Summary
Diffuse large B cell lymphoma (DLBCL) is a heterogeneous disease and response to therapy is difficult to predict. An algorithm to sort DLBCL cases using a series of five immunohistochemical markers (GCET1, CD10, BCL6, MUM1, FOXP1) accurately predicts survival in patients treated with current chemotherapeutic regimens.
In this issue of Clinical Cancer Research, Choi et al(1) refine prior attempts to use immunohistochemistry (IHC) to predict survival in DLBCL. Over the past decade, numerous groups have used gene expression profiles (GEPs) to identify distinctive molecular subtypes of a variety of human tumors, including DLBCL, an aggressive, common form of non-Hodgkin lymphoma. Morphologic appearance is not a reliable predictor of outcome in DLBCL, and overall roughly 50% of patients are cured and 50% die of their disease. In 2001, Staudt and colleagues identified two major subtypes of DLBCL, one with a GEP resembling normal germinal center B cells (the GC subtype), and a second with a GEP resembling activated B cells (the ABC subtype)(2); subsequent work from the same group identified a third, less common, “unclassified” group(3, 4). It is now well established that when classified by GEP, the prognosis of the GC subtype is significantly better than that of the ABC subtype, independent of the long-relied upon International Prognostic Index (5), when patients are treated with CHOP or, more recently, Rituxan(R)-CHOP chemotherapy.
An unsettled issue emerging from these and other GEP studies is how best to reduce discovery to practice, both within the context of clinical trials and the everyday diagnosis of cancer in academic and community-based hospitals. One approach is to “translate” GEPs into protein based tests such as IHC that can be performed routinely and relatively cheaply on formalin-fixed paraffin-embedded (FFPE) tissue biopsies. One of the earliest examples of this approach was an IHC algorithm developed by Hans et al(6), which relied on stains for 3 markers, MUM1/IRF4, CD10, and BCL6 (Figure 1), that are differentially expressed in GC and ABC subtypes of DLBCL. The “Hans” algorithm has proved useful in some studies (for example, in predicting response of DLBCL to certain chemotherapy regimens (7)), but not others (e.g., see ref. (8)).
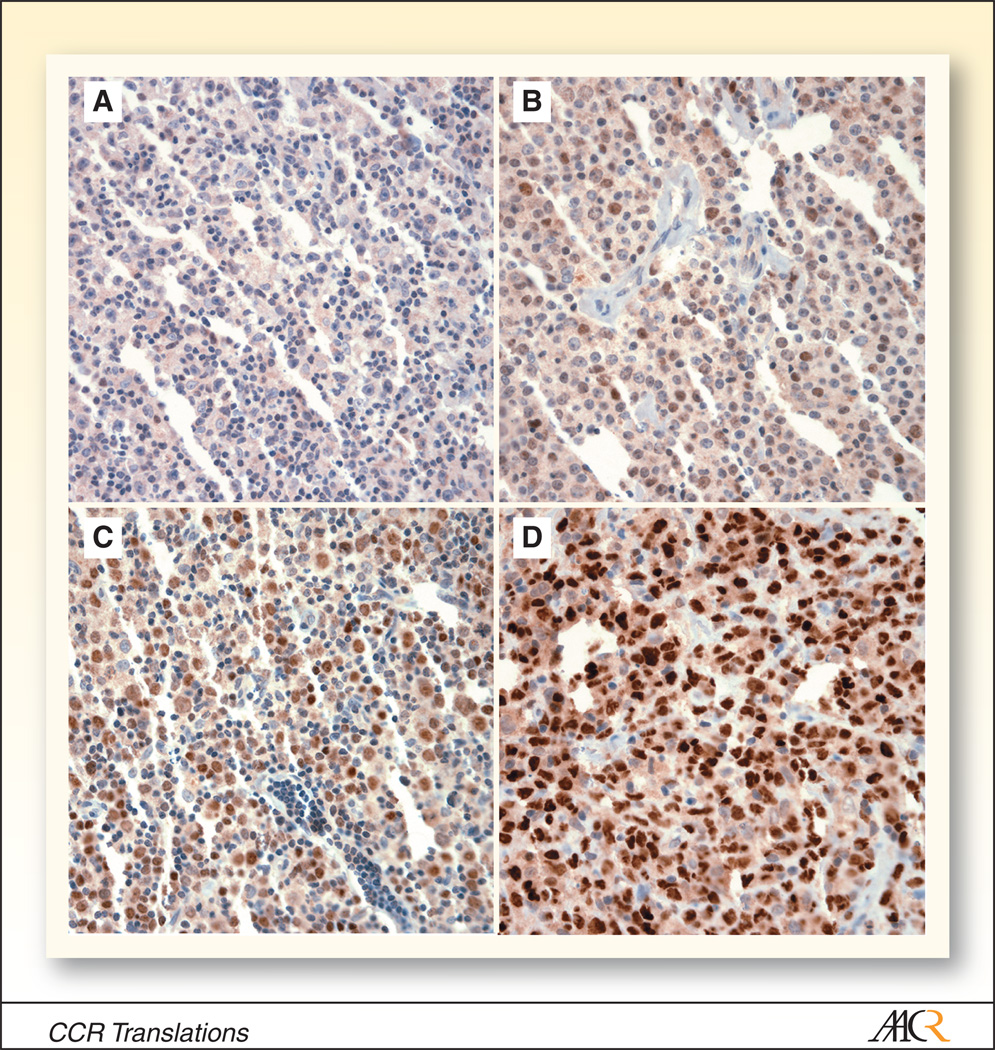
Figure 1.
Variable immunohistochemical staining for BCL6 in four diffuse large B cell lymphomas (A–D; original magnification 400x).
The new IHC algorithm reported here by Choi et al. builds on the “Hans” algorithm by incorporating stains for two additional markers, GCET1 and FOXP1, that are associated with the GC and ABC DLBCL subtypes, respectively (1). The new “Choi” algorithm correctly classified 93% of 63 cases of DLBCL into the GC or ABC subtypes (as compared to 86% for previous “Hans” algorithm(6)), and (as anticipated given this result) was effective at stratifying DLBCL patients treated with R-CHOP into good (GC) and bad (ABC) prognostic groups. Despite small numbers, it also appears to co-classify primary mediastinal large B cell lymphoma (PMLBCL) with the GC-type of DLBCL, which fits with the relatively good prognosis of PMLBCL.
These results are impressive and indicate that when applied by highly experienced hematopathologists with access to a state-of-the-art IHC laboratory the “Choi” algorithm can be used as a substitute for subclassification of DLBCL by GEP. However, it remains to be seen how well the new algorithm will perform in the hands of hematopathologists in other academic centers. Choi et al. stained DLBCL cores assembled in tissue microarrays, mitigating the inevitable slide-to-slide variation that is introduced when tumors are stained on individual slides as well as the complication of regional variation in staining intensity in large pieces of tissue. The latter is especially concerning when considering that the decisions points in the “Choi” algorithm are not positive or negative staining, but are based on cutoffs of 30% to 80% positivity in DLBCL cells for various markers. Pathologists are very good at pattern recognition, but are less reliable at estimating IHC staining intensity or percent-positive tumor cells by eye. It may be that this concern will prove to be unfounded, as Choi et al. have attempted to address the impact of intra- and inter-observer variations through a computer perturbation program that introduces random noise for each IHC marker. However, other sources of variation may also negatively affect the predictive value of the algorithm, most notably the absence of standard procedures for the fixation and processing of tissues and the performance of IHC across pathology departments. Thus, while there is little doubt that the algorithm can work, exporting its use to other academic groups will not be trivial, and without standardization, it will likely remain a research tool.
With these limitations in mind, it is worth considering other technologies that may ultimately supplant conventional IHC in the molecular subclassification of cancers. One group encompasses newer methods for determining GEPs using RNA retrieved from FFPE tissues, which have been successfully used in some instances to predict patient outcome (9). Another approach is to distill GEPs down to a limited number of genes that subclassify particular cancers; this approach has led to commercial tests using FFPE tissues involved by breast cancer(10), and it seems likely that similar classifiers will be developed for an array of cancers, including DLBCL. As a cautionary note, to date we are unaware of any direct comparisons of GEPs obtained with conventional platforms using RNAs prepared from fresh frozen samples and GEPs obtained on alternative platforms using RNAs prepared from paired FFPE samples; a formal proof of the interchangeability of these two approaches awaits further work.
A second group of nascent technologies rely on more sophisticated and reproducible methods for quantification of proteins of interest in FFPE tissue sections. Proteins are appealing as analytes, as they are generally well preserved in archival tissue blocks, and tissue-based analyses provide additional potentially valuable information about the expression of markers within both tumor and stromal cells. Quantification of proteins in situ in tissue sections requires that significant technical hurdles be surmounted (11, 12), but as disk storage space gets cheaper and computers become faster, it appears increasingly likely that automated measurement of proteins on scanned digitized slides will augment current scoring of IHC by eye. Initially, this will occur within the research realm, but it is notable that digital tissue image analysis platforms that perform FDA-approved biomarker tests in a HIPAA-compliant fashion have appeared in the last year, suggesting that these technologies may penetrate the clinical realm in the not-too-distant future. Some new tissue imaging technologies relying on fluorescence rather than bright-field microscopy can perform multi-parametric quantification of proteins in individual cells in tissue sections, bringing some of the advantages of flow cytometry to tissue sections while preserving the ability to assess tissue architecture simultaneously. Again, the strengths and potentials of such competing technologies will need to be thoroughly evaluated in the research laboratory, but it seems likely that one or another will emerge as a workhorse platform for accurate measurement of protein levels in FFPE tissues.
Beyond technological issues, it is also worth remembering that the prognostic utility of the Choi algorithm is tied to therapy and therefore may prove to be transient in nature. Experience has shown that when therapy changes, the significance of prognostic factors often changes as well. For example, in acute lymphoblastic leukemia (ALL) certain genomic aberrations or immunophenotype features that were once poor prognostic markers lost their predictive value with advances in therapy. One of the frustrations in DLBCL has been the failure to improve outcomes beyond those obtained with CHOP; rituximab is the first clear-cut example of therapeutic progress in this disease since CHOP was introduced in the mid-1970s. However, there is reason to hope that detailed genomic analyses will lead to the emergence of new rational therapeutic targets in the near future. As these present themselves, it can be imagined that the diagnostic emphasis will shift to a more focused analysis of the genetic lesions and/or protein markers that predict response to targeted therapies. Only then will the promise of expression profiling and other discovery-based approaches to understanding heterogeneity in DLBCL be truly fulfilled.
References
- 1.Choi WLW, Weisenburger DD, Greiner TC, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clinical Cancer Research. 2009 doi: 10.1158/1078-0432.CCR-09-0113. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Wright G, Tan B, Rosenwald A, Hurt EM, Wiestner A, Staudt L. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The International Non-Hodgkin's Lymphoma Prognostic Factors Project. A predictive model for aggressive non-Hodgkin's lymphoma. N Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 6.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 7.Dunleavy K, Pittaluga S, Czuczman MS, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood. 2009;113:6069–6076. doi: 10.1182/blood-2009-01-199679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moskowitz CH, Zelenetz AD, Kewalramani T, et al. Cell of origin, germinal center versus nongerminal center, determined by immunohistochemistry on tissue microarray, does not correlate with outcome in patients with relapsed and refractory DLBCL. Blood. 2005;106:3383–3385. doi: 10.1182/blood-2005-04-1603. [DOI] [PubMed] [Google Scholar]
- 9.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross JS, Hatzis C, Symmans WF, Pusztai L, Hortobagyi GN. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist. 2008;13:477–493. doi: 10.1634/theoncologist.2007-0248. [DOI] [PubMed] [Google Scholar]
- 11.Taylor CR, Levenson RM. Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49:411–424. doi: 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- 12.Walker RA. Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment I. Histopathology. 2006;49:406–410. doi: 10.1111/j.1365-2559.2006.02514.x. [DOI] [PubMed] [Google Scholar]