The salinosporamides comprise a natural product family of potent anticancer agents produced by the marine bacterium Salinispora tropica. This group of densely functionalized β-lactone-γ-lactam proteasome inhibitors is largely distinguished through structural differences at C-2 bearing methyl, ethyl, chloroethyl, and propyl substituents per salinosporamides D (1), B (2), A (3) and E (4), respectively (Scheme 1).1-3 The recent discovery of the related metabolite cinnabaramide A (5) from a terrestrial streptomycete,4 which instead harbors a C-2 hexyl chain, further extends the natural salinosporamide structural family. We recently reported that salinosporamides A and B are biosynthetic products derived from an unusual hybrid polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) pathway initiated by the chain elongation of acetyl-S~ACP by chloroethylmalonyl-CoA or ethylmalonyl-CoA,5,6 respectively, followed by the non-proteinogenic amino acid cyclohexenylalanine (Scheme 1).7 The selection of the PKS extender unit is controlled by the acyltransferase domain AT1 from the hexadomained SalA synthase. Herein we report that salinosporamides D and E are respectively accessed from methylmalonyl-CoA and propylmalonyl-CoA, the latter of which is a newly described PKS extender unit that belongs to a growing family of PKS substrates derived from α,β-unsaturated fatty acids.
Scheme 1.
Biosynthesis of salinosporamide A (3), its analogs and their substituted malonyl-CoA PKS building blocks (boxed). Abbreviations: ACP, acyl carrier protein; KS, ketosynthase; AT, acyltransferase; C, condensing domain; A adenylation domain; PCP, peptidyl carrier protein.
Based on the biosynthetic assembly of salinosporamide B from a butyrate building block via ethylmalonyl-CoA,5,6 we reasoned that the methyl analog salinosporamide D (1) would be similarly assembled from a propionate unit via the common PKS substrate (2S)-methylmalonyl-CoA (Scheme 1). We explored this assumption by administering [1-13C]propionate to the S. tropica salL-deficient mutant, which is specifically unable to synthesize the chlorinated salinosporamide A as the major product of the sal pathway due to inactivated 5'-chloro-5'-deoxyadenosine synthase SalL.8 Isolation and characterization of the resultant salinosporamide D by NMR revealed the specific 13C-enrichment (5%) at C-1, thereby confirming its assembly from propionate.
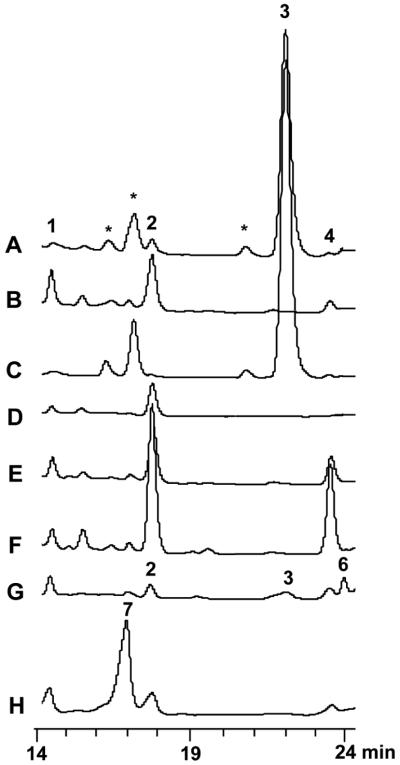
We next turned our attention to the propyl analog salinosporamide E (4), which would presumably derive from a pentanoate building block. If so, this would imply the prospect of a new PKS building block, namely propylmalonyl-CoA. Salinosporamide E was similarly isolated from the [1-13C]propionate feeding experiment, and NMR analysis confirmed 13C-enrichment (40%) at C-12, thereby suggesting an origin from pentanoate derived from propionate and acetate precursors (Scheme 1). While the administration of pentanoic acid to the S. tropica salL-deficient mutant resulted in a significant increase (~500%) in salinosporamide E titers, unsaturated trans-2-pentenoic acid had a much greater effect in enhancing its production at ~1500% (Figure 1). These observations suggested that the C5 substrate is limiting and that the α,β-unsaturated carboxylic acid is a more advanced biosynthetic intermediate.
Figure 1.
HPLC analysis at 210 nm of organic fractions of (A) wild-type S. tropica, (B) S. tropicasalL- mutant, (C) S. tropica Strop_3612- mutant (D) S. tropicasalG- mutant, (E) S. tropicasalL- + 0.8 mM pentanoic acid, (F) S. tropicasalL- + 0.8 mM trans-2-pentenoic acid, (G) S. tropicasalL- + 0.12 mM 4-bromocrotonoic acid, and (H) S. tropicasalL- + 0.15 mM 4-fluorocrotonoic acid. Salinosporamide analogs 1-4 and 6-7 are noted, while derivatives of 3 are marked with an asterisk.
The recent functional revision of crotonyl-CoA reductase (CCR) as a carboxylase that catalyzes the reductive carboxylation of crotonyl-CoA to (2S)-ethylmalonyl-CoA9 suggests the possibility of an analogous pathway to (2S)-propylmalonyl-CoA in S. tropica from 2-pentenyl-CoA (Scheme 1). Inspection of the complete genome sequence of S. tropica CNB-44010 revealed two CCR-encoding genes. We previously inactivated each by PCR-targeted mutagenesis and showed that Strop_3612 encodes a primary CCR involved in salinosporamide B biosynthesis while the homolog salG codes for a novel chlorocrotonyl-CoA reductase/carboxylase associated with salinosporamide A biosynthesis.6 Upon further analysis of the S. tropica CCR mutants, we observed that production of salinosporamide E was exclusively lost in the salG knockout mutant whereas it was maintained in the Strop_3612 mutant (Figure 1). Hence, this in vivo experiment suggests that SalG has relaxed substrate specificity towards 2-alkenyl-CoAs and is able to also reductively carboxylate 2-pentenyl-CoA in addition to 4-chlorocrotonyl-CoA as previously reported.6 In vitro analysis of recombinant octahistidyl-tagged SalG confirmed that it is able to reductively carboxylate 2-pentenyl-CoA (kcat 25.5 ± 0.7 min-1; Km 4.3 ± 0.5 μM) at comparable efficiency to 4-chlorocrotonyl-CoA (kcat 23.1 ± 2.6 min-1; Km 4.4 ± 1.8 μM), which is more efficient than that with crotonyl-CoA (kcat 15.4 ± 0.9 min-1; Km 20.7 ± 4.2 μM).6 Presumably, the CCR encoded by Strop_3612 has more substrate specificity towards crotonyl-CoA, although attempts to verify this hypothesis were unsuccessful due to issues with soluble expression of the protein. However, we did observe in parallel experiments that the CCR originally isolated from Streptomyces collinus11 uses crotonyl-CoA and not 2-pentenyl-CoA as a substrate for reductive carboxylation. Thus this CCR and SalG have markedly different substrate specificities yet likely have the same 2S-stereochemical outcome as in other homologous CCRs and medium-chain dehydrogenases/reductases.9
Given the relaxed in vitro and in vivo substrate specificity of SalG, we explored other substrates including 4-bromo- and 4-fluorocrotonate, which are anticipated substrates of natural bromosalinosporamide (6)3 and engineered fluorosalinosporamide (7),12 respectively (Scheme 1). Administration of the 4-halocrotonic acids to the S. tropicasalL-deficient mutant resulted in the production of 6 and 7 as anticipated (Figure 1), thereby establishing bromoethylmalonyl-CoA and fluoroethylmalonyl-CoA as additional PKS extender units unique to the salinosporamide biosynthetic pathway. While the 4-halo analogs were accepted as alternative in vivo substrates,13 elongated 2-alkenoates (C6 to C8) were not converted into new sal products as observed by HPLC-MS analysis. The capacity of a CCR homolog to preferentially accommodate a longer chain 2-alkenyl-CoA, however, is strongly suggestive in the biosynthesis of cinnabaramide A (5),4 which based on the salinosporamide biosynthetic model would incorporate (2S)-hexylmalonyl-CoA derived from the reductive carboxylation of 2-octenyl-CoA.
In conclusion, we discovered the new PKS extender unit propylmalonyl-CoA, in the context of salinosporamide E biosynthesis. It is rare for PKSs to incorporate pentyl building blocks in their polyketide products; to our knowledge only the macrolide immunosuppressant FK506,14 which carries an allyl side chain, and the acyl depsipeptide dentigerumycin15 may similarly incorporate propylmalonyl-CoA units. This discovery exemplifies a new strategy in PKS extender unit biochemistry in which α,β-unsaturated acyl-CoA thioesters are reductively carboxylated16 and furthermore suggests that CCR protein engineering may readily afford unnatural malonyl-CoA precursors for the bioengineering of novel polyketide molecules.
Supplementary Material
Acknowledgments
We gratefully acknowledge valuable discussions with Prof. Yoshihisa Kobayashi (UCSD) and financial support provided by the National Institutes of Health (CA127622 to B.S.M. and AI51629 to K.A.R.) and the Life Sciences Research Foundation via a Tularik postdoctoral fellowship to A.S.E.
Footnotes
Supporting Information Available: Experimental procedures, NMR data and SalG kinetic data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew. Chem. Int. Ed. 2003;115:369–371. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- (2).Williams PG, Buchanan GO, Feling RH, Kauffman CA, Jensen PR, Fenical W. J. Org. Chem. 2005;70:6196–6203. doi: 10.1021/jo050511+. [DOI] [PubMed] [Google Scholar]
- (3).Reed KA, Manam RR, Mitchell SS, Xu J, Teisan S, Chao TH, Deyanat-Yazdi G, Neuteboom ST, Lam KS, Potts BC. J. Nat. Prod. 2007;70:269–276. doi: 10.1021/np0603471. [DOI] [PubMed] [Google Scholar]
- (4).Stadler M, Bitzer J, Mayer-Bartschmid A, Müller H, Benet-Buchholz J, Gantner F, Tichy H-V, Reinemer P, Baco KB. J. Nat. Prod. 2007;70:246–252. doi: 10.1021/np060162u. [DOI] [PubMed] [Google Scholar]
- (5).Beer LL, Moore BS. Org. Lett. 2007;9:845–848. doi: 10.1021/ol063102o. [DOI] [PubMed] [Google Scholar]
- (6).Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. Proc. Natl. Acad. Sci. U.S.A. doi: 10.1073/pnas.0901237106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).McGlinchey RP, Nett M, Eustáquio AS, Asolkar RN, Fenical W, Moore BS. J. Am. Chem. Soc. 2008;130:7822–7823. doi: 10.1021/ja8029398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Eustáquio AS, Pojer F, Noel JP, Moore BS. Nat. Chem. Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Erb TJ, Berg IA, Brecht V, Müller M, Fuchs G, Alber BE. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Erb TJ, Brecht V, Fuchs G, Müller M, Alber BE. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8877–8882. doi: 10.1073/pnas.0903939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, Moore BS, Jensen PR. ISME J. 2009 doi: 10.1038/ismej.2009.58. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wallace KK, Bao ZY, Dai H, Digate R, Schuler G, Speedie MK, Reynolds KA. Eur. J. Biochem. 1995;233:954–962. doi: 10.1111/j.1432-1033.1995.954_3.x. [DOI] [PubMed] [Google Scholar]
- (12).Eustáquio AS, Moore BS. Angew. Chem. Int. Ed. 2008;47:3936–3938. doi: 10.1002/anie.200800177. [DOI] [PubMed] [Google Scholar]
- (13).Administration of 4-bromocrotonic acid to the S. tropica salL- mutant also provided the chlorinated 3 (Figure 1, trace G) as a minor product. Its production presumably derives from the transchlorination of the substrate in the seawater-based A1 medium.
- (14).Motamedi H, Shafiee A. Eur. J. Biochem. 1998;256:528–534. doi: 10.1046/j.1432-1327.1998.2560528.x. [DOI] [PubMed] [Google Scholar]
- (15).Oh D-C, Poulsen M, Currie CR, Clardy J. Nat. Chem. Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Chan YA, Podevels AM, Kevany BM, Thomas MG. Nat. Prod. Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.