Abstract
The bfrB gene from Pseudomonas aeruginosa was cloned and expressed in E. coli. The resultant protein (BfrB), which assembles into a 445.3 kDa complex0020from 24 identical subunits, binds 12 molecules of heme axially coordinated by two Met residues. BfrB, isolated with 5–10 iron atoms per protein molecule, was reconstituted with ferrous ions to prepare samples with a core mineral containing 600 ± 40 ferric ions per BfrB molecule and approximately one phosphate molecule per iron atom. In the presence of sodium dithionite or in the presence of P. aeruginosa ferredoxin NADP reductase (FPR) and NADPH the heme in BfrB remains oxidized and the core iron mineral is mobilized sluggishly. In stark contrast, addition of NADPH to a solution containing BfrB, FPR and the apo-form of P. aeruginosa bacterioferritin associated ferredoxin (apo-Bfd) results in rapid reduction of the heme in BfrB and in the efficient mobilization of the core iron mineral. Results from additional experimentation indicate that Bfd must bind to BfrB to promote heme mediation of electrons from the surface to the core to support the efficient mobilization of ferrous ions from BfrB. In this context, the thus far mysterious role of heme in bacterioferritins has been brought to the front by reconstituting BfrB with its physiological partner, apo-Bfd. These findings are discussed in the context of a model for the utilization of stored iron in which the significant upregulation of the bfd gene under low-iron conditions [Ochsner, U.A., Wilderman, P.J., Vasil, A.I., and Vasil, M.L. (2002) Mol. Microbiol. 45, 1277–1287] ensures sufficient concentrations of apo-Bfd to bind BfrB and unlock the iron stored in its core. Although these findings are in contrast to previous speculations suggesting redox mediation of electron transfer by holo-Bfd, the ability of apo-Bfd to promote iron mobilization is an economical strategy used by the cell because it obviates the need to further deplete cellular iron levels to assemble iron sulfur clusters in Bfd before the iron stored in BfrB can be mobilized and utilized.
Iron is the fourth most abundant element in Earth’s crust, after oxygen silicon and aluminum and the second most abundant metal. With a few exceptions, iron is a required nutrient because the metal ion is incorporated as cofactor in proteins and enzymes involved in a wide range of biological processes (1). Under biological conditions iron is present mainly in two oxidation states, the soluble Fe2+ which can react with O2 to form reactive oxygen species and the highly insoluble Fe3+ ion. These chemical properties present living organisms with enormous challenges for the acquisition and homeostasis of this nutrient. One mechanism whereby iron toxicity can be ameliorated or controlled is by storage of excess iron in ferritin and ferritin-like molecules. These molecules, found in prokaryotes and eukaryotes (2), likely evolved to combat the insolubility of Fe3+ and the toxicity of Fe2+ in aerobic environments, because they function by oxidizing Fe2+ using O2 and H2O2 as electron acceptors and by internalizing and compartmentalizing the resultant Fe3+ in the form of a mineral. Consequently, ferritin and ferritin-like molecules may also contribute to managing oxidative stress triggered by increased concentrations of “free” cellular Fe2+ (3). Intracellular reserves of iron in bacteria are placed in three types of ferritin-like molecules; the classic ferritins, which are also present in eukaryotes, the bacterioferritins, which are present only in eubacteria, and the Dps proteins, which are present only in prokaryotes (1). Ferritins are composed of 24 subunits that assemble into a spherical protein shell surrounding a central cavity where the iron mineral is stored. Bacterioferritins are also assembled from 24 subunits into a similar structure that harbors 12 heme molecules, and Dps proteins which assemble from 12 subunits to form a central cavity smaller than those formed by ferritins and bacterioferritins.
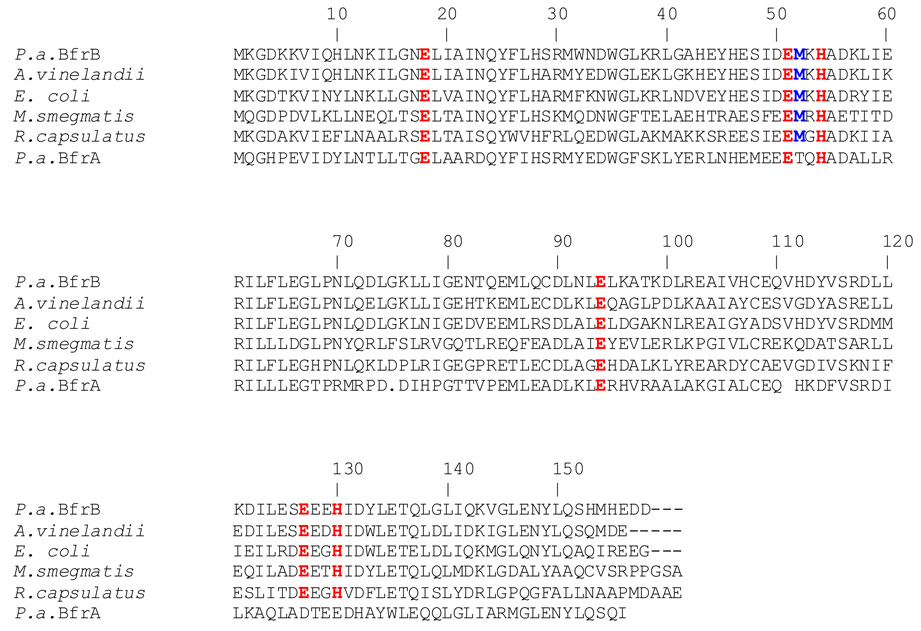
Early studies with bacterioferritin from P. aeruginosa suggested that the protein is composed of two subunits, termed α- and β-. Although the relative proportions of the two subunits varied widely from preparation to preparation, the α-subunit was the most abundant (4). More recent investigations have established that there are two genes coding for bacterioferritin molecules in P. aeruginosa, termed bfrA and bfrB (5). In Figure 1 the amino acid sequences of BfrA and BfrB from P. aeruginosa are compared with the sequences of structurally characterized bacterioferritins from E. coli (6), Azotobacter vinelandii (7) and Mycobacterium smegmatis (8). As indicated above, the structure of bacterioferritin is an assembly of 24 subunits that forms a nearly spherical, hollow molecule that binds 12 heme molecules, where each heme is coordinated by two Met residues (Met52) from a pair of subunits related to one another by two-fold symmetry. It is interesting that Met52 is absent from the sequence of BfrA, suggesting that the latter may not bind heme. In comparison, residues that act as ligands to the di-iron ferroxidase center are conserved in all the amino acid sequences shown in Figure 1, including that of BfrA. Thus, the amino acid sequence of BfrA, which indicates a full ferroxidase center but no heme-binding Met52 suggests that BfrA is a misnomer, as the bfrA gene likely codes for a bacterial ferritin and not a heme binding bacterioferritin. The bfrA gene is adjacent to the KatA gene, which codes for a catalase active in all growth phases, and is thought to be involved in imparting resistance to hydrogen peroxide. A bfrA mutant of P. aeruginosa was found to express only ~ 50% of the catalase activity observed in wild type cells due to a decrease in the synthesis of KatA. This observation, and the nearly complete restoration of catalase activity brought about by complementation with a plasmid expressing the bfrA gene, led to the suggestion that iron stored in BfrA is incorporated into protoporphyrin IX to make the heme cofactor of KatA (5).
Figure 1. Bacterioferritin sequences from different organisms aligned against P. aeruginosa BfrB (P.a. BfrB).
Residues involved in the ferroxidase center, as well as Met-52, which coordinates the heme axially, are highlighted. Note that Met52 is absent in the sequence of P. aeruginosa BfrA. The sequences were aligned with the aid of ClustalW (46).
The bfrB gene in P. aeruginosa and in several other bacteria is adjacent to a bfd gene, which codes for a small protein known as Bfd (bacterioferritin associated ferredoxin). Genes coding for Bfd in E. coli (9, 10) and in P. aeruginosa (11) have been cloned and their products shown to bind a [2Fe-2S] cluster. Although the function of Bfd is unknown, the ability of the recombinant protein to incorporate an iron sulfur cluster has been used to suggest that this small protein serves either as a scaffold for [2Fe-2S] cluster assembly and/or it aids in the uptake or release of iron from BfrB by mediating electron transport. An investigation of the global transcriptional response of iron-starved cultures of P. aeruginosa exposed to iron showed that mRNA levels of bfrB increased significantly (24-fold) when iron was made available, whereas bfrA mRNA levels remained unchanged (12). These observations are in agreement with previous findings indicating that in P. aeruginosa BfrA is expressed constitutively (13), whereas BfrB is expressed under high-iron conditions (13, 14). In comparison, the bfd gene is upregulated ~ 200-fold under low iron conditions (15). This differential regulation of the contiguous bfrB and bfd genes in response to cellular iron levels prompted us to hypothesize that Bfd synthesized under low-iron conditions functions to aid in the mobilization of iron stored in existing BfrB molecules.
Ferritin and bacterioferritin function by capturing excess cellular iron and storing it in the form of a ferric mineral. When cellular iron concentrations become limiting to metabolism, stored iron can be mobilized from ferritin or bacterioferritin to meet metabolic demands that involve incorporation of the nutrient in ubiquitous cofactors such as heme, non-heme iron centers and iron sulfur clusters. A significant volume of work has been carried out to understand the process of iron mineralization by ferritin and bacterioferritin, which starts with the binding of two ferrous ions to the highly conserved residues that act as ligands of the di-iron ferroxidase center (16–19). The two Fe2+ ions are oxidized by O2 and the resultant Fe3+ ions migrate to the cavity, where a ferrihydrite mineral is formed in the ferritins and a ferric phosphate mineral is formed in the bacterioferritins (20). In comparison, little detail is known about the process whereby mineralized iron is mobilized from the bacterioferritin cavity. Investigations conducted with ferritins from eukaryotes suggested that reduction of Fe3+ to Fe2+ by electrons shuttled from NADPH by way of a flavin-nucleotide is the more plausible mechanism for the mobilization of the iron stored in ferritin (21). Results from more recent work, also carried out with ferritins from eukaryotes, suggest that pores in the structure of ferritin reversibly unfold in the presence of millimolar concentrations of urea and largely increase the rate of iron release in vitro (22–24). These observations have led to the hypothesis that a yet unknown specific flavoprotein binds near the pores of ferritin, altering the dynamics of pore opening and closing and enhancing the rate of Fe3+ reduction and Fe2+ mobilization (25). In contrast, the issue of iron mobilization from bacterioferritins remains nearly completely enigmatic. Although X-ray crystal structures of bacterioferritins from different bacterial sources corroborated that these molecules bind 12 heme groups (8, 26–28), it is believed that the heme molecules do not play a role in iron mineralization, and their participation in iron mobilization from the core by facilitating Fe3+ reduction has been speculated but not demonstrated (29). In this report we present evidence that mobilization of iron stored in BfrB requires the presence of Bfd. Unexpectedly, the function of Bfd in iron mobilization is not the mediation of electrons from NADPH to the bacterioferritin; rather, apo-Bfd (devoid of its iron sulfur cluster) promotes the rapid mobilization of stored iron by binding to BfrB. Formation of a complex also affects the heme cofactor in BfrB, which is seen to mediate electrons from the surface to the core only if Bfd is bound to the bacterioferritin.
EXPERIMENTAL PROCEDURES
Cloning of Pseudomonas aeruginosa BfrB
The gene encoding for P. aeruginosa BfrB (PA3531) was synthesized, subcloned into a pET 11a vector and sequenced (GeneScript Corp., Pisctaway, NJ). The gene was engineered with silent mutations introducing codons favored by E. coli (30) and with Nde I and BamH I restriction enzyme sites at the 5’ and 3’ ends, respectively for subcloning into the pET 11a vector (Figure S1). The pET 11A vector harboring the bfrB gene was then transformed into E.coli Arctic express RIL competent cells (Stratagene).
Expression and purification of P. aeruginosa BfrB
A single colony of E. coli Arctic express RIL cells harboring the recombinant pET11-a/bfrB construct was cultured overnight at 37 °C in 50 mL of LB medium containing 100 µg/mL ampicilin and 20 µg/mL gentamicin. The cells were subsequently subcultured in 1 L of fresh LB containing no antibiotics and grown for 3 h at 37 °C. Cells were then transferred to a shaker incubator pre-equilibrated at 10 °C and incubated for 30 min before protein expression was induced by addition of isopropyl 1-thiol-D-galactopyranose (IPTG) to a final concentration of 1 mM. Cells were cultured for additional 20 h at 10 °C before they were harvested by centrifugation (4500 rpm for 10 min) and stored at −20 °C. The cell paste was resuspended in 50 mM Tris-HCl, pH 7.6 (2 mL/g cell paste) and PMSF (0.5 mM), DTT (5 mM), protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) and DNAse (Sigma-Aldrich, St. Louis, MO) were added before the cells were lysed by sonication on ice. Cell debris were pelleted by centrifugation at 4 °C and 23,500 rpm and the resultant supernatant was loaded onto a Q-Sepharose Fast Flow column (12 cm × 2.5 cm i.d.) equilibrated with 50 mM Tris-HCl (pH 7.6). The column was washed with three column volumes of 50 mM Tris-HCl (pH 7.6) before the protein was eluted using the same buffer and a linear gradient (0 mM – 600 mM) of NaCl. Fractions containing BfrB were pooled and reconstituted with heme by titrating with a solution of hemin (2 mg/mL in DMSO) until the absorbance of the Soret band (418 nm) no longer changed. The resultant protein solution was dialyzed against 4 L of 50 mM Tris-HCl (pH 7.6) for 12 h at 4 °C with at least two buffer changes. The resultant solution was then applied to a hydroxyapatite Bio-Gel column (10 cm × 2.5 cm i.d.) and washed with approximately 5 column volumes of 50 mM Tris-HCl (pH 7.6). BfrB was eluted with a linear gradient of potassium phosphate (50 mM–800 mM, pH 7.6). Fractions containing BfrB (red) were pooled and dialyzed overnight against 4 L of 50 mM Tris-HCl (pH 7.6) at 4 °C, with at least two buffer changes; the resultant solution was loaded to a second Q-Sepharose Fast Flow column (12 cm × 2.5 cm i.d.) and eluted as described above for the first ion exchange column. Fractions containing BfrB were loaded onto a second hydroxyapatite Bio-Gel column (10 cm × 2.5 cm i.d.) and the protein eluted using conditions identical to those described above for the first hydoxyapatite column. Fractions with absorbance ratio A280/A418 < 0.7 were pooled and dialyzed against 100 mM potassium phosphate (pH 7.6) and determined to be pure by SDS-PAGE.
Determination of molecular weight
The molecular weight of BfrB and that of the BfrB:Bfd complex were estimated with the aid of a high resolution size exclusion column (Superdex 200 16/60 Prep, GE Healthcare) equilibrated with 100 mM potassium phosphate (pH 7.6) and 1 mM tris (2-carboxyethyl)-phosphine hydrochloride (TCEP). The column was calibrated with a set of molecular weight standards (GE Healthcare) that included ferritin (440,000 Da), aldolase (158,000 Da), conalbumin (75,000 Da) and ovalbumin (43,000 Da).
Mineralization of iron in the BfrB cavity
As isolated, BfrB contains only a small amount of iron in its core (~ 5 – 10 iron atoms per molecule). Stocks of protein harboring ~ 600 iron atoms per molecule were prepared as follows: A solution of ferrous ammonium sulfate was prepared inside an anaerobic chamber (Coy Labs, Grass Lake, MI), placed in a container capped with a rubber septum and removed from the anaerobic chamber. Concentrated HCl was added to the ferrous ammonium sulfate solution (50 µL/100 mL) through the septum with the aid of a Hamilton micro syringe and a needle. The resultant solution was added to a stirred solution of 0.003 mM BfrB and 1.0 mM TCEP in 100 mM potassium phosphate buffer pH 7.6, in aliquots delivering 10% of the total iron necessary to load each BfrB molecule with 600 iron atoms. Fifteen minutes after the addition of an aliquot the resultant solution was transferred to an Eppendorf tube and centrifuged for 10 min at 4000 rpm. The supernatant was carefully transferred back into the reaction beaker before the following aliquot was added. The mineralization process was monitored spectrophotometrically using the 280 absorbance corresponding to the iron mineral (31) with the aid of a USB 2000 spectrophotometer (Ocean Optics, Dunedin, FL). The content of iron in the core was determined using previously reported methodology (32). In short, one mL of concentrated HCl was added to 1 mL of BfrB (0.003 mM) and the mixture incubated for 15 min, followed by addition of 1 mL of ascorbic acid (25 g/L) and 5 mL of saturated aqueous sodium acetate. A purple color developed upon the addition of 1 mL of ferrozine (5 g/L) and after 20 min the concentration was determined from the electronic absorption spectrum using the absorbance at 562 nm (ε562 = 27.9 mM−1 cm−1).
Mobilization of core iron from BfrB
Experiments to investigate the mobilization of core iron from BrfB were carried out in an anaerobic chamber. Reactions were conducted in a 1.0 cm path-length cuvette equipped with a magnetic stirring bar and monitored by electronic absorption spectroscopy using an Ocean Optics (Dunedin, FL) 2000 spectrophotometer. A few µL from stock solutions were added to a cuvette containing 3 mM 2,2, bipyrydil (bipy) in 1.5 mL of 20 mM potassium phosphate buffer (pH 7.6), to make a solution 15 µM in Bfd, 15 µM in FPR and 0.375 µM in BfrB. The reaction was initiated by addition of NADPH to a final concentration of 1.5 mM and the progress monitored by following the time-dependent changes in intensity of the band at 523 nm that is formed when bipy binds Fe2+ to form the [Fe(bipy)3]2+ complex. Experiments with apo-Bfd were conducted with a simple modification of the above procedure: Apo-Bfd was prepared in situ from holo-Bfd. To this end, holo-Bfd (15 µM final concentration) was added to a stirred cuvette containing 3 mM bipy in 20 mM potassium phosphate buffer (pH 7.6). Sodium dithionite (5 mM) was added to stoichiometrically reduce the Fe3+ in holo Bfd to Fe2+. Capturing of the ferrous ion by bipy was monitored by the time-dependent increase in the 523 nm absorbance, which was followed until it reached a plateau with intensity corresponding to the theoretical value calculated from the amount of holo Bfd placed in the cuvette. At this point, the solution containing apo-Bfd was reconstituted with FPR and BfrB (final concentrations of 15 µM and 0.375 µM, respectively) and the iron mobilization reaction initiated by the addition of NADPH to a final concentration of 1.5 mM.
RESULTS
Expression and purification of P. aeruginosa FPR, wild type Bfd and Cys43Ser Bfd
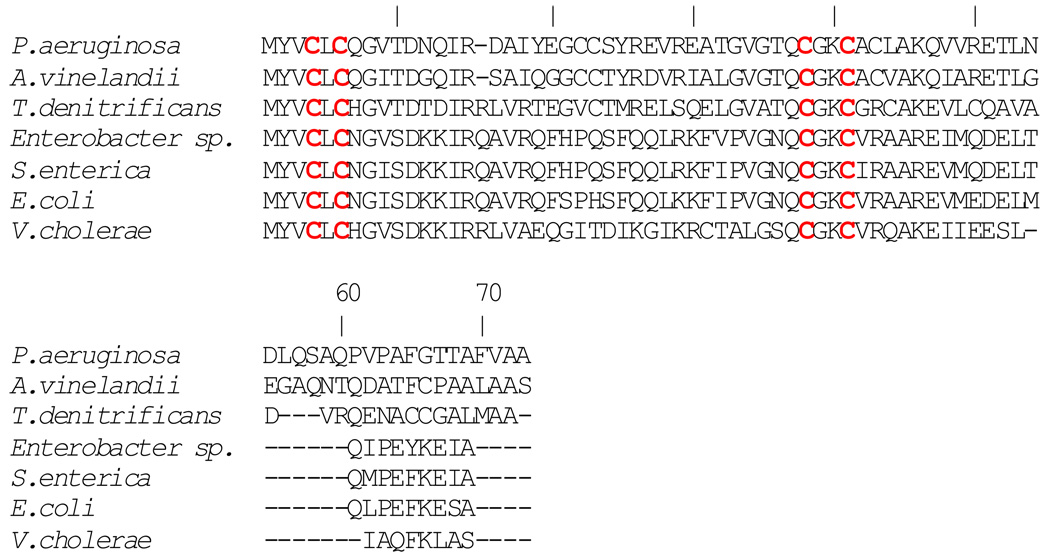
Ferredoxin NADP reductase from Pseudomonas aeruginosa (FPR) was expressed and purified as described previously (11, 33). Expression and purification of P. aeruginosa Bfd was carried out according to a previously described procedure (11) with the following modifications: In the previously described procedure Bfd was expressed in E. coli BL21(DE3) cells at 30 °C, which produced apoprotein encapsulated in inclusion bodies. Purification required denaturation with urea to solubilize the protein, in vitro reconstitution of the [2Fe-2S] cluster and subsequent chromatographic purification. In the present investigation the preparation of Bfd was facilitated by expressing it in E. coli Arctic Express RIL cells which produced soluble Bfd with an intact iron sulfur cluster. Although the preparation of P. aeruginosa Bfd was facilitated by use of the Arctic Express cells, the relatively low yield of protein (1–2 mg/L) was similar to that obtained previously; poor yields have also been reported for the expression of recombinant E. coli Bfd (10). The iron sulfur cluster in wild type Bfd is fragile and it tends to be lost or “damaged” during purification and sometimes during cryogenic storage. Inspection of the amino acid sequence of P. aeruginosa Bfd (Figure 2) shows that the 73-residue long protein contains 7 Cys, four of which are conserved; the latter are highlighted in red in the sequences corresponding to Bfd proteins from different organisms. Each of the three non-conserved Cys residues in P. aeruginosa Bfd was mutated to Ser in an attempt to increase the yield of expression and to stabilize the iron sulfur cluster of the recombinant protein. Mutation of Cys 22 or Cys 23 appears to have a negative impact on the fold of the protein because expression of these mutants did not yield detectable amounts of polypeptide, as judged by polyacrylamide gel electrophoresis. In contrast, the C43S mutant of Bfd can be expressed in yields (~ 4 mg/mL) that are at least two fold higher than those obtained with wild type Bfd. In addition, the [2Fe-2S] cluster in the C43S mutant, which is more resistant to purification, manipulation and to storage, exhibits an electronic absorption spectrum identical to that exhibited by wild type Bfd (Figure S2).
Figure 2. Bacterioferritin-associated ferredoxin sequences from different organisms aligned against that from P. aeruginosa Bfd.
Conserved cysteines forming the unique C-X-C-X31–32-CX2-C arrangement (highlighted) are thought to bind iron in the [2Fe-2S] cluster of the holo-protein. The sequences were aligned with the aid of ClustalW (46).
Overexpression, Purification and Characterization of BfrB
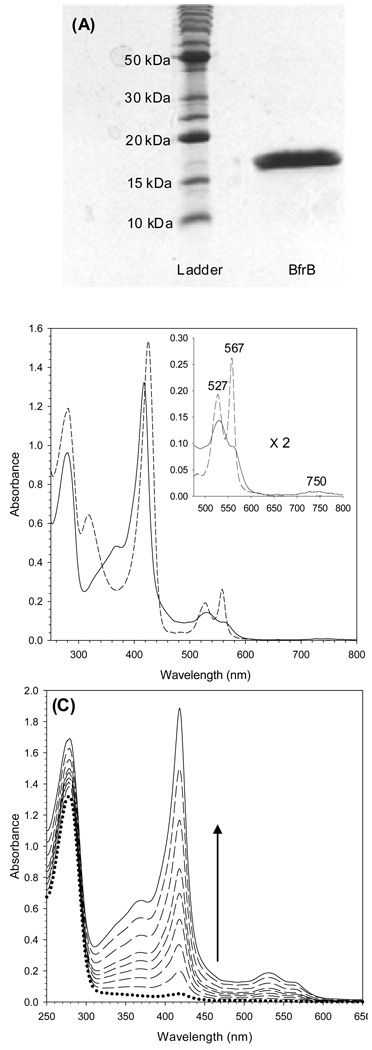
The BfrB protein expressed in E.coli Arctic express RIL cells is soluble and can be purified to homogeneity, as judged by the presence of a single band in the SDS-PAGE gel shown in Figure 3-A. The band corresponds to an approximate MW of 18,000 Da., which is in agreement with the molecular weight calculated from the amino acid sequence of a BfrB subunit (18,553 Da). The molecular weight of BfrB, estimated from its average elution volume (Ve = 58.9 ± 0.05 mL) from a Superdex 200 column calibrated as described in Experimental Methods is 447 ± 1 kDa (see Figure 8-A). This estimated MW indicates that it is composed of 24 subunits of MW 18.6 kDa, a value that is in good agreement with the 18.553 kDa per subunit value calculated from the amino acid sequence. The electronic absorption spectrum of pure BfrB with its heme-iron in the Fe3+ oxidation state is reminiscent of spectra obtained from other bacterioferritins, with a Soret band maxiumum at 418 nm, and α and β bands at 567 and 527 nm, respectively (solid trace in Figure 3-B). In addition, the spectrum displays the characteristic weak band at 740 nm (inset) indicative of heme coordinated by two axial methionine ligands (34). The spectrum of BfrB with its heme iron in the Fe2+ oxidation state, obtained upon addition of sodium dithionite (segmented trace in Figure 3-B), shows the Soret band at 425 nm, the α and β bands at 567 and 527 nm, respectively and no absorption near 740 nm, as expected. It is important to note that the spectrum of reduced heme was obtained with a sample of BfrB with a core devoid of iron mineral (~ 10 Fe atoms/BfrB). When the core is reconstituted with iron, on the other hand, the heme cannot be reduced, even if a large excess of dithionite is added under strict anaerobic conditions.
Figure 3. Characterization of recombinant BfrB.
(A) 15 % SDS-PAGE analysis of BfrB purified to homogeneity. (B) Electronic absorption spectra of oxidized-heme (full line) and reduced-heme (dashed line) BfrB in 100 mM potassium phosphate, pH 7.6. The inset shows the corresponding bands in the visible region. Note the 750 nm band in the oxidized-heme spectrum, which is characteristic of bis-methionine ligation. (C) Spectral changes observed upon reconstituting recombinant BfrB with exogenous heme.
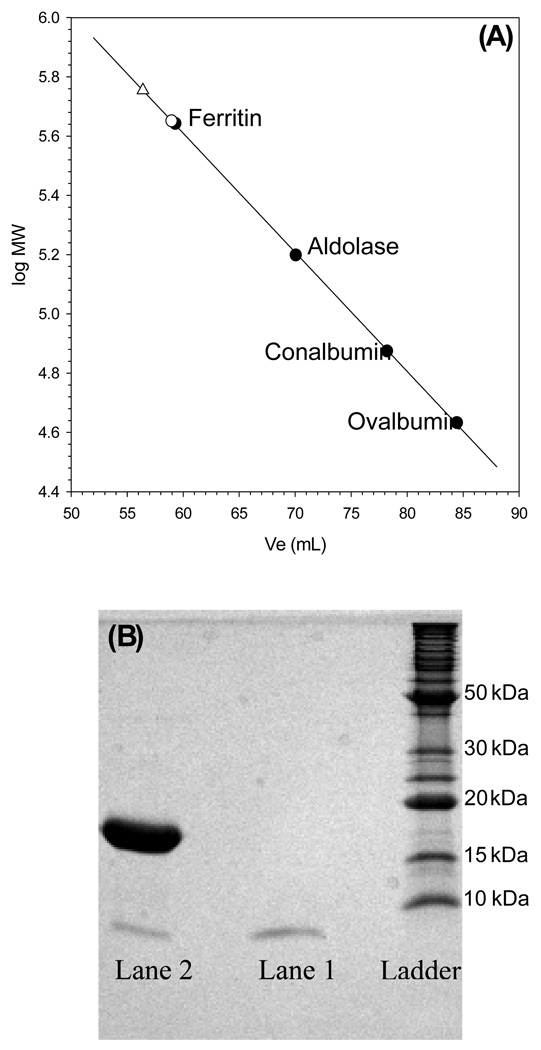
Figure 8. Bfd binds to BfrB.
(A) The calibration curve obtained to estimate the molecular weight of BfrB (○) and that of the BfrB-Bfd complex (Δ) was constructed from the elution volume (Ve) obtained for each of the following standard molecular weight markers: (●), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (44 kDa), loaded onto a Superdex 200 column. The average Ve (two measurements) obtained for pure BfrB (58.9 ± 0.05 mL) corresponds to a MW of 447 ± 1 kDa. (B) Two peaks were obtained upon loading the Superdex 200 column with a mixture of Bfd:BfrB in a mole ratio of 24: The peak with Ve = 57.0 ± 0.6 mL (MW ~ 538 kDa ±) contains a mixture of BfrB and Bfd (lane 2), whereas the peak with Ve = 96.3 mL contains only Bfd (lane 1). The presence of both Bfd and BfrB in peak with Ve = 57.0 mL indicates the formation of a stable complex between Bfd and BfrB and the difference in average Ve obtained from loading pure BfrB and a Bfrb:Bfd mixture is ~ 91 kDa, suggesting that ~ 12 molecules of Bfd bind a BfrB molecule.
In the process of establishing the purification protocol it was observed that BfrB-containing fractions eluting from the first ion exchange column (see Experimental) exhibited a faint red color, despite the fact that the protein concentration was high, as judged by the relative intensity of the subunit band in SDS-PAGE gels. This observation, which suggested the possibility that BfrB does not incorporate a full complement of heme when expressed in E. coli, was followed by reconstituting protein eluting from the first ion exchange column with heme. In the family of electronic absorption spectra obtained during the reconstitution process (Figure 3-C) the dotted trace, obtained before heme was added, shows an A280/A417 ratio > 10. Reconstitution with heme causes a large increase in the intensity of the Soret band, and a final A280/A417 ratio < 1 (solid trace). The large increase in the intensity of the Soret band upon reconstituting BfrB with heme confirms that the protein as isolated from the E. coli expressing strain has a low content of heme. Analysis of heme content in protein purified to homogeneity after in vitro reconstitution with the cofactor showed that BfrB binds 12 heme groups. Hence, BfrB is similar to other structurally characterized bacterioferritins in that it consists of 24 subunits that incorporate 12 heme molecules axially coordinated by two Met ligands. Analysis of the iron content in the core of BfrB as isolated from E. coli revealed the presence of 5–10 atoms of iron per protein molecule.
In attempts to circumvent the low iron and low heme content in as-isolated recombinant BfrB the expression medium was supplemented with (a) Fe(SO4)2 (4 mg/L) and δ-aminolevulinic acid (ALA) (17 mg/L) or (b) Fe(SO4)2 (4 mg/L) and hemin (5 mg/L). ALA was utilized because it has been shown that culture medium supplemented with the latter increases the yield of heme-bound recombinant proteins (35, 36). The content of heme and iron in recombinant BfrB expressed in E. coli cultured in LB medium supplemented with iron and ALA (or hemin) remained similar to those obtained from expressing BfrB in non-supplemented LB medium. The low content of iron in the core and the limited incorporation of heme in as isolated recombinant BfrB may be a consequence of overexpressing (40 mg/L) a protein that binds several heme molecules and is avid for iron, which places strain on the iron uptake and iron homeostasis processes of the host E. coli strain. Alternatively, it is possible that in the P. aeruginosa cell chaperones deliver and facilitate the binding of heme to BfrB. It is also interesting that heme appears to play an important role in the long-term stability of BfrB because attempts to purify it without reconstituting it with heme did not produce properly assembled dodecameric protein: BfrB devoid of heme was purified to homogeneity as judged by the presence of a single band, corresponding to a BfrB subunit (~ 18,000 Da), in an SDS PAGE gel. Determination of molecular weight in a Superdex 200 column, however, revealed the absence of a peak with elution volume corresponding to a dodecamer (MW 447 kDa). Instead, three main peaks were observed with Ve corresponding to molecular weights of approximately 30 kDa, 92 kDa and 254 kDa.
Preparation of iron loaded BfrB
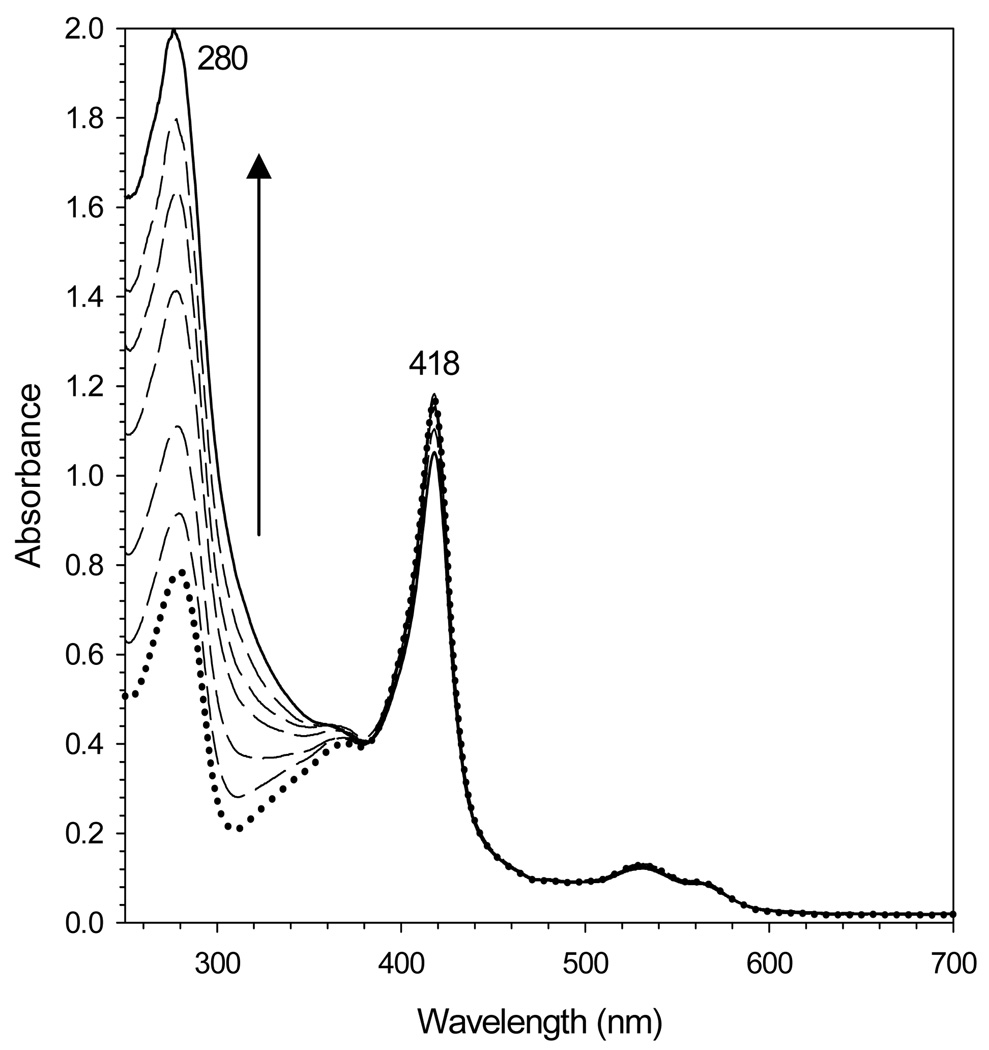
Protein containing ~ 600 iron atoms per BfrB molecule was prepared by titrating an anaerobic solution of Fe2+ into a solution of BfrB in air. Ferrous ions added with each aliquot are oxidized by two competing reactions: In the dominant process, Fe2+ is oxidized by BfrB using dissolved O2 as an electron acceptor and then incorporated into the Fe3+ mineral in the core of the bacterioferritin. In the competing process Fe2+ is oxidized by dissolved O2 without the participation of BfrB; the resultant Fe3+ is rapidly hydrolyzed to an insoluble colloidal hydrous oxide. Formation of the latter can result in significant loss of protein, presumably electrostatically associated to the colloid. To minimize the loss of protein during the preparation of iron-loaded BfrB, aliquots of Fe2+, each containing ~ 10% of the total iron were added approximately 15 min apart to allow for uptake, oxidation and mineralization, before the small amounts of colloidal iron oxide formed with each addition of Fe2+ were separated by centrifugation. The dotted trace in Figure 4, obtained before the addition of Fe2+ to a solution of BfrB harboring 10 iron atoms per protein molecule, displays A280/A418 ratio of 0.69. Incorporation of iron into the cavity induces a gradual increase in the intensity of the absorption ca. 280 nm because mineralized iron gives rise to absorption similar to band-edge bands displayed by polycrystalline semiconductors (31). The family of spectra in Figure 4 also shows that as the intensity of the band near 280 nm increases, the intensity of the Soret band remains almost unchanged; the small decrease in Soret band intensity is due to small losses of protein, presumably precipitated by small amounts of colloidal hydrous oxide formed with each addition of Fe2+. That the small amounts of colloid formed after each addition of Fe2+ are removed efficiently by the centrifugation step is evident in the preservation of baseline in the family of spectra; in the presence of colloids the baseline would drift toward higher absorbance values. This approach allowed for a relatively high recovery of protein, typically ~85%, with a consistent content of iron, 600 ± 40 iron atoms per protein molecule and phosphate to iron ratio of ~ 1.3. This relatively high phosphate content is typical of bacterioferritins isolated from native sources (19, 20, 37–39).
Figure 4. Reconstitution of core iron mineral in recombinant BfrB.
The family of electronic absorption spectra illustrates the intensity changes brought about by reconstituting the core of BrfB with iron. Note the large increase in the intensity of the band near 280 nm as the iron mineral grows in the core of the bacterioferritin. The small change in the intensity of the Soret absorbance is due to small loses of protein due to the formation of small amounts of colloidal hydrous oxide (see text). The spectrum with a dotted line was acquired before the addition of ferrous ammonium sulfate, and the spectrum with the full line, after the addition of the equivalent to 650 iron atoms per BfrB molecule.
Interactions with apo-Bfd unlock the iron stored in BfrB
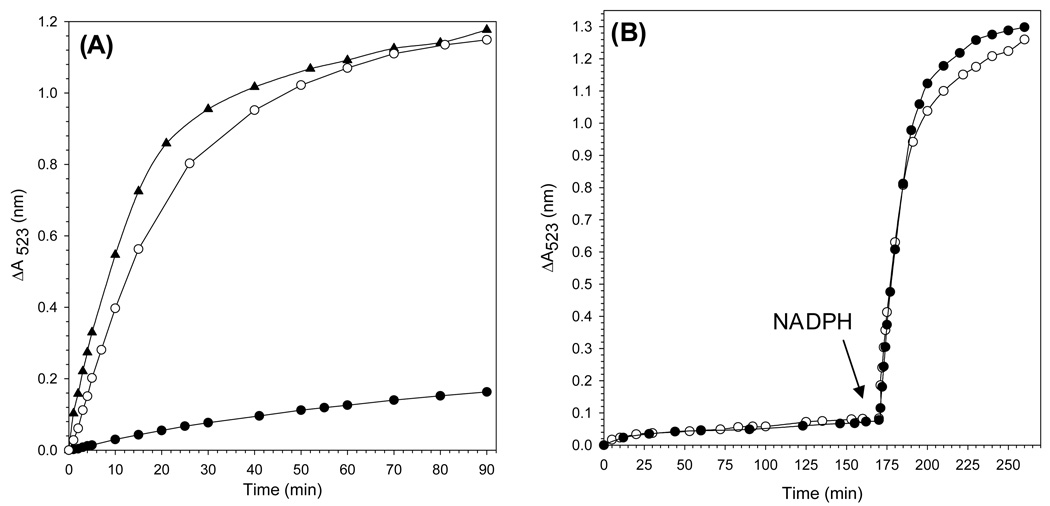
All experiments aimed at probing the mobilization of core iron from BfrB were carried out under anaerobic conditions. Iron mobilization was initiated by addition of NADPH (1.5 mM final concentration) to a cuvette containing a solution of the appropriate protein mixture and bipy (3 mM). The release of core iron was monitored by following the time-dependent formation of the ferrous bipyridine complex, [Fe(bipy)3]2+, which exhibits absorption maximum at 523 nm. The black circles in Figure 5-A correspond to the time dependent changes in absorbance at 523 nm following addition of NADPH to a solution containing BfrB (0.375 µM) and FPR (15 µM). The open circles correspond to the time dependent growth of the 523 nm band upon addition of NADPH to a solution containing wild type holo-Bfd (15 µM) in addition to BfrB (0.375 µM) and FPR (15 µM). It is evident that the presence of Bfd causes a significant acceleration in the rate of iron release, indicating that this small protein plays an important role in the mobilization of core iron from BfrB. It is also noteworthy that in the experiments conducted in the absence of Bfd (black circles) the Soret band remains at 417 nm (oxidized heme) throughout the experiment, which is in contrast with the rapid shift of the Soret band toward 425 nm that is observed when wild type holo-Bfd is present.
Figure 5. Bfd accelerates the mobilization of core iron from BfrB.
(A) Time dependent increase in the intensity of the 523 nm absorbance due to the formation of Fe[(bipy)3]2+ upon addition of NADPH (1.5 mM final concentration) to a solution containing BfrB (0.375 µM), FPR (15 µM), bipy (3 mM) and (●) no Bfd, (○) 15 µM wild type holo-Bfd, and (▲) wild type apo-Bfd. (B) Before the arrow: time dependent absorption changes in the intensity of the 523 nm caused by sequestration of the iron in the [2Fe-2S] cluster of (●) 15 µM wild type holo-Bfd and (○) 15 µM holo-C43S Bfd by bipy (3 mM); the solution contained 15 µM sodium dithionite to reduce the two iron ions in holo-Bfd. After the arrow: the resultant solution containing 15 µM apo-Bfd (●) or apo-C43S-Bfd (○) was made 0.375 µM in BfrB and 15 µM in FPR, before addition of NADPH (1.5 mM) started the rapid time dependent increase in the 523 nm absorption.
From the above observations it is tempting to assume that the [2Fe-2S] cluster of holo-Bfd catalyzes iron mobilization by shuttling electrons from FPR to BfrB. Results from an experiment in which wild type Bfd devoid of its [2Fe-2S] cluster (apo-Bfd) was used in lieu of holo-Bfd, however, indicate that Bfd does not participate in core iron mobilization by mediating electron transfer to BfrB. In these experiments wild type apo-Bfd was prepared in situ by incubating the holo protein with dithionite and bipy in a cuvette with constant stirring, in order to capture the iron in the [2Fe-2S] cluster as the [Fe(bipy)3]2+ complex. This process, although relatively slow (~ 170 min), is obvious in the appearance and growth of the characteristic 523 nm band (black circles in Figure 5-B). Evidence for the quantitative formation of apo-Bfd was obtained from the yield of [Fe(bipy)3]2+ calculated from the maximum intensity of the 523 nm band when it no longer changed with time, which is in excellent agreement with the amount of [Fe(bipy)3]2+ predicted to form if all the iron in the [2Fe-2S] cluster is captured by bipy. BfrB and FPR were subsequently added to the solution containing apo-Bfd, and the iron mobilization reaction started by the addition of NADPH (marked by an arrow). The fast growth in the intensity of the 523 absorbance that ensues upon addition of NADPH is also accompanied by rapid reduction of heme. To facilitate comparison, the time-dependent changes in the intensity of the 523 absorbance promoted by the presence of wild type apo-Bfd are shown with triangles in Figure 5-A, where it can be seen that the rate of iron release observed in the presence of apo-Bfd is slightly faster than that observed in the presence of holo-Bfd. These findings demonstrate that the [2Fe-2S] cluster of holo-Bfd is not necessary for the rapid mobilization of core iron and that Bfd does not function by shuttling electrons to BfrB. Rather, the effect exerted by Bfd on core iron mobilization appears to be brought about by binding to BfrB, presumably promoting conformational changes in the latter that enable the release of stored iron mineral.
As indicated above, the yield of recombinant wild type Bfd is significantly lower than that of the C43S mutant. In addition, the [2Fe-2S] cluster in the wild type protein has a tendency to degrade during protein purification and occasionally during protein storage. Cluster degradation leads to oligomerization of Bfd., presumably via formation of disulfide linkages. In an attempt to circumvent the limitations imposed by the low yields and fragility of wild type Bfd, the non-conserved Cys43 was mutated to a Ser. The mutant can be isolated pure in reasonable yield (4 mg per liter of culture) and is relatively more stable to manipulation and storage. The electronic absorption spectrum is identical to that of wild type Bfd (Figure S1), indicating that the mutation did not impair the fold of the protein or its ability to bind a [2Fe-2S] cluster. Apo-C43S Bfd was prepared in a cuvette in a manner similar that described above for the wild type protein. When the iron in the [2Fe-2S] cluster had been quantitatively captured as the [Fe(bipy)3]2+ complex, BfrB, FPR and NADPH were added to the cuvette and the iron mobilization reaction was followed by monitoring the growth in the intensity of the 523 nm absorbance. The time-dependent release of core iron promoted by the apo-form of the C43S mutant of Bfd (open circles in Figure 5-B) is nearly identical to the time-dependent release of iron promoted by apo-wild type protein. Consequently, subsequent studies involving Bfd were conducted with the C43S mutant to capitalize from its higher expression yields and enhanced stability. For simplicity, hereafter the C43S mutant of Bfd will be referred to as Bfd.
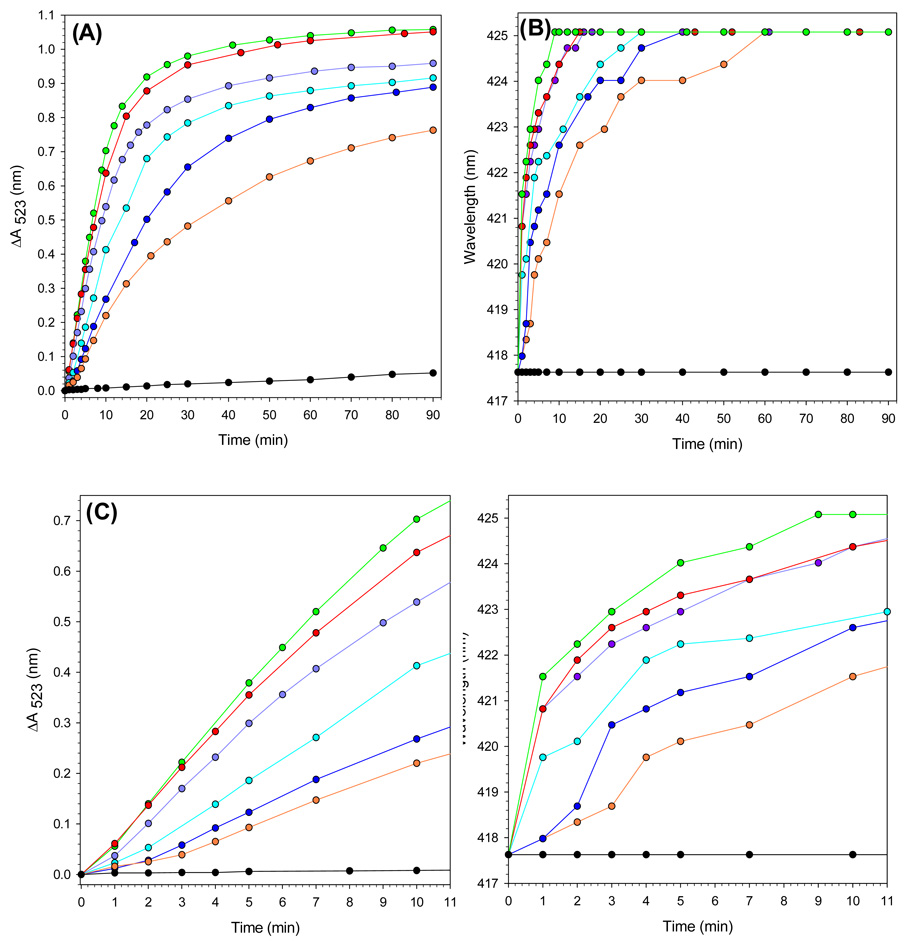
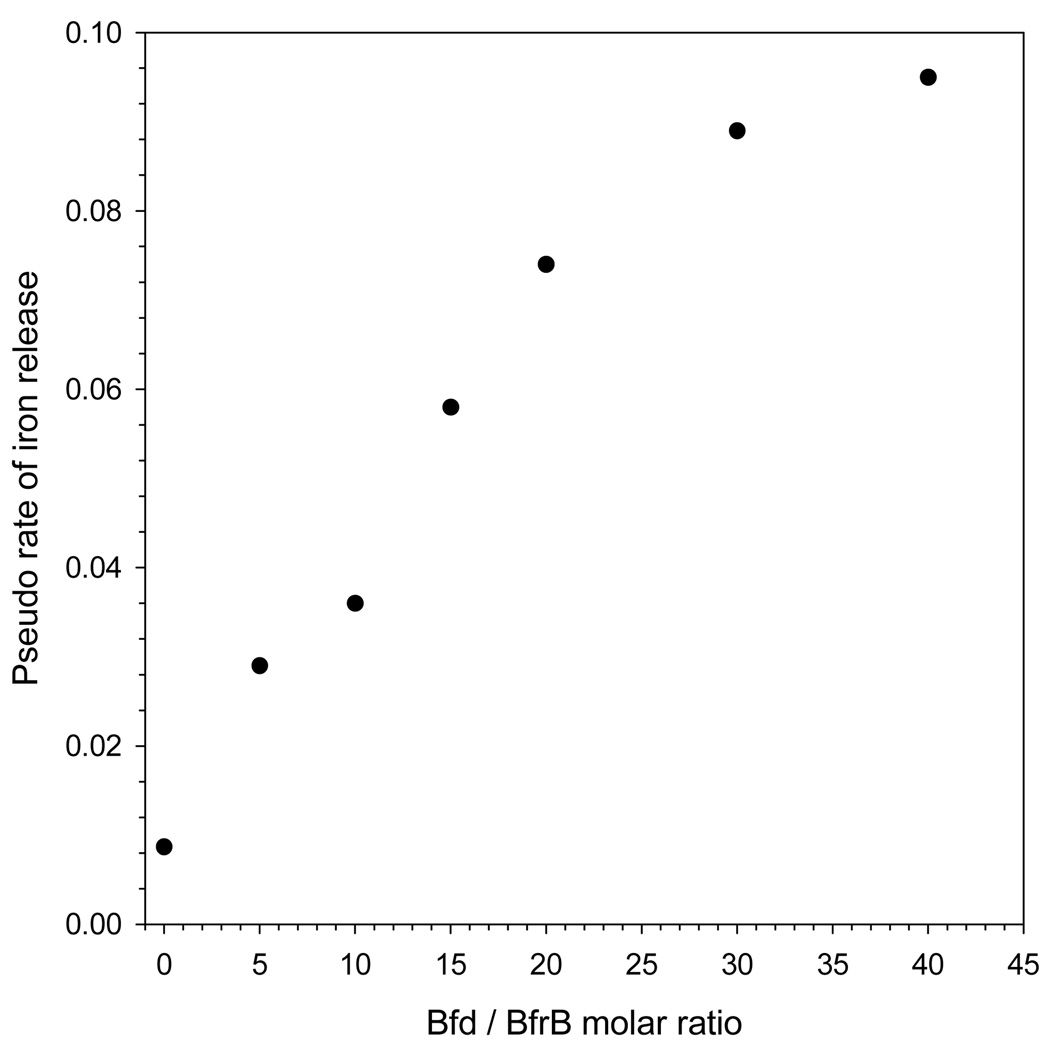
Results obtained from probing the effect of apo-Bfd concentration on the mobilization of core iron are summarized in Figure 6-A. In these experiments the concentrations of BfrB (0.375 µM) and FPR (15 µM) were maintained constant, while the concentration of Bfd was changed to explore conditions encompassing Bfd/BfrB ratios of 40:1 (green), 30:1 (red), 20:1 (purple), 15:1 (cyan), 10:1 (blue), 5:1 (orange) and 0:1 (black) . Lowering the Bfd/BfrB ratio from 40 to 5 causes a periodic slowing of iron release, followed by a pronounced change when the ratio is changed from five to zero. The accompanying plots in Figure 6-B, color coded as in Figure 6-A, show the time-dependent shift of the Soret band from 417 nm (ferric heme) to 425 nm (ferrous heme). It is apparent that when the Bfd/BfrB mole ratios are >5 the heme is reduced in an initial “fast phase” followed by a “slower phase” that continues until the Soret band has shifted to 425 nm (100 % reduction). A progressive slowing of each of the two phases is observed as the Bfd/FPR ratio is decreased from 40 to 5, while in stark contrast, in the absence of Bfd the Soret band remains at 417 throughout the duration of the experiment. Closer examination of the first 10 min of the iron mobilization reaction (Figure 6-C) and of the progress of heme reduction (Figure 6-D) reveals a lag phase lasting approximately 1–3 min (Figure 6-C) when the Bfd/BfrB ratio is 20 or lower. The fact that the lag in heme reduction tracks with the lag in iron mobilization suggests that electrons leaving the reductase (FPR) are channeled into the mineral core via the heme. Additional evidence supporting this notion is presented later in this report. In order to facilitate a more quantitative comparison of the rates of iron mobilization at varying concentrations of apo-Bfd, a pseudo rate was estimated by fitting the curves to an exponential function; data points corresponding to the lag phase observed when Bfd/BfrB < 20 were not included in the fit. The results, summarized in Table 1 and Figure 7, show a clear Bfd-dependent acceleration of core iron mobilization from BfrB.
Figure 6. Effect of Bfd concentration on the time-dependent processes of iron mobilization and heme reduction.
(A) Time dependent plot of absorption intensity change measured at 523 nm upon addition of NADPH (1.5 mM ) to solutions containing 3.0 mM bipy, 15 µM FPR, 0.375 µM BfrB and apo-Bfd with Bfd/Bfrb molar ratio of 40 (green), 30 (red), 20 (purple), 15 (cyan), 10 (blue), 5(orange) and 0 (black). (B) Time dependent changes in the position of the Soret band brought about by the addition of NADPH (1.5 mM) to the solutions in figure A; 417.5 nm and 425 nm correspond to fully oxidized and fully reduced heme, respectively. (C) Zoomed view (initial 11 min) of the time dependent absorption changes corresponding to the plots in A. (D) Zoomed view of the time dependent changes in the position of the Soret band corresponding to the plots in B.
Table 1.
Pseudo rates for the Bfd-dependent release of core iron mineral from BfrB.
| Bfd / BfrB molar ratio |
Pseudo rate of iron release (min−1)a,b |
|---|---|
| 0 | 0.0087 ± 0.0002 |
| 5 | 0.029 ± 0.001 |
| 10 | 0.036 ± 0.002 |
| 15 | 0.058 ± 0.008 |
| 20 | 0.074 ± 0.008 |
| 30 | 0.089 ± 0.007 |
| 40 | 0.095 ± 0.004 |
The standard deviations were obtained from triplicate measurements using proteins obtained from two independent preparations.
Measurements were carried out at 25 °C
Figure 7.
The release of core iron from BfrB is Bfd-dependent.
Bfd binds to BfrB
The observations described above strongly suggest that Bfd binds to BfrB. Additional evidence supporting the formation of a complex was obtained with the aid of size exclusion chromatography: The average volume of elution (Ve) corresponding to the peak obtained upon loading a Superdex 200 column with a 1.0 mL solution containing 0.0036 µmol of BfrB is 58.9 ± 0.05 mL (open circle in Figure 8-A). This value, which is nearly identical that obtained upon loading the column with the horse spleen ferritin standard, corresponds to an estimated molecular weight of 447 ± 1 kDa. In comparison, the avergae Ve corresponding to the peak obtained upon loading the column with 1.0 mL of a mixture containing 0.0036 µmol of BfrB and 0.0864 µmol of Bfd is 57.0 ± 0.6 mL and corresponds to an estimated molecular weight of 538 ± 30 kDa (open triangle in Figure 8-A). Excess Bfd elutes from the column significantly later (Ve = 96.3 ± 0.3 mL). The 91 kDa difference in the molecular weights estimated from the average elution volumes of pure BfrB and a BfrB-Bfd mixture corresponds to approximately 12 Bfd molecules, each with a molecular weight of 7.82 kDa. This observation suggests that 12 molecules of Bfd can bind a molecule of BfrB, which makes it tempting to speculate that one molecule of Bfd binds per subunit dimer of BfrB. In order to obtain additional evidence corroborating that Bfd elutes from the column together with BfrB, solutions corresponding to each peak in the chromatogram were analyzed by SDS page. The results presented in Figure 8-B show that the solution corresponding to the peak with elution volume 57.0 ± 0.6 mL (lane 2) is indeed a mixture of BfrB and Bfd, whereas the solution corresponding to the peak with elution volume 96.3 ± 0.3 mL (lane 1) contains only Bfd.
Binding of Bfd to BfrB enables heme mediation of electron transfer to the core
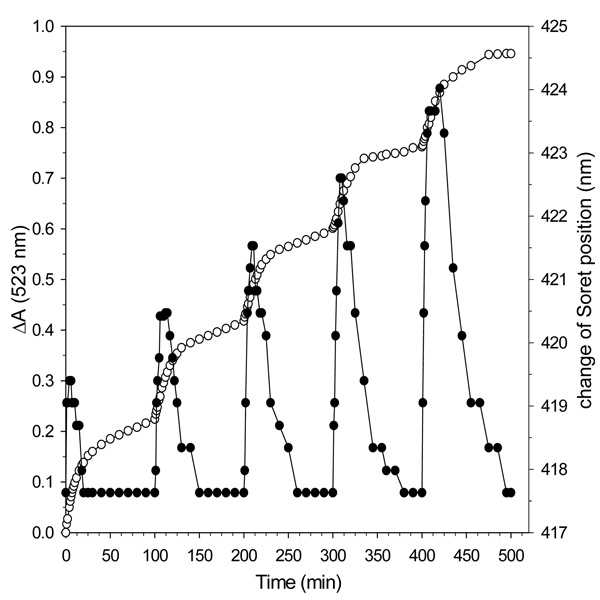
The idea that electron transfer from the reductase to the core mineral in BfrB is mediated by heme was further tested in an experiment where a limiting amount of NADPH, 15% of the total reducing equivalents needed to mobilize all the core iron, was added to a solution containing apo-Bfd, FPR and BfrB in molar proportion 40:40:1. These conditions were chosen because when the proportion of Bfd to BfrB is larger than 20, no lag phase is observed for iron mobilization or heme reduction (see Figure 6). The results, summarized in Figure 9, show rapid release of iron (open circles, left axis) with the addition of each aliquot of NADPH. The amount of Fe2+ released (0.043 ± 0.02 µmol) with each aliquot of NADPH is consistent with the number of electron equivalents (0.044 µmol) delivered, indicating quantitative utilization of NADPH. The plot of Figure 9 also shows that following each addition of NADPH the position of the Soret band shifts rapidly from 417 nm toward 425 nm and then back to its original position (closed circles, right axis), indicating a transient buildup of reduced heme. This transient buildup, which is indicative of a balance between reduction and subsequent oxidation of heme, strongly suggests that heme is reduced by electrons from the reductase faster than it is reoxidized by transferring those electrons to the core mineral. This idea finds additional support in the fact that the transient concentration of reduced heme increases with each addition of NADPH because as the size of the core mineral becomes progressively smaller, the electron demand decreases. Hence, if the electron flux from the surface to the heme is similar after each addition of NADPH but the electron flux from heme to the core decreases as the mineral shrinks, the transient concentration of reduced heme should be larger with each addition of NADPH, a prediction that is mirrored in the experimental observations summarized in Figure 9.
Figure 9. Binding of Bfd to BfrB promotes heme mediation of electron transfer to the core iron.
Open circles, left axis: time dependent changes in the intensity of absorption measured at 523 nm upon addition of aliquots of NADPH containing 15% of the total electron equivalents needed to reduce all the ferric ions in the core of BfrB (600 Fe3+ ions/BfB) to a solution containing BfrB (0.375 µM), Fpr (15 µM), apo-Bfd (15 µM ) bipy (3 mM). Dark circles, right axis: time dependent changes in the position of the heme Soret band following addition of each NADPH aliquot. The measurements were conducted at 22 °C.
DISCUSSION
The mobilization of ferritin iron in the cytosol of hepatic cells was shown to be promoted by electrons from NADH or NADPH which are shuttled to the ferritin via flavin- or xanthine-containing oxidoreductases (21). Consequently, work aimed at studying iron mobilization from mammalian ferritins utilizes FADH2 as an electron donor. In early work FADH2 was generated by NADH-flavin oxidoreductase (40) while in more recent studies FADH2 is generated by the beam of a spectrophotometer in the presence of NADH (41). In comparison, investigations of core iron mobilization from bacteriferritin have employed small molecule reductants such as dithionite or methyl viologen (42) because previous to this report virtually nothing was known about the process of electron delivery to the core iron of bacterioferritins or about the nature of the molecules that participate in the mobilization of stored iron. The present investigations, which were conducted with the aim of bridging these gaps, build from information obtained by probing the global genetic response of P. aeruginosa to iron levels (15). Amongst the genes reported to be positively regulated by low iron conditions, we previously noted those coding for Bfd and FPR because of their probable function in electron transfer (11). Biochemical and structural characterization of the Bfd and FPR proteins from P. aeruginosa allowed us to establish that FPR supports the catalytic activity of heme oxygenase by transferring the 7 electrons needed by this enzyme to degrade heme and release its iron (11). Hence, the NADPH dependent FPR was used in this work because it is the product of one of the two positively regulated genes coding for an electron transfer protein under iron starvation conditions and because it utilizes FADH2 as cofactor, like the flavin oxidoreductases implicated in delivering electrons to mammalian ferritin. The other positively regulated gene coding for a putative electron transfer protein is termed bacterioferritin associated ferredoxin (Bfd). The Bfd proteins from E. coli and from P. aeruginosa have been characterized and shown to bind a [2Fe-2S] cluster (9–11), observations that led to the hypothesis that Bfd may function in iron mobilization by mediating electron transfer to the iron core of BfrB (9, 10) or alternatively, function in iron mineralization by mediating electron transfer from the core to an external oxidant (10). In this context, it is significant that conditions of low-iron stress cause the bfrB gene to be down regulated (12) and the bfd gene to be strongly up regulated (15), because it suggests that Bfd is synthesized to aid in the mobilization of iron stored in BfrB, with the purpose of sustaining metabolic activity while additional mechanisms, also activated in response to iron starvation, procure the nutrient from extracellular sources. In the context of this hypothesis, holo Bfd would be expected to mediate electron transfer from NAD(P)H to the core iron in BfrB. Results from experiments aimed at testing the hypothesis showed that holo-Bfd promotes the efficient mobilization of core iron from BfrB, which would seem to support the notion that Bfd participates in iron mobilization by shuttling electrons to BfrB. Results obtained from additional experimentation aimed at testing the hypothesis, however, did not support this idea. Instead, these experiments conclusively demonstrated that apo-Bfd is effective in promoting the release of Fe2+ from BfrB, therefore demonstrating that assembly of a [2Fe-2S] cluster is not necessary for Bfd to exert its influence in iron mobilization. The collective evidence indicates that protein-protein interactions between apo-Bfd and BfrB promote the effective mobilization of iron stored in bacterioferritin.
Although these findings may appear at first to be surprising in light of previous expectations implicating the [2Fe-2S] cluster of Bfd, additional consideration makes it evident that bypassing the assembly of an iron-containing cofactor to mobilize and utilize iron stored in BfrB is an economical strategy when cellular levels are low. Hence, when these new observations are placed in the context of the known genetic response of P. aeruginosa to high- and low-iron conditions, a model for the efficient utilization of stored iron emerges: Down regulation of the bfrB gene clearly indicates that synthesis of additional BfrB protein is not necessary during periods of iron starvation. Iron stored in BfrB, however, is likely utilized for cell survival while additional mechanisms of iron acquisition are deployed to procure the nutrient from extracellular sources. Efficient release of iron from BfrB requires formation of a BfrB:Bfd complex with probable stoichiometry 1:12, which is in agreement with the strong positive regulation of the bfd gene by low-iron conditions. An electron transfer role for Bfd in the mobilization of stored iron, however, would place additional stress on the cell because assembly of [2Fe-2S] clusters in Bfd molecules would further deplete the levels of available iron. In contrast, a structural role in which apo-Bfd participates by binding to BfrB has the clear advantage of enabling utilization of stored iron without taxing the very resources that may allow cell survival under adverse conditions.
Binding of Bfd to BfrB enables heme mediation of electron transfer
It is noteworthy that reduction of the heme in P. aeruginosa BfrB occurs only in the presence of P. aeruginosa Bfd, which suggests that formation of a BfrB-Bfd complex enables the heme in the bacterioferritin to mediate electrons from the reductase to the core. Mediation of electron transfer is clearly manifested when limiting concentrations of NADPH (Figure 9) cause a transient buildup of reduced heme. Although heme mediation of electron transfer has previously been speculated (42) the function of heme in bacterioferritins has been difficult to establish. Findings from the present investigation make it apparent that the function of the heme in core iron mobilization can only be properly examined by reconstituting BfrB with Bfd because binding of the two physiological partner proteins is a prerequisite for making the heme “conductive”. The specific mechanism whereby heme is made “conductive” upon binding of Bfd is not understood at the moment, although some possible scenarios follow: (a) Formation of a complex with Bfd positively shifts the reduction potential of the heme in BfrB and enables it to accept electrons from FPR, (b) binding of Bfd causes conformational changes in BfrB that enable an electron transfer path from the surface to the heme; that is, acceleration of the electron transfer rate without a change in the driving force for the reaction, (c) the conformational changes brought about by binding Bfd both, alter the heme redox potential and enable a path for electron conduction and (d) binding of Bfd to Bfr may facilitate productive binding of the reductase, thus accelerating electron transfer to BfrB that is channeled to the core via the heme. Although additional work aimed at unraveling these possibilities is necessary to gather mechanistic information regarding the process of iron mobilization from bacterioferritin, it is clear that future studies directed at probing the mechanism of iron mobilization from bacterioferritins have to be conducted in the presence of the appropriate physiological partner protein. In this context, it is noteworthy that a recent study demonstrated that Bfd is necessary to recycle iron from Bfr in Erwinia chrysanthemi cells (43).
The rate of iron release from mammalian ferritins is thought to be controlled by “pore-gates” that open and close pores used for transporting Fe2+ ions from the core to the surface; “pore gates” are thought to regulate the outflow of Fe2+ by melting/ordering local structure (24). Given that very low concentrations of urea can melt the “pore gates”, it has been suggested that physiological regulators recognize them and protect the stored iron from leaking simply by exposure to physiological reductants in the cell (41). Peptides selected from a combinatorial library were also shown to unfold pores in the structure of mammalian ferritin and accelerate the mobilization of core iron (44). The presence of similar “pore gates” has not yet been demonstrated in bacterioferritins, although structural investigations suggest the presence of pores for the intake and release of iron into and out of the core (7, 8, 26, 27). Structural work with Azotobacter vinelandii Bfr, which exhibits the closest amino acid sequence homology to P. aeruginosa BfrB (Figure 1), showed redox-dependent conformational changes of several important residues in or near the ferroxidase center and the observation of iron at the four-fold pores (27). These observations, which are consistent with the idea that iron may enter (and perhaps leave) ferritin via these pores (7), support the notion of gated traffic of iron into and out of the bacterioferritin core. Hence, it is tempting to hypothesize that binding of Bfd to BfrB causes structural rearrangements that not only enable the heme to mediate electron transfer to the core mineral, but also open pore gates that enable the outflow of Fe2+. The amino acid sequence of Bfd does not hold any relationship to the sequence of the combinatorial peptides known to accelerate iron mobilization from mammalian ferritins. Thus, it is clear that additional investigations aimed at structurally characterizing BfrB and the Bfd-BfrB complex are necessary to dissect the effect exerted by Bfd on the structure of BfrB.
Bfd is likely not a classic ferredoxin
Early work with E. coli Bfd showed that the recombinant protein can be isolated from E. coli host cells with an intact [2Fe-2S] cluster analogous to that contained by bacterial and plant ferredoxins (9, 10). These observations together with the conservation of four Cys residues in the sequence of several Bfd genes, led to its classification as a ferredoxin, and the position of the bfd gene, upstream of the bfrB gene, resulted in the name bacterioferritin associated ferredoxin. Observations made during the present investigations show that apo-Bfd is fully functional in the mobilization of stored iron, demonstrating that assembly of an iron sulfur cluster is not necessary, at least for the purpose of mobilizing iron from BfrB. These findings strongly suggest that Bfd does not function in the classical electron transfer role of ferredoxins. In fact, inspection of the amino acid sequence alignment in Figure 2 shows that the four conserved cysteine residues thought to act as ligands for the iron in the [2Fe-2S] cluster are organized in a C-X-C-X31–32-C-X2-C arrangement. This unique arrangement and a relatively short peptide, which is approximately 50 residues shorter than typical [2Fe-2S] ferredoxins of bacteria, plants, fungi and vertebrates, lend support to the notion that Bfds do not function in the classical electron transfer role of ferredoxins. The resonance Raman spectral features of oxidized Bfd are nearly identical to those of the [2Fe-2S] cluster in the second domain of NifU (9, 11), a multi-domain protein required for the assembly of the metallocluster in nitrogenase (45). This has led to the suggestion that Bfds may function as a scaffold for the assembly of [2Fe-2S] clusters for subsequent assembly into other proteins and enzymes that utilize iron sulfur clusters (9–11). Assembly of a transient [2Fe-2S] cluster in Bfds is consistent with the fragility of this center and is not in disagreement with the role played by apo-Bfd in the mobilization of iron stored in BfrB, because it is possible that iron released from BfrB is coordinated by the four conserved Cys residues in Bfd to form a mononuclear iron center. Iron coordinated in this manner can be transported and delivered to where is needed, or alternatively, with the aid of proteins involved in the iron sulfur cluster assembly machinery (Isc), be used to form a transient [2Fe-2S] cluster in Bfd that can be transferred to a nascent ferredoxin. The ability to accommodate a tetracoordinated mononuclear iron center that can be converted into a [2Fe-2S] cluster may be built, at least partially, in the C-X-C motif, because its amino terminal position (see Figure 2) likely facilitates the conformational freedom that allow the corresponding Cys ligands to adapt to a mononuclear or dinuclear iron center. Clearly additional investigations with Bfd are needed to shed light on this issue.
Supplementary Material
Footnotes
This work was supported by grants from the National Institute of Health, GM-50503 (M.R.).
SUPPORTING INFORMATION AVAILABLE
DNA sequence of BfrB from P. aeruginosa engineered with silent mutations to include codons favored by E. coli and electronic absorption spectra of oxidized wild type and C43S Bfd.
REFERENCES
- 1.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial Iron Homeostasis. FEMS Microbiology Reviews. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Smith L. The Physiological Role of Ferritin-Like Compounds in Bacteria. Crit. Rev. Microbiol. 2004;30:173–185. doi: 10.1080/10408410490435151. [DOI] [PubMed] [Google Scholar]
- 3.Orino K, Lehman L, Tsuji Y, Ayaki H, Torti SV, Torti FM. Ferritin and the Response to Oxidative Stress. Biochem J. 2001;357:241–247. doi: 10.1042/0264-6021:3570241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore GR, Kadir HA, Al-Massad K, Le Brun NE, Thomson AJ, Greenwood C, Keen JN, Findlay JBC. Structural Heterogeneity of Pseudomonas aeruginosa Bacterioferritin. Biochem J. 1994;304:493–497. doi: 10.1042/bj3040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma J-F, Ochsner UA, Klotz MG, Nanayakkara VK, Howell ML, Johnson Z, Posey JE, Vasil ML, Monaco JJ, Hassett DJ. Bacterioferritin A Modulates Catalase A (KatA) Activity and Resistance to Hydrogen Peroxide in Pseudomonas aeruginosa. J. Bacteriol. 1999;181:3730–3742. doi: 10.1128/jb.181.12.3730-3742.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frolow F, Kalb AJ, Yariv J. Structure of a Unique Twofold Symmetric Haem-Binding Site. Nat. Struct. Biol. 1994;1 doi: 10.1038/nsb0794-453. [DOI] [PubMed] [Google Scholar]
- 7.Liu H-L, Zhou H-N, Xing W-M, Zhao J-F, Li S-X, Huang J-F, Bi R-C. 2.6 Å Resolution Crystal Structure of the Bacterioferritin from Azotobacter vinelandii. FEBS Lett. 2004;573:93–98. doi: 10.1016/j.febslet.2004.07.054. [DOI] [PubMed] [Google Scholar]
- 8.Janowski R, Auerbach-Nevo T, Weiss MS. Bacterioferritin from Mycobacterium smegmatis Contains Zinc in its Di-Nuclear Site. Prot. Sci. 2007;17:1138–1150. doi: 10.1110/ps.034819.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quail MA, Jordan P, Grogan JM, Butt JN, Lutz M, Thomson AJ, Andrews SC, Guest JR. Spectroscopic and Voltammetric Characterization of Bacterioferritin-Associated Ferredoxin of Escherichia coli. Biochem. Biophys. Res. Commun. 1996;229:635–642. doi: 10.1006/bbrc.1996.1856. [DOI] [PubMed] [Google Scholar]
- 10.Garg RP, Vargo CJ, Cui X, Kurtz DMJ. A [2Fe-2S] Protein Encoded by an Open Reading Frame Upstream of the Escherichia coli Bacterioferritin Gene. Biochemistry. 1996;35:6297–6301. doi: 10.1021/bi9600862. [DOI] [PubMed] [Google Scholar]
- 11.Wang A, Zeng Y, Han H, Weeratunga S, Morgan BN, Moënne-Loccoz P, Schönbrunn E, Rivera M. Biochemical and Structural Characterization of Pseudomonas aeruginosa Bfd and FPR: Ferredoxin NADP+ Reductase and Not Ferredoxin is the Redox Partner of Heme Oxygenase under Iron-Starvation Conditions. Biochemistry. 2007;46:12198–12211. doi: 10.1021/bi7013135. [DOI] [PubMed] [Google Scholar]
- 12.Palma M, Worgall S, Quadri LEN. Transcriptome Analysis of the Pseudomonas aeruginosa Response to Iron. Arch. Microbiol. 2003;180:374–379. doi: 10.1007/s00203-003-0602-z. [DOI] [PubMed] [Google Scholar]
- 13.Vasil ML, Ochsner UA. The Response of Pseudomonas aeruginosa to Iron: Genetics, Biochemistry and Virulence. Mol. Microbiol. 1999;34:399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- 14.Wilderman PJ, Sowa NA, FitzGerald DJ, FitGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of Tandem Duplicate Regulatory Small RNAs in Pseudomonas aeruginosa Involved in Iron Homeostasis. Proc. Natl. Acad. Sci. USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip Expression Analysis of the Iron Starvation Response in Pseudomonas Aeruginosa: Identification of Novel Pyoverdine Biosynthesis Genes. Mol. Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 16.Pereira AS, Small W, Krebs C, Tavares P, Edmondson DE, Theil EC, Huynh BH. Direct Spectroscopic and Kinetic Evidence for the Involvement of Peroxodiferric Intermediate during the Ferroxidase Reaction in Fast Ferritin Mineralization. Biochemistry. 1998;37:9871–9876. doi: 10.1021/bi980847w. [DOI] [PubMed] [Google Scholar]
- 17.Le Brun NE, Wilson MT, Andrews SC, Guest JR, Harrison PM, Thomson AJ, Moore GR. Kinetic and Structural Characterization of an Intermediate in the Biomineralization of Bacterioferritin. FEBS Lett. 1993;333:197–202. doi: 10.1016/0014-5793(93)80404-i. [DOI] [PubMed] [Google Scholar]
- 18.Carrondo MA. Ferritins, Iron Uptake and Storage from the Bacterioferritin Viewpoint. EMBO J. 2003;22:1959–1968. doi: 10.1093/emboj/cdg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews SC. Iron Storage in Bacteria. Advan. Microbial Physiol. 1998;40:281–351. doi: 10.1016/s0065-2911(08)60134-4. [DOI] [PubMed] [Google Scholar]
- 20.Mann S, Williams JM, Treffry A, Harrison PM. Reconstituted and Native Iron-Cores of Bacterioferritin and Ferritin. J. Mol. Biol. 1987;198:405–416. doi: 10.1016/0022-2836(87)90290-7. [DOI] [PubMed] [Google Scholar]
- 21.Topham R, Goger M, Pearce K, Schultz P. The Mobilization of Ferritin Iron by Liver Cytosol. Biochem J. 1989;261:137–143. doi: 10.1042/bj2610137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagi H, Shi D, Ha Y, Allewell NM, Theil EC. Localized Unfolding at the Junction of Three Ferritin Subunits. J. Biol. Chem. 1998;273 doi: 10.1074/jbc.273.30.18685. 18685-18618-18688. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Jin W, Theil EC. Opening Protein Pores with Chaotropes Enhances Fe Reduction and Chelation of Fe from the Ferritin Biomineral. Proc. Natl. Acad. Sci. USA. 2003;100:3653–3658. doi: 10.1073/pnas.0636928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin W, Takagi H, Pancorbo B, Theil EC. "Opening" the Ferritin Pore for Iron Release by Mutation of Conserved Amino Acids at Interhelix and Loop Sites. Biochemistry. 2001;40:7525–7532. doi: 10.1021/bi002509c. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Theil EC. Ferritins: Dynamic Management of Biological Iron and Oxygen Chemistry. Acc. Chem. Res. 2005;38:167–175. doi: 10.1021/ar0302336. [DOI] [PubMed] [Google Scholar]
- 26.Macedo S, Romão CV, Mitchell E, Matias PM, Liu MY, Xavier AV, LeGall J, Teixeira M, Lindley P, Carrondo MA. The Nature of the Di-Iron Site in the Bacterioferritin from Desulfovibrio desulfuricans. Nat. Struct. Biol. 2003;10:285–290. doi: 10.1038/nsb909. [DOI] [PubMed] [Google Scholar]
- 27.Swartz L, Kuchinskas M, Li H, Poulos TL, Lanzilotta WN. Redox-Dependent Structural Changes in the Azotobacter vinelandii Bacterioferritin: New Insights into the Ferroxidase and Iron Transport Mechanism. Biochemistry. 2006;45:4421–4428. doi: 10.1021/bi060146w. [DOI] [PubMed] [Google Scholar]
- 28.van Eerde A, Wolterink-van Loo S, van der Oost J, Dijkstra BW. Fortituous Structure Determination of "As-Isolated" Escherichia coli Bacterioferritin in a Novel Crystal Form. Acta. Cryst. F. 2006;62:1061–1066. doi: 10.1107/S1744309106039583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews SC, Le Brun NE, Barynin V, Thomson AJ, Moore GR, Guest JR, Harrison PM. Site-Directed Replacement of the Coaxial Heme Ligands of Bacterioferritin Generates Heme-free Variants. J Biol Chem. 1995;270:23268–23274. doi: 10.1074/jbc.270.40.23268. [DOI] [PubMed] [Google Scholar]
- 30.Ikemura T. Codon Usage and tRNA Content in Unicellular and Multicellular Organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 31.Yariv J, Kalb AJ, Sperling R, Bauminger ER, S.G. C, Ofer S. The Composition and Structure of Bacterioferritin of Escherichia coli. Biochem J. 1981;197:171–175. doi: 10.1042/bj1970171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ringeling PL, Davy SL, Monkara FA, Hunt C, Dickson DPE, McEwan AG, Moore GR. Characterization of Bacterioferritin and Formation of Non-Haem Iron Particles in Intact Cells. Eur. J. Biochem. 1994;223:847–855. doi: 10.1111/j.1432-1033.1994.tb19061.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang A, Rodríguez JC, Han H, Schönbrunn E, Rivera M. X-Ray Crystallographic and Solution State Nuclear Magnetic Resonance Spectroscopic Investigations of NADP+ Binding to Ferredoxin NADP Reductase from Pseudomonas aeruginosa. Biochemistry. 2008;47:8080–8093. doi: 10.1021/bi8007356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheesman MR, Le Brun NE, Kadir FH, Thomson AJ, Moore GR, Andrews SC, Guest JR, Harrison PM, Smith JA, Yewdall SJ. Haem and Non-Haem Iron Sites in Escherichia coli Bacterioferritin: Spectroscopic and Model Building Studies. Biochem J. 1993;292:47–56. doi: 10.1042/bj2920047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivera M, Walker FA. Biosynthetic Preparation of Isotopically Labeled Heme. Anal. Biochem. 1995;230:295–302. doi: 10.1006/abio.1995.1477. [DOI] [PubMed] [Google Scholar]
- 36.Rivera M, Caignan GA. Recent Developments in the 13C NMR Spectroscopic Analysis of Paramagnetic Hemes and Hemoproteins. Anal. Bioanal. Chem. 2004;378:1464–1483. doi: 10.1007/s00216-003-2340-0. [DOI] [PubMed] [Google Scholar]
- 37.Watt GD, Frankel RB, Jacobs D, Huang H. Fe2+ and Phosphate Interactions in Bacterial Ferritin from Azotobacter vinelandii. Biochemistry. 1992;31:5672–5679. doi: 10.1021/bi00139a035. [DOI] [PubMed] [Google Scholar]
- 38.Watt GD, Frankel RB, Papaefthymiou GC, Spartalian K, Stiefel EI. Redox Properties and Mössbauer Spectroscopy of Azotobacter vinelandii Bacterioferritin. Biochemistry. 1986;1986:4330–4336. [Google Scholar]
- 39.Aitken-Rogers H, Singleton C, Lewin a, Taylor-Gee A, Moore GR, Le Brun NE. Effect of Phosphate on Bacterioferritin-Catalysed Iron(II) Oxidation. J. Biol. Inorg. Chem. 2004;9:161–170. doi: 10.1007/s00775-003-0504-1. [DOI] [PubMed] [Google Scholar]
- 40.Jones T, Spencer R, Walsh C. Mechanism and Kinetics of Iron Release from Ferritin by Dihydroflavins and Dihydroflavin Analogues. Biochemistry. 1978;17:4011–4017. doi: 10.1021/bi00612a021. [DOI] [PubMed] [Google Scholar]
- 41.Theil EC, Liu XS, Tosha T. Gated Pores in the Ferritin Protein Nanocage. Inorg. Chim. Acta. 2008;361:868–874. doi: 10.1016/j.ica.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richards TD, Pitts KR, Watt GD. A Kinetic Study of Iron Release from Azotobacter Vinelandii Bacterial Ferritin. J. Inorg. Biochem. 1996;61:1–13. doi: 10.1016/0162-0134(95)00029-1. [DOI] [PubMed] [Google Scholar]
- 43.Expert D, Boughammoura A, Franza T. Sidreophore-Controlled Iron Assimilation in the Enterobacterium Erwinia chrysanthemi. J. Biol. Chem. 2008;283:36564–36572. doi: 10.1074/jbc.M807749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu XS, Patterson LD, Miller MJ, Theil EC. Peptides Selected for the Protein Nanocage Pores Change the Rate of Iron Recovery from the Ferritin Mineral. J. Biol. Chem. 2007;282:31821–31825. doi: 10.1074/jbc.C700153200. [DOI] [PubMed] [Google Scholar]
- 45.Dos Santos PC, Smith AD, Frazzon J, Cash VL, Johnson MK, Dean DR. Iron-Sulfur Cluster Assembly: NifU-Directed Activation of the Nitrogenase Fe Protein. J. Biol. Chem. 2004;279:19705–19711. doi: 10.1074/jbc.M400278200. [DOI] [PubMed] [Google Scholar]
- 46.Larkin MA, Blackshields G, Brown NP, McGettigan PR, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X Version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.