycopersicon esculentum) anther development. Journal of Horticultural Science and Biotechnology. 2006;81:341–348. [Google Scholar]
- Pressman E, Moshkovitch H, Rosenfeld K, Shaked R, Gamliel B, Aloni B. Influence of low night temperatures on sweet pepper flower quality and the effect of repeated pollinations, with viable pollen, on fruit setting. Plant Physiology and Biochemistry. 1998;73:131–136. [Google Scholar]
- Pressman E, Peet MM, Pharr MD. The effect of heat-stress on tomato pollen characteristics is associated with changes in carbohydrate concentration in the developing anthers. Annals of Botany. 2002;90:631–636. doi: 10.1093/aob/mcf240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressman E, Shaked R, Firon N. Tomato response to heat stress: focus on pollen grains. Plant Stress. 2007;1:216–227. [Google Scholar]
- Queitsch C, Suk-Whan Hong, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. The Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Panshyshyn JA, Ward JM. Analysis of transport activity of Arabidopsis sugar alcohol permease homolog AtPLT5. Journal of Biological Chemistry. 2005;280:1594–1602. doi: 10.1074/jbc.M410831200. [DOI] [PubMed] [Google Scholar]
- Saini HS, Aspinall D. Abnormal sporogenesis in wheat (Triticum aestivum L) induced by short periods of high-temperature. Annals of Botany. 1982;49:835–846. [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proceedings of the National Academy of Sciences, USA. 2006;103:18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Peet MM, Thomas JF. Physiological factors limit fruit set of tomato (Lycopersicon esculentum Mill.) under chronic, mild heat stress. Plant, Cell and Environment. 2000;23:719–726. [Google Scholar]
- Sato S, Peet MM, Thomas JF. Determining critical pre- and post-anthesis periods and physiological processes in Lycopersicon esculentum Mill. exposed to moderately elevated temperatures. Journal of Experimental Botany. 2002;53:1187–1195. doi: 10.1093/jexbot/53.371.1187. [DOI] [PubMed] [Google Scholar]
- Scharf KD, Rose S, Zott W, Schoffl F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO Journal. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Rose S, Zott W, Thierfelde J, Nover L. Two cDNAs for tomato heat stress transcription factors. Plant Physiology. 1993;102:1355–1356. doi: 10.1104/pp.102.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, Koskull-Doring P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Molecular Biology. 2006;60:759–772. doi: 10.1007/s11103-005-5750-x. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Speranza A, Calzoni GL, Pacini E. Occurrence of mono- or disaccharides and polysaccharide reserves in mature pollen grains. Sexual Plant Reproduction. 1997;10:110–115. [Google Scholar]
- Stone P. The effects of heat stress on cereal yield and quality. In: Basra AS, editor. Crop responses and adaptations to temperature stress. Binghampton, NY: Food Products Press; 2001. pp. 243–291. [Google Scholar]
- Sung DY, Kaplan F, Lee KJ, Guy LC. Acquired tolerance to temperature extremes. Trends in Plant Science. 2003;8:179–187. doi: 10.1016/S1360-1385(03)00047-5. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Bajad S, Shuman J, Shulaev V, Mittler R. The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. Journal of Biological Chemistry. 2008;283:9269–9275. doi: 10.1074/jbc.M709187200. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Mittler R. Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiologia Plantarum. 2006;126:45–51. [Google Scholar]
- Suzuki N, Rizhsky L, Hongjian Liang, Shuman J, Shulaev V, Mittler R. Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiology. 2005;139:1313–1322. doi: 10.1104/pp.105.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov RA, Panchuk, Mullineaux PM, Schoffl F. Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Molecular Biology. 2006;61:733–746. doi: 10.1007/s11103-006-0045-4. [DOI] [PubMed] [Google Scholar]
- Volkov RA, Panchuk, Schoffl F. Small heat shock proteins are differentially regulated during pollen development and following heat stress in tobacco. Plant Molecular Biology. 2005;57:487–502. doi: 10.1007/s11103-005-0339-y. [DOI] [PubMed] [Google Scholar]
- Wien HC. Peppers. In: Wien HC, editor. The physiology of vegetable crops. Wallingford, UK: CAB International; 1997. pp. 259–293. [Google Scholar]
- Wing RA, Yamaguchi J, Larabell SK, Ursin VM, McCormick S. Molecular and genetic characterization of two pollen-expressed genes that have sequence similarity to pectate lyases of the plant pathogen Erwinia. Plant Molecular Biology. 1990;14:17–28. doi: 10.1007/BF00015651. [DOI] [PubMed] [Google Scholar]
- Yoon GM, Dowd PE, Gilroy S, McCubbin AG. Calcium-dependent protein kinase isoforms in Petunia have distinct functions in pollen tube growth, including regulating polarity. The Plant Cell. 2006;18:867–878. doi: 10.1105/tpc.105.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Jones B, Frasse P, Marty C, Maitre B, Latch A, Pech JC, Bouzayen M. Ethylene-regulated gene expression in tomato fruit: characterization of novel ethylene-responsive and ripening-related genes isolated by differential display. The Plant Journal. 1999;18:589–600. doi: 10.1046/j.1365-313x.1999.00483.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.