Abstract
Objectives/Hypothesis
The avian cochlea regenerates hair cells following aminoglycoside treatment through supporting cell proliferation. Immunocytochemical labeling of BrdU, a thymidine analog, is a popular nonradioactive marker for identifying cells in the DNA Synthesis (S phase) of the cell cycle. However, it requires harsh treatments to denature double-stranded DNA for the antibody to bind BrdU. We explored a new method using EdU as a thymidine analog and a non-antibody azide/alkyne reaction between the EdU and the fluorescent probe. We propose that EdU is as effective as BrdU but without the requirement for harsh denaturation or the use of antibodies for detection.
Study Design
Two week-old chicks received a single gentamicin injection followed by a single EdU injection 72h later. Cochleae were extracted 4-8h later, fixed, and processed for fluorescent detection of EdU.
Methods
Cochleae were processed for detection of incorporated EdU using the Click-iT™ Imaging Kit (Invitrogen) and co-labeled with Sox2, myosin VI, or myosin VIIa antibodies. Whole-mount cochlear preparations were examined with confocal microscopy.
Results
Supporting cells incorporated EdU into their newly synthesized DNA during the 4-8h following the EdU injection and were readily detected with little background signal. The intensity and quantity of cells labeled were similar to or better than that seen for BrdU.
Conclusions
The EdU method is as effective as BrdU without requiring harsh denaturation or secondary antibodies to identify proliferating cells. Thus, the non-antibody EdU system allows more flexibility by enabling co-labeling with multiple antibodies to other cellular proteins involved in regeneration.
Introduction
Hearing loss is a significant problem in the United States today. Nearly 35 million Americans suffer from measurable hearing impairment and related speech disorders. Hearing loss affects approximately 17 in every 1,000 children under the age 18. The incidence increases with age: approximately 314 in 1,000 people over age 65 have a hearing loss and 40 to 50 percent of people 75 and older have a hearing loss1. Hearing loss affects more people than epilepsy, multiple sclerosis, spinal injury, stroke, Huntington’s, and Parkinson’s diseases combined2, and although it is rarely life-threatening, it has a huge financial impact on our economy and lifestyle. Two million Americans are completely deaf and two-to-three out of every thousand children born are severely to profoundly deaf, half of those due to hereditary causes1. The primary cause of these hearing impairments is thought to be damage to the sensory cells, supporting cells and neurons in the cochlea, and is referred to clinically as sensorineural or “nerve” deafness, as opposed to conductive hearing loss. These hearing deficits can be caused by administration of ototoxic drugs, exposure to intense work-related or recreational noise, genetic mutations, or as a consequence of the aging process. The loss of hair cells in the mammalian cochlea leads to permanent hearing loss because these cells are generated only during embryonic development3 and must last throughout a person’s lifetime. However, it was recently discovered that birds are able to rapidly and repeatedly produce new hair cells and supporting cells in their cochleae following hair cell damage which leads to a significant recovery of hearing4,5,6.
The primary mechanism for regeneration in the bird cochlea is the proliferation of supporting cells in the damaged region of the sensory epithelium that results in the generation of new hair cells and supporting cells. In the normal bird cochlea, both the hair cells and supporting cells are post-mitotic and remain in a state of quiescence known in the cell cycle field as G07. But once the dying hair cells are ejected from the epithelium, the adjacent supporting cells are stimulated to leave quiescence, re-enter the cell cycle (the G1 phase), double their DNA content (the DNA synthesis, or S phase), generate proteins needed to divide (G2 phase), and finally split into two identical daughter cells (the Mitosis, or M phase). In the avian cochlea, the daughter cells will go on to differentiate into new hair cells or supporting cells replacing those that were lost. At the time of our original regeneration discovery in the bird, there was morphological evidence that new stereociliary bundles were appearing in the region of hair cell loss within 4-6 days after the trauma8. However, we were unsure as to whether this was from the repair of surviving hair cells or the generation of new ones.
In order to test whether new hair cells were being produced by cell divisions, we needed to label the tissues with markers for evidence of the production of new cells. At that time, the standard technique was to inject radioactive (tritiated, or 3H) thymidine, one of the four nucleotides required to duplicate DNA. Tritiated thymidine is detected by the radioactive decay of the 3H tag, however, this weak decay can only travel through tissues over a short distance. Thus, the method for detecting tritiated thymidine is to slice the tissue into thin (1-30 um) sections, cover the sections with a photographic emulsion, expose the tissues in the dark for several hours to days in order to detect exposed silver grains in the overlying emulsion. While this is a very powerful technique, it has several drawbacks, such as dealing with radioactive isotopes, having to section tissues, and the fact that the signal is only detected in the overlying emulsion, not in the tissue itself. Plus, the necessity of excluding extraneous light that would ruin the emulsion during the exposure can be frustrating.
In the mid-1980′s a new technique was developed for detecting DNA synthesis using a non-radioactive analog of thymidine, 5-bromo-2′-deoxyuridine (BrdU). This analog is incorporated into DNA just as readily as thymidine during S phase and can be detected with a monoclonal antibody that binds directly to the BrdU molecule within the DNA9. The BrdU technique to label mitotically active cells is quite beneficial, as one can sidestep the significant dangers, regulations, and permitting associated with the radioactive isotope tritiated thymidine. Moreover, since the antibody binds directly to the DNA within the cell, it does not require sectioning and overlaying with a photographic emulsion. The detecting signal can be a stain such as diaminobenzidine (DAB), for brightfield histological analysis or a fluorescent probe attached to a secondary antibody or even directly to the primary antibody for immunofluorescent detection. Finally, detection of the signal within the tissue, rather than in an overlying emulsion, and penetration of the antibody label through thick sections of tissue or even whole-mount preparation of tissues like the cochlear sensory epithelium, enables three dimensional localization of the labeled nuclei with tools such as the confocal laser scanning microscope or computer-aided 3D reconstruction programs coupled with standard light microscopy. While these benefits of BrdU as a method for detecting proliferating cells offer several major advantages, there are also a few significant drawbacks. Detection of BrdU incorporated into the DNA requires harsh denaturation techniques to give the primary antibody access to the BrdU molecule. This harsh denaturation also tends to affect many protein epitopes and significantly hinders the ability of many standard antibodies to detect their target proteins. Moreover, the requirement of a monoclonal antibody to detect the BrdU limits the ability to detect other proteins of interest within the tissue that are normally also labeled with monoclonal antibodies.
A new technique for detecting DNA synthesis in proliferating cells in vivo and in vitro has been developed within the last two years10. The thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) has a terminal alkyne group replacing a methyl group at the 5 position of the pyrimidine ring and can be readily incorporated into DNA during synthesis. The incorporated EdU molecule can be detected by a reaction of the terminal alkyne group with fluorescent azides, in a Cu(I)-catalyzed [3 + 2] cycloaddition “click” chemistry (Click-iT™, Invitrogen/Molecular Probes, Carlsbad)10. Because the Click-iT™ reagents are significantly smaller than antibody molecules, they can penetrate much more easily through tissues and the incorporated EdU can be detected without the necessity of DNA denaturation. This leads to a greater sensitivity of detection and retains the availability of other proteins for double-labeling using standard immunocytochemistry. In this study, we have examined the ability of EdU incorporation and detection by Click-iT™ chemistry to identify proliferating supporting cells in the regenerating chick cochlea following gentamicin injection. We have shown that EdU is as sensitive, if not more so, than traditional BrdU labeling with less background labeling and that several antibodies to other proteins involved in regeneration can be used in conjunction with EdU.
Materials & Methods
Animals
White Leghorn chickens (Gallus domesticus) were obtained at one week of age from Specific Pathogen-Free Avian Supply (Charles River SPAFAS, Wilmington, MA). The chicks were housed in communal brooders with ad libitum access to food and water. All animal procedures were approved by the Boston University Medical Center Institutional Animal Care and Use Committee (IACUC).
Gentamicin injections
To create cochlear hair cell death, 11-16 day-old chicks were given a single, subcutaneous injection of gentamicin sulfate (300 mg/kg, Sigma, St. Louis, MO), with the time of injection designated as time “0 hours” after injection (AI). Injections were performed either in the morning or early afternoon, so that the nephrotoxic effects of gentamicin could pass before the evening11,12. An additional group of age-matched birds did not receive gentamicin injections and served as unmanipulated experimental controls.
EdU injections
In order to label mitotically active supporting cells, a single, subcutaneous injection of EdU (50 mg/kg) in sterile, phosphate buffered saline (PBS, pH 7.4) was administered to each bird 72h after the gentamicin injection.
Harvesting and fixation of the cochleae
Four to 8 hours after the EdU was injected, birds were euthanized with an intracardiac injection of Fatal Plus (390 mg/mL pentobarbital, Vortech Pharmaceuticals, Dearborn, MI) and the heads were removed for cochlear dissection. To harvest cochlear tissues, the tympanic membrane and columella were removed and the lagena was exposed. Forceps were used to chip away at the bones surrounding the middle ear to provide direct access to the oval window. The bony spur separating the round and oval windows was removed and bone was chipped away, exposing the entire length of the cochlea. Using forceps to grasp the lagena, the cochlea was gently lifted out of the temporal bone and placed into chilled Hank’s Balanced Salt Solution (HBSS, pH 7.4; Gibco, Grand Island, NY). Once in HBSS, the tissues of the tegmentum vasculosum and the lagena were carefully dissected away leaving the basilar papilla suspended between the superior and inferior cartilaginous plates. In most cases, the tectorial membrane overlying the basilar papilla was left in place and tissue was fixed in chilled 4% paraformaldehyde in PBS for 1h. Cochleae were then rinsed three times in PBS for 5 minutes each rinse and stored in PBS at 4°C until time of EdU labeling.
EdU Click-iT™ reaction
EdU incorporation into DNA was detected using the Click-iT™ EdU Alexa Fluor® Imaging kit (Invitrogen/Molecular Probes, Eugene, OR). All steps of the Click-iT™ reaction were performed at room temperature. Cochlear tissue was permeabilized in 1.0% Triton ×-100 for 25 minutes and rinsed three times in PBS for 10 minutes each rinse. During the permeabilization step, the Click-iT™ kit azide and buffer additive were removed from -20°C storage and allowed to thaw in a light-protected box. The buffer additive is stored as a 10X solution; once it is thawed, it must be diluted with dH2O to make a working 1X solution. During the third PBS rinse, the reaction cocktail is made, adding the azide last. The reaction cocktail must be used within 15 minutes of creation. The amount of reaction cocktail needed was determined, and made according to the chart below:
| TOTAL VOLUME | 200 μL | 400 μL | 600 μL | 800 μL | 1000 μL |
|---|---|---|---|---|---|
| Reaction Buffer | 175 μL | 350 μL | 525 μL | 700 μL | 875 μL |
| Buffer Additive | 20 μL | 40 μL | 60 μL | 80 μL | 100 μL |
| CuSO4 | 4 μL | 8 μL | 12 μL | 16 μL | 20 μL |
| Azide | 1 μL | 2 μL | 3 μL | 4 μL | 5 μL |
Cochleae were then incubated in the EdU cocktail for 30 minutes and rinsed three times in PBS for 15 minutes per rinse. The azides used were coupled to either an Alexa Fluor® 488 (green) or Alexa Fluor® 594 (red) fluorophore. Upon completion of the EdU Click-iT™ reaction, cochleae were mounted in Vectashield mounting medium onto glass slides and sealed with clear fingernail polish.
Immunocytochemistry for other proteins
All steps of the immunocytochemistry procedure were performed at room temperature, unless otherwise noted. Cochleae were rinsed in PBS three times for 10 minutes per rinse between each step. Cochleae were permeabilized in 1.0% Triton ×-100 for 25 minutes and blocked in 10% normal goat serum (myosin VI or myosin VIIa) or 10% normal donkey serum (Sox2) for 10 minutes. Following incubation in primary antibody solution (rabbit anti-myosin VI or VIIa, 1:200 in PBS, Proteus Biosciences, Ramona, CA; or goat anti-Sox2 (Y-17), 1:500 in PBS, Santa Cruz Biotechnology, Santa Cruz, CA) for 2h, cochlear tissue was incubated in secondary antibody solution (Alexa Fluor® 568 goat anti-rabbit IgG, 1:1000 in PBS or Alexa Fluor® 568 donkey anti-goat IgG, 1:5000 in PBS; Invitrogen/Molecular Probes, Eugene, OR) for 2h. Cochleae were then run through the EdU Click-iT™ reaction cocktail, as described above and mounted in Vectashield mounting medium onto glass slides and sealed with clear fingernail polish.
Whole-mount analysis by light microscopy or confocal microscopy
Fluorescent digital images of the whole-mount cochlea were obtained using either a Nikon TE2000-U inverted microscope (Nikon Instruments, Melville, NY) equipped with a Spot RT CCD (Diagnostic Imaging, Sterling Heights, MI) or a Zeiss 510 Meta confocal laser-scanning microscope (Carl Zeiss MicroImaging, Thornwood, NY). On the light microscope, images were obtained using a 20x (N.A. 0.75) water immersion lens and images were captured on a Spot RT digital camera interfaced with a Power Mac G4 (Apple, Cupertino, CA) workstation. For the confocal, a z-series of scans through the basilar papilla was made using a 20X (N.A. 0.75) immersion objective. Z-series scans were compressed to produce a single digital image used to visualize labeling in cochlear hair cells following gentamicin treatment. All images were digitally processed using Adobe Photoshop CS for generating publication quality micrographs. The tissues in all micrographs were oriented so that the basal (proximal) end of the cochlea appeared on the left and the apical (distal) end on the right.
Results
EdU labeling of S phase nuclei
When basilar papillae are examined at 76h, four hours after the EdU injection, numerous brightly labeled nuclei of supporting cells are apparent in the basal third of the sensory epithelium (Fig. 1). This is the region of the cochlea where hair cell death is induced following a single gentamicin injection11,13. No EdU labeling of supporting cell nuclei is seen in the remainder of the sensory epithelium apical to the region of hair cell loss. The EdU results are qualitatively equivalent to what was seen with our previous BrdU injections. However, the EdU-Click iT™ labeling is much brighter than the BrdU labeling and there is much less background labeling, as the signal-to-noise ratio is much better for the EdU technique. We were able to label EdU-incorporating nuclei using either the Alexa Fluor® 488 (green) fluorophore or the Alexa Fluor® 594 (red) fluorophore, although the 594 fluorophore seemed somewhat brighter, especially when contrasted against a blue DAPI nuclear counterstain. There is a faint level of fluorescence seen in the cytoplasm of both the normal and dying hair cells within the sensory epithelium. However, this label is not nuclear and is seen in both the green and red channels, thus it may be nonspecific autofluorescence generated by the dense, mitochondrial and ribosomal rich pericuticular cytoplasm of the hair cells.
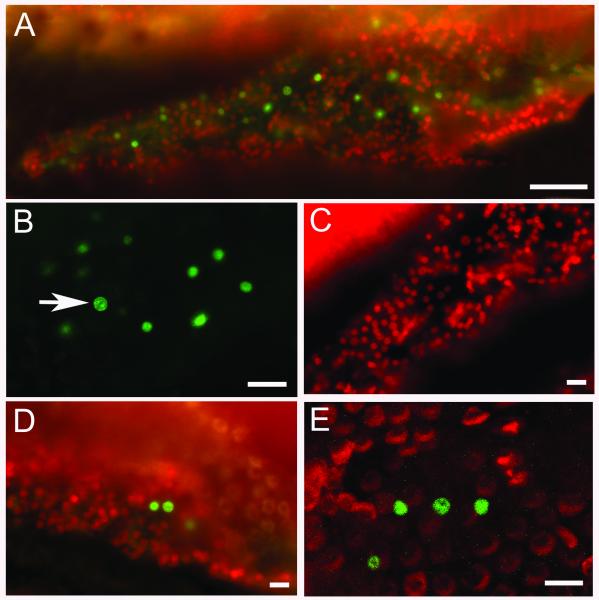
Figure 1.
EdU Click-iT™ labeling of EdU incorporated into supporting cell nuclei of the regenerating chick cochlea 76h after gentamicin injection. The EdU was injected at 72h. A) A low magnification light micrograph of EdU-labeled nuclei (green) in the basal region of the cochlea. Other nuclei in the sensory epithelium (red) are made fluorescent by the Click-iT™ processing. Bar = 50 μm. B) A higher magnification light micrograph of EdU+ supporting cells in S phase showing both large (arrow) and small SC nuclei. Ba r= 20 μm. C) A higher magnification light micrograph of supporting cell nuclei made fluorescent (red) by the Click-iT™ process when viewed in the 350nm (DAPI) channel. Bar = 20μm. D) A higher magnification light micrograph showing supporting cell nuclei double labeled for EdU (green) and Sox2 (red). Bar = 20μm. E) A higher magnification confocal micrograph of cochlear tissues double labeled for EdU (green) and myosin VI (red). Bar = 20μm.
Detection of EdU labeling indicates that numerous supporting cell nuclei are present at various levels within the sensory epithelium. Small, labeled nuclei, similar in size to the normal supporting cell nuclei are seen in the deepest layers of the epithelium, close to the basilar membrane (Fig. 1b). Large labeled nuclei can be identified higher up in the epithelium, close to the luminal surface (Fig 1, arrow). These nuclei are presumably ones that are in the later stages of DNA synthesis or in G2 preparing for mitosis and contain twice the normal DNA content. Previous light microscopic and transmission electron microscopic studies have shown that the nuclei of cells in S phase progressively move up to the luminal surface of the epithelial layer where they subsequently complete the cell cycle and undergo mitosis14,15.
When EdU is injected at 72h and allowed to incorporate for 4h, some of the supporting cells that first entered S phase around 65h are labeled with EdU at the end of their S phase. They are able to reach the mitotic phase and some even divide during this time. By 76h, we are able to identify a few cells in the midst of mitosis and a few that have already divided and exist as small, adjacent pairs of labeled nuclei. However, many more EdU-positive, post-mitotic paired nuclei are seen at 80h (data not shown). This occurs because the supporting cells have completed M phase and divided into two new daughter cells.
One interesting side effect of the Click-It™ technique is that it seems to make all of the nuclei in the tissue fluorescent with an orange-to-yellow emission when viewed through the DAPI (350nm) filters (Fig. 1c). This fluorescence of the nuclei is not visible in the green or the red channels. While this may be problematic in some situations, it can actually be beneficial for localizing the EdU-labeled nuclei within the population of surrounding unlabeled supporting cell nuclei.
EdU/Sox2 double labeling
Sox2 is a developmental transcription factor in the cochlea that labels the entire presumptive sensory epithelium during embryonic development but is restricted to only the nuclei of supporting cells in the mature mammalian and avian cochlea16,17.
Double-labeling studies of EdU incorporation and Sox2 protein expression in supporting cells at 76h of regeneration (72h EdU injection, cochleae harvested at 76h) indicate that the cells taking up EdU during S phase continue to label with Sox2 (Fig 1d). However, in those EdU labeled cells undergoing mitosis at 76h we see a loss of Sox2 labeling in the nuclei that have reached the metaphase stage of mitosis (midline alignment of the chromosomes) (data not shown). This loss of Sox2 labeling of dividing nuclei continues through anaphase and telophase.
EdU/Myosin VI & VIIa double labeling
Myosins VI and VIIa are hair cell specific markers in the vertebrate cochlea and normally have a characteristic pericuticular and stereociliar localization within the hair cells18. Our studies of regeneration in the bird cochlea have demonstrated that both myosins VI and VIIa exhibit a change in localization and an increase in labeling intensity as gentamicin-damaged hair cells undergo apoptosis12. Moreover, we have shown that myosin VI first appears in the newly regenerating hair cells as early as 78h after the gentamicin injection and that myosin VIIa first appears at 90h12.
When the chick cochleae are double labeled with EdU and either myosin VI or VIIa polyclonal antibodies at 76h after gentamicin, we can clearly identify the nuclei of supporting cells that have entered S phase within the previous 4h by EdU labeling. Also, while the myosin VI or VIIa prominently labels the dying and ejected hair cells trapped in the tectorial membrane overlying the damaged region, the new hair cells cannot yet be detected at this early time point. But our results for the 76h cochleae do show that both EdU and myosin antibodies can readily be used together to double label regenerating cochlear tissues (Fig 1e).
Discussion
The results of our experiments indicate that EdU effectively labels cochlear supporting cells undergoing DNA synthesis during avian hair cell regeneration. The Click-iT™ technique is a simple, quick procedure that provides a very intense fluorescent label for proliferating cell nuclei that have incorporated EdU. In addition, it has a very low background signal, so it gives a very high signal-to-noise ratio in the whole-mount cochlear tissues. Surprisingly, we found that the Click-iT™ procedure also causes all the cell nuclei in the tissue to fluoresce in an orange-yellow color that was restricted to the 350nm excitation wavelength. Thus, we can identify not only the nuclei that are undergoing DNA synthesis, we can also localize these nuclei relative to other supporting cell and hair cell nuclei within the sensory epithelium.
Our previous studies using BrdU have determined that the first cells enter S phase around 65h after a single gentamicin injection and the peak of DNA synthesis occurs between 72h and 96h12. Comparison of the extent of labeling at 76h for both BrdU and EdU (4h after their injection) indicates that the two probes identify an equivalent number of supporting cells in S phase within the gentamicin-damaged region of the sensory epithelium. Thus, our initial experiments suggest that the EdU Click-iT™ technique can be used in place of the BrdU technique and give essentially identical results.
An additional benefit to the EdU Click-iT™ technique over BrdU is that the Alexa Fluor® azide used to detect the incorporated EdU is much smaller than the BrdU antibody (1/500th the size10) and can readily penetrate thick tissues like our whole mount preparations of the cochlea. Moreover, its small size allows the fluorophore to reach the incorporated EdU molecules without needing to harshly denature the DNA double strands first. This allows the tissues to remain structurally intact and it retains important protein epitopes for many of the most popularly utilized monoclonal and polyclonal antibodies. We have been able to double-label EdU-treated cochleae with monoclonal and polyclonal antibodies to myosins VI and VIIa, Sox2, and Math1 to determine the expression patterns of these proteins relative to DNA synthesis and hair cell regeneration. However, we have not yet been able to establish a protocol that enables EdU to co-label with Phalloidin, a very popular marker for labeling F-actin in hair cells and supporting cells of the cochlea. It is well known that the BrdU procedure destroys the ability of Phalloidin to bind to actin filaments in cochlear tissues and this was assumed to be due to the harsh HCl treatment used for denaturing the DNA. We had hoped that since EdU labeling does not require denaturation, it might allow Phalloidin to bind to F-actin in cochlear tissues. But it appears that components in the azide-alkyne reaction also prohibit Phalloidin from binding after the Click-iT™ processing. We are continuing to explore alterations in the procedure that would enable Phalloidin to successfully double label with the EdU-tagged tissues.
Conclusions
This research has been able to demonstrate that the EdU Click-iT™ method for labeling proliferating cells is as effective as the standard BrdU technique without requiring harsh denaturation or secondary antibodies to identify proliferating cells. As of now, the EdU technique does not allow co-labeling with Phalloidin, although alterations in the procedure are currently being explored. Despite this minor drawback, the non-antibody EdU Click-iT™ detection system allows more flexibility than BrdU immunocytochemistry by enabling co-labeling of EdU-labeled sensory epithelia with multiple antibodies to other cellular proteins involved in regeneration.
Supplementary Material
Acknowledgments
This work was supported by research grants DC01689 (DAC) and DC008235 (CLK) from NIH/NIDCD.
Financial Support: NIH/NIDCD Grants DC01689 (DAC) and DC008235 (CLK)
References
- 1. [Accessed January 9, 2009];Statistics about Hearing, Balance, Ear Infections, and Deafness [NIDCD website] 2008 June 11; Available at: http://www.nidcd.nih.gov/health/statistics/hearing.asp.
- 2.Hudspeth AJ. How hearing happens. Neuron. 1997;19:947–950. doi: 10.1016/s0896-6273(00)80385-2. [DOI] [PubMed] [Google Scholar]
- 3.Ruben RJ. Development of the inner ear of the mouse. A radioautographic study of terminal mitosis. Acta Otolaryngol. 1967;220(Suppl.):1–44. [PubMed] [Google Scholar]
- 4.Corwin JT, Cotanche DA. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240:1772–1774. doi: 10.1126/science.3381100. [DOI] [PubMed] [Google Scholar]
- 5.Ryals BM, Rubel EW. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240:1774–1776. doi: 10.1126/science.3381101. [DOI] [PubMed] [Google Scholar]
- 6.Tucci DL, Rubel EW. Physiological status of regenerated hair cells in the avian inner ear following aminoglycoside ototoxicity. Otolaryngol Head Neck Surg. 1990;103:443–450. doi: 10.1177/019459989010300317. [DOI] [PubMed] [Google Scholar]
- 7.Baserga R. The Biology of Cell Reproduction. Harvard University Press; Cambridge, MA: 1985. [Google Scholar]
- 8.Cotanche DA. Regeneration of hair cell stereociliary bundles in the chick cochlea following severe acoustic trauma. Hear Res. 1987;30:181–196. doi: 10.1016/0378-5955(87)90135-3. [DOI] [PubMed] [Google Scholar]
- 9.Gratzner HG. Monoclonal antibody to 5-bromo-2-iododeoxyuridine: a new reagent for detection of DNA replication. Science. 1982;218:474–475. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- 10.Salic A, Mitchison T. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Nat Acad Sci. 2008;105:2415–20. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberson DW, Alosi JA, Messana EP, Cotanche DA. Effect of violation of the labyrinth on the sensory epithelium in the chick cochlea. Hear Res. 2000;141:155–164. doi: 10.1016/s0378-5955(99)00218-x. [DOI] [PubMed] [Google Scholar]
- 12.Duncan L, Mangiardi DA, Matsui J, Anderson J, McLaughlin-Williamson K, Cotanche DA. Differential expression of unconventional myosins in apoptotic and regenerating hair cells confirms two regeneration mechanisms. J Comp Neurol. 2006;499:691–701. doi: 10.1002/cne.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangiardi DA, McLaughlin-Williamson K, May K, Messana EP, Mountain DC, Cotanche DA. Progression of hair cell ejection and molecular markers of apoptosis in the avian cochlea following gentamicin treatment. J Comp Neurol. 2004;475:1–18. doi: 10.1002/cne.20129. [DOI] [PubMed] [Google Scholar]
- 14.Raphael Y, Adler HJ, Wang Y, Finger PA. Cell cycle of transdifferentiating supporting cells in the basilar papilla. Hear Res. 1994;80:53–63. doi: 10.1016/0378-5955(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 15.Tsue TT, Watling DL, Weisleder P, Coltrera MD, Rubel EW. Identification of hair cell progenitors and intermitotic migration of their nuclei in the normal and regenerating avian inner ear. J Neuroscience. 1994;14:140–152. doi: 10.1523/JNEUROSCI.14-01-00140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiernan A, Pelling A, Leung K, et al. Sox2 is required for sensory organ development in the mammalan inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- 17.Neves J, Kamaid A, Alsina B, Giraldez F. Differential expression of Sox2 and Sox3 in neural and sensory progenitors of the de3veloping inner ear of the chick. J Comp Neurol. 2007;503:487–500. doi: 10.1002/cne.21299. [DOI] [PubMed] [Google Scholar]
- 18.Hasson T, Gillespie P, Garcia J, et al. Unconventional myosins in inner-ear sensory epithelia. J Cell Biol. 1997;137:1287–1307. doi: 10.1083/jcb.137.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.