Abstract
Macroautophagy (autophagy) is a lysosomal degradation pathway for the breakdown of intracellular proteins and organelles. Although, constitutive autophagy is a homeostatic mechanism for intracellular recycling and metabolic regulation, autophagy is also stress responsive where it is important for the removal of damaged proteins and organelles. Autophagy thereby confers stress tolerance, limits damage and sustains viability under adverse conditions. Autophagy is a tumor suppression mechanism yet it enables tumor cell survival in stress. Reconciling how loss of a prosurvival function can promote tumorigenesis, emerging evidence suggests that preservation of cellular fitness by autophagy may be key to tumor suppression. As autophagy is such a fundamental process, establishing how the functional status of autophagy influences tumorigenesis and treatment response is important. This is especially critical as many current cancer therapeutics activate autophagy. Therefore, efforts to understand and modulate the autophagy pathway will provide new approaches to cancer therapy and prevention.
Autophagy is a lysosomal degradation pathway for intracellular digestion
Stress stimuli activate cellular pathways for adaptation that are crucial for cells to either tolerate adverse conditions, or to trigger cell suicide mechanisms such as apoptosis to eliminate damaged and potentially dangerous cells (1). Metabolic stress, including starvation, increases the cellular requirement for energy production and damage mitigation, and catabolic cellular self-digestion by autophagy plays a critical role in both instances. Stress activates autophagy where double membrane vesicles form and engulf proteins, cytoplasm, protein aggregates and organelles that are then delivered to lysosomes where they are degraded (2). This serves to maintain cellular metabolism through recycling of cellular components when the availability of external nutrient sources is limited. Autophagy-deficient mice have tissues with low ATP levels and fail to survive the neonatal starvation period, providing a clear example of autophagy-mediated management of energy homeostasis (3). Stress, particularly that resulting form oxidative damage due to aging or hypoxic conditions, damages proteins and organelles that require autophagy for elimination. Mice with autophagy defects accumulate cells with polyubiqutinated, p62 (sequestosome1)-containing protein aggregates, and damaged mitochondria and show elevated oxidative stress and cell death (4-8). Thus, autophagy is important for the degradative turnover of damaged proteins and organelles during stress, the failure of which is toxic to cells and tissues and can be pro-inflammatory. Peptides generated from proteins degraded by autophagy can also be utilized for antigen presentation to T cells for regulation of immunity and host defense (9). The importance of autophagy as a homeostatic and survival-promoting mechanism is underscored by the association of autophagy defects in the etiology of many diseases, including neurodegeneration, steatosis, Crohn's disease, infection, aging and cancer (2, 10).
Autophagy localizes to metabolically stressed tumor regions
As in normal cells, autophagy is activated in tumor cells by stress including starvation, hypoxia, and factor deprivation (2, 10). Tumor cells experience elevated metabolic stress from nutrient, factor and oxygen deprivation caused by inadequate blood supply resulting from deficient angiogenesis (11-13). This environmental metabolic stress in tumors is compounded by cell intrinsic metabolic stress derived from the high metabolic demand of cell proliferation and altered metabolism (aerobic glycolysis) where ATP production is inefficient. Autophagy localizes to hypoxic regions of tumors most distal to blood vessels where is supports tumor cell survival (14-17). Thus, autophagy plays a similar role in tumor cells as it does in normal cells, but because the inherent stress tumor cells encounter is greater, the dependence on autophagy may be more substantial. This difference between normal and tumor cells with respect to autophagy dependence may be useful for exploiting autophagy modulation in cancer therapy by providing a therapeutic window. Additionally, tumor masses have heterogeneous areas of vessel and nutrient supply, and tumor cells residing in hypoxic tumor regions undergoing autophagy are the tumor cells that resist radiation and chemotherapy. Knowing that autophagy supports survival of this important subpopulation of tumor cells provides an opportunity to target these resistant cells to improve cancer therapy.
Autophagy promotes tumor cell survival to metabolic stress
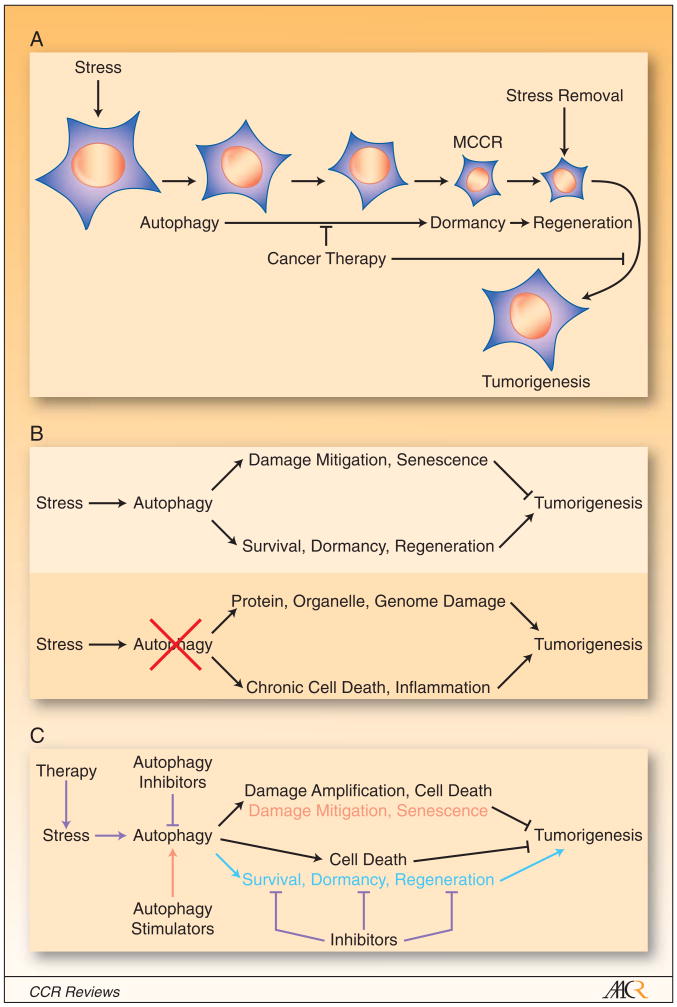
One of the most remarkable feats within the tumor cells' repertoire is to activate autophagy in response to stress that enables long-term survival, particularly when apoptosis is defective (14, 18). Normally apoptosis would eliminate tumor cells enduring unrelenting stress as a tumor suppression mechanism. Tumor cells frequently evolve defects in apoptosis, allowing autophagy to sustain survival for weeks in conditions of depravation (10, 11, 14, 18). Tumor cells can progressively eat themselves under prolonged stress, becoming less than one-third their normal size (Figure 1A). During the process of cellular consumption through autophagy, cell division and motility are suppressed which may represent and energy conservation effort (14). These apparently “dormant” tumor cells represent the minimal cells capable of recovery (MCCRs) that retain the capacity to return to their normal size and resume cell proliferation within 24 hours of having normal growth conditions restored (10, 11) (Figure 1A). Autophagy must be a highly selective process to permit extensive cellular degradation while retaining functional integrity. Establishing how cellular components are selected and directed to the autophagy pathway for degradation will be important, but likely involves specialized processes such as mitophagy (19) and p62 protein aggregate formation (20). Establishing dormancy with the capacity for regeneration are highly dependent on autophagy as tumor cells with autophagy defects are less efficient at achieving dormancy and regenerating. Thus, autophagy confers tumor cells with superior stress tolerance that limits damage, maintains viability, sustains dormancy, and facilitates recovery (Figure 1B).
Figure 1. Strategies for modulation of autophagy for cancer therapy.
(A), Autophagy-mediated survival and regeneration in tumor cells. Stress activates autophagy in tumor cells and in those with apoptotic defects this allows sustained, progressive autophagy and selective cellular self-consumption. This produces small cells that can remain in a dormant state in the presence of stress but when the stress is removed these minimal cells capable of recovery (MCCRs) regenerate and resume cellular proliferation. Autophagy thereby affords cancer cells with the flexibility to tolerate stress, even therapeutic stress, and resume growth when conditions are more favorable. Cancer therapy directed at blocking autophagy-mediated survival with autophagy inhibitors or specific inhibitors targeting the dormancy and recovery process may be extremely valuable.
(B), The double-edged sword of autophagy in tumor suppression. Stress activates autophagy, which mitigates damage and promotes senescence that limit tumorigenesis. Autophagy also enables tumor cells to survive metabolic stress, become dormant and regenerate with stress relief that can promote tumorigenesis. In tumors with autophagy defects, damage mitigation is impaired but the surviving, damaged tumor cells, particularly those with genome damage, can promote tumorigenesis. The impaired survival and induction of chronic cell death in tumors can also stimulate inflammation and tumorigenesis. Although autophagy-defective cells may have reduced fitness, these other factors compensate and overall tumorigenesis is stimulated.
(C), Autophagy modulation in cancer therapy. Cytotoxic, targeted and radiation therapy amplifies stress and autophagy in addition to the inherent metabolic stress in the tumor microenvironment. Some therapeutics such as angiogenesis inhibitors and 2-deoxyglucose may specifically amplify metabolic stress. Autophagy inhibitors such as HCQ block autophagy, which amplifies damage and cell death while also impairing dormancy and regeneration tilting the balance in favor of tumor regression (blue type). Specific inhibitors of survival such as apoptosis-inducing Bcl-2 inhibitors may work in part by preventing the downstream consequence of productive autophagy. Similarly, dormancy and regeneration inhibitors may be additionally useful anti-cancer therapeutics (blue type). Mechanisms to amplify tumor cell damage by specifically blocking autophagy of proteins or organelles or using proteasome inhibitors in conjunction with autophagy inhibitors should be explored. Alternatively, autophagy stimulators may be useful for cancer prevention by enhancing damage mitigation and senescence (red type).
A major aspect of cancer treatment is infliction of damage and stress on tumor cells sufficient to kill them by apoptosis, necrosis or alternate forms of cell death. Nonetheless, a small number of remaining tumor cells is too often sufficient for tumors to reoccur, often years later with deadly consequences. Remaining tumor cells that manage to tolerate treatment and persist only to re-emerge at a later time is a fundamental barrier to successful cancer treatment. The mechanism by which tumor cells achieve dormancy and regenerate needs to be established, and the precise role that autophagy plays in these processes needs to be defined. Therapeutic targeting of autophagy to impair dormancy and regeneration is worthwhile, but there may also be additional opportunities targeting the dormancy and regeneration pathways specifically (Figure 1C).
Autophagy is suppressed in many human tumors
Although autophagy is a survival pathway utilized by both normal and tumor cells to survive starvation and stress, paradoxically, autophagy defects are found in many human tumors. Allelic loss of the essential autophagy gene beclin1 is frequent in human breast, ovarian and prostate cancers (21). This limited assessment requires further substantiation along with evaluation of other autophagy genes coupled to functional analysis of autophagy in tumors. Allelic loss of beclin1 also renders mice prone to hepatocellular carcinoma (HCC), lung adenocarcinoma, mammary hyperplasia and lymphoma (22, 23). Defective autophagy through allelic loss of beclin1 or deficiency in the essential autophagy gene atg5 promotes tumorigenesis of immortal epithelial kidney and mammary cell lines (15, 17, 24). Loss of other autophagy regulators such as bif-1 (25) and atg4C (26) also renders mice tumor prone. The most common mutational event influencing autophagy in tumors, however, may be constitutive activation of the PI-3 kinase pathway.
PI-3 kinase and mammalian target of rapamycin (mTOR) activation are one of the most common events in human cancers and mTOR inhibits autophagy (2). The PI-3 kinase pathway functions to integrate growth factor and nutrient availability with biosynthetic processes such as protein translation and anabolic metabolism to favor cell growth and proliferation (27). This enables cells to manage activity and metabolic demand with the availability of external nutrient resources. When nutrients and growth factors are readily available, the demand for the catabolic activity of autophagy is reduced by mTOR. Under starvation conditions, the activity of the PI-3 kinase pathway and mTOR is suppressed, which down-regulates cellular biosynthetic processes and cellular proliferation but de-represses autophagy enabling catabolism. This permits cells to adapt to environmental fluctuations by adjusting behavior, utilization and consumption. The problem arises in cancer cells where the PI-3 kinase pathway in constitutively activated by mutations. This causes unrelenting cell growth signals uncoupled from nutrient and growth factor availability (11). While constitutive growth signals drive tumor cell proliferation, this also renders tumor cells less able to induce autophagy or to suppress consumption in response to stress that can lead to metabolic catastrophe where energetic demand exceeds production (11). This metabolic fragility of tumor cells has suggested therapeutic starvation as an approach to cancer therapy to exploit an inherent difference between normal and tumor cells (28). Collectively these findings suggest that although autophagy supports tumor cell survival, many tumors may paradoxically have autophagy suppressed. In the case of constitutive activation of the PI-3 kinase pathway, the advantage of deregulation of cell growth may be greater than the survival disadvantage conferred by suppressed autophagy. Alternatively, there may be additional aspects of autophagy defects that promote oncogenesis that compensate for survival deficit.
Physiological context of autophagy in cancer
A remaining question is the role that autophagy plays in oncogene activation and tumor suppressor gene inactivation that are forms of stress that may elevate the requirement for autophagy. Hypoxia induces hypoxia inducible transcription factor 1α (Hif-1α), which activates autophagy as part of a stress responsive and adaptive transcription program that also promotes angiogenesis and alters metabolism (29). Induction of Hif-1α due to inactivation of the von Hippel-Lindau (Vhl) tumor suppressor protein occurs with high frequency in renal clear cell carcinoma, but the contribution of autophagy in this setting is not known. Loss of the retinoblastoma (Rb) tumor suppressor protein de-represses the HIF-1α target Bnip3, and promotes autophagy and cell death (30). This suggests that autophagy may protect against Rb inactivation by enabling cellular preservation. Loss of checkpoint regulation may also increase tumor cell damage that may be counteracted by autophagy-mediated protein and organelle quality control surveillance to limit tumor progression. Indeed, deficiency or mutation in the p53 tumor suppressor gene promotes autophagy. This may be due to damage resulting from loss of the p53 DNA damage checkpoint or mechanisms not yet determined (31). Induction of wild type p53 also activates autophagy that could be due to direct transcriptional activation of downstream pro-autophagy regulators such as DRAM (32) or perhaps is an indirect consequence of p53 modulating cellular metabolism (33). A small mitochondrial form of the p19Arf tumor suppressor induces autophagy and cell death but whether this is related to Arf tumor suppression is not clear (34). Finally, endoplasmic reticulum localized Bcl-2 interacts with Beclin1 and suppresses autophagy; however, starvation disrupts this interaction allowing autophagy to take place (35). Bcl-2 blocks apoptosis and promotes tumorigenesis even in autophagy-defective cells suggesting that suppression of autophagy by Bcl-2 is not likely a critical factor regulating tumorigenesis (14). Thus, the role of autophagy in cancer needs to be assessed in context with the genetic makeup and environment of the tumor, and the guiding principals here are currently lacking.
Autophagy limits cell death and inflammation
One consequence of autophagy defects in tumors is impaired survival in stress, which results in chronic tumor cell death (14). Superficially, stimulation of cell death in tumors may appear to be a desirable outcome. However, persistent chronic cell death elicits an inflammatory response that can be pro-tumorigenic (36). Dead cells, particularly apoptosis-defective cells that undergo necrotic cell death releasing cellular contents, potently activating a pro-inflammatory immune response (37, 38). The nuclear protein high mobility group protein B1 (HMGB1) is released from necrotic cells where it is a ligand for the cell surface receptor for advanced glycation endproducts (RAGE) that is a potent activator of NF-κB. Similarly, nucleic acids released from necrotic cells can stimulate inflammation through activation of Toll-like receptors. The presence of these damage-associated molecular patterns (DAMPs) signals tissue damage and inflammation. Thus, tumors can appear as wounds that do not heal, which benefit from the persistent presence of inflammatory cells and cytokines meant to repair tissue damage. In the case of chronic cell death in a tumor, however, the wound does not heal, inflammation does not resolve, and instead tumor growth is enhanced (Figure 1B).
Stimulation of apoptotic cell death in tissues can also be pro-inflammatory and oncogenic. Chronic apoptotic cell death in the liver can trigger inflammation, more tissue damage and an increased risk of HCC (39). Hepatocyte cell death activates resident macrophages (Kupffer cells) to produce hepatomitigens that stimulate compensatory proliferation. This is a normal reaction to repair tissue damage, but when the underlying cause is persistent (hepatitis virus infection, alcohol consumption, toxins, perhaps defective autophagy), this chronic inflammation promotes tumorigenesis. Thus, acute cell death may be required for tumor eradication while chronic cell death can promote tissue damage, inflammation and tumorigenesis. Interestingly, autophagy defects in mice cause hepatocyte toxicity, liver damage and HCC (6, 8, 22, 23).
Autophagy-defective embryonic tissues are impaired for the removal of cell corpses (40), which raises the potential to prolong pro-inflammatory stimuli through both increased cell death and the failure to eliminate dead cells. Immortal mouse mammary epithelial cells with allelic loss of beclin1 accumulate cell corpses when grown as three-dimensional mammospheres, but if this contributes to inflammation and increased tumorigenesis is not known (15). Apoptosis-defective tumors with autophagy defects display chronic necrosis and inflammation with dramatic macrophage infiltration, NF-κB activation and cytokine production compared to tumors where autophagy is intact (14). These findings are consistent with autophagy promoting tumor cell survival and limiting inflammation as a non-cell autonomous means to suppress tumorigenesis. These contrasting tumor cell survival promoting and tumor suppressing activities contribute to autophagy acting as a double-edged sword in the cancer setting (Figure 1B).
Autophagy limits genome damage
Cells in tissues of mice with autophagy defects accumulate damaged mitochondria and p62- and ubiquitin-containing protein aggregates, suggesting a general role for autophagy in maintenance of cellular health through damage mitigation. Immortal epithelial cell lines from autophagy-defective mice manifest genome damage, indicating that in checkpoint-defective cells that failure of damage mitigation by autophagy may ultimately result in DNA mutations and chromosome instability (15, 17). Since an elevated mutation rate and genome instability promote cancer, this raises the possibility that damage mitigation by autophagy and protection of the genome is a possible tumor suppression mechanism. How autophagy protects the genome from damage is not yet clear but may result from clearance of damaged proteins and organelles and the suppression of oxidative stress. Accumulation of either protein aggregates or damaged mitochondria are associated with increased production of reactive oxygen species (ROS) (8). How damaged protein accumulation causes oxidative stress is not clear but may result from increased protein reduction and isomerization during re-folding, which is an oxidative reaction. Degradation of denatured or unfolded proteins through autophagy may eliminate the need for excessive protein folding activity and oxidative stress.
Damaged mitochondria are a well-known source of ROS emanating from disruption of electron transport. ROS, in turn, mediate further organelle, protein damage and DNA damage. The failure of protein and organelle quality control in autophagy-defective cells may thereby set up a downward spiral where persistence of damaged proteins and organelles causes ROS that further damages proteins and organelles and ultimately the genome. If this is the case then acceleration of oxidative stress may ultimately be the route cause of cell damage that renders autophagy-defective cells more tumor prone and illustrates the protective function of autophagy. If so, this would be a novel mechanism of tumor suppression and would implicate anti-oxidants as a protective measure where autophagy defects are predicted to predispose to cancer. Ultimately, identification of the source of ROS and whether unfolded protein or damaged organelles contribute to ROS production will be informative.
Role of autophagic cell death
In contrast to the survival-promoting function autophagy that is supported by substantial evidence, induction autophagic cell death has also been proposed as a possible tumor suppression mechanism. This comes from the observation that cell death can occur concomitant with features of autophagy (41), and that excessive stimulation of autophagy through over-expression of beclin1, suppresses tumorigenesis (42). Prolonged stress and progressive autophagy can also eventually lead to cell death (10). Excessive cellular damage may lead to cell death by over-stimulating autophagy and cellular self-consumption. Small molecules that promote autophagic cell death in Vhl-negative renal cell carcinoma have been identified in a screen for synthetic lethality with loss of Vhl (43). Whether the approach of induction of autophagic tumor cell death induction can work in the clinic remains to be tested. In vivo evidence in mammals to support these concepts has so far been limited (41). One obstacle to validating the concept of autophagic cell death is that the only marker is improved cellular survival when autophagy is inhibited. The identification of biochemical markers for, and definition of, the process of autophagic cell death will be valuable to establish how it may be distinct from apoptosis (44), necrosis (45), entosis (46, 47), necroptosis (48, 49), or other forms of cell death. Moreover, many of the studies reporting induction of autophagic cell death have been performed with non-specific pharmacological inhibitors of autophagy or apoptosis, or with RNAi-mediated knockdown of autophagy regulators (50). These approaches have been problematic due to off-target effects.
In contrast, autophagic cell death in Drosophila has been shown to be an important tissue remodeling and resource reutilization process is larval morphogenesis (51). A potentially similar role for autophagy in mammalian cells during tissue remodeling will be important to investigate. An example where stress orchestrates cellular resource reutilization in mammals is senescence. The stress of oncogene activation triggers oncogene-induced senescence (OIS), a tumor suppression mechanism that diverts emerging tumor cells to cell cycle exit, cellular remodeling and secretion of inflammatory mediators (52). Autophagy is induced during and promotes senescence including the secretory phenotype (53, 54). The ability of autophagy to turnover intracellular components may facilitate senescence by intracellular recycling and remodeling during acquisition of the senescence phenotype. It will be of interest to test the role that autophagy-enabled senescence plays in tumor suppression (53).
Cancer is a proteinopathy
Autophagy plays a role in the degradation of mutant proteins, and by preventing their accumulation, may provide protection against degenerative conditions such as Parkinson's and Huntington's disease (55-57). Accumulation of mutant huntingtin protein and neurodegeneration is accelerated by defective autophagy and suppressed by autophagy stimulation. The accumulation of polyubiquitinated protein aggregates and neurodegeneration in the course of normal aging is also accelerated by autophagy defects. Progression of diseases of mutant protein accumulation or proteinopathies, in general, may benefit from autophagy as a mechanism to facilitate their degradation and limit accumulation. Cancer is also a disease of mutant or over-expressed protein accumulation and can be considered a form of proteinopathy where overexpressed mutated oncoproteins (receptor tyrosine kinases, for example) or tumor suppressor proteins (p53, for example) are prominent features of many tumors. Whether autophagy suppresses the accumulation of these proteins as a means of limiting oncogenesis is not known.
Defects in autophagy and impairment of protein degradation cause the abnormal accumulation of endoplasmic reticulum (ER) chaperones and p62 that functions to direct polyubiqutinated proteins to autophagosomes for degradation (6, 8, 20). Cancer may place a greater burden on the protein quality control system through the overproduction of proteins, mutant protein expression or through a high rate of protein synthesis brought about by constitutive growth. Some cancers such as multiple myeloma are sensitive to proteasome inhibitors, which may result from a high rate of immunoglobulin production and increased generation of unfolded proteins. Proteaseome- and autophagy-mediated protein degradation may serve overlapping and complementary roles in maintaining protein quality control and cancer may increase the demand for these activities to limit damage, preserving viability.
Double-edged sword of autophagy and cancer: Damage mitigation vs. survival promotion
The emerging role of autophagy in cancer is one of a double-edged sword (Figure 1B). On the one hand, autophagy enables tumor cells to tolerate stress including a hypoxic microenvironment, starvation and probably some forms of therapy. Even with prolonged stress, autophagy can allow prolonged survival, generating dormant tumor cells that have the capacity to resume growth when conditions are more favorable. This process of stress survival, dormancy and regeneration afforded by autophagy may be a major obstacle to achieving successful cancer treatment. On the other hand, autophagy plays an important role in damage mitigation in response to stress that can limit tumorigenesis (Figure 1B). By clearing away damaged proteins and organelles, and perhaps by maintaining energy homeostasis through intracellular recycling, autophagy can ultimately prevent genome damage that drives tumorigenesis. Damage mitigation can also suppress tumorigenesis by limiting chronic cell death and inflammation that can promote tumorigenesis. With this in mind, it is easier to visualize how disruption of autophagy, which reduces cellular fitness, actually promotes tumorigenesis. It is the capacity of tumor cells to adapt and evolve in response to selective pressure and become progressively more deleterious to their human host that makes cancer difficult to treat. Autophagy may suppress this evolution by maintaining homeostatic conditions. Although survival of tumor cells may be increased by autophagy, this may be compensated for by the damage mitigation function (Figure 1B). In tumor cells with autophagy inactivated, the reduction in survival may be inconsequential in the presence of enhanced genome damage and chronic inflammation to promote tumor growth and progression (Figure 1B). The next challenge will be to use these guiding principals to test autophagy modulation in the setting of cancer therapy, bearing in mind that tumors with autophagy intact may respond differently from those with autophagy suppressed. An additional challenge will be to recognize functional autophagy status in tumors to direct the appropriate therapy.
Strategies for targeting the autophagy pathway for cancer therapy
Blocking survival to metabolic stress with autophagy inhibitors
As autophagy is a survival pathway utilized by tumor cells to tolerate metabolic stress, autophagy inhibitors are expected to by useful for cancer therapy (Figure 1C) (10-13, 58-60). Autophagy inhibitors are particularly attractive because they can target those tumor cells in hypoxic tumor regions, which are therapy, particularly radiation, resistant. Additionally, tumor cells in the process of metastasizing may be particularly dependent on autophagy, supporting approaches to abrogate autophagy in early progression and the adjuvant setting. While some death my result from autophagy inhibitors producing a survival disadvantage in stress, they are also expected to impair dormancy and recovery.
Combining autophagy inhibitors with inducers of metabolic stress
It is unlikely that autophagy inhibitors will be useful for cancer therapy as single agents because only a subpopulation of tumor cells undergo autophagy. Since many cancer therapeutics induce autophagy (Table 1), because they induce damage (cytotoxic chemotherapy), metabolic stress (angiogenesis inhibitors, 2-deoxyglucose), or block growth signaling pathways (targeted non-cytotoxics, kinase inhibitors) mimicking factor deprivation or starvation, the addition of autophagy inhibitors would be expected to enhance cytotoxicity of these agents. mTOR inhibitors, in particular, induce autophagy and if this provides stress protection, then their full therapeutic advantage will be realized in combination with autophagy inhibitors. The approach here will be to block stress adaptation and amplify damage by inhibiting autophagy (Figure 1C). It will be important to induce acute rather than chronic cell death and inflammation that can be counterproductive. The downside to this approach is the loss of the damage mitigation and potentially tumor suppression activity of autophagy (Figure 1C). This may not be an issue if the induction of stress in combination with inhibition of autophagy augments toxicity sufficient to kill all tumor cells. Potential collateral damage to normal tissue will need to be addressed, although the inherent metabolic stress in tumors may provide a therapeutic window.
Table 1. Therapeutic agents demonstrated to induce autophagy.
| Agent | Drug Class | Reference |
|---|---|---|
| Endostatin | Anti-angiogenesis | (82) |
| Sorafenib | TKI, VEGF inhibitor, Raf | (83) |
| SAHA | Histone deacetylase inhibitor | (68) |
| Farnesyltransferase inhibitors | Farnesyltransferase inhibitors | (84) |
| Temsirolimus | mTOR inhibitor | (85) |
| Everolimus | mTOR inhibitor | (86) |
| Deoxyglucose | Glycolysis inhibitor | (28) |
Inhibition of apoptosis, dormancy and regeneration
An alternate approach would be to specifically target the survival, dormancy and regeneration mechanisms acting in concert with or downstream of autophagy. Disabling apoptosis, for example, should decrease acquisition of dormancy and regeneration (Figure 1C). Although autophagy inhibition stimulates apoptosis, there may be a specific apoptotic mechanism responsible for the survival and regeneration of dormant cells that can be identified. An anti-apoptotic Bcl-2 inhibitor is in clinical trials to promote tumor cell apoptosis (61). Metabolic stress triggers apoptosis that is inhibited by Bcl-2, requires proapoptotic Bim and is signaled through the core proapoptotic regulators Bax and Bak (62, 63). Whether Bcl-2 is also essential for the viability of dormant and regenerating tumor cells should be assessed. Defining the mechanisms governing dormancy and regeneration may reveal novel targets downstream of autophagy and inhibiting these processes is potentially valuable. Dormancy- and regeneration-specific inhibitors may have the advantage of retaining the protective, damage mitigation function of autophagy.
Development and assessment of autophagy inhibitors in cancer therapy
Development of specific autophagy inhibitors for cancer therapy should be possible as there are kinases (Atg1/Unc-51-like kinase 1/2/3, Vps34), proteases (Atg4) and two ubiquitin-like conjugation systems that regulate the activation of autophagy and autophagosome formation (64). There are also signaling pathways, both mTOR-dependent and –independent, that regulate activation of the autophagy that can be targeted. Elongation factor-2 kinase (eEF-2 kinase), for example, which is downstream of mTOR, promotes autophagy-mediated survival of glioblastoma, and the expectation is that eEF-2 kinase inhibitors will be therapeutically useful (65).
There are poorly understood mechanisms for targeting proteins and organelles to autophagosomes that are likely to contain additional targets. Signaling pathways that promote autophagy are also good candidates for inhibitor development. Currently, hydroxychloroquine (HCQ), which blocks lysosome acidification and autophagosome degradation, is available as an autophagy inhibitor that will enable assessment of the utility of this approach. A number of clinical trials have been initiated and are currently accruing patients with solid and hematopoietic tumors to test this (Table 2). Most of these trials are combinations of HCQ with cytotoxic chemotherapy, inducers of metabolic stress or targeted therapies with the overall hypothesis that autophagy is a mechanism of therapeutic resistance and HCQ will increase cytoxicity by abrogation of autophagy. As shown in Table 2, most of these studies combine HCQ with a more standard cancer therapy expected to induce autophagy (some of these potential agents or classes of agents are shown in Table 1). These trials should begin to reveal if there is therapeutic benefit to blocking autophagy for cancer therapy. Although HCQ is not an exclusive autophagy inhibitor, it is relatively non-cytotoxic and blocks flux through the autophagy pathway by inhibiting the terminal lysosome degradation step. It remains to be demonstrated if HCQ can block autophagy in human tumors in vivo, and if or how the genetic makeup of tumors influences this response. To this end, the identification of biomarkers and signatures that reflect functional autophagy status and that detect therapeutic modulation of autophagy in human tumors will require development.
Table 2. Selected Clinical Trials of Hydroxychloroquine and Modulation of Autophagy*.
| Condition | Intervention | Phase | Sponsors Collaborators | ClinicalTrials.gov Identifier | Title |
|---|---|---|---|---|---|
| Prostate Cancer | docetaxel hydroxy-chloroquine | II | The Cancer Institute of New Jersey (CINJ), The National Cancer Institute (NCI) | NCT00786682 | A Phase II Study of Docetaxel and Modulation of Autophagy With Hydroxychloroquine for Metastatic Hormone Refractory Prostate Cancer |
| Prostate Cancer | hydroxy-chloroquine | II | CINJ, NCI | NCT00726596 | Autophagic Cell Death in Patients With Hormone-Dependent Prostate-Specific Antigen Progression After Local Therapy for Prostate Cancer |
| Multiple Myeloma, Plasma Cell Neoplasm | bortezomib hydroxy-chloroquine | I/II | U. Pennsylvania, NCI | NCT00568880 | A Phase I/II Trial of Hydroxychloroquine Added to Bortezomib for Relapsed/Refractory Myeloma |
| Brain, Central Nervous System Tumors | hydroxy-chloroquine temozolomide | I/II | U. Pennsylvania, NCI | NCT00486603 | A Phase I/II Trial of Hydroxychloroquine in Conjunction With Radiation Therapy and Concurrent and Adjuvant Temozolomide in Patients With Newly Diagnosed Glioblastoma Multiforme |
| Breast Cancer | hydroxy-chloroquine ixabepilone | I/II | CINJ, NCI | NCT00765765 | Phase I/II Study of Ixabepilone in Combination With the Autophagy Inhibitor Hydroxychloroquine for the Treatment of Patients With Metastatic Breast Cancer |
| Lung Cancer | bevacizumab carboplatin hydroxy-chloroquine paclitaxel | I/II | CINJ, NCI | NCT00728845 | Modulation of Autophagy With Hydroxychloroquine in Combination With Carboplatin, Paclitaxel and Bevacizumab in Patients With Advanced/Recurrent Non-Small Cell Lung Cancer - A Phase I/II Study |
| Adult Solid Tumors | hydroxy-chloroquine temozolomide | I | U. Pennsylvania, NCI | NCT00714181 | A Phase I Study of Hydroxychloroquine in Combination With Temozolomide in Patients With Advanced Solid Tumors |
| Non-Small Cell Lung Cancer | gefitinib, hydroxy-chloroquine | I/II | National University Hospital, Singapore, Massachusetts General Hospital, AstraZeneca | NCT00809237 | A Phase II With a Lead in Phase I Study to Examine the Tolerability, Safety Profile and Efficacy of Hydroxychloroquine and Gefitinib in Advanced Non-Small Cell Lung Cancer |
| Cancer | hydroxy-chloroquine sunitinib malate | I | CINJ, NCI | NCT00813423 | Autophagic Modulation With Anti-Angiogenic Therapy in Patients With Advanced Malignancies: A Phase I Trial of Sunitinib and Hydroxychloroquine |
| B-Cell Chronic Lymphocytic Leukemia | hydroxy-chloroquine | II | North Shore Long Island Jewish Health System | NCT00771056 | Phase II Study to Evaluate the Tolerability and Efficacy of Treatment of Previously Untreated B-Cell Chronic Lymphocytic Leukemia (B-CLL) Patients With Hydroxychloroquine. |
Study citations were obtained form the NIH/NCI clinicaltrials.gov website
Efficacy of HCQ and chloroquine (CQ) in preclinical models
Efficacy of autophagy inhibition with HCQ and CQ has begun to be assessed in animal models and human cancer cells and lines and the results are encouraging. In a mouse model for Myc-driven lymphoma, HCQ and autophagy inhibition enhances the ability of either p53 or alkylating agents to promote tumor cell death (66). CQ also impairs spontaneous lymphomagenesis in Atm-deficient mice that model ataxia telangiectasia and in Myc-transgenic mice that model human Burkitt lymphoma, although CQ did not prevent spontaneous lymphomagenesis in p53-deficient mice (67). The mechanism of CQ-mediated tumor cell toxicity was consistent with promotion of lysosomal stress and p53-dependent and apoptosis-independent tumor cell death. Thus CQ can enhance the death-promoting activity of a common tumor suppression pathway to prevent cancer development. CQ also promotes cancer cell death in cooperation with the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) in chronic myelogenous leukemia (CML) cell lines and primary CML cells expressing wild type and imatinib-resistant mutant forms of Bcr-Abl (68). Synergy of CQ and SAHA was associated with induction of, and was enhanced by, the lysosomal protease cathepsin D, suggesting that CQ and autophagy inhibition promotes tumor cell death by a lysosome-driven process. As CQ and HCQ block flux through the autophagy pathway at the lysosomal degradation step, it will be of interest to compare and contrast this with the effectiveness of blocking autophagy initiation. Collectively, these findings suggest that autophagy inhibition can augment the anti-cancer activity of both chemotherapy and endogenous tumor suppressor mechanisms to promote tumor regression and to limit spontaneous tumor development.
Compromising protein degradation pathways for cancer chemotherapy
There are two main protein degradation systems in cells, autophagy, which is a mechanism for bulk protein degradation in lysosomes, and the proteasome pathway, which is a mechanism for degradation of individual proteins tagged with polyubiquitin in proteasomes. These two pathways for protein degradation may be partly complementary or interdependent in that inhibition of the proteasome pathway induces autophagy (69) and inhibition of autophagy increases the accumulation of polyubiqutinated proteins (4-7). Moreover, autophagy rescues toxicity (70) whereas autophagy defects confer sensitivity to proteasome inhibition (8), supporting the compensatory nature of these protein degradation pathways. There is also evidence that polyubiquitin-containing protein aggregates can block proteasome-mediated degradation by clogging up proteasomes (71). Predictions are that inhibiting both the proteasome and autophagy degradation pathways may be more toxic to cancer cells, particularly those with a high rate of protein synthesis such as those secreting immunoglobulin (multiple myeloma). Indeed, multiple myeloma is sensitive to the proteasome inhibitor Velcade, which is FDA approved for treatment of this cancer (72). The validity of combining HCQ to augment Velcade in multiple myeloma is being assessed in the clinic (Table 2). Similarly the histone deacetylase HDAC6, which binds polyubiquitinated proteins and promotes autophagy-mediated protein degradation, and inhibition of HDAC activity with SAHA synergizes with Velcade to kill multiple myeloma (73). Thus, inhibiting both the proteasome and autophagy pathways may be an important approach to cancer treatment.
Stimulation of autophagy for cancer chemoprevention
Since autophagy defects predispose to cancer and other diseases, the prospect of stimulating autophagy as a disease prevention measure is promising (Figure 1C). In models of neurodegeneration, inhibiting autophagy accelerates disease progression whereas stimulating autophagy with mTOR inhibitors or other autophagy stimulators or by augmentation HDAC6 expression delays disease progression (55-57, 70, 74-78). Whether this strategy will be effective for cancer prevention remains to be investigated. Non-specific means for stimulating autophagy such as caloric restriction and fasting may improve human health and suppress cancer, whereas excess nutritional consumption that should repress autophagy is considered deleterious (79). It will be of interest to test whether these activities can be attributed to modulation of autophagy.
Autophagy defects in mice cause liver disease resembling steatohepatitis and HCC (6, 8, 22, 23). Patients with steatohepatitis are at risk for developing HCC and are a candidate group to test autophagy stimulation for cancer prevention. CQ can suppress spontaneous, oncogene activation-driven lymohomagenesis in mice suggesting that inhibition of autophagy may be a prevention strategy (67). Augmentation of tumor suppressor activity, such as that of p53, with autophagy inhibitors may be more advantageous in this setting. In contrast, steatohepatitis is symptomatic of protein quality control failure may rather benefit from autophagy stimulation. With steatohepatitis, tissue damage and the resulting inflammation promote HCC (39) that may be suppressed by autophagy stimulation. Indeed, stimulation of autophagy eliminates p62-containing Mallory Bodies induced by proteasome inhibition (80), p62 accumulation is responsible for liver toxicity (6) and tumorigenesis (8) in autophagy-defective mice, and low Beclin1 levels and reduced autophagy may be associated with poor prognosis in HCC (81). It is clear that combination cancer therapy targeting multiple, distinct pathways is important to achieve cures and the autophagy pathway provides novel means to augment therapy.
Acknowledgments
We thank Drs. Robin Mathew, Cristina Karp, Anne Strohecker, Vassilki Karantza, Mark Stein, and Janice Menhert for helpful comments and discussion and Tami Sharkey for assistance with preparation of the manuscript. Support from the National Institutes of Health (R37CA53370 and RO1CA130893 to E. W.) and Department of Defense (W81XWH06-1-0514 and W81XWH05 to R. S. D. and E. W.) is gratefully acknowledged.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 4.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 5.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 6.Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 7.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009 doi: 10.1016/j.cell.2009.03.048. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crotzer VL, Blum JS. Autophagy and its role in MHC-mediated antigen presentation. J Immunol. 2009;182:3335–41. doi: 10.4049/jimmunol.0803458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–7. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin S, DiPaola RS, Mathew R, White E. Metabolic catastrophe as a means to cancer cell death. J Cell Sci. 2007;120:379–83. doi: 10.1242/jcs.03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin S, White E. Tumor suppression by autophagy through the management of metabolic stress. Autophagy. 2008;4:563–6. [PMC free article] [PubMed] [Google Scholar]
- 14.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karantza-Wadsworth V, Patel S, Kravchuk O, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathew R, Karantza-Wadsworth V, White E. Assessing metabolic stress and autophagy in epithelial tumors. Methods Enzymol. 2009;453:51–78. doi: 10.1016/S0076-6879(08)04004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Kundu M, Ney PA. Mitophagy in mammalian cells: the reticulocyte model. Methods Enzymol. 2009;452:227–45. doi: 10.1016/S0076-6879(08)03615-X. [DOI] [PubMed] [Google Scholar]
- 20.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 21.Aita VM, Liang XH, Murty VV, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 22.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Degenhardt K, Chen G, Lindsten T, White E. BAX and BAK mediate p53-independent suppression of tumorigenesis. Cancer Cell. 2002;2:193–203. doi: 10.1016/s1535-6108(02)00126-5. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 27.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 28.DiPaola RS, Dvorzhinski D, Thalasila A, et al. Therapeutic starvation and autophagy in prostate cancer: a new paradigm for targeting metabolism in cancer therapy. Prostate. 2008;68:1743–52. doi: 10.1002/pros.20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Bosch-Marce M, Shimoda LA, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–42. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crighton D, Wilkinson S, O'Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 33.Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 34.Reef S, Zalckvar E, Shifman O, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–75. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–6. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 37.Coffelt SB, Scandurro AB. Tumors sound the alarmin(s) Cancer Res. 2008;68:6482–5. doi: 10.1158/0008-5472.CAN-08-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lotze MT, Zeh HJ, Rubartelli A, et al. The grateful dead: damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol Rev. 2007;220:60–81. doi: 10.1111/j.1600-065X.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 39.Sun B, Karin M. NF-kappaB signaling, liver disease and hepatoprotective agents. Oncogene. 2008;27:6228–44. doi: 10.1038/onc.2008.300. [DOI] [PubMed] [Google Scholar]
- 40.Qu X, Zou Z, Sun Q, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 41.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–10. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 43.Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams JM, Cory S. Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr Opin Immunol. 2007;19:488–96. doi: 10.1016/j.coi.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zong WX, Thompson CB. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- 46.Overholtzer M, Mailleux AA, Mouneimne G, et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–79. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 47.White E. Entosis: it's a cell-eat-cell world. Cell. 2007;131:840–2. doi: 10.1016/j.cell.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008;135:1161–3. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Hitomi J, Christofferson DE, Ng A, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–23. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White E. Autophagic cell death unraveled: Pharmacological inhibition of apoptosis and autophagy enables necrosis. Autophagy. 2008;4:399–401. doi: 10.4161/auto.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–48. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chien Y, Lowe SW. Secreting tumor suppression. Cell. 2008;132:339–41. doi: 10.1016/j.cell.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White E, Lowe SW. Eating to exit: autophagy-enabled senescence revealed. Genes Dev. 2009;23:784–7. doi: 10.1101/gad.1795309. [DOI] [PubMed] [Google Scholar]
- 54.Young AR, Narita M, Ferreira M, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 56.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–12. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 57.Williams A, Jahreiss L, Sarkar S, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 58.Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 2007;13:7271–9. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 59.Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbamcr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dang CV. Antimalarial therapy prevents Myc-induced lymphoma. J Clin Invest. 2008;118:15–7. doi: 10.1172/JCI34503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tse C, Shoemaker AR, Adickes J, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–8. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 62.Tan TT, Degenhardt K, Nelson DA, et al. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7:227–38. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 63.White E. Role of the metabolic stress responses of apoptosis and autophagy in tumor suppression. Ernst Schering Found Symp Proc. 2007:23–34. doi: 10.1007/2789_2008_087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 65.Wu H, Yang JM, Jin S, Zhang H, Hait WN. Elongation factor-2 kinase regulates autophagy in human glioblastoma cells. Cancer Res. 2006;66:3015–23. doi: 10.1158/0008-5472.CAN-05-1554. [DOI] [PubMed] [Google Scholar]
- 66.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carew JS, Nawrocki ST, Kahue CN, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–22. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding WX, Ni HM, Gao W, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pandey UB, Nie Z, Batlevi Y, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 71.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 72.Chauhan D, Bianchi G, Anderson KC. Targeting the UPS as therapy in multiple myeloma. BMC Biochem. 2008;9 1:S1. doi: 10.1186/1471-2091-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nawrocki ST, Carew JS, Maclean KH, et al. Myc regulates aggresome formation, the induction of Noxa, and apoptosis in response to the combination of bortezomib and SAHA. Blood. 2008;112:2917–26. doi: 10.1182/blood-2007-12-130823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 75.Sarkar S, Perlstein EO, Imarisio S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–8. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shibata M, Lu T, Furuya T, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–85. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 77.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–31. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang L, Yu J, Pan H, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104:19023–8. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singletary K, Milner J. Diet, autophagy, and cancer: a review. Cancer Epidemiol Biomarkers Prev. 2008;17:1596–610. doi: 10.1158/1055-9965.EPI-07-2917. [DOI] [PubMed] [Google Scholar]
- 80.Harada M, Hanada S, Toivola DM, Ghori N, Omary MB. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47:2026–35. doi: 10.1002/hep.22294. [DOI] [PubMed] [Google Scholar]
- 81.Ding ZB, Shi YH, Zhou J, et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008;68:9167–75. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- 82.Ramakrishnan S, Nguyen TM, Subramanian IV, Kelekar A. Autophagy and angiogenesis inhibition. Autophagy. 2007;3:512–5. doi: 10.4161/auto.4734. [DOI] [PubMed] [Google Scholar]
- 83.Park MA, Zhang G, Martin AP, et al. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide-dependent CD95 and PERK activation. Cancer Biol Ther. 2008;7:1648–62. doi: 10.4161/cbt.7.10.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pan J, Chen B, Su CH, et al. Autophagy induced by farnesyltransferase inhibitors in cancer cells. Cancer Biol Ther. 2008;7:1679–84. doi: 10.4161/cbt.7.10.6661. [DOI] [PubMed] [Google Scholar]
- 85.Yazbeck VY, Buglio D, Georgakis GV, et al. Temsirolimus downregulates p21 without altering cyclin D1 expression and induces autophagy and synergizes with vorinostat in mantle cell lymphoma. Exp Hematol. 2008;36:443–50. doi: 10.1016/j.exphem.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 86.Cao C, Subhawong T, Albert JM, et al. Inhibition of mammalian target of rapamycin or apoptotic pathway induces autophagy and radiosensitizes PTEN null prostate cancer cells. Cancer Res. 2006;66:10040–7. doi: 10.1158/0008-5472.CAN-06-0802. [DOI] [PubMed] [Google Scholar]