Abstract
Nicotianamine (NA), a small molecule ubiquitous in plants, is an important divalent metal chelator and the main precursor of phytosiderophores. Nicotianamine synthase (NAS) is the enzyme catalyzing NA synthesis by the condensation of three aminopropyl moieties of S-adenosylmethionine (SAM) and the cyclization of one of them to form an azetidine ring. Here we report five crystal structures of an archaeal NAS from Methanothermobacter thermautotrophicus, either free or in complex with its product(s) and substrate(s). These structures reveal a two-domains fold arrangement of MtNAS, a small molecule related to NA (named here thermoNicotianamine or tNA), and an original mechanism of synthesis in a buried reaction chamber. This reaction chamber is open to the solvent through a small inlet, and a single active site allows the selective entrance of only one substrate at a time that is then processed and translocated stepwise.
Keywords: metal homeostasis, reaction mechanism
Iron is an essential element for almost all living species. Although abundant in the earth's crust, the availability of iron is limited as the insoluble ferric (Fe3+) form predominates. Plants use two strategies to obtain iron from the soil (1–3). The first one (strategy I) is by reducing the ferric ion pool into its more soluble Fe2+ form before transport through the roots. The second (strategy II) is by the roots excreting small molecules called phytosiderophores, chelators belonging to the mugineic acid family, which have a very high affinity for Fe3+. Once they are formed, the phytosiderophore-Fe3+ complexes are absorbed by the root system of the plant.
Nicotianamine (NA) is a ubiquitous metabolite in plants that is able to bind heavy metals both in vitro and in vivo and that is crucial to the homeostasis of essential metals such as iron and copper (4–6). It is the main precursor in phytosiderophore synthesis and therefore contributes to strategy II iron acquisition. In both strategies, it is also an important metal chelator allowing long distance iron transport and sequestration. The role of NA in the redistribution of iron in plants is illustrated by the phenotype of a tomato mutant called chloronerva, which, unable to synthesize NA, develops interveinal chlorosis despite high iron concentrations in its leaves (7, 8). The NA level in plants can be manipulated through genetic engineering to produce crops with higher iron contents or with increased tolerance to low iron availability (9, 10). The action of NA is not restricted to iron homeostasis, but extends to that of other metal ions such as Cu2+, Zn2+, Mn2+, and Ni2+ (10–15). For example, one of the most pronounced defects in the chloronerva mutant is a lack of root-to-shoot transport of Cu that induces a severe shortage in leaves with the consequence that Cu-containing enzymes are less active. NA has also been shown to be the main metabolite in Cd2+ and Zn2+ hyperaccumulation in Arabidopsis haleri (16) and largely contributes to Ni2+ chelation and long distance transport in Thlaspi caerulescens, a natural hyperaccumulator plant (12, 17). Aside from its roles in metal homeostasis, NA inhibits the angiotensin I-converting enzyme in spontaneously hypertensive rats and may therefore reduce high blood pressure in humans (18, 19).
The biosynthesis of NA is catalyzed by nicotianamine synthase (NAS; EC 2.5.1.43) and consists in the condensation of three aminopropyl moieties of S-adenosylmethionine (SAM), with the cyclization of one of them to form an azetidine ring (Fig. S1) (20). In barley, NAS is a single polypeptide whose functional form has been reported to be a monomer (20) or a trimer (21). The trimer form was taken as an indication that the catalytic mechanism involves the cooperation of three monomers to achieve the triple condensation of aminopropyl moieties. Despite this controversy, an intriguing feature of NAS, when compared to other aminopropyltranferase enzymes, is that it uses three molecules of SAM without any other aminopropyl acceptor. By comparison, spermidine synthase and spermine synthase, two enzymes belonging to the aminopropyltransferase family, use putrescine or spermidine as their respective acceptors and are limited to only one aminopropyl transfer (22, 23).
The NAS gene family has long been considered to be plant-specific. However, recent sequencing projects have revealed nas-like genes in the genome of various organisms including plants, fungi, and archaea (21, 24). Apart from in plants, both NA and a functional NAS have only been detected in Neurospora crassa, a filamentous fungus (24). On the contrary, nothing is known about the presence of NA, its putative role, and the functional relevance of a nas-like gene in archaea.
Structure of MtNAS.
Here we report the high-resolution crystal structures of (i) the native protein from Methanothermobacter thermautotrophicus (MtNAS) as a complex with one thermoNicotianamine (tNA) or two catalytic products [tNA and 5′-methylthioadenosine (MTA)] and (ii) a mutant protein in its apo-form or as a complex with one (glutamate) or two substrates (SAM and glutamate). The crystal structure of the selenomethionine-labeled native MtNAS was first determined by the multiple-anomalous-dispersion (MAD) method. The structures of all of the other complexes were subsequently solved by molecular replacement and then refined at high resolution (Table S1).
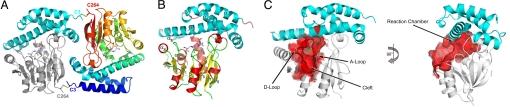
In all of the crystal forms, MtNAS is a homodimer stabilized by two intermolecular disulfide bridges linking the first (C3) and last (C264) cysteines of each monomer (Fig. 1A). This is consistent with the redox-dependent dimerization previously revealed by size exclusion chromatography (25). Many disulfide bonds were identified and found to contribute significantly to the protein thermostability in some archaea (26). However, upon dimerization, only about 720 Å2 of the surface is buried, equivalent to 5.6% of the solvent-accessible surface of each monomer. With 10 intermolecular hydrogen bonds and nine salt bridges located at the dimeric interface, this dimer does not appear to be strongly stabilized, and it is not evident that the functional form in vivo is a dimer.
Fig. 1.
Overall structure of MtNAS. (A) View of the dimer. The monomers are head-to-tail and linked by two disulfide bridges between C3 and C264. In the monomer on the left the α-helical N-terminal domain is colored in cyan and the Rossman fold C-terminal domain in gray. The monomer on the right is colored in a rainbow gradient from blue (N-terminal) to red (C-terminal). (B) Ribbon diagram of one monomer with helices of the N-terminal domain colored in cyan and helices and strands of the C-terminal domain colored in red and yellow, respectively. (C) Representation of the reaction chamber of MtNAS at the interface between the N- and C-terminal domains. The cleft accommodates the base moiety in proteins adopting the Rossmann fold. The reaction chamber is large, extends deep inside the protein, and communicates with the solvent through a small inlet located just above the cleft.
The MtNAS monomer consists of a five-helix bundle N-terminal domain (residues 2–101) on top of a classic Rossmann fold C-terminal domain (residues 102–264) (Fig. 1 A and B). A structural homology search using DALI (27) revealed that the N-terminal domain is specific to the NAS family, whereas the C-terminal domain is highly homologous to the class I family of SAM-dependent methyltransferases, as previously predicted by the level of sequence similarity (28). A large internal catalytic cavity (hereafter called the reaction chamber) with dimensions of 16 × 8 × 8 Å is located at the interface of the two domains (Fig. 1C) and delineated by residues scattered along the primary structure [helix α4 and helix α6, loops connecting β1 to α7 (called the D-Loop), β4 to α10 (called the A-Loop), β6 to β7 and strand β5]. This reaction chamber is open to the solvent by a small inlet restricted by residues belonging to helix α4 and residues from both the A- and D-Loops. At the entrance of this reaction chamber on the solvent exposed side, a cleft is clearly visible that corresponds to the nucleotide binding site such as is found in other proteins adopting a Rossmann fold (29, 30).
The Reaction Product Is Co-Purified and Trapped in the Reaction Chamber.
In the course of refining the MtNAS model, an unexplained electron density was observed in the reaction chamber (Fig. 2 A and B and Fig. 3C). Since no substrate, product, or inhibitor was added to the crystallization medium, this molecule must have been co-purified with the protein. Because of the location in the reaction chamber, we attempted to fit it to one molecule of NA, but although the two aminopropyl moieties fit the observed electron density well, the azetidine ring does not. However, a glutamate moiety matches the observed density perfectly (Fig. 2A). We then confirmed the structure of this compound by using noncovalent mass spectrometry (MS) and electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis (Figs. S2 and S3). The bound molecule is therefore composed of three moieties, a glutamate (called Glu1) and two (central and terminal) aminopropyl moieties (called AP2 and AP3, respectively). We propose naming this compound thermoNicotianamine or tNA. This molecule has never been described earlier, and its properties are still unknown. It is most probably able to bind to various divalent metals with high affinity, but this remains to be evaluated. The same holds true for its physiological role in the archaea that is still unknown.
Fig. 2.
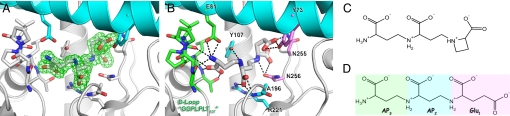
Identification and environment of tNA. (A) Electron density map (Fo-Fc contoured at 2.5σ) around the tNA molecule. The residues involved in the binding of tNA are shown in stick form and colored according to the domain to which they belong (cyan, N-terminal domain; white, C-terminal domain). (B) As in A, hydrogen bonds are shown as dotted lines. Residues stabilizing the AP3, AP2, and Glu1 moieties are colored in green, blue, and magenta, respectively. Note the seven hydrogen bonds involving the terminal aminopropyl moiety (AP3). (C–D) Chemical structures of NA (C) and tNA (D).
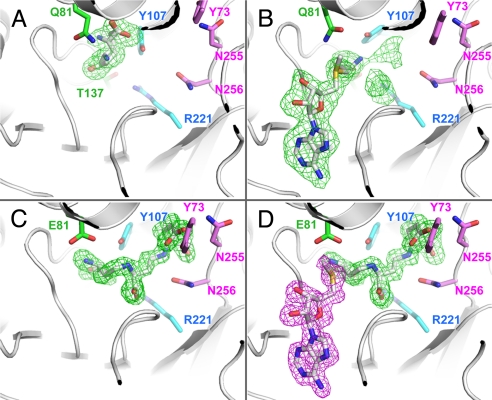
Fig. 3.
Unbiased difference Fourier maps of wild-type and variant MtNAS co-crystallized or soaked with substrate and/or products. All of the maps are contoured at 2.5σ. Residues from MtNAS lining the reaction chamber are color-coded as for Fig. 2B. (A) E81Q-MtNAS co-crystallized in the presence of 5 mM glutamate. The glutamate substrate is in the donor site and adopts two alternative positions. (B) E81Q-MtNAS co-crystallized with 5 mM glutamate and soaked in a solution containing 5 mM SAM. The aminopropyl moiety of the SAM takes the position previously occupied by the glutamate substrate. The glutamate position is however disordered and contributes to the bulky electron density in the reaction chamber. (C) Electron density map of native MtNAS co-purified with tNA. (D) Co-crystallization of native MtNAS with 5 mM MTA (the electron density associated with MTA is colored in magenta).
In the three-dimensional structure of the MtNAS-tNA complex, the tNA molecule is buried in the reaction chamber with only one aminopropyl moiety (AP3) exposed to the solvent (Fig. 1C). A total of 16 hydrogen bonds contribute to the stabilization of the tNA molecule, not including water molecules (Fig. 2B). The residues involved in the binding of tNA belong to both the N- and C-terminal domains, and most of them are conserved in sequence alignments (Fig. S4). It is conceivable that the interaction between the N- and C-terminal domains is not only important for the formation of the reaction chamber but also for the interdomain dynamics, as the mutation in the chloronerva mutant (replacing a hydrophobic phenylalanine residue with a polar serine) is located at the interface of these two domains in a hydrophobic “hotspot” that contributes to the interdomain cohesion (Fig. S5). The AP3 moiety is the most stabilized part of tNA, with numerous hydrogen bonds mainly with residues located in the conserved D-Loop (G131-T137), but also with one hydrogen bond with the side chain of E81 in helix α4 from the N-terminal domain (Fig. 2B). In the middle of the cavity, the amino group of AP2 is bound to the protein via one hydrogen bond with the side chain of Y107, whereas the carboxyl moiety is stabilized by two hydrogen bonds with the main chain of A196 and the side chain of R221 Finally, Glu1 is the most buried part of tNA, and it interacts with the side chains of two residues of the loop connecting β6 to β7 (N255-N256) and one residue from helix α4 (Y73). The chemical similarities between tNA and NA, together with the location in a reaction chamber mostly lined with conserved residues, strongly suggested that tNA had been synthesized by MtNAS in Escherichia coli and that it remained trapped in its active site. Co-purification of an enzyme with its catalytic product is unusual but is corroborated by the strong stabilization of tNA observed in the MtNAS-tNA complex.
tNA Is the Catalytic Product of MtNAS.
From the product-bound complex of MtNAS with tNA, we propose the reaction catalyzed by MtNAS to be: Glu + 2 × SAM → tNA + 2 × 5′-MTA.
This reaction can be compared with the one catalyzed by eukaryotic NAS that uses three SAM molecules to produce NA and three MTA. The chemical similarities between tNA and NA suggest that the azetidine ring is replaced by a glutamate moiety in archaea. This difference in the overall reaction provided us with an opportunity to characterize the successive steps of the reaction (see below). We assayed the reaction catalyzed by MtNAS in vitro using the purified protein (monomer or dimer) and its two substrates, SAM and glutamate. The reaction was followed by ESI-MS, and we observed a slow accumulation of tNA, the rate depending on the temperature (Fig. S2 B and C). The optimal temperature might be above 55 °C (the optimal growth temperature for M. thermoautotrophicus is around 65 °C), but at these higher temperatures, the degradation of SAM prevented tNA formation. At 45 °C, SAM did not degrade, and the enzyme steadily synthesized tNA for at least 1 h. However, although we did observe net tNA synthesis in MS experiments, this corresponds to only a few turnovers of the enzyme over several tens of minutes, even at temperatures close to the optimum growth conditions for M. thermoautotrophicus. Some additional unknown mechanisms are therefore likely to occur in vivo to free the product.
Structure of the Enzyme with Its Second Product.
MTA is another product of the reaction, and the structure of the MtNAS-tNA-MTA complex was solved at 1.66 Å resolution by co-crystallizing the MtNAS protein with MTA. In addition to the tNA molecule, the resulting high-resolution electron density map clearly showed the presence of one MTA molecule bound to the enzyme in the solvent-exposed cleft (Fig. 3D). The nucleoside of MTA is sandwiched between the A-Loop and the loop connecting strand β2 to helix α8. The side chain of E153 stabilizes the ribose through two H-bonds, and the main chain carbonyl of E199 is hydrogen-bonded to the N6 and N7 atoms of the base (Fig. S6). The nucleoside binding site is completed by residues from the D-Loop and the loop connecting β3 to α9 with the side chain of E181 hydrogen bonded to the N6 atom of the base. Most importantly, the sulfur atom of MTA and the C3 atom of tNA, i.e., the two atoms of the bond broken in the last step of the reaction catalyzed by MtNAS, are in Van der Waals contact (3.5 Å; ideal distance would be 3.55 Å for C-S). This supports the view that MTA and tNA are the natural catalytic products of MtNAS.
The Catalytic Product Does Not Co-Purify with the E81Q Mutant Form.
Since MtNAS always co-purified with tNA, the crystal structures of the enzyme either free or in complex with its substrates (glutamate and/or SAM) could not be obtained. We therefore attempted to design a mutant that would allow expression of the apo-enzyme in E. coli. We based our analysis on the aminopropyl transfer mechanism proposed for spermidine and spermine synthase (22, 23, 31). In these mechanisms, the transfer of the aminopropyl group from dcAdoMet to putrescine (or spermidine) was shown to occur via nucleophilic attack of putrescine (or spermidine), a reaction facilitated by its amino group interacting with an acidic residue of the enzyme (D170 for Thermotoga maritima spermidine synthase and D173 and D167 for human spermidine and spermine synthase, respectively). In the structure of MtNAS, the only possible candidate was E81, even though this residue lies in a completely different location when the MtNAS structure is superimposed on either spermidine or spermine synthase structures (Fig. S7). Although the catalytic mechanism of MtNAS turned out to be quite different from both those of spermine and spermidine synthase, the E81Q mutant was nevertheless free of tNA in the reaction chamber as found by solving its structure in the apo form. This was also confirmed by mass spectrometry. The absence of the catalytic product therefore allowed us to try co-crystallizing the E81Q mutant with its substrates (SAM and Glu).
Structures of Enzyme-Substrate Complexes.
We next obtained crystals of the MtNAS-E81Q mutant co-crystallized with glutamate and solved its structure at 1.8 Å resolution. The glutamate is bound at the entrance of the reaction chamber, a position occupied by the AP3 moiety of tNA in the MtNAS-tNA structure (Fig. 3A). This position will be called the donor site hereafter. The electron density map revealed that glutamate adopts two alternative orientations. In one conformation, the side chain of the glutamate is hydrogen-bonded to Y107 OH and its main chain carbonyl is hydrogen-bonded to T137 OH in the D-Loop. In the second conformation, the positions of the two carboxylate groups of the glutamate substrate are exchanged. In both cases an ion (attributed to Na+) contribute to the stabilization of the glutamate. Comparison of the structures of the MtNAS-tNA complex with that of the E81Q-Glu complex reveals two main differences: (i) A small rotation of Y73 around the Cα-Cβ bond and importantly because it is involved in the binding of the glutamate; (ii) a displacement of the Y107 ring (the position of Y107 OH in the two structures differs by 2.4 Å; compare Fig. 3 A and B). The position of the glutamate at the donor site suggests that upon binding of one SAM substrate molecule, it would translocate to the acceptor site for the formation of the AP2-Glu1 reaction intermediate. To test this hypothetical mechanism, we solved the structure of the E81Q mutant co-crystallized with glutamate and SAM. However and contrary to our expectations, we obtained the structure of a ternary complex with bound products (E81Q-MtNAS-tNA-MTA). This means that although this mutant does not co-purify with the catalytic products when expressed in E. coli, it is still able to complete tNA synthesis in the course of the crystallization process. We then tried to trap an intermediate by soaking a crystal of the MtNAS-E81Q mutant protein co-crystallized with glutamate in a solution containing 5 mM SAM. The structure of this complex, solved by molecular replacement and refined to 1.8 Å resolution, differs from the previous one, as the SAM molecule is clearly and entirely visible in the experimental electron density map (Fig. 3B). The base and the ribose moieties of the SAM adopt the same conformation as the one observed for the MTA product in the structure of the MtNAS-tNA-MTA ternary complex. In addition, the aminopropyl moiety of SAM extends through the entrance of the reaction chamber and occupies the glutamate binding site as in the single substrate structure previously described. The carboxylate group of SAM is hydrogen-bonded to T137 OH in the donor site, whereas the amino group is hydrogen-bonded to Y107 OH in the acceptor site. The binding of SAM therefore affects the position of the glutamate substrate, which moves away in the reaction chamber. Its release into the solvent is however prevented by the position of the SAM molecule that “closes the door” of the reaction chamber. The electron density map in this reaction chamber indicates the glutamate substrate is disordered. Indeed, there are a few bulky electron densities localized over all of the carboxylates found in the MtNAS-tNA complex, as if the glutamate adopted a few discrete positions. We believe that this disorder is due to the E81Q mutation, which likely destabilizes the proper binding of the glutamate substrate. As to why the glutamate does not occupy its final position in the back of the reaction chamber, primary amines are not favored at this position mostly because of the presence of a conserved hydrophobic residue (L110) that cannot engage in a hydrogen bond (Fig. S8).
Crystallographic Snapshot of the Catalytic Mechanism.
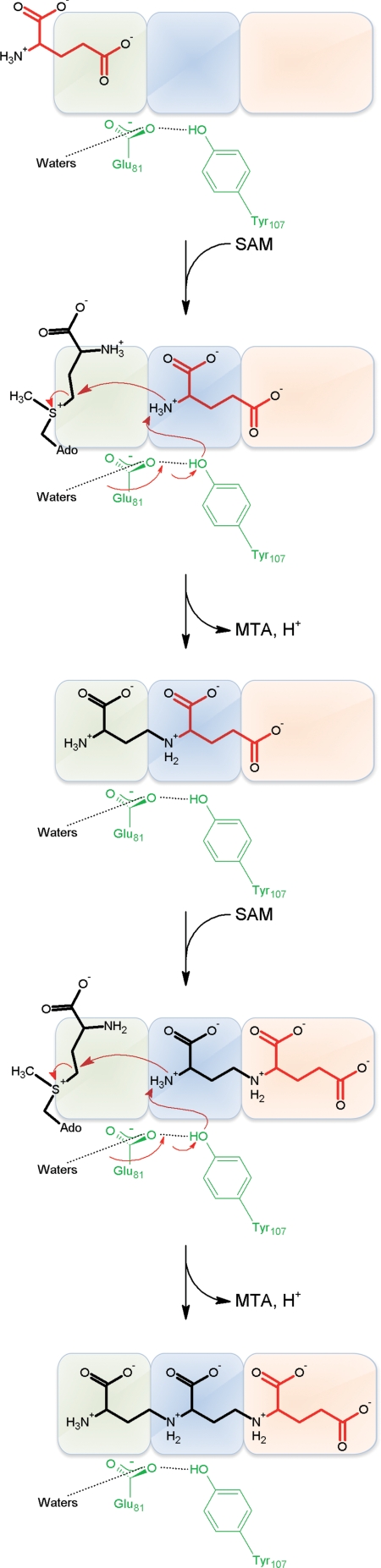
Taken together, the crystal structures of MtNAS bound to its different substrates or products allow us to propose a realistic reaction mechanism for the synthesis of tNA and, by analogy, for the synthesis of NA (Fig. 4). In tNA synthesis, the first step is the binding of the glutamate substrate in the donor site. Upon binding of SAM, the glutamate is translocated to the acceptor site. Deprotonation of the free amino group of glutamate, assisted by the side chains of Y107 and E81, triggers the nucleophilic attack onto the methylene carbon of SAM. The importance of these residues is confirmed by site-directed mutagenesis. Indeed, although mutation of either one of these residues (E81Q or Y107F) does not completely abolish the catalytic activity, the double mutant E81Q/Y107F is completely inactive (Fig. S9). The next steps are the release of one molecule of MTA and the release of one proton. The chain of water molecules connecting the Y107 OH to E81 Oδ1 and to the bulk solvent, clearly visible in the highest resolution crystal structure obtained for the ternary complex between MtNAS-tNA-MTA (Fig. S9), creates a proton wire that permits a rapid release of protons into the solvent (32). In turn, the reaction intermediate that formed, AP2-Glu1, would be translocated deeper into the catalytic chamber to allow the binding of another molecule of SAM in the donor site and the positioning of the free amino group of the AP2 moiety in the acceptor site. A second catalytic reaction then occurs identical to the first one, taking place at the same catalytic site. The release of another proton and a molecule of MTA accompany this process, while a complete molecule of tNA is assembled. With regard to the difference between tNA and NA synthesis, only the first step would differ and would consist in the azetidine ring formation by autocyclization of the aminopropyl moiety of one molecule of SAM. Sequence divergences between archaeal and eukaryotic enzymes are likely responsible for the difference in the reaction catalyzed. From our sequence alignment (Fig. S4), we found that the main difference between the two types of enzymes is in the A-Loop region, and we therefore propose that variation in this region determines the capacity to form the azetidine ring. The two subsequent aminopropyl transfers would then occur similarly in eukaryotic and archaeal enzymes.
Fig. 4.
Proposed reaction mechanism of the synthesis of tNA by MtNAS. The reaction chamber is schematized by three boxes depicting the donor site (green), acceptor site (blue), and the final site of the glutamate moiety of tNA (magenta). See the text for details of the reaction.
Overall, these first crystal structures of a NAS family member represent a major advance in understanding the chemistry used for NA synthesis and provide the structural basis for understanding an unusual polymerization mechanism that takes place in a reaction chamber with ordered substrate binding and sequential substrate and reaction intermediate translocations.
Materials and Methods
MtNAS was cloned, expressed, and purified as described by Dreyfus et al. (25). MtNAS mutants were constructed using the QuikChange II XL Site-Directed Mutagenesis kit from Stratagene following the manufacturer's manual. Crystals of MtNAS-tNA, MtNAS-tNA-MTA, and mutant protein in complex with substrates were obtained from 0.1 mol/L HEPES buffer, pH 7.5, 22% (wt/vol) PEG3350, and 0.4 mol/L NaBr. The structure of the native dimeric protein was solved using selenomethionine-substituted protein and the MAD phasing method. Noncovalent MS measurements were taken with a MicroTOF-Q (Bruker) equipped with an ESI source. MS/MS fragmentation was done directly on the eluting peptide. Wild-type MtNAS (5 μM of the dimer in 20 mM Tris buffer, pH 9.0) and mutants activity were measured by following the production of tNA by mass spectrometry. Please see SI Materials and Methods for more details.
Supplementary Material
Acknowledgments.
We thank Edward L. Bolt (University of Nottingham, Nottingham) for the gift of M. thermoautotrophicus genomic DNA; Dr. J. Lavergne [Commissariat à l'Energie Atomique Cadarache, France] for critical reading of the manuscript; and ID14, ID23, ID-29, and BM-30 staff [European Synchrotron Radiation Facility, Grenoble] for technical assistance in synchrotron data collection. This work was supported by National Agency for Research project HEMOLI, National Agency for Research Grant ANR-07-BLAN-0115-01, and the Commissariat à l'Energie Atomique.
Footnotes
The authors declare no conflicts of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [ID codes 3FPE (MtNAS-tNA), 3FPF (MtNAS-tNA-MTA), 3FPG (E81QMtNAS), 3FPH (E81QMtNAS-Glu), and 3FPJ (E81QMtNAS-SAM-Glu)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0904439106/DCSupplemental.
References
- 1.Curie C, Briat JF. Iron transport and signaling in plants. Annu Rev Plant Biol. 2003;54:183–206. doi: 10.1146/annurev.arplant.54.031902.135018. [DOI] [PubMed] [Google Scholar]
- 2.Hell R, Stephan UW. Iron uptake, trafficking, and homeostasis in plants. Planta. 2003;216:541–551. doi: 10.1007/s00425-002-0920-4. [DOI] [PubMed] [Google Scholar]
- 3.Mori S. Iron acquisition by plants. Curr Opin Plant Biol. 1999;2:250–253. doi: 10.1016/S1369-5266(99)80043-0. [DOI] [PubMed] [Google Scholar]
- 4.Colangelo EP, Guerinot ML. Put the metal to the petal: Metal uptake and transport throughout plants. Curr Opin Plant Biol. 2006;9:322–330. doi: 10.1016/j.pbi.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Curie C, et al. Metal movement within the plant: Contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot (Lond) 2009;103:1–11. doi: 10.1093/aob/mcn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haydon MJ, Cobbett CS. Transporters of ligands for essential metal ions in plants. New Phytol. 2007;174:499–506. doi: 10.1111/j.1469-8137.2007.02051.x. [DOI] [PubMed] [Google Scholar]
- 7.Ling HQ, Koch G, Baumlein H, Ganal MW. Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA. 1999;96:7098–7103. doi: 10.1073/pnas.96.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling HQ, Pich A, Scholz G, Ganal MW. Genetic analysis of two tomato mutants affected in the regulation of iron metabolism. Mol Gen Genet. 1996;252:87–92. doi: 10.1007/BF02173208. [DOI] [PubMed] [Google Scholar]
- 9.Cheng L, et al. Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol. 2007;145:1647–1657. doi: 10.1104/pp.107.107912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi M, et al. Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell. 2003;15:1263–1280. doi: 10.1105/tpc.010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Wiren N, et al. Nicotianamine chelates both FeIII and FeII. Implications for metal transport in plants. Plant Physiol. 1999;119:1107–1114. doi: 10.1104/pp.119.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vacchina V, et al. Speciation of nickel in a hyperaccumulating plant by high-performance liquid chromatography-inductively coupled plasma mass spectrometry and electrospray MS/MS assisted by cloning using yeast complementation. Anal Chem. 2003;75:2740–2745. doi: 10.1021/ac020704m. [DOI] [PubMed] [Google Scholar]
- 13.Pianelli K, Mari S, Marques L, Lebrun M, Czernic P. Nicotianamine over-accumulation confers resistance to nickel in Arabidopsis thaliana. Transgenic Res. 2005;14:739–748. doi: 10.1007/s11248-005-7159-3. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, et al. Increased nicotianamine biosynthesis confers enhanced tolerance of high levels of metals, in particular nickel, to plants. Plant Cell Physiol. 2005;46:1809–1818. doi: 10.1093/pcp/pci196. [DOI] [PubMed] [Google Scholar]
- 15.Koike S, et al. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004;39:415–424. doi: 10.1111/j.1365-313X.2004.02146.x. [DOI] [PubMed] [Google Scholar]
- 16.Weber M, Harada E, Vess C, Roepenack-Lahaye E, Clemens S. Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J. 2004;37:269–281. doi: 10.1046/j.1365-313x.2003.01960.x. [DOI] [PubMed] [Google Scholar]
- 17.Mari S, et al. Root-to-shoot long-distance circulation of nicotianamine and nicotianamine-nickel chelates in the metal hyperaccumulator Thlaspi caerulescens. J Exp Bot. 2006;57:4111–4122. doi: 10.1093/jxb/erl184. [DOI] [PubMed] [Google Scholar]
- 18.Kinoshita E, Yamakoshi J, Kikuchi M. Purification and identification of an angiotensin I-converting enzyme inhibitor from soy sauce. Biosci Biotechnol Biochem. 1993;57:1107–1110. doi: 10.1271/bbb.57.1107. [DOI] [PubMed] [Google Scholar]
- 19.Shimizu E, Hayashi A, Takahashi R, Aoyagi Y, Murakami T, Kimoto K. Effects of angiotensin I-converting enzyme inhibitor from Ashitaba (Angelica keiskei) on blood pressure of spontaneously hypertensive rats. J Nutr Sci Vitaminol (Tokyo) 1999;45:375–383. doi: 10.3177/jnsv.45.375. [DOI] [PubMed] [Google Scholar]
- 20.Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S. Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 1999;119:471–480. doi: 10.1104/pp.119.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbik A, et al. Isolation, characterization, and cDNA cloning of nicotianamine synthase from barley. A key enzyme for iron homeostasis in plants. Eur J Biochem. 1999;265:231–239. doi: 10.1046/j.1432-1327.1999.00717.x. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, et al. Crystal structure of human spermine synthase: Implications of substrate binding and catalytic mechanism. J Biol Chem. 2008;283:16135–16146. doi: 10.1074/jbc.M710323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, et al. Structure and mechanism of spermidine synthases. Biochemistry. 2007;46:8331–8339. doi: 10.1021/bi602498k. [DOI] [PubMed] [Google Scholar]
- 24.Trampczynska A, Bottcher C, Clemens S. The transition metal chelator nicotianamine is synthesized by filamentous fungi. FEBS Lett. 2006;580:3173–3178. doi: 10.1016/j.febslet.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 25.Dreyfus C, Pignol D, Arnoux P. Expression, purification, crystallization and preliminary X-ray analysis of an archaeal protein homologous to plant nicotianamine synthase. Acta Crystallogr F. 2008;64:933–935. doi: 10.1107/S1744309108027796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallick P, Boutz DR, Eisenberg D, Yeates TO. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc Natl Acad Sci USA. 2002;99:9679–9684. doi: 10.1073/pnas.142310499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 28.Kozbial PZ, Mushegian AR. Natural history of S-adenosylmethionine-binding proteins. BMC Struct Biol. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin JL, McMillan FM. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 30.Schluckebier G, O'Gara M, Saenger W, Cheng X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J Mol Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- 31.Korolev S, et al. The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor. Nat Struct Biol. 2002;9:27–31. doi: 10.1038/nsb737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.