Abstract
Purpose
microRNAs have been shown to be involved in different human cancers. We therefore have performed expression profiles on a panel of pediatric tumors to identify cancer-specific microRNAs. We also investigated if microRNAs are co-regulated with their host gene.
Experimental Design
We performed parallel microRNAs and mRNA expression profiling on 57 tumor xenografts and cell lines representing 10 different pediatric solid tumors using microarrays. For those microRNAs that map to their host mRNA, we calculated correlations between them.
Results
We found that the majority of cancer types clustered together based on their global microRNA expression profiles by unsupervised hierarchical clustering. Fourteen microRNAs were significantly differentially expressed between rhabdomyosarcoma and neuroblastoma, and 8 of them were validated in independent patient tumor samples. Exploration of the expression of microRNAs in relationship with their host genes demonstrated that the expression for 43 (63%) of 68 microRNAs located inside known coding genes were significantly correlated with that of their host genes. Among these 43 microRNAs, 5 out of 7 microRNAs in the OncomiR-1 cluster correlated significantly with their host gene MIRHG1 (P<0.01). In addition, high expression of MIRHG1 was significantly associated with high stage and MYCN-amplification in neuroblastoma tumors; and the expression level of MIRHG1 could predict the outcome of neuroblastoma patients independently from the current neuroblastoma risk-stratification in two independent patient cohorts.
Conclusion
Pediatric cancers express cancer-specific microRNAs. The high expression of the OncomiR-1 host gene MIRHG1 correlates with poor outcome for patients with neuroblastoma, indicating important oncogenic functions of this microRNA cluster in neuroblastoma biology.
Keywords: microRNA, gene expression profiling, microarray, pediatric cancer, neuroblastoma, cancer classification, OncomiR-1, MIRHG1, prognosis
Introduction
MicroRNAs are small, non-coding RNA molecules encoded in the genomes of plants and animals. These highly conserved, ~21-neucleotide RNAs regulate the expression of genes by binding to the 3'-untranslated regions (3'-UTR) of specific mRNAs, causing translational inhibition or mRNA degradation (1). As many mRNAs may share this short sequence, microRNAs are capable of simultaneously influencing the expression of large sets of genes. It is estimated that each microRNA can target hundreds of genes (2), and conversely multiple microRNAs can target a single gene. So far 701 microRNAs (version 12.0) have been reported to be expressed in human cells (http://microrna.sanger.ac.uk/). Due to their regulatory roles in gene expression, there is increasing evidence that microRNAs are directly involved not only in normal embryogenesis, metabolism, cell growth, differentiation, and apoptosis, but also in pathogenesis of human cancers (3–6). Because most pediatric malignancies are developmental tumors arising from aberrant differentiation, we hypothesized that pediatric tumors will exhibit cancer and tissue-specific microRNA expression profiles associating with development and the tumorigenic process, which can be used in classification and prognosis of cancers.
To test this hypothesis, we investigated the expression profiles of microRNAs for a panel of 57 pediatric cell lines and human tumor xenografts for which mRNA profiles were available, and the majority of which are currently used as pediatric pre-clinical models for drug screening (7, 8). Using this panel of samples representing 10 different types of pediatric tumors (Table 1), we explored whether pediatric tumors differentially express microRNAs according to their diagnosis using microarray technology. A machine learning algorithm and statistical analysis was applied to the microRNA expression data to identify tumor-specific profiles for the two major subgroups of cancers (neuroblastoma and rhabdomyosarcoma) represented in our data set, and we validated these findings on independent neuroblastoma and rhabdomyosarcoma patient tumor samples. We explored if the microRNAs that map within host messenger RNAs are co-regulated with their host mRNAs. Finally we investigated if the expression of MIRHG1 gene (formally C13orf25), which hosts the oncogenic microRNA OncomiR-1 (miR-17-92 cluster), correlated with aggressive disease and poor outcome for patients with neuroblastoma.
Table 1.
Summary of Samples
| Sample Name | Type | Diagnosis |
|---|---|---|
| D212-X | xenograft | Glioblastoma |
| D456-X | xenograft | Glioblastoma |
| SJBT39-X | xenograft | Glioblastoma |
| SJBT56-X | xenograft | Glioblastoma |
| SJGBM2-X | xenograft | Glioblastoma |
| BT45-X | xenograft | BT other (Medulloblastoma) |
| BT46-X | xenograft | BT other (Medulloblastoma) |
| BT50-X | xenograft | BT other (Medulloblastoma) |
| BT36-X | xenograft | BT other (Ependymoma) |
| BT41-X | xenograft | BT other (Ependymoma) |
| ASLuc-C | cell line | Neuroblastoma |
| BE2-C | cell line | Neuroblastoma |
| CHP134-C | cell line | Neuroblastoma |
| GILIN-C | cell line | Neuroblastoma |
| IMR32-C | cell line | Neuroblastoma |
| IMR5-C | cell line | Neuroblastoma |
| KCNR-C | cell line | Neuroblastoma |
| LAN1-C | cell line | Neuroblastoma |
| LAN5-C | cell line | Neuroblastoma |
| NB1691-C | cell line | Neuroblastoma |
| NBEB-C | cell line | Neuroblastoma |
| SKNAS-C | cell line | Neuroblastoma |
| SKNDZ-C | cell line | Neuroblastoma |
| SKNFI-C | cell line | Neuroblastoma |
| SKNSH-C | cell line | Neuroblastoma |
| SY5Y-C | cell line | Neuroblastoma |
| CHLA79-X | xenograft | Neuroblastoma |
| NB-1382-X | xenograft | Neuroblastoma |
| NB1643-X | xenograft | Neuroblastoma |
| NB1691-X | xenograft | Neuroblastoma |
| NB1771-X | xenograft | Neuroblastoma |
| NBEBc1-X | xenograft | Neuroblastoma |
| NBSD-X | xenograft | Neuroblastoma |
| SKNAS-X | xenograft | Neuroblastoma |
| OS1-X | xenograft | Osteosarcoma |
| OS17-X | xenograft | Osteosarcoma |
| OS2-X | xenograft | Osteosarcoma |
| OS21-X | xenograft | Osteosarcoma |
| SKNEP-X | xenograft | Other (Diffuse anaplastic Wilm's) |
| EW5-X | xenograft | Other (Ewings) |
| EW8-X | xenograft | Other (Ewings) |
| BT29-X | xenograft | Other (Rhabdoid tumor of brain) |
| KT12-X | xenograft | Other (Rhabdoid tumor of kidney) |
| KT16-X | xenograft | Other (Rhabdoid tumor of kidney) |
| Unknown1-X | xenograft | Other (Unknown) |
| Unknown2-X | xenograft | Other (Unknown) |
| Rh28-X | xenograft | Rhabdomyosarcoma |
| Rh30-X | xenograft | Rhabdomyosarcoma |
| RH30R-X | xenograft | Rhabdomyosarcoma |
| Rh36-X | xenograft | Rhabdomyosarcoma |
| Rh41-X | xenograft | Rhabdomyosarcoma |
| Rh65-X | xenograft | Rhabdomyosarcoma |
| KT10-X | xenograft | Wilm's tumor |
| KT11-X | xenograft | Wilm's tumor |
| KT13-X | xenograft | Wilm's tumor |
Materials and Methods
Cell lines, Xenografts and Primary Tumor Samples
Neuroblastoma cell lines (n=16) were cultured as described (9). Xenograft samples (n=41) were described elsewhere (7, 8), and obtained through the Pediatric Preclinical Testing program (PPTP) established by the National Cancer Institute (NCI). Anonymous primary snap-frozen neuroblastoma (n=6) and rhabdomyosarcoma tumors (n=6) were acquired from Corporative Human Tissue Network (CHTN) and were deemed exempt from NCI institutional review board for this study. The clinical characteristics of these primary samples are described in the Supplemental Table 1.
Microarray and TaqMan Real-Time RT-PCR assays for microRNA Expression
Small RNA (<200bp) was purified using a previously published protocol (10). MicroRNA expression profiling was performed on our in-house printed microarrays. Synthetic DNA probes were designed using the sequences available from the Sanger miRBase Sequence Database (11), and custom-made by Sigma (St. Louis, MO). Each probe contained two tandem complementary sequences against each mature microRNA or its counterpart strand in the hairpin stem-loop structure, and there were 521 unique probes for human microRNAs on our in-house microRNA microarrays. An amine group tag was added on the 5’ of each probe for tethering on Nexterion epoxy glass slides (Schott, Louisville, KY). The reference synthetic DNA oligos complementary to the probes were labeled with Cy3 dye and the sample with Cy5 dye using miRVana microRNA labeling kit (Ambion, Austin, TX). Hybridization was performed on the MAUI hybridization systems (BioMicro System, Salt Lake City, UT) at 52°C with mixing for overnight. Slides were then washed in 2XSSC with 0.2%SDS for 15 minutes at 42°C, 2XSSC at room temperature for 10 minutes, and 0.2XSSC at room temperature for 10 minutes. Finally, slides were dried by centrifugation and scanned in an Agilent microarray scanner (Agilent, Santa Clara, CA).
TaqMan microRNA RT-PCR assays (Applied Biosystems, Foster City, CA) were performed according to the manufacturer’s protocol as previously described (10).
MicroRNA Microarray Data Filtering and Normalization
The Cy3 reference channel was first normalized using quantile normalization. Then, low-quality probes were removed using the Cy5 sample channel with a criterion that required raw intensity of the probe to be larger than 128 fluorescent units for at least four samples. Two hundred seven probes passed this quality filter. After quality filtering, the log2 ratios (log2(Cy5/Cy3)) were calculated, and were subsequently normalized by subtracting the average log2 ratio of the internal control probes. Then, we added a constant value to get positive values. Every probe was printed in duplicate on the array and the average of these duplicates was used to represent the final expression measurements. All of the quality-filtered microRNA and parallel mRNA data can be found on our website (http://pob.abcc.ncifcrf.gov/cgi-bin/JK).
mRNA Microarray Experiments
Gene expression profiling was performed on Affymetrix U133 Plus 2.0 arrays according to the manufacturer's instruction (Affymetrix, Santa Clara, CA). We obtained the gene expression profiling data of 38 xenograpfts from the study by Neale et al. (7), and profiled the rest samples in our lab. Relative expression values were obtained using Affymetrix PLIER algorithm through Affymetrix Power Tools version 1.8.5 and further log-tranformed to base 2.
Hierarchical Clustering, Nearest Centroid Classifiers, Statistical, and Genomic Location Analyses
We performed two-way hierarchical clustering of 57 pediatric cancer samples using all expression values from microRNA after quality filtering. The Cluster 3.0 software (12) was utilized, wherein expression values were median- centered per gene, and clustered using Pearson correlation distance and average linkage. The result was visualized using TreeView (13).
Nearest centroid classifiers (NCC) (14) were trained to separate neuroblastoma from rhabdomyosarcoma samples. In the NCC, the centroid for each class was calculated as the average profile across samples. For a test sample, the prediction output was calculated as the Pearson correlation against the rhabdomyosarcoma centroid minus the Pearson correlation against the neuroblastoma centroid. Therefore a large prediction output suggests that the sample is a rhabdomyosarcoma sample. Prediction accuracy was evaluated using a leave-one-out scheme, in which all but one samples were used for training and the status of the left out sample was predicted by the trained classifier. We used a permutation test to estimate statistical significance of prediction accuracy. Sample labels were randomly permuted 100,000 times, and for each randomization the leave-one-out procedure was repeated, and a P value was calculated corresponding to the probability to obtain perfect predictions for random sample labels.
To identify the host genes for microRNAs, genomic locations for microRNAs and mRNA probe sets were retrieved from Sanger miRBase (version 10.1) and Affymetrix respectively, and were mapped to the UniGene. The host genes (represented by probe sets) were identified for each microRNA by mapping them in the same UniGene with the same orientation. We calculated the Pearson correlation across all 57 samples. If a microRNA was matched to several probe sets, the largest absolute correlation was used.
Survival Analysis
Cox regression analyses and Log-rank tests were performed using the survival R package (http://cran.r-project.org/web/packages/survival/index.html). Expression values of a gene were dichotomized into high- and low- expression using the median as cutoffs.
Results
microRNA Expression Profiling of Pediatric Cancers and Classification of Cancers using microRNA Profiles
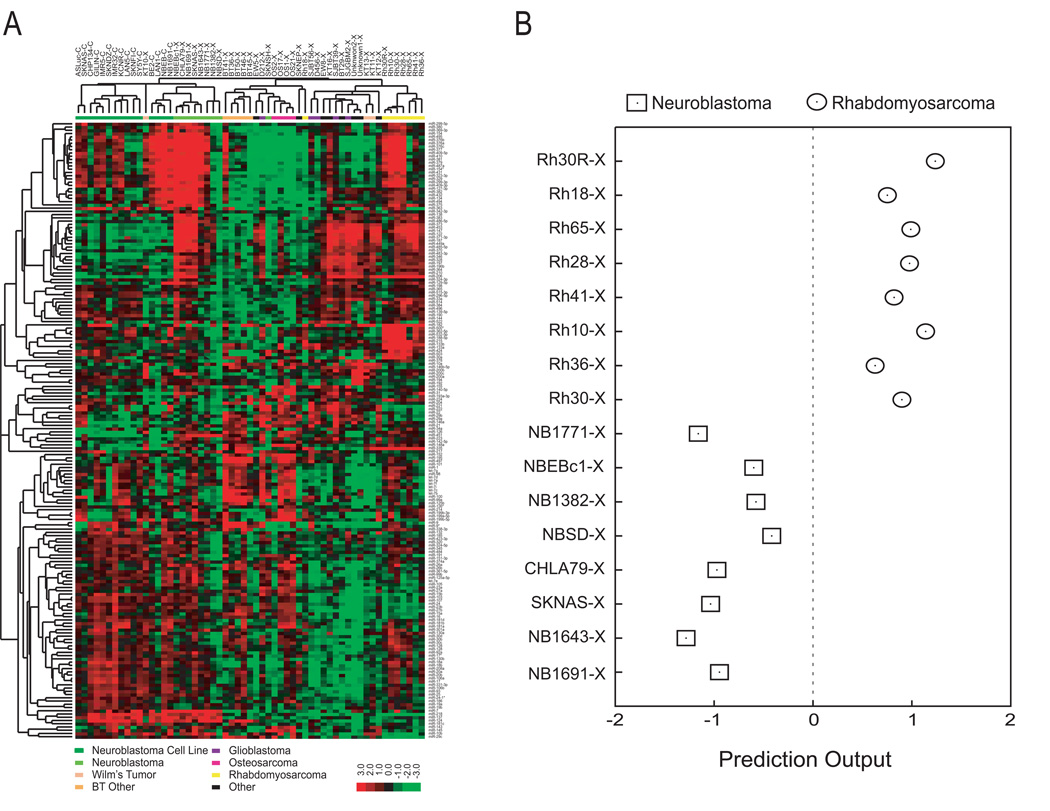
We first hypothesized that the microRNA expression profiles would reflect the cancer type for pediatric malignancies. We performed microRNA microarray analysis of 16 neuroblastoma cell lines and 41 xenografts including brain tumor (n=10), neuroblastoma (n=8), rhabdomyosarcoma (n=8), osteosarcoma (n=4), Wilms’ tumor (n=3), and others (n=8) (Table 1). An unsupervised hierarchical clustering analysis using all 207 microRNA probes with good quality demonstrated that microRNA expression profiles can separate these samples according to their diagnosis (Figure 1A). One of the two major branches consists of all except one neuroblastoma samples indicating that these samples have a neuroblastoma-specific microRNA expression profile. The other major branch contains non-neuroblastoma samples formed clusters primarily of the same diagnostic categories. For example, seven of eight rhabdomyosarcomas form a tight sub-cluster, as did four of four osteosarcomas (Figure 1A). The property of the sample clustering shows that there is a clear tumor-specific microRNA expression profile in these pediatric cancer samples.
Figure 1. MicroRNAs are diagnostic for pediatric tumor samples.
(A) Unsupervised hierarchical clustering demonstrated clustering of the samples according to the tumor types. We performed two-way hierarchical clustering of 57 pediatric cancer samples using all microRNA expression values after quality filtering. The Cluster 3.0 software (12) was utilized, wherein expression values were median-centered per gene and clustered using Pearson correlation distance and average linkage. (B) Nearest centroid classifiers were trained to separate neuroblastoma from rhabdomyosarcoma xenograft samples. The centroid for each diagnostic category was calculated as the average profile across samples. The prediction output of a sample was calculated as the Pearson correlation against the rhabdomyosarcoma centroid minus the Pearson correlation against the neuroblastoma centroid. Prediction accuracy was evaluated using a leave-one-out scheme, and all samples were correctly classified (P=1.6×10−4).
To further examine if microRNAs can be used to classify cancers, we applied a machine learning algorithm to the microRNA expression data for the two major subgroups of cancers (neuroblastoma and rhabdomyosarcoma) represented in our data set using all high-quality probes. To avoid classifications heavily driven by cell line-specific signatures, we used only xenograft samples in this analysis. We built nearest centroid classifiers (14) to separate the 8 neuroblastoma xenografts from the 8 rhabdomyosarcoma xenografts using a leave-one-out scheme. All 16 samples were perfectly diagnosed (P=1.6×10−4) (Figure 1B). Therefore, these experiments showed that neuroblastoma and rhabdomyosarcoma differentially express tumor-specific microRNAs.
Differential Expression of microRNAs Distinguishes Neuroblastoma vs. Rhabdomyosarcoma
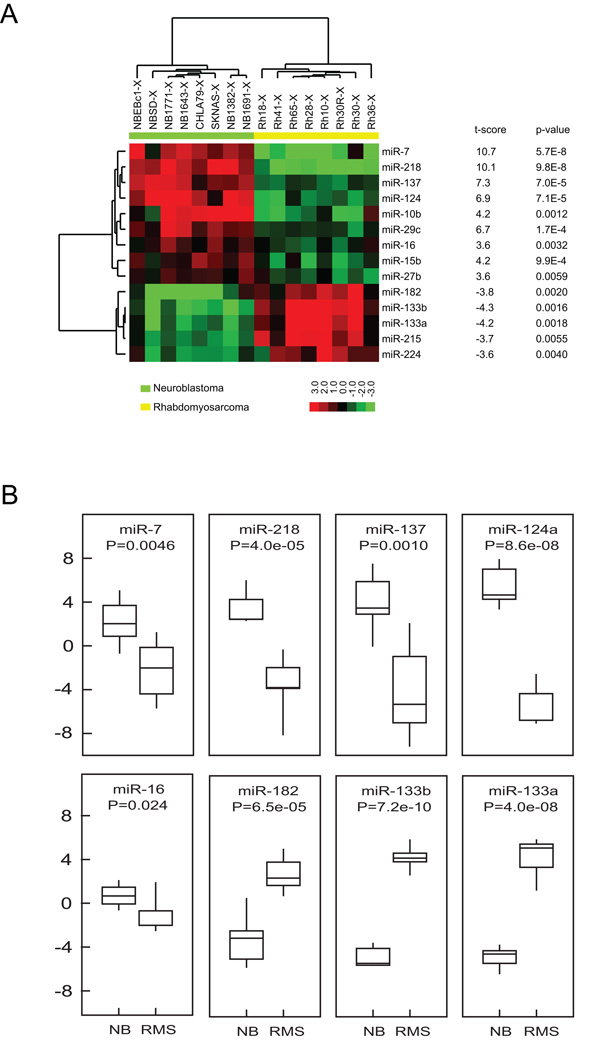
To identify the tumor-specific microRNAs which may contribute to the biology of these cancers, we performed a t-statistical test between the two major cancer types, neuroblastoma and rhabdomyosarcoma, using the microRNA expression profiles of 8 neuroblastoma and 8 rhabdomyosarcoma xenograft samples. We found 14 microRNAs significantly differentially expressed in these two cancer types (P<0.01; FDR=15%) (Figure 2A). To validate if these microRNAs are differentially expressed in human primary tumor samples, we performed Taqman® real-time RT-PCR using an independent set of primary neuroblastoma (n=6) and rhabdomyosarcoma (n=6) tumors from patient biopsies. We found 8 of the 14 microRNAs to be significantly differentially expressed in the patient tumor samples (P<0.05; Figure 2B), demonstrating the potential of using these microRNAs as biomarkers to distinguish these two cancer types.
Figure 2. Differentially expressed cancer-specific microRNAs for Neuroblastoma and rhabdomyosarcoma.
(A) Comparing the microRNA expression in rhabdomyosarcoma (n=8) and neuroblastoma (n=8) xenograft samples using a t-test, we identified 14 differentially expressed microRNAs (P<0.01). Samples and microRNAs were hierarchically clustered using Pearson correlation distance and average linkage. Data were centralized prior clustering such that median expression of each microRNA was zero. (B) The expression of 8 differentially expressed microRNAs was validated using Taqman® real-time RT-PCR in an independent set of primary neuroblastoma and rhabdomyosarcoma tumors (P<0.01) indicating the value of microRNA expression levels in distinguishing these cancers.
Co-regulation of microRNA with Host Gene
Currently, the control of microRNA expression is largely unclear, and we hypothesized that the expression level of microRNAs mapping within coding genes is controlled by the promoter of the host gene. We therefore investigated how many of the microRNAs on our array map within and also correlate with their host mRNAs. Out of 207 microRNAs of good quality detected by our arrays, 189 unique microRNAs can be mapped to the human genome with their genomic coordinates. Among them, 68 (36%) are located within known coding genes, and also have the same orientation with their host genes. We calculated correlations of the expression between these 68 microRNAs and their corresponding mRNAs, and found that 43 (63%) were significantly co-expressed (r>0.34; P<0.01) with their host genes (Figure 3 and Supplemental Table 2). OncomiR-1, also known as the miR-17-92 cluster, has been reported to be associated with multiple human malignancies (15, 16). Among these 43 microRNAs, we found that 5 out of 7 microRNAs in the OncomiR-1 cluster significantly correlated with the expression level of its host gene MIRGH1 (r=0.37– 0.49; P<0.01) (Figure 3), indicating that the level of MIRGH1 transcript directly affected the level of these oncogenic microRNAs.
Figure 3. MicroRNA expression correlates with their host gene expression.
Genomic locations for microRNAs and probe sets were retrieved from Sanger miRBase (version 10.1) and Affymetrix respectively. Genomic locations were mapped to UniGene clusters. Off 189 microRNAs with known locations, 68 (36%) were located within a UniGene with the same orientation. For each pair of microRNA and its host gene located within the same UniGene, the Pearson correlation was calculated across all 57 samples. Of 68 microRNAs located within a UniGene, 43 (63%) were found to be significantly (P<0.01) positively correlated with a probe set from its host gene. Colors in the heatmap represent z-score normalized (zero mean and unity variance per microRNA/gene) expression. Samples (columns) are sorted with respect to diagnosis, and mRNAs or microRNAs (rows) are sorted with respect to genomic location. Chr, chromosome; Start, the start coordinate of microRNA.
High MIRGH1 Expression Level is Associated with Aggressive Behavior and Poor Outcome in Neuroblastoma Patients
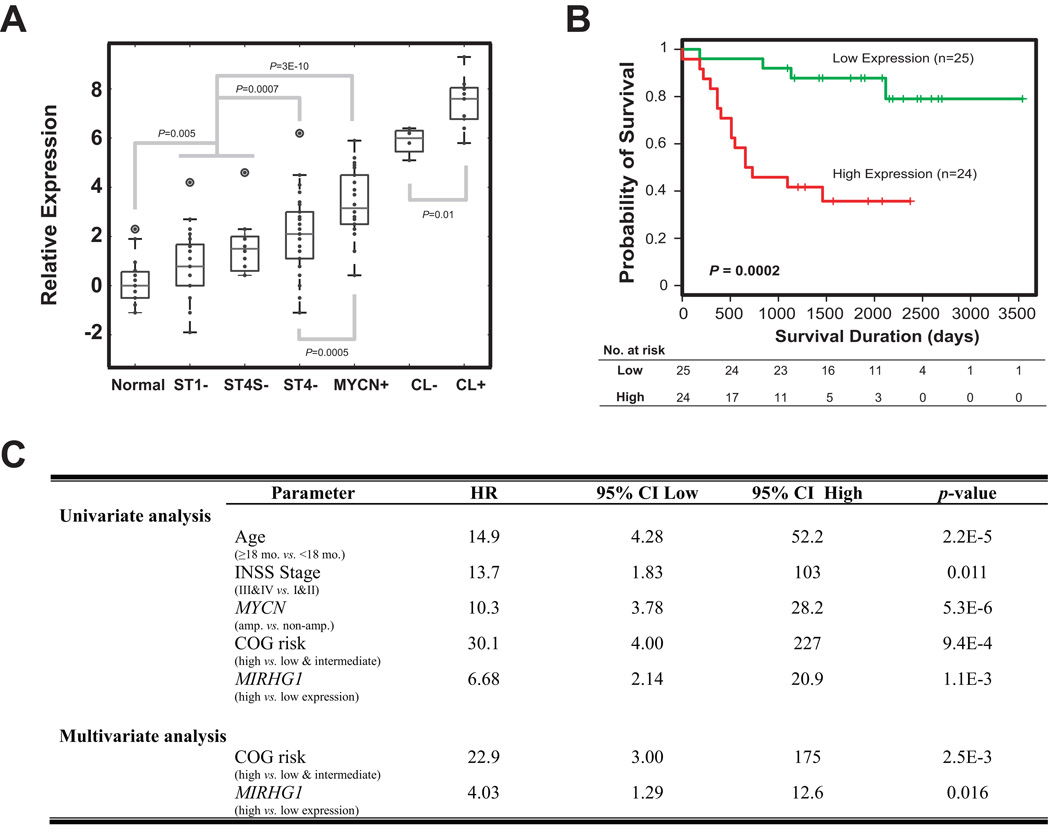
Since OncomiR-1 has been reported to be directly downstream of MYC (17–20), and MYCN is often highly expressed in pediatric cancers including neuroblastoma and rhabdomyosarcoma (21, 22), we explored if the expression of OncomiR-1 host gene MIRHG1 was associated with more aggressive phenotype in our neuroblastoma tumor gene expression database (23, 24) (http://pob.abcc.ncifcrf.gov/cgi-bin/JK). We found a significantly higher expression level in MYCN-amplified tumors and cell lines, as well as in higher stage (Stage 4) compared with lower stage and more benign tumors (stage 1 and 4S) (Figure 4A). The highest expression was in cell lines particularly if the MYCN was amplified; and the lowest expression in normal human tissues (n=19) (Figure 4A). Furthermore, we found that high expression of MIRHG1 is significantly associated with poor outcome of neuroblastoma patients in our published cohort consisting of patients of all major stages, with or without MYCN-amplification (NCI cohort) (P=0.0002 in a log-rank test or P=0.0011 in an univariate Cox model ) (Figure 4B and C) (23). A multivariate analysis showed that the prognostic power of MIRHG1 expression is independent of current COG risk-stratification (P<0.05, Figure 4C).
Figure 4. High MIRHG1 expression is significantly correlated with poor clinical outcome in neuroblastoma patients.
(A) A box plot of MIRHG1 expression in neuroblastoma primary tumors and cell lines demonstrates that MIRHG1 is expressed at a higher level in high-stage (P=0.0007) and MYCN-amplified (P=0.0005) neuroblastoma tumors as well as in the cell lines (P=0.01). Normal samples express lower level of MIRHG1 (P=0.005). Normal, normal tissues; ST, stage; CL, cell line; “+”, MYCN-amplification; “−”, MYCN not-amplified. (B) A Kaplan-Meier curve for survival probability using the mRNA level of MIRHG1 in a published NCI neuroblastoma patient cohort (23) demonstrates MIRHG1 expression level can predict the outcomes of neuroblastoma patients. Median expression of MIRHG1 was used as the cutoff, and P value is calculated using a Log-rank test. (C) The expression of MIRHG1 predicts the outcome of neuroblastoma patients independently from current COG risk-stratification in the NCI cohort (23). Cox regression models were used in both univariate and multivariate analyses.
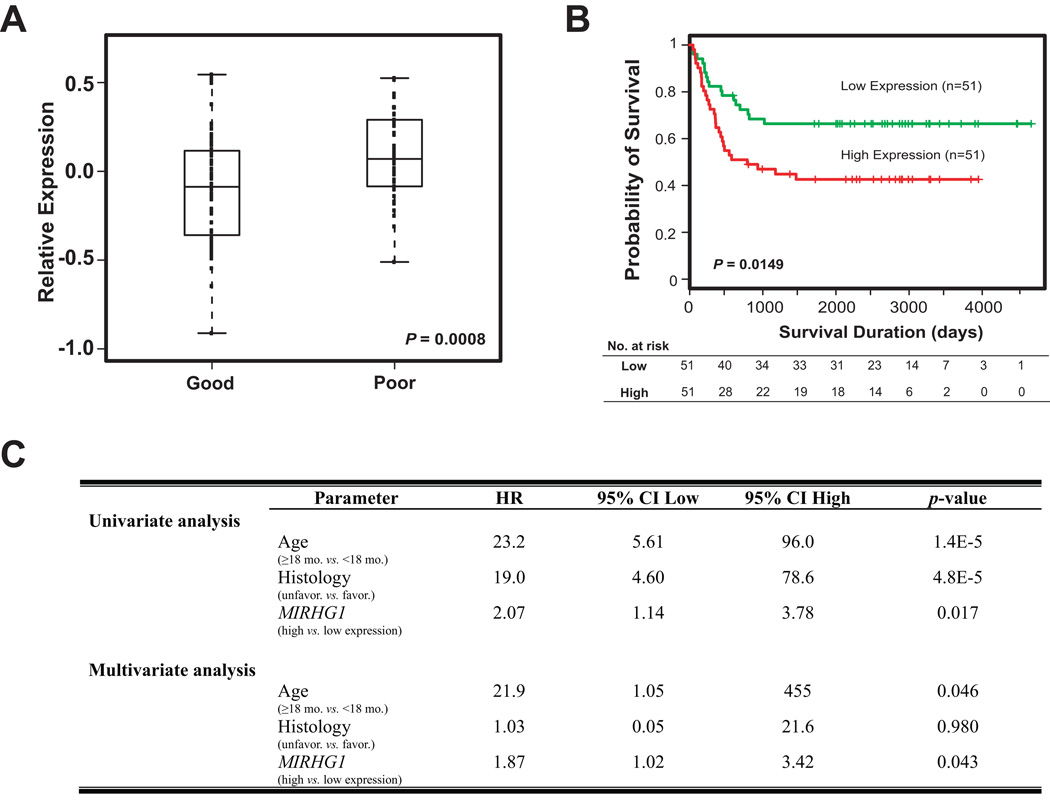
Finally, we examined if our findings could be validated in another independent neuroblastoma patient cohort reported by Asgharzadeh et al., which only included neuroblastoma patients with stage 4 diseases without MYCN-amplification (CHLA cohort) (25). Indeed, we observed higher MIRHG1 expression in the patients with poor outcome in this dataset (P=0.0008) (Figure 5A), confirming that high expression level of MIRHG1 was significantly associated with poor prognosis (P=0.0149) (Figure 5B). Furthermore, we examined if the expression level of MIRHG1 added any predictive power to the current Children’s Oncology Group (COG) risk-stratification in this dataset. Because the intermediate-risk patients in CHLA cohort all survived, we could not build a Cox regression model using the COG-risk stratification criteria in the multivariate analysis. Instead we used the available risk factors (age and histology) in this analysis (Figure 5C). Multivariate analysis in this dataset again demonstrated that MIRHG1 expression is a significant prognostic marker independent from COG risk-stratification factors (Figure 5C).
Figure 5. The predictive power of MIRHG1 expression is validated in an independent CHLA neuroblastoma patient cohort.
(A) In an independent published CHLA patient cohort which only consists of neuroblastoma patients with stage 4 diseases and without MYCN-amplification (25), the high expression level of MIRHG1 is also observed in the patients of poor outcome (P=0.0008). The expression of MIRHG1 significantly correlated with adverse outcome in a Kaplan-Meier plot (P=0.0149) (B) and Cox models (P<0.05) (C). Median expression of MIRHG1 was used as the cutoff.
Discussion
The expression patterns of microRNA represent novel methods for categorizing pediatric cancers, and have potential advantages over mRNA profiling. First, the relatively smaller number of microRNA in the human genome (~1000 for microRNA vs. tens of thousands for mRNA) makes their profiles less complex. Second, because of the short length of microRNAs, they are better preserved in clinical samples such as tumor specimen and paraffin sections (26, 27). Reports have suggested that microRNA expression profiles may be better predictors of diagnosis and clinical outcome of human diseases than mRNA based methods (28, 29), however, a recent study on NCI 60 cell lines showed that mRNA profiles are more informative for discriminating tissue types than microRNA profiles (30). In our study, the 57 samples clustered according to the major cancer types using the microRNA expression profiles (Figure 1). However, the microRNA expression profiles did not perform as well as mRNA expression profiles (7, 8) in separating these samples into their cancer types by hierarchical clustering (data not shown). Therefore, the value of using global microRNAs profiles in cancer classification is still unclear.
We identified 14 significantly differentially expressed microRNAs distinguishing neuroblastoma from rhabdomyosarcoma in these xenografts. Of these 8 were validated by Taqman RT-PCR in an independent cohort of primary human tumor samples indicating the potential utility of these microRNAs as tumor-specific biomarkers for tumor classification. However, due to the rarity of pediatric solid tumors, this study focused on validation of microRNAs only in neuroblastoma and rhabdomyosarcoma. Although the xenografts and cell lines are kept in an artificial environment, studies have shown that they express a large panel of genes resembling their corresponding human tumors (7, 8). Furthermore, studies in the xenografts and cell lines have yielded valuable information in a pre-clinical setting such as the Pediatric Preclinical Testing Program (PPTP, http://pptp.stjude.org/) to identify effective agents for these tumors (7, 8). Therefore, we believe our approach in this study is valid and the results from this study showed that we identified cancer-specific microRNAs. However, these microRNA signatures need to be validated in a much larger study with human tumors before clinical use as diagnostic markers.
In addition to potential use a diagnostic biomarkers, these differentially expressed microRNAs may shed lights to our understanding of the biology of these pediatric tumors, because microRNA expression has been reported to associate with tissue differentiation (31). Thus, tumor-specific microRNA profiles may reveal not only the tissue origin of cancers, but also its biology in tumorigenesis. For example, miR-133a is expressed during normal muscle development (31), and was found by us to be expressed abundantly in rhabdomyosarcoma. Recently miR-133a has been shown to play a critical role in the regulation of myocyte growth (32, 33). Similarly, we found that miR-7, 124a, 137 and 218 were expressed at a high level in neuroblastoma samples, and their expression has been reported to be specific in neural tissues during zebra fish development (34). The correlation between the expression levels of miR-218 and its host gene, SLIT3, is the highest in our analysis (Supplemental Table 2). SLIT family members have been implicated to play a critical role in the formation of central nervous system (35). This indicates that the expression of miR-218 from the SLIT3 transcript may also play a role in the differentiation of neural tissues. Intriguingly, we have previously reported that SLIT3 is over-expressed in the poor-prognosis neuroblastoma (23). The high correlation between SLIT3 and miR-218 expression suggests that expression of miR-218 may also predict poor prognosis. Recently, Makeyev et al. have reported that miR-124 promotes neuronal differentiation through inducing nerve system specific alternative splicing (36) demonstrating the importance of this microRNA during neural development. Therefore, differentially expressed microRNAs are likely to play important roles in the normal tissue development as well as in the tumorigenesis of pediatric cancers.
Despite the increasing knowledge of microRNA expression patterns in different biological systems including cancers, the regulation of microRNA expression is largely unknown. We attempted to determine if the genomic location of microRNAs in relationship to their host genes affected the expression of microRNAs. Using this pediatric tumor dataset containing both gene and microRNA expression profiles, and a low stringent cut-off (r>0.34), surprisingly we observed that only 63% of microRNAs residing within host genes in the same orientation showed expression patterns that correlated with their host genes (Figure 3). Therefore, a gene unit is a more complicated functional transcription unit than the protein coding gene itself, indicating there are multiple mechanisms to regulate microRNA levels other than sharing the common promoter with their host genes.
Among all the microRNAs and their host gene with highly correlated expression levels, OncomiR-1 (miR-17-92 cluster) and its non-protein coding host gene, MIRHG1 (also known as C13orf25), are of particular interest due to its oncogenic potential in human cancers (16, 17, 20). Although MIRHG1 and the miRNAs in OncomiR-1 are correlated, the putative promoter of MIRGH1 is about 2000 bp upstream of the E-boxes at the OncomiR-1 locus (18, 20). It is therefore still possible that MIRHG1 and OncomiR-1 are transcribed from different promoters, but are still co-regulated. OncomiR-1 has been demonstrated to be directly transactivated by an important oncogene c-Myc (18), and MYC oncogene family members are important transcription factors often hyper-activated in many human cancers (37). Therefore, OncomiR-1 can mediate at least some of the oncogenic functions of MYC. Several recent studies have indicated that MYCN, another MYC family member, can up-regulate OncomiR-1 (19, 20, 38). Fontana et al. have demonstrated that MYCN activates OncomiR-1 cluster by directly binding to its promoter (20). This is of particular interest because the MYCN gene is frequently amplified in neuroblastoma and rhabdomyosarcoma (21, 22), and this molecular characteristic is used in clinic to stratify treatment for patients with neuroblastoma. In this study, we have demonstrated that the high expression of the OncomiR-1 host gene, MIRHG1, is correlated with tumors with not only MYCN-amplification but also higher stages and poor prognosis. In addition, Fontana et al have shown the tumorigenic role of Oncomicr-1 cluster in neuroblastoma cells by promoting cell growth (20). Therefore, these studies indicate an important biological role of OncomiR-1 cluster in the aggressive form of neuroblastoma. These findings warrant future studies to characterize the oncogenic mechanisms of individual microRNA encoded in this cluster and explore the potential targeted therapies against these microRNAs.
In summary, we have shown that pediatric cancers cluster according to their diagnosis based on microRNA expression profiles, and we identified 8 tumor specific microRNAs for rhabdomyosarcoma and neuroblastoma. In addition, we have shown evidence that the expression of microRNAs located within protein coding genes are co-regulated with their host transcripts. Finally, we demonstrated that the high expression of a microRNA cluster host gene MIRHG1 is significantly associated with aggressive neuroblastoma. Our results indicate that MIRHG1 may play an important biological role in aggressive neuroblastoma, and the predictive value of MIRHG1 expression in neuroblastoma patients should be further validated in a much larger cohort in future studies.
Statement of Translational Relevance
microRNAs are small, non-coding regulatory RNAs that are implicated in cancer development. Because most pediatric malignancies are developmental tumors arising from aberrant differentiation, we hypothesized that pediatric tumors will exhibit cancer and tissue-specific microRNA expression profiles associating with development and tumorigenic process, which can be used in diagnosis and prognosis. Here we have performed microRNAs and mRNA expression profiling on a panel of 57 tumor xenografts and cell lines representing 10 different pediatric solid tumors using microarrays. We showed that pediatric cancers differentially express microRNAs specific to their origins and types. In addition, we showed evidence that the expression of microRNAs located within protein coding genes are co-regulated with their host gene transcripts. Finally, we demonstrated that the high expression of the OncomiR-1 host gene, MIRHG1, is significantly associated with aggressive neuroblastoma. This finding warrants further studies of the role of the OncomiR-1 in neuroblastoma patients with adverse outcomes.
Supplementary Material
Acknowledgments
This manuscript is dedicated to our friend and colleague Dr. Stephen Qualman (Cooperative Human Tissue Network, Columbus, OH) who fought a valiant battle against pancreatic cancer. We would like to thank Dr. Ernest Kawasaki, David Petersen, Jonathon Paarlberg, and Temitope Fadiran for their excellent technical support; and Dr. Thomas Badgett for his insightful comments on this manuscript. We also thank CHTN for providing human tumor samples. This study is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neale G, Su X, Morton CL, et al. Molecular characterization of the pediatric preclinical testing panel. Clin Cancer Res. 2008;14:4572–4583. doi: 10.1158/1078-0432.CCR-07-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiteford CC, Bilke S, Greer BT, et al. Credentialing preclinical pediatric xenograft models using gene expression and tissue microarray analysis. Cancer Res. 2007;67:32–40. doi: 10.1158/0008-5472.CAN-06-0610. [DOI] [PubMed] [Google Scholar]
- 9.Khan J, Wei JS, Ringner M, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei JS, Song YK, Durinck S, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 13.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 14.Dabney AR. Classification of microarrays to nearest centroids. Bioinformatics. 2005;21:4148–4154. doi: 10.1093/bioinformatics/bti681. [DOI] [PubMed] [Google Scholar]
- 15.Ota A, Tagawa H, Karnan S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 16.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 17.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 19.Schulte JH, Horn S, Otto T, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 20.Fontana L, Fiori ME, Albini S, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwab M, Ellison J, Busch M, Rosenau W, Varmus HE, Bishop JM. Enhanced expression of the human gene N-myc consequent to amplification of DNA may contribute to malignant progression of neuroblastoma. Proc Natl Acad Sci U S A. 1984;81:4940–4944. doi: 10.1073/pnas.81.15.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williamson D, Lu YJ, Gordon T, et al. Relationship between MYCN copy number and expression in rhabdomyosarcomas and correlation with adverse prognosis in the alveolar subtype. J Clin Oncol. 2005;23:880–888. doi: 10.1200/JCO.2005.11.078. [DOI] [PubMed] [Google Scholar]
- 23.Wei JS, Greer BT, Westermann F, et al. Prediction of clinical outcome using gene expression profiling and artificial neural networks for patients with neuroblastoma. Cancer Res. 2004;64:6883–6891. doi: 10.1158/0008-5472.CAN-04-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krasnoselsky AL, Whiteford CC, Wei JS, et al. Altered expression of cell cycle genes distinguishes aggressive neuroblastoma. Oncogene. 2005;24:1533–1541. doi: 10.1038/sj.onc.1208341. [DOI] [PubMed] [Google Scholar]
- 25.Asgharzadeh S, Pique-Regi R, Sposto R, et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006;98:1193–1203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Smyth P, Flavin R, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 29.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 30.Blower PE, Verducci JS, Lin S, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6:1483–1491. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 31.Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 32.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 34.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 35.Itoh A, Miyabayashi T, Ohno M, Sakano S. Cloning and expressions of three mammalian homologues of Drosophila slit suggest possible roles for Slit in the formation and maintenance of the nervous system. Brain Res Mol Brain Res. 1998;62:175–186. doi: 10.1016/s0169-328x(98)00224-1. [DOI] [PubMed] [Google Scholar]
- 36.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.