Abstract
Plasminogen activator inhibitor type I (PAI-1) is a marker of the fibrinolytic system, and serves as a possible predictor for hepatic metabolic syndromes. Fenofibrate, a peroxisome proliferator-activated receptor α (PPARα) agonist, is a drug used for treatments of hyperlipidemia. Orphan nuclear receptor small heterodimer partner (SHP) plays a key role in transcriptional repression of crucial genes involved in various metabolic pathways. In this study, we show that fenofibrate increased SHP gene expression in cultured liver cells and in the normal and diabetic mouse liver by activating the AMP-activated protein kinase (AMPK) signaling pathway in a peroxisome proliferator-activated receptor α (PPARα)-independent manner. Administration of transforming growth factor β (TGFβ) or a methionine and choline deficient (MCD) diet to induce the progressive fibrosing steatohepatitis model in C57BL/6 mice was significantly reversed by fenofibrate via AMPK-mediated induction of SHP gene expression with a dramatic decrease in PAI-1 mRNA and protein expression along with other fibrotic marker genes. No reversal was observed in SHP null mice treated with fenofibrate. Treatment with another PPARα agonist, WY14643, showed contrasting effects on these marker genes expressions in wild type and SHP null mice, demonstrating the specificity of fenofibrate in activating AMPK signaling. Fenofibrate exhibited a differential inhibitory pattern on PAI-1 gene expression depending on the transcription factors inhibited by SHP.
Conclusion
By demonstrating that a PPARα-independent fenofibrate-AMPK-SHP regulatory cascade can play a key role in PAI-1 gene down-regulation and reversal of fibrosis, our study suggests that various AMPK activators regulating SHP might provide a novel pharmacologic option in ameliorating hepatic metabolic syndromes.
Keywords: SHP, AMPK, PAI-1, Fenofibrate
Introduction
Small heterodimer partner (SHP; NR0B2) is an atypical orphan nuclear receptor lacking the classical DNA binding domain (DBD) but is classified as a nuclear receptor due to the presence of a putative ligand binding domain (LBD) [1]. SHP is predominantly expressed in liver and functions mainly as a transcriptional corepressor of a large variety of nuclear receptors and other transcription factors and regulates key enzyme genes involved in various metabolic processes as well as in ameliorating hepatic fibrosis and insulin sensitivity [2, 3]. Inducers of SHP such as bile acids and AMPK activators have been shown to down-regulate cholesterol 7α-hydroxylase (CYP7A1), the rate-limiting enzyme in cholesterol biosynthesis, and key gluconeogenic genes phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase), respectively, via repression of transcription factors regulating these gene promoters [2-5], thereby highlighting its significance in maintenance of metabolic homeostasis.
Plasminogen activator inhibitor-1 (PAI-1), a serine proteinase inhibitor, is the main physiological inhibitor of the fibrinolytic system regulating intravascular fibrinolysis and thereby controlling thrombus dissolution along with invasion and migration of cells through the extracellular matrix [6-9]. Studies with transgenic mice suggest a major role of PAI-1 in metabolic disturbances such as obesity and insulin resistance and metabolic syndrome [6]. Recent reports suggest a direct correlation between plasma PAI-1 levels and steatosis in human subjects [10]. Various reports demonstrate induction and transcriptional regulation of the PAI-1 gene by cytokines and inflammatory mediators such as endotoxins, interleukin-1 (IL-1), transforming growth factor-β (TGFβ), insulin, tumor necrosis factor α (TNFα), hepatocyte growth factor (HGF), and phorbol 12-myristate 13-acetate (PMA) [11].
Fenofibrate, an agonist of the nuclear receptor peroxisome proliferator-activated receptor α (PPARα), is mainly used to treat hyperlipidemia in patients at risk of cardiovascular disease [12]. Recent studies suggest beneficial effects of fenofibrate on the insulin resistance [13] and in normalizing biochemical abnormalities associated with the metabolic syndromes more effectively when used in combination with metformin [14]. AMP-activated protein kinase (AMPK) is a ubiquitous, heterotrimeric, serine/threonine protein kinase, mediating cellular adaptation to nutritional and environmental variations/stress by acting as a “metabolic switch” in regulating key genes involved in glucose and lipid metabolism [15]. Because of its favorable widespread metabolic effects in the activated state, AMPK stands out to be a tempting therapeutic target in prevention of metabolic syndromes related to hepatic steatosis, insulin resistance and other hepatic disorders.
Here, we elucidate a novel role of fenofibrate in activating the AMPK signaling pathway to induce SHP gene expression and to regulate PAI-1 gene expression. Using in vivo and in vitro studies, we demonstrate that SHP functions as a mediator of fenofibrate inhibition of PAI-1 gene expression and possibly functions as a key negative regulator of liver fibrosis and steatohepatitis, suggesting that targeting SHP with AMPK activators might provide us with a new therapeutic approach for the treatment of metabolic syndromes.
Materials and Methods
Animals and Experimental Protocols
Male C57BL/6J, congenic SHP null mice (B6.129/Sv-Shptm1, described in Supplementary ref. 1) at 12 weeks of age, were injected intraperitoneally with 25 μg/kg of TGFβ or fed with MCD diet for 4 weeks. C57BL/6J and SHP null mice were then divided into 5 groups for each model and fed 1) MCD diet for 3 days, 2) MCD diet with fenofibrate (100 mg/kg/day) for 3 days, 3) MCD diet with WY14643 (50 mg/kg/day) for 3 days, 4) TGFβ for 4 hours and 5) TGFβ with fenofibrate (100 mg/kg/day) for 3 days. Another group was fed the control chow diet for 4 weeks. Male C57BLKS-leptin receptor deficient mice (BKS-Leprdb/db) and PPARα null mice (B6.129/Sv-Pparαtm1Gonz described in Supplementary reference 2) at 8−9 weeks of age were fed with fenofibrate (100 mg/kg/day) for the indicated time period. Animal studies were conducted in accordance with the institutional guidelines for care and use of laboratory animals.
Materials and Methods
Information on the materials and methods used in this study is given in the Supplementary Methods section.
Results
Fenofibrate induces SHP gene expression in hepatic cell lines and rat primary hepatocytes
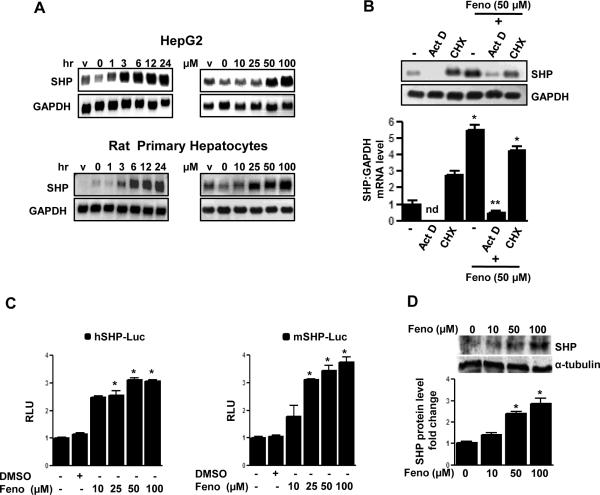
Previous reports suggest repressive effects of fenofibrate [16] and SHP [17] on PAI-1 gene expression. To understand the molecular mechanism of fenofibrate action on PAI-1 gene expression, the effect of fenofibrate on SHP gene expression was assessed. Treatment of various hepatoma cell lines (HepG2 and H4IIE), non-transformed mouse hepatocytes (AML12) and rat primary hepatocytes with fenofibrate (50 μM) for different times (1 to 24 hrs) and different doses of fenofibrate (10−100 μM) for 24 hours caused a significant induction of SHP mRNA expression by approximately 5−10 fold in all hepatic cell lines tested and primary hepatocytes (Fig.1A and Supplementary Fig.1A). In an attempt to determine whether the increase of SHP mRNA level by fenofibrate treatment was due to the increase in transcription or protein synthesis, HepG2 cells were pretreated with the transcription inhibitor, actinomycin D (Act D), alone or preceding fenofibrate treatment resulting in a drastic decrease in SHP mRNA levels (Fig.1B). However, the protein synthesis inhibitor, cycloheximide (CHX), showed no significant effect on fenofibrate-induced SHP mRNA levels, thereby suggesting that fenofibrate induces SHP gene expression at the transcriptional level and does not require de novo protein synthesis. Finally, to determine whether fenofibrate treatment increases SHP gene promoter activity and SHP protein synthesis, transient transfection assays and immunoblotting analysis were performed. Fenofibrate treatment resulted in a 3−4 fold increase in both human and mouse SHP gene promoter activity, in HepG2 cells and AML12 cells, respectively, (Fig. 1C) and the SHP protein level was significantly increased in a dose dependent manner in HepG2 cells (Fig.1D). Taken together, these results indicate that fenofibrate induces SHP gene expression in hepatic cell lines and primary hepatocytes.
Figure 1. Induction of SHP gene expression by fenofibrate.
A: HepG2 cells and rat primary hepatocytes were treated with fenofibrate (50μM) or vehicle (DMSO) for the time indicated and in the concentrations indicated for 24 h. Total RNA was isolated for Northern blot analysis of SHP mRNA expression and was normalized to GAPDH expression. Data represent mean ± SD of 3 individual experiments. B: HepG2 cells were treated with Actinomycin D (Act D) or Cycloheximide (CHX) alone for 1 h or pretreated following fenofibrate treatment for 24 h at the indicated concentration. Total RNA was isolated for Northern blot analysis of SHP mRNA expression and was normalized to GAPDH expression. Data represent mean ± SD of 3 individual experiments. nd: not detectable. *P < 0.05 and **P < 0.01 compared to untreated control and fenofibrate treated cells. C: The wild-type human (h) and mouse (m) SHP-luciferase reporters were transfected into HepG2 and AML12 cells respectively and treated with fenofibrate (Feno, 50 μM) or vehicle (DMSO) for 24 h under serum-starved conditions. All experiments were done in triplicate, and data are expressed in relative luciferase units (RLU) and as the fold activation relative to the control, representing mean ± SD of 3 individual experiments.*P < 0.05 compared to untreated control. D: HepG2 cells were treated with fenofibrate at the indicated concentrations for 6 h and whole cell extracts (50 μg/lane) were analyzed by immunoblotting with SHP and α-tubulin antibodies. Data represent mean ± SD of 3 individual experiments. *P < 0.05 compared to untreated control.
PPARα-independent induction of SHP gene expression by fenofibrate is mediated by the AMPK-signaling pathway
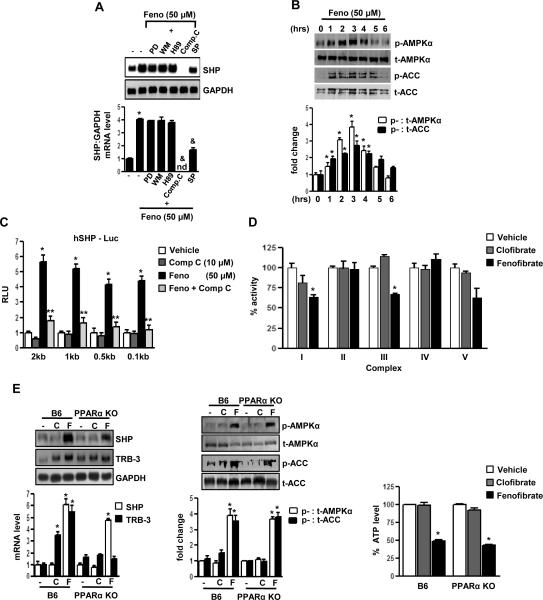
To evaluate the potential signaling pathways involved in the induction of SHP gene expression by fenofibrate, HepG2 cells were pretreated with several specific protein kinase inhibitors followed by fenofibrate treatment for 24 hours. Northern blot analysis indicated that pretreatment of Compound C (Comp C, an AMPK inhibitor) completely abolished fenofibrate-mediated SHP induction and SP600125 (SP, a JNK inhibitor) showed significant repressive effects. However, there was no significant effect of PD98059 (PD, a MAPK inhibitor), Wortmannin (WM, a PI3 kinase inhibitor) and H-89 (H89, a PKA inhibitor) (Fig.2A). To confirm the involvement of the AMPK signaling pathway in fenofibrate-mediated SHP gene regulation, the phosphorylation levels of AMPK and its direct downstream target Acetyl CoA carboxylase (ACC) by fenofibrate was assessed using immunoblotting analysis with antibodies specifically detecting the phosphorylated as well as the total AMPK (t-AMPKα) and ACC (t-ACC) levels in HepG2 cells (Fig.2B). Fenofibrate treatment phosphorylated AMPK (p-AMPKα) and ACC (p-ACC) at 3 hours, followed by a gradual decrease to the basal levels at the 6 hour time point, confirming that fenofibrate activated the AMPK signaling pathway. To elucidate further the mechanism of AMPK-mediated induction of SHP by fenofibrate, transient transfection assays were performed in HepG2 cells using serial deletion construct of the human SHP gene promoter (Fig.2C). Fenofibrate treatment significantly induced SHP promoter activity and pretreatment with Compound C dramatically abolished the fenofibrate-induced SHP promoter activity in all these constructs, suggesting that the fenofibrate and AMPK-responsive factors involved in SHP gene regulation reside within the 100 base pair (bp) region upstream of the SHP gene promoter.
Figure 2. AMPK mediates PPARα-independent induction of SHP gene expression by fenofibrate.
A: HepG2 cells were pretreated with protein kinase inhibitors PD98059 (PD, 20 μM), Wortmannin (WM, 0.1 μM), H-89 (H89, 10 μM), Compound C (Comp C, 10 μM) and SP600125 (SP, 25 μM) for 1 h (left) or with Comp C alone (middle) and then treated with fenofibrate (Feno, 50 μM) for 24 h. Total RNA was isolated for Northern blot analysis of SHP mRNA expression and was normalized to GAPDH expression. HepG2 cells were treated with fenofibrate at the indicated time (right) under serum starved conditions and whole cell extracts (50 μg/lane) were analyzed by immunoblotting with phospho-AMPKα (p-AMPKα), total-AMPKα (t-AMPKα), phospho-ACC (p-ACC) and total-ACC (t-ACC) antibodies. Data represent mean ± SD of 3 individual experiments. *P < 0.05 and &P < 0.005 compared to untreated control and fenofibrate treated cells. nd; not detectable. B: HepG2 cells were treated with fenofibrate under serum-starved conditions for the indicated time period and concentration. Whole cell extracts (50 μg/lane) were analyzed by immunoblotting with phospho-AMPKα (p-AMPKα), total-AMPKα (t-AMPKα), phospho-ACC (p-ACC) and total-ACC (t-ACC) antibodies. Data represent mean ± SD of 3 individual experiments. *P < 0.05 compared to untreated control. C: The human (h) SHP-luciferase reporter constructs were transfected into HepG2 cells and treated with compound C alone for 1 h or pretreated with compound C preceding treatments with fenofibrate or vehicle (DMSO) for 24 h under serum-starved conditions. All experiments were done in triplicate, and data are expressed in relative luciferase units (RLU) and as the fold activation relative to the control, representing mean ± SD of 3 individual experiments.*P < 0.05 and **P < 0.05 compared to vehicle and fenofibrate treated cells. bp: basepair D: Mitochondrial respiratory enzyme activities were performed as described in Materials and Methods. Fenofibrate (50 μM) and clofibrate (50 μM) were mixed with the mitochondrial fraction in each reaction buffers and enzymatic activities were calculated. Data were normalized with respect to the specific activities in corresponding DMSO control and presented as mean ± SD of 3 independent experiments. .*P < 0.05 and **P < 0.001 compared to control. E: C57BL/6 mice (B6, n=3 per group) and PPARα null mice (PPARαKO, n=3 per group) were fed with fenofibrate (100mg/kg/day) or clofibrate (500mg/kg/day) for the 2 days and liver samples were obtained for total RNA isolation for Northern blot analysis of SHP and Trb3 mRNA expression and normalized to GAPDH expression. Tissue extracts (100 μg/lane) were analyzed by immunoblotting with p-AMPKα, t-AMPKα, p-ACC and t-ACC antibodies. Total intracellular ATP was quantified by luminescence according to manufacturer's protocol. Results are expressed as the % decrease in levels of intracellular ATP, setting control liver extract samples as 100, and are normalized to the total protein level. Data represent mean ± SD. *P < 0.005 compared to control.
To elucidate whether fenofibrate lowers cellular ATP levels, the mitochondrial respiratory chain complex (I – V) activity was assessed after fenofibrate and clofibrate treatment of mitochondrial fractions from normal mice liver homogenates (Fig.2D). Clofibrate showed no significant effect on any of the complexes, whereas fenofibrate treatment significantly lowered respiratory chain complex I and III activity and thereby decreased cellular ATP levels. Treatments with other fibrates or synthetic PPARα agonists showed no significant change in SHP gene expression or AMPK activity in HepG2 cells (Supplementary Fig.1B). Fenofibrate alone showed a significant decrease in cellular ATP levels, compared with other PPARα agonists (Supplementary Fig. 1C). However, TRB-3, a PPARα target gene (Supplementary ref.14), was significantly induced by all PPARα agonists, highlighting the specificity of fenofibrate in activating AMPK and inducing SHP gene expression. MTT assay in HepG2 cells demonstrated that fenofibrate or clofibrate have no significant effect on cell viability at these concentrations (Supplementary Fig.1D), as was reported previously [Supplementary ref.13].
Finally, we confirmed the PPARα-independent effect of fenofibrate in vivo, using fenofibrate and clofibrate treatments to wild type (B6) and PPARα null mice (PPARαKO) (Fig.2E). Fenofibrate showed a significant induction of SHP mRNA levels (left) along with activation of AMPK (middle) and a decrease in ATP levels (right) in vivo. Treatment with clofibrate showed no significant change in any of these parameters ascertained. However, TRB-3 mRNA levels were significantly induced upon treatments with either fenofibrate or clofibrate in wild type mice and expectedly, no changes in TRB-3 mRNA levels were observed in PPARα null mice upon these treatments. Collectively, these results suggest that fenofibrate decreases cellular ATP levels via inhibition of mitochondrial respiratory chain complex I and III, consequently activating the AMPK signaling to induce SHP gene expression in a PPARα-independent manner both in vitro and in vivo.
Fenofibrate represses TGFβ-induced PAI-1 gene expression via SHP in vivo
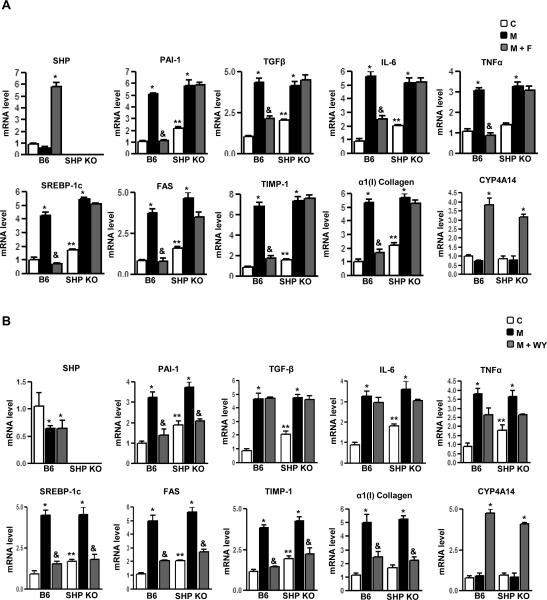
To confirm in vitro observations in the animal model, wild type mice fed with fenofibrate clearly demonstrated a significant induction of SHP mRNA levels from 12 hours to 7 days (Fig.3A left). Immunoblotting analysis of AMPK and ACC activity (Fig.3A middle) from liver extracts of these animals demonstrated a significant increase in phosphorylation of AMPK and ACC on fenofibrate treatment and this pattern of AMPK activation was inversely correlated to the cellular ATP levels which showed significant decrease during fenofibrate treatment (Fig.3A right). Fenofibrate feeding for 3 days showed a similar pattern of increase in SHP gene expression and subsequent decrease in PAI-1 mRNA levels (left), along with a significant decrease in PAI-1 intracellular protein levels (middle) and an increase in AMPK and ACC phosphorylation (right) in the BKS-Leprdb/db (db/db) mice model, a unique model of diabetic dyslipidemia (Supplementary Fig.2A). PAI-1 levels are significantly elevated by transforming growth factor β (TGFβ) treatments [18]. The effect of TGFβ injection (T) in wild type mice and SHP null mice (SHPKO) in the presence or absence of fenofibrate treatments (Fig.3B) was studied. Wild type mice injected with TGFβ for 4 hours showed a dramatic increase in PAI-1 mRNA and protein levels as well as hepatic collagen α1(I) gene expression. The expression of various other marker genes which were previously reported to be elevated in fibrosis conditions was checked. Treatment with fenofibrate for 3 days (T+F) was associated with significant reduction of PAI-1 mRNA and protein levels (Supplementary Fig.2B left) (by 80−90%), collagen α1 (I), TGF-β, IL-6 and TIMP-1 mRNA levels along with a subsequent increase in SHP gene expression (Fig.3B). However, TNFα gene expression showed no significant changes either upon TGFβ injection or with fenofibrate treatments. Interestingly, SHP null mice showed increased basal mRNA levels of all genes involved in fibrosis in comparison to wild type (C) and did not respond to fenofibrate treatments, suggesting the role of SHP in mediating effects of fenofibrate. As expected, AMPK and ACC phosphorylation status is similar in both wild type and SHP null mice (Supplementary Fig.2B). Overall, these results suggest that activation of SHP by fenofibrate may reverse TGFβ-induced hepatic fibrosis directly via repression of PAI-1 and other pro-inflammatory cytokine and profibrogenic marker genes expression.
Figure 3. SHP null mice reverse inhibitory effects of fenofibrate on TGFβ-induced PAI-1 gene expression.
A: C57BL/6 mice (B6, n=4 per group) were fed with fenofibrate (100mg/kg/day) for the indicated time period and liver samples were obtained for total RNA isolation for Northern blot analysis of SHP mRNA expression and normalized to GAPDH expression. Tissue extracts (100 μg/lane) were analyzed by immunoblotting with phospho-AMPKα (p-AMPKα), total-AMPKα (t-AMPKα), phospho-ACC (p-ACC) and total-ACC (t-ACC) antibodies. Total intracellular ATP was quantified by luminescence according to manufacturer's protocol. Results are expressed as the % decrease in levels of intracellular ATP, setting control liver extract samples as 100, and are normalized to the total protein level. Data represent mean ± SD.*P < 0.05 compared to control. B: C57BL/6 mice (B6, n=4 per group) and SHP null mice (SHPKO, n=4 per group) were fed with chow (C) or injected with TGFβ (T, 25 μg/kg) intraperitoneally followed by treatment with fenofibrate (100mg/kg/day) (T+F) for an additional 3 days. Liver samples were obtained for total RNA isolation for semi-quantitative RT-PCR analysis of PAI-1, α(1) I collagen, TNFα, IL-6, TIMP-1, TGFβ and SHP mRNA expression and normalized to actin expression. Data represent mean ± SD. *P < 0.05, &P < 0.005 and **P < 0.005 compared to chow-fed mice, TGFβ fed mice and B6 chow-fed mice.
Fenofibrate represses MCD diet induced PAI-1 gene expression via SHP in vivo
Next, wild type and SHP null mice were treated with a methionine and choline deficient diet (MCD), which causes progressive liver fibrosis, pathologically similar to human metabolic steatohepatitis [19]. Wild type mice fed with MCD diet (M) for 4 weeks showed a dramatic increase in PAI-1 mRNA and protein levels (Supplementary Fig.2C) along with collagen α1 (I) gene expression (Fig. 4A-B). Expression of genes involved in fatty acid synthesis (SREBP-1c and FAS), proinflammatory cytokines (TNFα and IL-6), profibrogenic marker (TGFβ) and TIMP-1 mRNA levels (Fig.4A-B) were significantly increased in MCD diet fed wild type mice. Interestingly, treatment with fenofibrate (M+F) (Fig.4A) showed significant induction of SHP gene expression whereas WY (M+WY, Fig. 4B) had no significant effect on SHP gene expression in wild type mice. Both fenofibrate and WY significantly repressed the PAI-1 mRNA levels in wild type mice but fenofibrate failed to repress PAI-1 in SHP null mice. However, WY repressed PAI-1 gene expression in SHP null mice, suggesting a PPARα-dependent mechanism of this action. Interestingly, fenofibrate significantly repressed expression of proinflammatory cytokine genes (TGFβ, TNFα and IL-6) in wild type mice and this repression was abrogated in SHP null mice. In contrast, WY had no significant effect on these cytokines gene expression in both wild type and SHP null mice. These results suggest a SHP-dependent specific repressive effect of fenofibrate on proinflammatory cytokine gene expression.
Figure 4. SHP null mice reverse inhibitory effects of fenofibrate on MCD diet-induced PAI-1 gene expression.
A-B: C57BL/6 mice (B6, n=4 per group) and SHP null mice (SHPKO, n=4 per group) were fed with chow (C) or with a methionine and choline deficient diet (M) for 4 weeks and compared to MCD diet-fed mice followed by treatment with fenofibrate (100mg/kg/day) (M+F) or WY14643 (50mg/kg/day) (M+WY) for an additional 3 days. Liver samples were obtained for total RNA isolation for semi-quantitative RT-PCR analysis of PAI-1, SREBP-1c, FAS, α(1) I collagen, TNFα, IL-6, TIMP-1, TGFβ, CYP4A14 and SHP mRNA expression and normalized to actin expression. Data represent mean ± SD. *P < 0.05, &P < 0.005 and **P < 0.005 compared to chow-fed mice, MCD-diet fed mice and B6 chow-fed mice. nd, not-detectable.
Furthermore, Wy significantly repressed the expression of genes involved in fatty acid synthesis (SREBP-1c and FAS) along with fibrotic marker genes (collagen α1 (I), TIMP-1) in both wild type and SHP null mice, confirming the previously reported effect of WY on fibrotic marker genes expression [19]. Fenofibrate showed similar repressive effects in wild type mice but failed to repress these genes in SHP null mice. This implicated a predominantly PPARα-dependent action in the case of WY, whereas regulation of these genes expression by fenofibrate occurs in a SHP-dependent manner. Interestingly, both fenofibrate and WY induced expression of PPARα-target gene CYP4A14 (fatty acid ω oxidation marker gene) (Fig.4A-B) in a SHP-independent manner. As expected, AMPK and ACC were activated in a similar manner in both wild type and SHP null mice on fenofibrate treatment (Supplementary Fig.2C). Overall, these results demonstrate that fenofibrate repression on various target genes expression upregulated during the diet-induced progressive fibrosis model (MCD diet) is mediated by the induction of SHP in a PPARα-independent manner, thereby highlighting the significance of the fenofibrate-AMPK-SHP pathway in partial reversal of progressive liver fibrosis.
Fenofibrate differentially regulates stimulatory factor-induced PAI-1 gene expression via AMPK and SHP
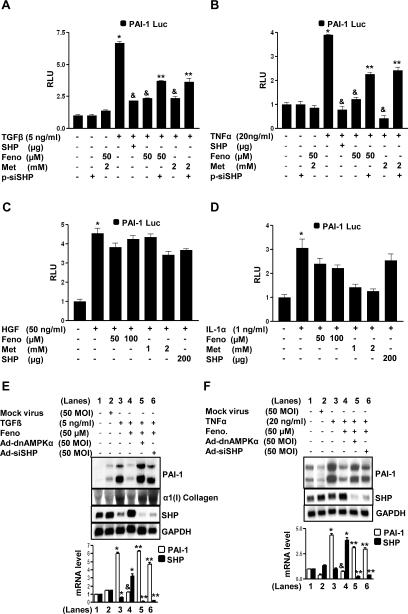
The PAI-1 gene has been reported to be regulated at the transcriptional level by various physiologically important factors [7-11]. To determine the involvement of fenofibrate-mediated induction of SHP gene expression in the regulation of various stimulatory factor-induced PAI-1 gene expression, the effects of TGFβ, tumor necrosis factor α (TNFα), hepatocyte growth factor (HGF) and interleukin-1α (IL-1α) on PAI-1 gene expression in HepG2 cells (Fig.5) was examined. Adenoviral overexpression of SHP (Ad-SHP) or AMPKα (Ad-AMPKα) showed significant repression of TGFβ and TNFα–induced PAI-1 mRNA levels (Fig.5A-B) consistent with dose-dependent fenofibrate treated HepG2 cells (Supplementary Fig.3A-B). However, fenofibrate, Ad-SHP or Ad-AMPKα showed no significant effect on HGF and IL-1α-mediated induction of PAI-1 mRNA (Fig.5C-D). Interestingly, HGF treatments showed a significant increase in SHP mRNA levels (Fig.5C). Similar effects were observed with mouse AML12 cells treated with either TGFβ or HGF (Supplementary Fig.3C-D). These results suggest that fenofibrate differentially inhibits PAI-1 gene expression by repressing specific stimulatory factor effects via AMPK mediated up-regulation of SHP gene expression.
Figure 5. Inhibition of cytokine-induced PAI-1 mRNA levels by fenofibrate is mediated by AMPK-dependent induction of SHP.
A-D: HepG2 cells were treated with mock virus, adenovirus SHP (Ad-SHP) and adenovirus AMPKα (Ad-AMPKα) at indicated concentrations for 24 h after which cells were treated with TGFβ (panel A), TNFα (panel B), HGF (panel C) or IL-1α (panel D) at indicated concentrations for another 4 h under serum starved conditions. Total RNA was isolated for Northern blot analysis of PAI-1 and SHP mRNA expression and was normalized to GAPDH expression. Data represent mean ± SD of 3 individual experiments. *P < 0.05 and &P < 0.005 compared to untreated control and cytokine treated cells.
Fenofibrate represses cytokine-induced PAI-1 promoter activity via SHP
Next, to elucidate the differential regulation of PAI-1 gene expression by fenofibrate via SHP at the promoter level, the effects of TGFβ, TNFα, HGF and IL-1α on human PAI-1 promoter activity in HepG2 cells (Fig.6A-D) was examined. Fenofibrate significantly repressed TGFβ (Fig.6A) and TNFα (Fig.6B) induced PAI-1 promoter (−800bp) activity but consistent with the previous observations (Fig.5C-D), failed to repress HGF (Fig.6C) and IL-1α (Fig.6D) induced activation of the PAI-1 promoter. The repression pattern of fenofibrate was significantly similar to cells cotransfected with the SHP expression vector and treatment with metformin, a known activator of SHP [5], for all four factors tested. Endogenous knockdown of AMPK and SHP expression by adenovirus dominant negative AMPK (Ad-dnAMPKα) or the expression vector for siRNA SHP (p-siSHP) and adenovirus siRNA SHP (Ad-siRNA SHP) significantly reversed the fenofibrate-mediated repression of PAI-1 promoter activity and mRNA levels in the case of TGFβ (Fig.6A, E) and TNFα (Fig.6B, F) treated cells. α1 (I) Collagen was used as a positive control for TGFβ treatment (Fig.6E). These results further confirmed that SHP-mediated repression of PAI-1 gene transcription by fenofibrate is a stimulatory factor-specific (TGFβ and TNFα) phenomena rather than a general inhibition of various other factors (HGF and IL-1α) regulating PAI-1 gene transcription.
Figure 6. Inhibition of cytokine-induced PAI-1 promoter activity by fenofibrate is SHP-dependent.
A-D: HepG2 cells were transfected with the human PAI-1 gene promoter (−800bp) for 18 h followed by treatments with TGFβ (panel A), TNFα (panel B), HGF (panel C) and IL-1α (panel D) at indicated concentrations for 24 h in the presence or absence of SHP (200 μg), pSuper siRNA SHP (p-siSHP, 200 μg), fenofibrate and metformin for another 24 h under serum-starved conditions. All experiments were done in triplicate, and data are expressed in relative luciferase units (RLU) and as the fold activation relative to the control, representing mean ± SD of 3 individual experiments.*P < 0.05, &P < 0.05 and **P < 0.005, compared to untreated control, individual cytokine treatments and fenofibrate or metformin treated cells. E-F: HepG2 cells were treated with mock virus, adenovirus siRNA SHP (Ad-siSHP) and adenovirus dominant negative AMPKα (Ad-dnAMPKα) for 48 h, followed by fenofibrate (Feno) treatment at indicated concentration for an additional 24 h, after which cells were treated with TGFβ (panel E), TNFα (panel F), at indicated concentrations for another 4 h under serum starved conditions. Total RNA was isolated for Northern blot analysis of PAI-1, Collagen type I (Col I) and SHP mRNA expression and was normalized to GAPDH expression. Data represent mean ± SD of 3 individual experiments. *P < 0.05, &P < 0.005 and **P < 0.005 compared to untreated control, cytokine treated cells and fenofibrate treated cells.
Fenofibrate represses transcription factor-mediated activation of PAI-1 promoter via AMPK and SHP
Finally, to elucidate the underlying molecular mechanism of fenofibrate-mediated repression of PAI-1 gene promoter activity, we tried to determine the role of SHP on various transcription factors mediating the effect of TGFβ, TNFα, HGF and IL-1α on PAI-1 promoter transactivation in HepG2 cells. Transient transfection assays demonstrated that cotransfections with Smad7 (iSmad, inhibitor of Smad activity), dominant negative Nur77, dominant negative upstream stimulatory factor-1 (USF-1) and inhibitory kinase of NF-κB (IκBα) significantly repressed the PAI-1 promoter activity by TGFβ, TNFα, HGF and IL-1α respectively (Supplementary Fig.4A). Next, the effect of fenofibrate on these transcription factors and the role played by the SHP and AMPK signaling pathway (Fig.7A-E) was assessed. Fenofibrate treatment or overexpression of constitutively active AMPK (ca-AMPK) and SHP significantly repressed the PAI-1 promoter activity by Smad3 and Smad4 (TGFβ) and Nur77 (TNFα), whereas overexpression of a dominant negative AMPK (dn-AMPK) significantly reversed the inhibitory effects of fenofibrate on the PAI-1 promoter activity (Fig.7A-B). Furthermore, SHP competes with coactivator p300 for Smad3 and Smad4, and with CREB binding protein (CBP) for Nur77 mediated activation of the PAI-1 gene promoter activity (Fig.7C-D). These results are in accordance with in vivo results demonstrating that the inhibitory effect of fenofibrate on fibrotic genes expression in TGFβ-treated mice was mediated by SHP (Fig.3B) as well as treatments with either TGFβ or TNFα in HepG2 cells to induce PAI-1 gene expression was repressed by fenofibrate and knockdown of AMPK activity or endogenous SHP expression significantly reversed this inhibitory effect of fenofibrate (Fig. 6E-F). As expected, fenofibrate or metformin treatments or overexpression of SHP was unable to repress the transcriptional activation of the PAI-1 promoter by either USF-1 (HGF) or NF-κB p65 subunit (IL-1α) (Fig.7E). Fenofibrate also exhibited a similar selective repressive effect on the transcriptional activity of liver receptor homolg-1 (LRH-1), a transcription factor repressed by SHP, but not on steroidogenic factor-1 (SF-1), which is not repressed by SHP, in a multicopy LRH-1/SF-1 binding reporter (Sft4-Luc) (Supplementary Fig.4B-C). Overall, these results demonstrate that inhibition of PAI-1 promoter activity by fenofibrate depends on the nature of transcription factors mediating the stimulatory factor effect, essentially repressing those effects which involve transcription factors that are repressed by SHP via AMPK activation (Fig.7F) and a selective but highly effective role of the fenofibrate-AMPK-SHP cascade in repressing major inflammatory and fibrogenic stimuli related to progressive hepatic fibrosis.
Figure 7. Inhibition of transcription factor-induced PAI-1 promoter activity by fenofibrate is SHP-dependent.
A-E: HepG2 cells were cotransfected with the human PAI-1 gene promoter (−800bp), and Smad3, Smad4 (panel A,C), Nur77 (panel B,D), USF1 (panel E) and NF-kB p65 (panel E) in the presence or absence of cotransfected constitutively active AMPK (ca-AMPK), dn-AMPK (panel A-B, D-E) or with p300 (panel C), CBP (panel D) and SHP (panel A-E) at indicated concentrations for 18 h or p-siSHP for 48hrs and fenofibrate (Feno) treatment for an additional 24 h at indicated concentrations. All experiments were done in triplicate, and data are expressed in relative luciferase units (RLU) and as fold activation relative to the control, representing mean ± SD of 3 individual experiments. *P < 0.05, &P < 0.05, **P < 0.05 and #P < 0.05 compared to untreated control, individual transcription factor cotransfection, fenofibrate treatment and coactivator cotransfection. F: Schematic model depicting differential regulation of hepatic PAI-1 gene expression by fenofibrate in a SHP-dependent manner. Fenofibrate represses hepatic PAI-1 transcriptional activity and expression in animals through AMPK-dependent induction of SHP gene expression. Fenofibrate decreases the cellular ATP/AMP ratio subsequently leading to AMPK activation and induction of SHP gene expression in liver. Consequently, SHP selectively inhibits transcription factors mediating various stimulatory signaling pathways which induces PAI-1 gene expression, and as a result can reverse steatohepatitis and liver fibrosis in animals.
Discussion
The pathogenesis of steatohepatitis, which is correlated with a set of metabolic syndromes such as insulin resistance, diabetes and obesity or non-alcoholic fatty liver disease (NAFLD), and the underlying molecular mechanisms related to the regulation of genes involved in this process are yet to be elucidated fully. In this study, we demonstrate that reversing the PAI-1 gene expression pattern in liver with fenofibrate via SHP in an AMPK-dependent pathway was rapid and effective both in vitro as well as fibrosis- induced in vivo murine models and provides a new insight at the molecular etiology of steatohepatitis and liver fibrosis at a broader level with possible therapeutic significance.
Fenofibrate has beneficial effects in amelioration of metabolic syndromes via down-regulation of multiple target genes involved in the pathogenesis of steatohepatitis and fibrosis [12-14], including PAI-1 gene expression, which is critical for ameliorating fibrosis and steatohepatitis [16]. Reports from our group have suggested previously the inhibitory role of SHP in TGFβ-mediated PAI-1 gene transcription [17]. However, the exact molecular mechanism of antifibrotic effects of fenofibrate remained unclear. Our study demonstrates that fenofibrate induces SHP gene expression both in vitro and in vivo via AMPK signaling. Our results in wild type and PPARα null mice clearly suggest that the induction of SHP by fenofibrate was PPARα-independent. Recent reports demonstrated AMPK activation by fenofibrate in a similar PPARα-independent manner [20]. Our results demonstrate that in comparison to other PPARα agonists, fenofibrate specifically activates AMPK by lowering the cellular ATP levels via inhibition of mitochondrial respiratory complex I and III in PPARα null mice. This result reconfirmed previous reports suggesting inhibition of mitochondrial respiratory chain complexes by fenofibrate and to a significantly lesser extent by clofibrate and the mechanism of inhibition as well as the inhibitory function differed considerably [21]. We previously demonstrated that metformin also activates AMPK and induces SHP expression [5]. Thus, it can be suggested that activation of AMPK by both drugs leads to induction of SHP gene expression, which may be the plausible mechanism underlying the effects.
PAI-1 is a marker of hyperfibrinolysis, and plasma PAI-1 levels are correlated to PAI-1 expression in the liver [22] and are directly correlated to the features observed with insulin resistance and visceral obesity as well as steatohepatitis and liver fibrosis [10]. By inducing severe and progressive liver fibrosis using TGFβ injection in mice, we elucidated the protective role of fenofibrate via SHP. As previously demonstrated [23], we observed auto-induction of TGFβ expression itself upon this treatment as well as an increase in gene expression of α1 (I) collagen IL-6, TIMP-1 and PAI-1 and fenofibrate significantly reversed these TGFβ-mediated effects via SHP. This reversal was completely abolished in SHP null mice, thereby establishing a positive correlation between SHP and its ameliorative effects in liver fibrosis. Furthermore, using the nutritional fibrosis model (MCD diet), we observed that treatment with fenofibrate significantly abrogated mRNA levels of genes involved in fatty acid synthesis (FAS and SREBP-1c), inflammatory response (TNFα and IL-6), inhibitor of fibrosis reversal (TIMP-1) along with PAI-1 and profibrogenic markers (α1 (I) collagen and TGFβ). This inhibitory effect of fenofibrate was significantly abrogated in SHP null mice. However, treatments with another PPARα agonist WY14643 did not show any significant increase of SHP expression in normal mice, consistent with a recent report suggesting no effect of WY on SHP gene expression [24]. The effect of WY treatment on the marker genes reflected a pattern independent of SHP. Surprisingly, WY treatment shows no significant effect on the expression of any proinflammatory genes (TGFβ, TNFα and IL-6) we tested, whereas fenofibrate significantly represses these gene expressions in a SHP-dependent manner. Our results illustrate a significant difference in mechanism of fenofibrate and WY action in ameliorating hepatic fibrosis. A previous study showed a similar role of SHP in ameliorating bile duct ligated fibrosis model in rodents [25]. Importantly, the fenofibrate-AMPK-SHP cascade provides a broader range of inhibitory effect in the development of fibrosis and signifies the antifibrogenic effects of fenofibrate via induction of SHP.
Our current study demonstrates a selective inhibitory effect of fenofibrate via AMPK and SHP on Smad3 and Smad4 [17] and Nur77 [9] mediated transactivation on PAI-1 promoter activity via competition with coactivators p300 and CBP, respectively, but no effect on USF-1 [26] or NF-κB p65 subunit [11] mediated transactivation on PAI-1 promoter (Fig 7E). Interestingly, our previous studies demonstrated an inhibitory effect of SHP on Smad3 and Smad4 [17] and Nur77 transactivation [Supplementary ref. 4] but stimulatory effect on NF-kB p65 in RAW264.7 cells, thus supporting our current observations. However, TGFβ signaling has also been demonstrated to involve transcription factors from the AP-1 family and SHP also inhibits AP-1 transactivity [23, 25], thereby suggesting the possibility of other transcription factors that might be repressed by the fenofibrate-AMPK-SHP signaling cascade. We are currently investigating the mechanism and possible transcription factors involved in AMPK mediated induction of SHP gene expression. The effect of fenofibrate on the AMPK mediated SHP gene expression leads to the possibility that the effects of several other anti-diabetic drugs and adipokines, such as thiazolidinediones, adiponectin and leptin [27] might be mediated by SHP. Currently, we are investigating the effect of several hepatokines on AMPK signaling and SHP gene regulation. Overall this selective inhibitory effect of fenofibrate on PAI-1 gene expression provides a possible molecular mechanism for beneficial effects of fenofibrate in the treatment of hepatic metabolic syndromes in addition to lipid-lowering.
To correlate the possible significance of our results in humans, it is important to consider species differences in the case of PPARα activation in rodents and humans. PPARα is expressed at a much lower level in humans compared to rodents and this is likely to be of functional relevance for fibrate therapy [19]. By demonstrating a PPARα-independent mechanism of fenofibrate action and the predominance of SHP gene expression in mammalian species, our study signifies the importance of SHP in liver-related metabolic syndrome. In summary, the present study provides an in-depth analysis of the molecular etiology underlying the regulation of PAI-1, a key gene involved in various metabolic syndromes. The pharmacologic modulation of SHP gene expression by fenofibrate, metformin and other AMPK activators in the treatment of metabolic syndromes associated to liver is therefore worthy of further research.
Supplementary Material
Supplementary Data
Supplementary Materials and Methods
Reagents, Plasmids and Adenovirus vectors. The reagents fenofibrate, bezafibrate, clofibrate, GW7647, WY14643, Wortmannin and H89 were obtained from Sigma; PD98059, SP600125 and Compound C from Calbiochem; TGFβ, TNFα, HGF and IL-1α from R&D systems; and metformin from Wako Chemicals. Human and mouse SHP promoter luciferase reporter, human PAI-1 promoter luciferase reporter, Sft4 luciferase reporter and PPRE luciferase reporter constructs were described previously [4, 5, 16]. Smad 3, Smad 4, LRH-1, SF-1, PPARα, SHP, Nur77, dn-Nur77, ca-AMPK, dn-AMPK, dn-USF-1, NF-κBp65 subunit, IκBα, p300 and CBP expression vectors were described previously [4, 5, 17 and Supplementary ref. 3−5]. The Smad7 (inhibitor of Smad, iSmad) expression vector was a kind gift from Dr CH Heldin. The USF-1 expression vector was a kind gift from Dr. IK Chung, the pSuper vector and pSuper siSHP were kind gifts from Dr. JK Kemper. Ad-SHP, Ad-AMPKα, Ad-siSHP and Ad-dnAMPKα constructs were described [4, 5].
Cell Culture and transient transfection assays. HepG2, H4IIE and AML12 cells were obtained from the American Type Culture Collection. Maintenance of cell lines and transient transfections were performed as described [4, 5].
RNA isolation and analysis. Total RNA was isolated for Northern blot analysis and RT-PCR analysis using primers for PAI-1, SHP, α(1) I collagen, TNFα, IL-6, TIMP-1, TGFβ, SREBP-1c, FAS and β-actin as described [4, 5, 23, 26 and Supplemetary ref. 6, 7].
Immunoblotting analysis. Cell lysate preparation and immunoblotting analysis were performed as described [4]. Mouse monoclonal anti-PAI-1 antibody and rabbit polyclonal SHP antibody (H-160) was from SantaCruz (Santa Cruz, CA, USA).
Measurement of ATP concentration. ATP concentrations were estimated via luciferase acitivity using ATP bioluminescence assay kit (Roche Applied Bioscience, Switzerland) as described previously [4, Supplementary ref. 12].
Isolation of mitochondria. Healthy C57BL/6 mice were asphyxiated and liver tissues were rinsed twice with ice-cold buffer A (320mM sucrose, 1mM EDTA, 10mM Tris, pH 7.5). Livers were finely minced and homogenized in 4 ml of buffer AT (75mM sucrose, 225mM mannitol, 1mM EGTA, 0.01% BSA, pH 7.4) per gram of liver using a glass-teflon homogenizer. The resultant homogenate was centrifuged at 1,000 × g for 5 min at 4 °C.
The 1,000 × g supernatant was centrifuged twice at 13,000 × g for 10 min [Supplementary ref. 8]. The mitochondria-enriched pellet was used for measuring mitochondrial respiratory chain activity.
Assay of respiratory enzyme activities. To prepare submitochondrial particles, mitochondrial pellets were suspended in buffer AT, then freezen and thawed three times. Mitochondrial protein concentration was measured by using BSA as standard [Supplementary ref. 9]. Complex I activity (NADH CoQ oxidoreductase) was measured in the presence of decylubiquinone as the rotenone-sensitive decrease in NADH at 340nm. The activity of complex II (succinate: DCIP oxireductase) was measured in the presence decylubiquinone plus rotenone as the antimycin A-sensitive reduction of 2,6-DCIP at 600nm with 520nm as reference wavelength. Complex III activity (ubiquinol: cytochrome c oxireductase) was measured in the presence of rotenone and decylubiquinone following the rate of reduction of cytochrome c at 550nm with 580nm as the reference wavelength. Complex IV activity (cytochrome c oxidase) was measured as the disappearance of reduced cytochrome c at 550nm [Supplementary ref. 10−11]. All absorbance measurements were performed in a Beckman DU650 (Beckman coulter fullerton, CA) spectrophotometer.
MTT cell viability assay. Cell viability was evaluated in HepG2 cells as described previously [Supplementary ref. 14].
Statistical analyses. Data are means ± SD. Analysis of variance (ANOVA) was used to determine significant differences, followed by Duncan's multiple comparison tests. All experiments were performed at least three times. Data calculation and statistical analysis were performed using GraphPad Prism 4.0 software. Two-way ANOVA analysis for repeated measures and Student's t-test for unpaired data were used as appropriate to detect any significant differences. Significance was accepted at the P < 0.05 level.
Supplementary Figure Legends
Supplementary Figure 1. Induction of SHP and activation of AMPK by fenofibrate.
A: H4IIE and AML12 cells were treated with fenofibrate (50μM) or vehicle (DMSO) for the time indicated and in the concentrations indicated for 24 h. Total RNA was isolated for Northern blot analysis of SHP mRNA expression and was normalized to GAPDH expression. Data represent mean ± SD of 3 individual experiments. B: HepG2 cells were treated with various PPARα agonists, bezafibrate (Beza, 100 μM), clofibrate (Clo, 250 μM), WY14643 (WY, 100 μM) and fenofibrate (Feno, 50 μM) for 12 h (left) or for 3 h (right) under serum-starved conditions. Total RNA was isolated for Northern blot analysis of SHP and Trb3 mRNA expression and was normalized to GAPDH expression or whole cell extracts (50 μg/lane) were analyzed by immunoblotting with phospho-AMPKα (p-AMPKα), total-AMPKα (t-AMPKα), phospho-ACC (p-ACC) and total-ACC (t-ACC) antibodies. Data represent mean ± SD of 3 individual experiments. *P < 0.05 compared to untreated control. C: HepG2 cells were treated with fenofibrate (50μM) for the indicated time and at indicated concentrations for 3 h and with various PPARα agonists for 3 h under serum-starved conditions. Total intracellular ATP was quantified by luminescence according to manufacturer's protocol. Results are expressed as the % decrease in levels of intracellular ATP, setting untreated cells as 100, and are normalized to the total protein level. Data represent mean ± SD of 3 individual experiments. *P < 0.05 compared to untreated cells. D: HepG2 cells were treated with fenofibrate or clofibrate at indicated concentrations for 24 h and MTT assay was performed according to the manufacturer's protocol to determine cell viability. Data represent mean ± SD of 3 individual experiments.
Supplementary Figure 2. Inhibition of PAI-1 gene expression and activation of AMPK signaling by fenofibrate in vivo.
A: C57BLKS-Leptin receptor deficient db/db mice (BKS-Leprdb/db, n=4 per group) were fed with fenofibrate (100mg/kg/day) for the indicated time period and liver samples were obtained for semiquantitative RT-PCR analysis of PAI-1 and SHP mRNA expression and was normalized to actin expression (left) and tissue extracts (100 μg/lane) were analyzed by immunoblotting with PAI-1 and α-tubulin antibodies (middle) or with phospho-AMPKα (p-AMPKα), total-AMPKα (t-AMPKα), phospho-ACC (p-ACC) and total-ACC (t-ACC) antibodies (right). Data represent mean ± SD.*P < 0.05 compared to untreated control. B-C: C57BL/6 mice (B6, n=4 per group) and SHP null mice (SHPKO, n=4 per group) were fed with chow (C) or injected with TGFβ (T, 25 μg/kg) intraperitoneally followed by treatment with fenofibrate (100mg/kg/day) (T+F) for an additional 3 days (panel B) or a methionine and choline deficient diet (M) for 4 weeks and compared to diet-fed mice treated with fenofibrate (100mg/kg/day) (M+F) for the final 3 days of the experimental period (panel C). Liver tissue extracts (100 μg/lane) were analyzed by immunoblotting with PAI-1, α-tubulin, phospho-AMPKα (p-AMPKα), total-AMPKα (t-AMPKα), phospho-ACC (p-ACC) and total-ACC (t-ACC) antibodies. Data represent mean ± SD. *P < 0.05 compared to chow-fed mice.
Supplementary Figure 3. Inhibition of cytokine-induced PAI-1 mRNA levels by fenofibrate is mediated by AMPK-dependent induction of SHP in AML12 cells.
A-B: HepG2 cells were treated with fenofibrate (Feno) at indicated concentrations for 24 h after which cells were treated with TGFβ (panel A) and TNFα (panel B) at indicated concentrations for further 4 h under serum starved conditions. Total RNA was isolated for Northern blot analysis of PAI-1 and SHP mRNA expression and was normalized to GAPDH expression. Data represent mean ± SD of 3 individual experiments. *P < 0.05 and &P < 0.005 compared to untreated control and cytokine treated cells. nd, non-detectable. C-D: AML12 cells were treated with mock virus, adenovirus siRNA SHP (Ad-siSHP) or adenovirus dominant negative AMPKα (Ad-dnAMPKα) for 48 h, followed by fenofibrate (Feno) treatment at indicated concentrations for an additional 24 h, after which cells were treated with TGFβ (panel A) or HGF (panel B) at indicated concentrations for further 4 h under serum starved conditions. Total RNA was isolated for semiquantitative RT-PCR analysis of PAI-1 and SHP mRNA expression and was normalized to actin expression. Data represent mean ± SD of 3 individual experiments. *P < 0.05, &P < 0.005 and **P < 0.005, compared to untreated control, cytokine treatment and fenofibrate treated cells.
Supplementary Figure 4. Repression of LRH-1 but not SF-1 transcriptional activity by fenofibrate. A: HepG2 cells were transfected with the human PAI-1 gene promoter (−800bp) for 18 h followed by treatments with TGFβ, TNFα, HGF and IL-1α at indicated concentrations for 24 h in the presence or absence of cotransfected Smad7 (200 μg), dominant negative Nur77 (dn-Nur77, 200 μg), dominant negative USF1 (dn-USF1, 200 μg) or IκB respectively under serum-starved conditions. All experiments were done in triplicate, and data are expressed in relative luciferase units (RLU) and as the fold activation relative to the control, representing mean ± SD of 3 individual experiments.*P < 0.05, &P < 0.05, compared to untreated control and individual cytokine treatments. B: HepG2 cells were transfected with pSuper vector (pSuper only, 200μg) or pSuper siRNA SHP (p-siSHP, (200μg) for 48 h. Total RNA was isolated for semiquantitative RT-PCR analysis of SHP mRNA expression and was normalized to actin expression. Data represent mean ± SD of 3 individual experiments. .*P < 0.05 compared to untreated control and pSuper only treated cells. C: HepG2 cells were cotransfected with Sft4-luc along with LRH-1 (200 μg) or SF-1 (200 μg) in the presence or absence of SHP (200 μg), pSuper siRNA SHP (p-siSHP, 200 μg) for 18 h followed by fenofibrate and metformin treatments as indicated for a further 24 h under serum-starved conditions. All experiments were done in triplicate, and data are expressed in relative luciferase units (RLU) and as the fold activation relative to the control, representing mean ± SD of 3 individual experiments. *P < 0.05, &P < 0.05 and **P < 0.005, compared to untreated control, individual transcription factor cotransfection and fenofibrate or metformin treatment.
Supplementary References
1. Park YJ, Qatanani M, Chua SS, LaRey JL, Johnson SA, Watanabe M, Moore DD, Lee YK. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology. 2008; 47(5):1578−86.
2. Kim BH, Won YS, Kim EY, Yoon M, Nam KT, Oh GT, Kim DY. Phenotype of peroxisome proliferator-activated receptor-alpha (PPARalpha)deficient mice on mixed background fed high fat diet. J Vet Sci. 2003; 4(3):239−44.
3. Kim JH, Kim K, Jin HM, Youn BU, Song I, Choi HS, Kim N. Upstream stimulatory factors regulate OSCAR gene expression in RANKL-mediated osteoclast differentiation. J Mol Biol. 2008;383(3):502−11.
4. Yeo MG, Yoo YG, Choi HS, Pak YK, Lee MO. Negative cross-talk between Nur77 and small heterodimer partner and its role in apoptotic cell death of hepatoma cells. Mol Endocrinol. 2005; 19(4): 950−63.
5. Kim YS, Han CY, Kim SW, Kim JH, Lee SK, Jung DJ, et al. The orphan nuclear receptor small heterodimer partner as a novel coregulator of nuclear factor-kappa b in oxidized low density lipoprotein-treated macrophage cell line RAW 264.7. J Biol Chem. 2001; 276(36): 33736−40.
6. Bergheim I, Guo L, Davis MA, Lambert JC, Beier JI, Duveau I, et al. Metformin prevents alcohol-induced liver injury in the mouse: Critical role of plasminogen activator inhibitor-1. Gastroenterology 2006; 130(7): 2099−112.
7. Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, et.al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004; 113: 1408−18.
8. Fernandez-Vizarra E, Fernandez-Silva P, Enriquez JA. Isolation of Mitochondria from Mammalian Tissues and Cultured Cells. Cell Biology- A laboratory handbook, 3rd edition (2), 69−77.
9. Lowry OH, Rosenbrough NJ, Farr A L, Randall RJ. Protein measurement with Folin phenol reagent. J Biol Chem. 1951; 193: 265−275.
10. Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods Cell Biology 2007; 80: 93−119.
11. López LC, Escames G, Tapias V, Utrilla P, León J, Acuña-Castroviejo D. Identification of an inducible nitric oxide synthase in diaphragm mitochondria from septic mice: its relation with mitochondrial dysfunction and prevention by melatonin. Int J Biochem Cell Biol. 2006; 38(2): 267−78.
12. Yu W, Gong JS, Ko M, Garver WS, Yanagisawa K, Michikawa M. Altered cholesterol metabolism in Niemann-Pick type C1 mouse brains affects mitochondrial function. J Biol Chem. 2005; 280(12):11731−9.
13. Jiao HL, Zhao BL. Cytotoxic effect of peroxisome proliferator fenofibrate on human HepG2 hepatoma cell line and relevant mechanisms. Toxicol Appl Pharmacol. 2002; 185(3):172−9.
14. Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004; 10(5):530−4.
Acknowledgements
We would like to thank Dr. David D. Moore (Baylor College of Medicine, Houston, TX) for permitting us to use SHP null mice and Dr. Keun-Gyu Park (Keimyung University School of Medicine, Daegu, South Korea) and Dr. Young-Joo Park (Seoul National University College of Medicine, Seongnam, Republic of Korea) for helpful discussions.
Financial Support: This work was supported by the KOSEF through the National Research Laboratory program (NRL-ROA-2005-000-10047-0) and by the KRF (2006-005-J03003) (HSC). This research was also supported by NIH grants DK44442 and DK58379 (JYLC) and KOSEF (M10753020001-07N5302-00110) (MS).
Abbreviations
- SHP
Small heterodimer partner
- PAI-1
Plasminogen activator inhibitor 1
- TGFβ
Transforming growth factor β
- TNFα
Tumor necrosis factor α
- HGF
Hepatocyte growth factor
- IL-1α
Interleukin-1α
- AMPK
AMP-activated protein kinase
- USF-1
Upstream stimulatory factor 1
- NF-κB
Nuclear factor kappa-B
Footnotes
Conflict of Interest: No conflict of interest.
References
- 1.Seol W, Choi HS, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 2.Chanda D, Park JH, Choi HS. Molecular basis of endocrine regulation by orphan nuclear receptor small heterodimer partner. Endocr J. 2008;55:253–268. doi: 10.1507/endocrj.k07e-103. [DOI] [PubMed] [Google Scholar]
- 3.Lee YS, Chanda D, Sim J, Park YY, Choi HS. Structure and function of the atypical orphan nuclear receptor small heterodimer partner. Int Rev Cytol. 2007;261:117–158. doi: 10.1016/S0074-7696(07)61003-1. [DOI] [PubMed] [Google Scholar]
- 4.Chanda D, Kim SJ, Lee IK, Shong M, Choi HS. Sodium arsenite induces orphan nuclear receptor SHP gene expression via AMP-activated protein kinase to inhibit gluconeogenic enzyme gene expression. Am J Physiol Endocrinol Metab. 2008;295(2):E368–79. doi: 10.1152/ajpendo.00800.2007. [DOI] [PubMed] [Google Scholar]
- 5.Kim YD, Park KG, Lee YS, Park YY, Kim DK, Nedumaran B, et al. Metformin inhibits hepatic gluconeogenesis through AMP-activated protein kinase dependent regulation of the orphan nuclear receptor SHP. Diabetes. 2007;57:306–314. doi: 10.2337/db07-0381. [DOI] [PubMed] [Google Scholar]
- 6.Loskutoff DJ, Quigley JP. PAI-1, fibrosis, and the elusive provisional fibrin matrix. J Clin Invest. 2000;106(12):1441–3. doi: 10.1172/JCI11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lijnen HR. Pleiotropic functions of plasminogen activator inhibitor-1. J Thromb Haemost. 2005;3:35–45. doi: 10.1111/j.1538-7836.2004.00827.x. [DOI] [PubMed] [Google Scholar]
- 8.Targher G, Bertolini L, Scala L, Zenari L, Lippi G, Franchini M, Arcaro G. Plasma PAI-1 levels are increased in patients with nonalcoholic steatohepatitis. Diabetes Care. 2007;30(5):e31–2. doi: 10.2337/dc07-0109. [DOI] [PubMed] [Google Scholar]
- 9.Gruber F, Hufnagl P, Hofer-Warbinek R, Schmid JA, Breuss JM, Huber-Beckmann R, et al. Direct binding of Nur77/NAK-1 to the plasminogen activator inhibitor 1 (PAI-1) promoter regulates TNF alpha -induced PAI-1 expression. Blood. 2003;101(8):3042–8. doi: 10.1182/blood-2002-07-2331. [DOI] [PubMed] [Google Scholar]
- 10.Van Gaal LF, Mertens IL, De Block CE. Mechanisms linking obesity with cardiovascular disease. Nature. 2006;14(7121):875–80. doi: 10.1038/nature05487. 444. [DOI] [PubMed] [Google Scholar]
- 11.Dawson SJ, Wiman B, Hamsten A, Green F, Humphries S, Henney AM. The two allele sequences of a common polymorphism in the promoter of the plasminogen activator inhibitor-1 (PAI-1) gene respond differently to interleukin-1 in HepG2 cells. J Biol Chem. 1993;268(15):10739–45. [PubMed] [Google Scholar]
- 12.Zambon A, Cusi K. The role of fenofibrate in clinical practice. Diab Vasc Dis Res. 2007;(Suppl 3):S15–20. doi: 10.3132/dvdr.2007.053. [DOI] [PubMed] [Google Scholar]
- 13.FIELD study investigators Effects of long-term fenofibrate therapy on cardiovascular events in 9795 people with type 2 diabetes mellitus (the FIELD study): randomised controlled trial. Lancet. 2005;366(9500):1849–61. doi: 10.1016/S0140-6736(05)67667-2. [DOI] [PubMed] [Google Scholar]
- 14.Nieuwdorp M, Stroes ES, Kastelein JJ. Normalization of metabolic syndrome using fenofibrate, metformin or their combination. Diabetes Obes Metab. 2007;6:869–78. doi: 10.1111/j.1463-1326.2006.00668.x. [DOI] [PubMed] [Google Scholar]
- 15.Hardie DG. The AMP-activated protein kinase pathway—new players upstream and downstream. J Cell Sci. 2004;117:5479–5487. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- 16.Chen LL, Zhang JY, Wang BP. Renoprotective effects of fenofibrate in diabetic rats are achieved by suppressing kidney plasminogen activator inhibitor-1. Vascul Pharmacol. 2006;44(5):309–15. doi: 10.1016/j.vph.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Suh JH, Huang J, Park YY, Seong HA, Kim D, Shong M, et al. Orphan nuclear receptor small heterodimer partner inhibits transforming growth factor-beta signaling by repressing SMAD3 transactivation. J Biol Chem. 2006;281(51):39169–78. doi: 10.1074/jbc.M605947200. [DOI] [PubMed] [Google Scholar]
- 18.Gauldie J, Bonniaud P, Sime P, Ask K, Kolb M. TGF-beta, Smad3 and the process of progressive fibrosis. Biochem Soc Trans. 2007;35:661–4. doi: 10.1042/BST0350661. [DOI] [PubMed] [Google Scholar]
- 19.Ip E, Farrell G, Hall P, Robertson G, Leclercq I. Administration of the potent PPARalpha agonist, Wy-14,643, reverses nutritional fibrosis and steatohepatitis in mice. Hepatology. 2004;39(5):1286–96. doi: 10.1002/hep.20170. [DOI] [PubMed] [Google Scholar]
- 20.Murakami H, Murakami R, Kambe F, Cao X, Takahashi R, Asai T, et al. Fenofibrate activates AMPK and increases eNOS phosphorylation in HUVEC. Biochem Biophys Res Commun. 2006;341(4):973–8. doi: 10.1016/j.bbrc.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 21.Nadanaciva S, Dykens JA, Bernal A, Capaldi RA, Will Y. Mitochondrial impairment by PPAR agonists and statins identified via immunocaptured OXPHOS complex activities and respiration. Toxicol Appl Pharmacol. 2007;223(3):277–87. doi: 10.1016/j.taap.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Alessi MC, Bastelica D, Mavri A, Morange P, Berthet B, Grino M, Juhan-Vague I. Plasma PAI-1 levels are more strongly related to liver steatosis than to adipose tissue accumulation. Arterioscler Thromb Vasc Biol. 2003;23(7):1262–8. doi: 10.1161/01.ATV.0000077401.36885.BB. [DOI] [PubMed] [Google Scholar]
- 23.Kim SJ, Angel P, Lafyatis R, Hattori K, Kim KY, Sporn MB, et al. Autoinduction of transforming growth factor beta 1 is mediated by the AP-1 complex. Mol Cell Biol. 1990;10(4):1492–7. doi: 10.1128/mcb.10.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gyamfi MA, Wan YJ. Mechanisms of Resistance of Hepatocyte Retinoid X Receptor a-null Mice to WY-14,643-induced Hepatocyte Proliferation and Cholestasis. J Biol Chem. 2009 Jan 27; doi: 10.1074/jbc.M808861200. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiorucci S, Antonelli E, Rizzo G, Renga B, Mencarelli A, Riccardi L, et al. The nuclear receptor SHP mediates inhibition of hepatic stellate cells by FXR and protects against liver fibrosis. Gastroenterology. 2004;127(5):1497–512. doi: 10.1053/j.gastro.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Imagawa S, Fujii S, Dong J, Furumoto T, Kaneko T, Zaman T, et al. Hepatocyte growth factor regulates E box-dependent plasminogen activator inhibitor type 1 gene expression in HepG2 liver cells. Arterioscler Thromb Vasc Biol. 2006;26(10):2407–13. doi: 10.1161/01.ATV.0000240318.61359.e3. [DOI] [PubMed] [Google Scholar]
- 27.Kola B, Boscaro M, Rutter GA, Grossman AB, Korbonits M. Expanding role of AMPK in endocrinology. Trends Endocrinol Metab. 2006;17(5):205–15. doi: 10.1016/j.tem.2006.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Materials and Methods
Reagents, Plasmids and Adenovirus vectors. The reagents fenofibrate, bezafibrate, clofibrate, GW7647, WY14643, Wortmannin and H89 were obtained from Sigma; PD98059, SP600125 and Compound C from Calbiochem; TGFβ, TNFα, HGF and IL-1α from R&D systems; and metformin from Wako Chemicals. Human and mouse SHP promoter luciferase reporter, human PAI-1 promoter luciferase reporter, Sft4 luciferase reporter and PPRE luciferase reporter constructs were described previously [4, 5, 16]. Smad 3, Smad 4, LRH-1, SF-1, PPARα, SHP, Nur77, dn-Nur77, ca-AMPK, dn-AMPK, dn-USF-1, NF-κBp65 subunit, IκBα, p300 and CBP expression vectors were described previously [4, 5, 17 and Supplementary ref. 3−5]. The Smad7 (inhibitor of Smad, iSmad) expression vector was a kind gift from Dr CH Heldin. The USF-1 expression vector was a kind gift from Dr. IK Chung, the pSuper vector and pSuper siSHP were kind gifts from Dr. JK Kemper. Ad-SHP, Ad-AMPKα, Ad-siSHP and Ad-dnAMPKα constructs were described [4, 5].
Cell Culture and transient transfection assays. HepG2, H4IIE and AML12 cells were obtained from the American Type Culture Collection. Maintenance of cell lines and transient transfections were performed as described [4, 5].
RNA isolation and analysis. Total RNA was isolated for Northern blot analysis and RT-PCR analysis using primers for PAI-1, SHP, α(1) I collagen, TNFα, IL-6, TIMP-1, TGFβ, SREBP-1c, FAS and β-actin as described [4, 5, 23, 26 and Supplemetary ref. 6, 7].
Immunoblotting analysis. Cell lysate preparation and immunoblotting analysis were performed as described [4]. Mouse monoclonal anti-PAI-1 antibody and rabbit polyclonal SHP antibody (H-160) was from SantaCruz (Santa Cruz, CA, USA).
Measurement of ATP concentration. ATP concentrations were estimated via luciferase acitivity using ATP bioluminescence assay kit (Roche Applied Bioscience, Switzerland) as described previously [4, Supplementary ref. 12].
Isolation of mitochondria. Healthy C57BL/6 mice were asphyxiated and liver tissues were rinsed twice with ice-cold buffer A (320mM sucrose, 1mM EDTA, 10mM Tris, pH 7.5). Livers were finely minced and homogenized in 4 ml of buffer AT (75mM sucrose, 225mM mannitol, 1mM EGTA, 0.01% BSA, pH 7.4) per gram of liver using a glass-teflon homogenizer. The resultant homogenate was centrifuged at 1,000 × g for 5 min at 4 °C.
The 1,000 × g supernatant was centrifuged twice at 13,000 × g for 10 min [Supplementary ref. 8]. The mitochondria-enriched pellet was used for measuring mitochondrial respiratory chain activity.
Assay of respiratory enzyme activities. To prepare submitochondrial particles, mitochondrial pellets were suspended in buffer AT, then freezen and thawed three times. Mitochondrial protein concentration was measured by using BSA as standard [Supplementary ref. 9]. Complex I activity (NADH CoQ oxidoreductase) was measured in the presence of decylubiquinone as the rotenone-sensitive decrease in NADH at 340nm. The activity of complex II (succinate: DCIP oxireductase) was measured in the presence decylubiquinone plus rotenone as the antimycin A-sensitive reduction of 2,6-DCIP at 600nm with 520nm as reference wavelength. Complex III activity (ubiquinol: cytochrome c oxireductase) was measured in the presence of rotenone and decylubiquinone following the rate of reduction of cytochrome c at 550nm with 580nm as the reference wavelength. Complex IV activity (cytochrome c oxidase) was measured as the disappearance of reduced cytochrome c at 550nm [Supplementary ref. 10−11]. All absorbance measurements were performed in a Beckman DU650 (Beckman coulter fullerton, CA) spectrophotometer.
MTT cell viability assay. Cell viability was evaluated in HepG2 cells as described previously [Supplementary ref. 14].
Statistical analyses. Data are means ± SD. Analysis of variance (ANOVA) was used to determine significant differences, followed by Duncan's multiple comparison tests. All experiments were performed at least three times. Data calculation and statistical analysis were performed using GraphPad Prism 4.0 software. Two-way ANOVA analysis for repeated measures and Student's t-test for unpaired data were used as appropriate to detect any significant differences. Significance was accepted at the P < 0.05 level.
Supplementary Figure Legends
Supplementary Figure 1. Induction of SHP and activation of AMPK by fenofibrate.
A: H4IIE and AML12 cells were treated with fenofibrate (50μM) or vehicle (DMSO) for the time indicated and in the concentrations indicated for 24 h. Total RNA was isolated for Northern blot analysis of SHP mRNA expression and was normalized to GAPDH expression. Data represent mean ± SD of 3 individual experiments. B: HepG2 cells were treated with various PPARα agonists, bezafibrate (Beza, 100 μM), clofibrate (Clo, 250 μM), WY14643 (WY, 100 μM) and fenofibrate (Feno, 50 μM) for 12 h (left) or for 3 h (right) under serum-starved conditions. Total RNA was isolated for Northern blot analysis of SHP and Trb3 mRNA expression and was normalized to GAPDH expression or whole cell extracts (50 μg/lane) were analyzed by immunoblotting with phospho-AMPKα (p-AMPKα), total-AMPKα (t-AMPKα), phospho-ACC (p-ACC) and total-ACC (t-ACC) antibodies. Data represent mean ± SD of 3 individual experiments. *P < 0.05 compared to untreated control. C: HepG2 cells were treated with fenofibrate (50μM) for the indicated time and at indicated concentrations for 3 h and with various PPARα agonists for 3 h under serum-starved conditions. Total intracellular ATP was quantified by luminescence according to manufacturer's protocol. Results are expressed as the % decrease in levels of intracellular ATP, setting untreated cells as 100, and are normalized to the total protein level. Data represent mean ± SD of 3 individual experiments. *P < 0.05 compared to untreated cells. D: HepG2 cells were treated with fenofibrate or clofibrate at indicated concentrations for 24 h and MTT assay was performed according to the manufacturer's protocol to determine cell viability. Data represent mean ± SD of 3 individual experiments.
Supplementary Figure 2. Inhibition of PAI-1 gene expression and activation of AMPK signaling by fenofibrate in vivo.
A: C57BLKS-Leptin receptor deficient db/db mice (BKS-Leprdb/db, n=4 per group) were fed with fenofibrate (100mg/kg/day) for the indicated time period and liver samples were obtained for semiquantitative RT-PCR analysis of PAI-1 and SHP mRNA expression and was normalized to actin expression (left) and tissue extracts (100 μg/lane) were analyzed by immunoblotting with PAI-1 and α-tubulin antibodies (middle) or with phospho-AMPKα (p-AMPKα), total-AMPKα (t-AMPKα), phospho-ACC (p-ACC) and total-ACC (t-ACC) antibodies (right). Data represent mean ± SD.*P < 0.05 compared to untreated control. B-C: C57BL/6 mice (B6, n=4 per group) and SHP null mice (SHPKO, n=4 per group) were fed with chow (C) or injected with TGFβ (T, 25 μg/kg) intraperitoneally followed by treatment with fenofibrate (100mg/kg/day) (T+F) for an additional 3 days (panel B) or a methionine and choline deficient diet (M) for 4 weeks and compared to diet-fed mice treated with fenofibrate (100mg/kg/day) (M+F) for the final 3 days of the experimental period (panel C). Liver tissue extracts (100 μg/lane) were analyzed by immunoblotting with PAI-1, α-tubulin, phospho-AMPKα (p-AMPKα), total-AMPKα (t-AMPKα), phospho-ACC (p-ACC) and total-ACC (t-ACC) antibodies. Data represent mean ± SD. *P < 0.05 compared to chow-fed mice.
Supplementary Figure 3. Inhibition of cytokine-induced PAI-1 mRNA levels by fenofibrate is mediated by AMPK-dependent induction of SHP in AML12 cells.
A-B: HepG2 cells were treated with fenofibrate (Feno) at indicated concentrations for 24 h after which cells were treated with TGFβ (panel A) and TNFα (panel B) at indicated concentrations for further 4 h under serum starved conditions. Total RNA was isolated for Northern blot analysis of PAI-1 and SHP mRNA expression and was normalized to GAPDH expression. Data represent mean ± SD of 3 individual experiments. *P < 0.05 and &P < 0.005 compared to untreated control and cytokine treated cells. nd, non-detectable. C-D: AML12 cells were treated with mock virus, adenovirus siRNA SHP (Ad-siSHP) or adenovirus dominant negative AMPKα (Ad-dnAMPKα) for 48 h, followed by fenofibrate (Feno) treatment at indicated concentrations for an additional 24 h, after which cells were treated with TGFβ (panel A) or HGF (panel B) at indicated concentrations for further 4 h under serum starved conditions. Total RNA was isolated for semiquantitative RT-PCR analysis of PAI-1 and SHP mRNA expression and was normalized to actin expression. Data represent mean ± SD of 3 individual experiments. *P < 0.05, &P < 0.005 and **P < 0.005, compared to untreated control, cytokine treatment and fenofibrate treated cells.
Supplementary Figure 4. Repression of LRH-1 but not SF-1 transcriptional activity by fenofibrate. A: HepG2 cells were transfected with the human PAI-1 gene promoter (−800bp) for 18 h followed by treatments with TGFβ, TNFα, HGF and IL-1α at indicated concentrations for 24 h in the presence or absence of cotransfected Smad7 (200 μg), dominant negative Nur77 (dn-Nur77, 200 μg), dominant negative USF1 (dn-USF1, 200 μg) or IκB respectively under serum-starved conditions. All experiments were done in triplicate, and data are expressed in relative luciferase units (RLU) and as the fold activation relative to the control, representing mean ± SD of 3 individual experiments.*P < 0.05, &P < 0.05, compared to untreated control and individual cytokine treatments. B: HepG2 cells were transfected with pSuper vector (pSuper only, 200μg) or pSuper siRNA SHP (p-siSHP, (200μg) for 48 h. Total RNA was isolated for semiquantitative RT-PCR analysis of SHP mRNA expression and was normalized to actin expression. Data represent mean ± SD of 3 individual experiments. .*P < 0.05 compared to untreated control and pSuper only treated cells. C: HepG2 cells were cotransfected with Sft4-luc along with LRH-1 (200 μg) or SF-1 (200 μg) in the presence or absence of SHP (200 μg), pSuper siRNA SHP (p-siSHP, 200 μg) for 18 h followed by fenofibrate and metformin treatments as indicated for a further 24 h under serum-starved conditions. All experiments were done in triplicate, and data are expressed in relative luciferase units (RLU) and as the fold activation relative to the control, representing mean ± SD of 3 individual experiments. *P < 0.05, &P < 0.05 and **P < 0.005, compared to untreated control, individual transcription factor cotransfection and fenofibrate or metformin treatment.
Supplementary References
1. Park YJ, Qatanani M, Chua SS, LaRey JL, Johnson SA, Watanabe M, Moore DD, Lee YK. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology. 2008; 47(5):1578−86.
2. Kim BH, Won YS, Kim EY, Yoon M, Nam KT, Oh GT, Kim DY. Phenotype of peroxisome proliferator-activated receptor-alpha (PPARalpha)deficient mice on mixed background fed high fat diet. J Vet Sci. 2003; 4(3):239−44.
3. Kim JH, Kim K, Jin HM, Youn BU, Song I, Choi HS, Kim N. Upstream stimulatory factors regulate OSCAR gene expression in RANKL-mediated osteoclast differentiation. J Mol Biol. 2008;383(3):502−11.
4. Yeo MG, Yoo YG, Choi HS, Pak YK, Lee MO. Negative cross-talk between Nur77 and small heterodimer partner and its role in apoptotic cell death of hepatoma cells. Mol Endocrinol. 2005; 19(4): 950−63.
5. Kim YS, Han CY, Kim SW, Kim JH, Lee SK, Jung DJ, et al. The orphan nuclear receptor small heterodimer partner as a novel coregulator of nuclear factor-kappa b in oxidized low density lipoprotein-treated macrophage cell line RAW 264.7. J Biol Chem. 2001; 276(36): 33736−40.
6. Bergheim I, Guo L, Davis MA, Lambert JC, Beier JI, Duveau I, et al. Metformin prevents alcohol-induced liver injury in the mouse: Critical role of plasminogen activator inhibitor-1. Gastroenterology 2006; 130(7): 2099−112.
7. Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, et.al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004; 113: 1408−18.
8. Fernandez-Vizarra E, Fernandez-Silva P, Enriquez JA. Isolation of Mitochondria from Mammalian Tissues and Cultured Cells. Cell Biology- A laboratory handbook, 3rd edition (2), 69−77.
9. Lowry OH, Rosenbrough NJ, Farr A L, Randall RJ. Protein measurement with Folin phenol reagent. J Biol Chem. 1951; 193: 265−275.
10. Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods Cell Biology 2007; 80: 93−119.
11. López LC, Escames G, Tapias V, Utrilla P, León J, Acuña-Castroviejo D. Identification of an inducible nitric oxide synthase in diaphragm mitochondria from septic mice: its relation with mitochondrial dysfunction and prevention by melatonin. Int J Biochem Cell Biol. 2006; 38(2): 267−78.
12. Yu W, Gong JS, Ko M, Garver WS, Yanagisawa K, Michikawa M. Altered cholesterol metabolism in Niemann-Pick type C1 mouse brains affects mitochondrial function. J Biol Chem. 2005; 280(12):11731−9.
13. Jiao HL, Zhao BL. Cytotoxic effect of peroxisome proliferator fenofibrate on human HepG2 hepatoma cell line and relevant mechanisms. Toxicol Appl Pharmacol. 2002; 185(3):172−9.
14. Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004; 10(5):530−4.