Abstract
Lamination of cortical regions of the vertebrate brain depends on glial-guided neuronal migration. The conserved polarity protein Par6α localizes to the centrosome and coordinates forward movement of the centrosome and soma in migrating neurons. The cytoskeletal components that produce this unique form of cell polarity and their relationship to polarity signaling cascades are unknown. We show that F-actin and Myosin II motors are enriched in the neuronal leading process and Myosin II activity is necessary for leading process actin dynamics. Inhibition of Myosin II decreased the speed of centrosome and somal movement, while Myosin II activation increased coordinated movement. Ectopic expression or silencing of Par6α inhibited Myosin II motors by decreasing Myosin Light Chain phosphorylation. These findings suggest leading process Myosin II may function to “pull” the centrosome and soma forward during glial-guided migration by a mechanism involving the conserved polarity protein Par6α.
Keywords: glial-guided migration, neuronal polarity, centrosome, Myosin II
Introduction
In recent years, remarkable progress has been made in understanding the general features of cell migration (Ridley et al., 2003). Fibroblasts or epithelial cells crawling on 2 dimensional substrates, the most common models for cell motility, possess lamellipodial actin-rich protrusions that advance a “leading edge” in the direction of movement. A variety of actin regulatory molecules; such as Arp2/3, ADF/cofilin or Ezrin-Radixin-Moesin (ERM) proteins organize lamellipodial actin cytoskeletal dynamics (Le Clainche and Carlier, 2008). Just behind the “leading edge”, Myosin II motors act in concert with integrin dependent adhesions to provide traction forces needed for protrusion and forward movement (Giannone et al., 2004; Gupton and Waterman-Storer, 2006; Hu et al., 2007; Vicente-Manzanares et al., 2009). Although many of these features are conserved, the morphology or mode of cell migrations can vary depending upon cell type or migratory pathway.
Directed migrations of neurons along glial fibers are essential for development of the laminar architecture of cortical regions of the mammalian brain and ultimately pattern synaptic connectivity (Hatten, 2002; Rakic, 1971). The cerebellar granule neuron (CGN) has long provided a model for understanding the molecular mechanism of glial-guided migration, including the first real time imaging of the mode of neuronal movement along the glial fiber (Edmondson and Hatten, 1987) and the identification of astrotactin (ASTN) as a neuron-glial ligand in the adhesion beneath the cell soma (Fishell and Hatten, 1991). A key difference between neuronal locomotion along glial fibers and the general crawling movement of metazoan cells is the absence of a “leading edge” in migrating neurons (Edmondson and Hatten, 1987; Gregory et al., 1988; Komuro and Rakic, 1998; O’Rourke et al., 1992; Solecki et al., 2004; Tanaka et al., 2004; Tsai et al., 2007; Umeshima et al., 2007). Migrating neurons extend an extremely thin (1–2 μm in diameter) leading process that wraps around the glial fiber in the direction of migration (Rivas and Hatten, 1995). Although the leading process is highly dynamic, extension of the tip of this process does not correlate with somal translocation (Edmondson and Hatten, 1987). Forward movement of the soma and nucleus occurs with the release of the neuron-glial adhesion beneath the migrating neuron, after which the cell glides along the glial fiber until a new adhesion forms (Gregory et al., 1988). In contrast to the crawling movements described above, integrin-based adhesions are not involved in neuronal migration along glial fibers (Belvindrah et al., 2007; Edmondson et al., 1988; Fishell and Hatten, 1991).
Over the last decade functional studies on signaling pathway components and cytoskeletal proteins has begun to define the molecular basis of glial-guided neuronal migration (Bellion et al., 2005; Komuro and Rakic, 1998; Schaar and McConnell, 2005; Solecki et al., 2004; Tanaka et al., 2004; Tsai et al., 2007; Umeshima et al., 2007). Live imaging and functional studies examining the activity of the Par6α signaling complex in neuronal migration reveal that the neuronal centrosome enters the leading process prior to forward movement of the neuronal soma and that the PAR complex coordinates centrosomal motility with forward locomotion (Solecki et al., 2004). Thus, the directed movement of the centrosome indicates the direction of migration, as does the direction of leading process extension and somal translocation (Schaar and McConnell, 2005; Solecki et al., 2004; Tanaka et al., 2004; Tsai et al., 2007; Umeshima et al., 2007). Studies by Vallee (Faulkner et al., 2000; Tsai et al., 2007; Tsai et al., 2005), Wynshaw-Boris (Hirotsune et al., 1998) and others (Feng et al., 2000; Gleeson et al., 1999; Niethammer et al., 2000; Smith et al., 2000; Tanaka et al., 2004) demonstrate that cytoplasmic dynein and its cofactors LIS1 and Doublecorti, organize the microtubule cytoskeleton of migrating neurons while actin based motors, e.g. Myosin II (Bellion et al., 2005; Ma et al., 2004; Schaar and McConnell, 2005), small GTPases (Guan et al., 2007; Kholmanskikh et al., 2006) and f-actin regulatory molecules (Bellenchi et al., 2007) control the microfilament system. In spite of these advances, an integrated view of the function of polarity signaling pathways and diverse cytoskeletal components function in living, migrating CNS neurons is lacking. In particular, although the dynamics and organization of f-actin have been examined in great detail in the leading edge of fibroblasts and neuronal growth cones (Hu et al., 2007; Ponti et al., 2004; Schaefer et al., 2002; Schaefer et al., 2008), little is known about f-actin organization and dynamics in the soma and leading process of migrating neurons.
In this report, we use time-lapse imaging to investigate the role of the actin cytoskeleton and Myosin II motor in organizing the coordinated movement of the centrosome and soma during CGN migration along glial fibers. We show that dynamic rearrangement of the actin cytoskeleton in the proximal region of the leading process, where the cell body tapers into the leading process, functions in centrosomal and somal motility. In addition, Myosin II motors are primarily localized to the neuronal leading process, suggesting that acto-myosin contractility in this region coordinates actin dynamics linked to migration. Our findings suggest a model where acto-myosin contractility in the leading process, rather than in a classical “leading edge” at the tip of the leading process, provides the traction force needed for forward movement. These studies further suggest that the conserved polarity protein Par6α regulates acto-myosin contractility during neuronal migration by modifying Myosin Light Chain phosphorylation.
Results
Organization of the actin cytoskeleton in migrating neurons
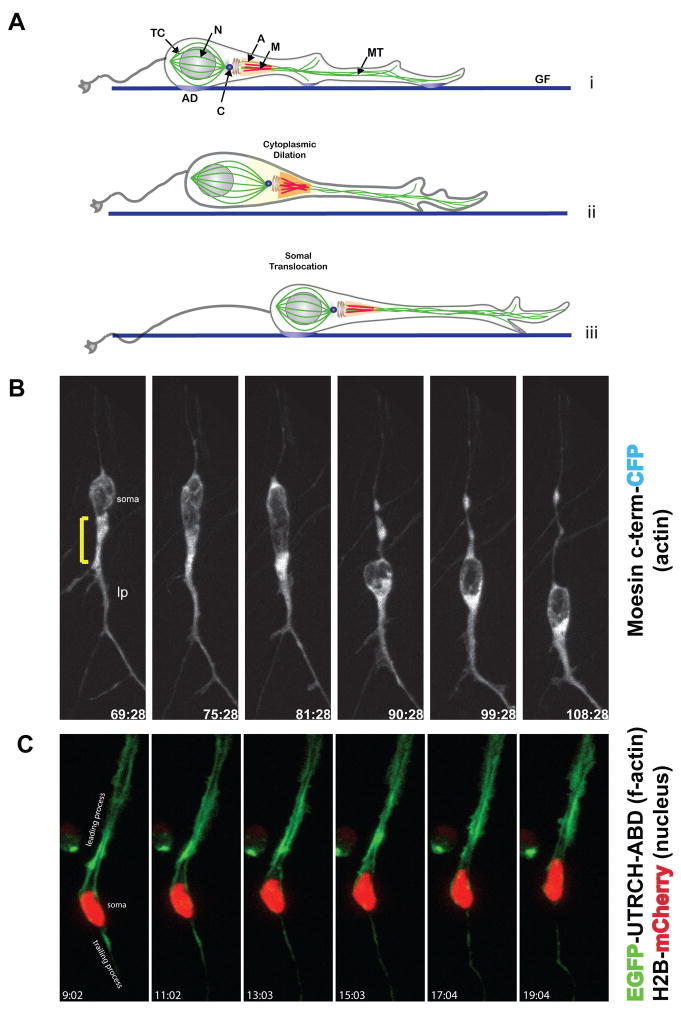
The actin cytoskeleton plays key roles in cell structure and motility in a variety of cell types and processes (Le Clainche and Carlier, 2008). We used actin-binding proteins as convenient reporters of actin localization in live CGNs migrating along Bergmann glial fibers. We first examined the spatial distribution of actin using CFP-tagged moesin C-terminus, a well characterized actin-binding reporter (Litman et al., 2000). During neuronal migration, high levels of moesin C-terminus-CFP were observed in the proximal region of the leading process (Figure 1B, Movie 1), which widens prior to somal translocation. This widening of the leading process during the initial phase of forward locomotion has been characterized as a cytoplasmic dilation by Schaar et al. (Schaar and McConnell, 2005). Interestingly, we noted that actin reporter fluorescence increases as the neuron takes a “step” along the glial fiber (Figure 1A, compare frame 75:28 to 81:28). To confirm and refine these observations, we utilized a reporter based on the actin-binding domain of the Utrophin protein (UTRCH-ABD). The UTRCH-ABD probe has been shown to faithfully report the presence of f-actin without altering f-actin concentrations in cells expressing the probe (Burkel et al., 2007). Examination of the localization of EGFP-UTRCH-ABD in live CGNs confirmed that f-actin is present in the leading process of migrating neurons (Figure 1C, Movie 2). These experiments establish the neuronal leading process as a major site for f-actin enrichment in neurons migrating along glial fibers.
Figure 1. Structure of the f-actin cytoskeleton of live cerebellar CGNs migrating on glial fibers.
(A) Model of the glial-guided neuronal migration cycle. Migrating CGNs extend a leading process in the direction of migration. Before somal translocation the centrosome enters the proximal leading process. Release of the glial-neuron adhesion junction occurs as the soma translocates along the glial fiber. TC Tubulin Cage, N Nucleus, A M Acto-Myosin, MT Microtubules, AD Adhesion Junction, C Centrosome and GF Glial Fiber. (B) F-actin (moesin-CFP) is concentrated in the proximal leading process in a live, migrating CGN. Bracket=proximal leading process. lp leading process. (C) The six panels depict the localization and intracellular movement of fluorescently labeled f-actin (EGFP-UTRCH) and nucleus (Histone 2B-Cherry). F-actin is heavily concentrated in the neuronal leading process.
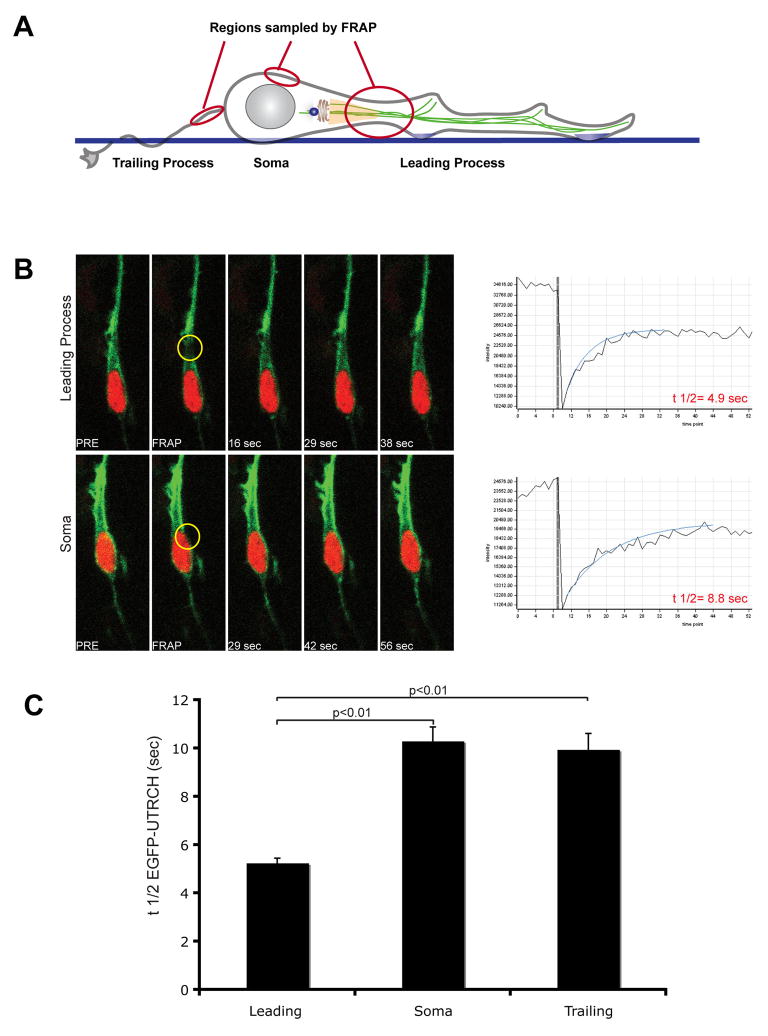
High levels of actin turnover are thought to remodel the actin cytoskeleton and facilitate cell motility and migration. To examine changes in f-actin dynamics during neuronal movement, we probed f-actin turnover in the cell soma, the proximal region of the leading process or in the trailing (axonal) process when present, by Fluorescent Recovery after Photo-bleaching (FRAP) using the EGFP-UTRCH-ABD probe (Figure 2A for a diagram). FRAP analysis demonstrates that the rate of f-actin turnover in the proximal leading process is two-fold faster than in other domains within migrating neurons (Figure 2B, C).
Figure 2. F-Actin turnover is more dynamic in the neuronal leading process.
(A) Regions probed by Fluorescence Recovery After Photobleaching (FRAP) are superimposed upon a schematic diagram of a migrating neuron. (B) FRAP profile of f-actin (EGFP-UTRCH). Regions of interest (gold circles) were photobleached and time-lapse imaging used to determine the recovery time of f-actin. Profiles compare leading process to somal recovery time. (C) Average half-life of f-actin in leading process (n=30), soma (n=15) and trailing process (n=30). The shorter recovery time of f-actin in the leading process indicates rapid actin turnover in this region of migrating neurons.
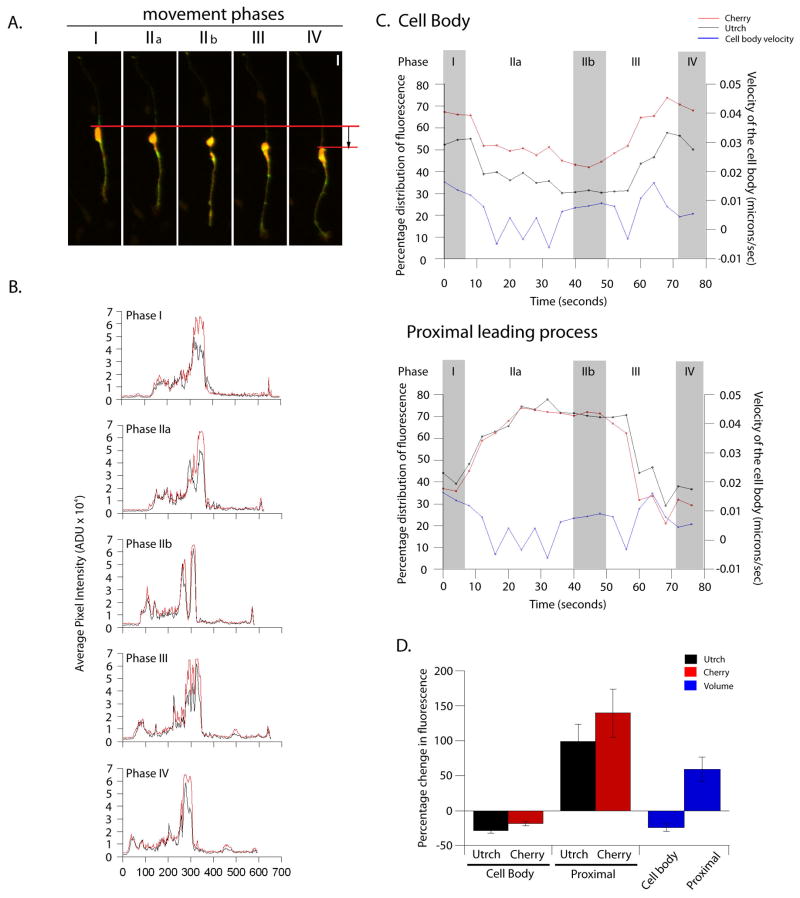
To uncover the particular changes in f-actin dynamics linked to migration, we utilized time-lapse microscopy to examine the fluorescence intensity of EGFP-UTRCH-ABD along the entire length of migrating neurons, i.e. leading process, trailing process and soma, which could be observed for at least one full motility cycle. We quantitated f-actin concentration in the different domains described above using a newly developed 4D volumetric analysis protocol that involved measuring the 3D intensity and volume of actin, normalized to cytoplasmic volume. These studies revealed a complex interplay between the f-actin in the soma and the proximal portion of the leading process, which we define as stages of the migratory cycle (Figure 3). Before movement, the cell soma possesses the highest actin concentration. Actin then accumulates in the proximal leading process with a concurrent decrease in somal actin. Finally, as the soma moves forward, sometimes entering the space previously occupied by the proximal leading process, actin concentration in the leading process and soma return to pre-movement levels. On average, there was a two-fold enrichment of f-actin in the proximal leading process before somal translocation. Additionally, line intensity scanning, normalized to a cytoplasmic marker, resolves the proximal leading process and somal f-actin as individual peaks that merge when the neuronal somal translocates during migration. Regional concentrations of f-actin within the entire trailing process and distal leading process remained stable for the entire length of the migratory cycle (data not shown). These studies show that changes in f-actin concentration during movement predominantly occur in the proximal leading process and soma of migrating neurons, and that f-actin becomes concentrated in the proximal portion of the leading process just prior to movement.
Figure 3. Line scan and 4D-volumetric analysis of f-actin content in migrating neurons.
(A) Representative movie frames demonstrating different movement cycle phases. Phase I is pre-movement. The cytoplasm begins to polarize in the proximal leading process in Phase IIa and reaches maximal polarization by Phase IIb. The cell soma then translocates forward in Phase III and comes to rest by Phase IV, at which point the cycle may start over again. Red bars indicate the distance moved by the rear of the soma during an 80 minute period. Scale bar = 10 μm. (B) Line scan of the images shown in A to illustrate the distribution of Utrch and the cytoplasm (as indicated by diffusible mCherry fluorophore). Most of the Utrch and mCherry are localized in the cell soma and the proximal region throughout the cycle, but note that the Utrch enriched in the proximal region of the leading process before mCherry (Phase IIa and Phase IIb respectively). Once two peaks of Utrch and cytoplasm localization are established in Phase IIb, the somal and leading process peaks merge in Phase III, coinciding with somal translocation. Phase IV is almost identical to Phase I. (C) Graphical representation of the change in percentage distribution of Utrch and cytoplasm during a movement cycle in the cell body (upper panel) and proximal leading process (lower panel). During Phase II (a and b), protein levels increase in the proximal region and decrease in the cell body, while remaining the same in the other parts of the cell (not shown). Before the cell translocates, as shown by the increase in velocity (blue line), protein distribution changes dramatically to increase in the cell body and decrease in the proximal leading process. (D) Summary of changes in protein/cytoplasmic concentration during Phase I and Phase IIb of the movement cycle (n = 7). On average there was a 28% (± 4%) decrease in Utrch concentration, an 18% (± 3%) decrease in cytoplasmic concentration and a 24% (± 5%) decrease in overall volume of the cell body during this period. In the proximal region of the leading process, the Utrch concentration increased by 98% (± 25%), accompanying a 139% (± 34%) increase in the cytoplasm concentration. This resulted in a 59% (± 17%) increase in the overall volume of the proximal region.
As FRAP analysis and 4D volumetric measurements implicate the proximal leading process as a major site for actin cytoskeletal remodeling preceding somal translocation, we also examined the fate of proximal leading process f-actin by photo-activation of Photo Activatible-EGFP (PA-EGFP) labeled UTRCH-ABD. Photoactivation allows one to perform “pulse-chase” time-lapse imaging analysis to examine the local dynamics of a sub-set of f-actin molecules (Burkel et al., 2007). Pulse-chase analysis of leading process f-actin shows for the first time that actin translocates forward from proximal to distal portions of the leading process and is a new indicator of the vector of polarization in migrating neurons. (Figure 5B). The average speed of forward actin translocation was 0.028±0.001 μm/sec (n=18). The concentration of f-actin in the leading process agrees with the localization of microfilaments observed in correlated electron micrographic studies of neuronal migration on glial fibers (Gregory et al., 1988) and the localization of actin-polymerizing agent Arp3 (Figure S2A). Taken together, FRAP, 4D volumetric measurements and pulse-chase imaging, show that fluctuations in f-actin concentration linked to somal movement predominantly occur in the proximal leading process and neuronal soma. Importantly, these previously unappreciated changes in actin concentration occur prior to the forward gliding movement of the soma.
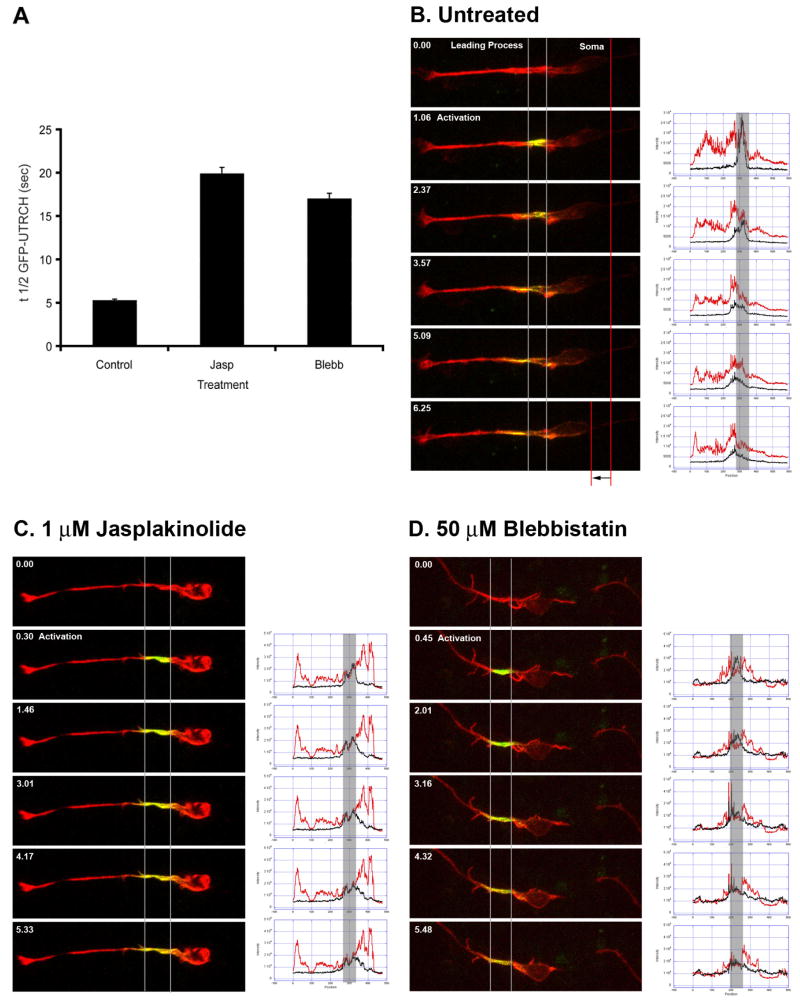
Figure 5. Myosin II motor activity is essential for leading process actin dynamics.
(A) FRAP analysis of the leading process of control, Jasplakinolide or Blebbistatin treated CGNs. EGFP-UTRCH was photobleached in regions of interest in the proximal leading process of neurons treated with 1 μM Jasplakinolide (n=15) or 50 μM Blebbistatin (n=15). Actin filament stabilization with Jasplakinolide or inhibition of Myosin II motor activity with Blebbistatin increased recovery time of EGFP-UTRCH 3-fold, indicating that Myosin II motors drive leading process f-actin dynamics. (B) Photo-activation dynamics in a control cell. CGNs were electroporated with two expression vectors: one encoding a photo-activatible EGFP f-actin reporter (PA-EGFP-UTRCH-ABD) and another encoding RFP-UTRCH as a counter stain. In the second frame of the sequence, the proximal leading process was exposed to 400 nm light to activate actin probe fluorescence. Time-lapse imaging was used to track the movement of photo-activated actin molecules in a pulse-chase experiment. Actin rapidly translocates towards distal portions of the leading process in migrating neurons. Adjacent to the time lapse images are line scan profiles of RFP-UTRCH-ABD (red) and PA-EGFP-UTRCH-ABD (black), the left of the curve is the tip of the leading process while right is the tip of the trailing process. Note that the PA-EGFP-UTRCH-ABD signal migrates to the left of its initial position. (C) Photo-activation dynamics in a Jasplakinolide treated cell. Stabilization of actin inhibits forward flow of actin. Adjacent line scan profiles show that the PA-EGFP-UTRCH-ABD signal barely exits its initial position marked. (D) Photo-activation dynamics in a Blebbistatin treated cell. Inhibiting Myosin II motors halts forward actin flow. Adjacent line scan profiles show that the PA-EGFP-UTRCH-ABD signal barely exits its initial position marked.
Myosin II motors are enriched along the length of the leading process in migrating neurons
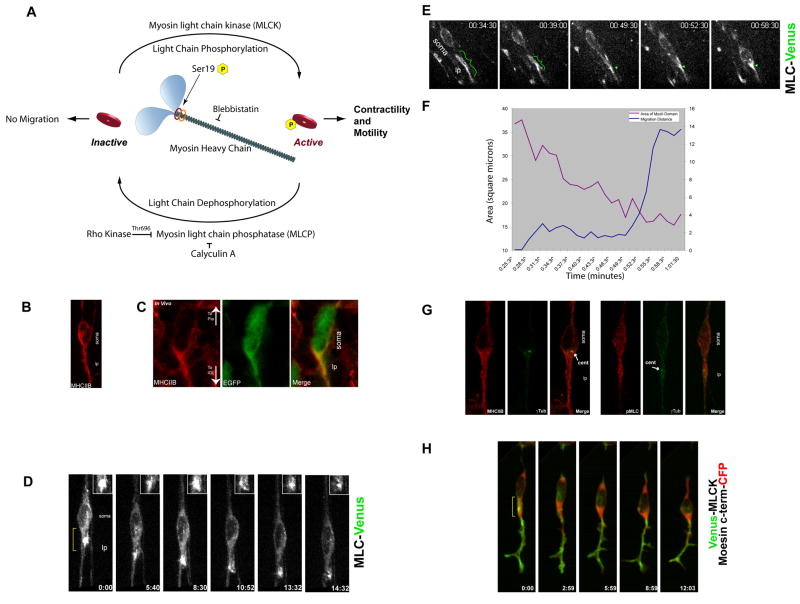
During general cell motility, the actin-based motor Myosin II contracts actin filaments to generate the force needed to power cell motility and turn over actin-based adhesions (Gupton et al., 2002; Gupton and Waterman-Storer, 2006; Vicente-Manzanares et al., 2009; Webb et al., 2004). We chose to study the role of Myosin IIB in neuronal polarization and motility, since Myosin IIB is the predominant Myosin II motor expressed in the nervous system (Kawamoto and Adelstein, 1991; Rochlin et al., 1995), and Myosin IIB activity is required for normal CGN migration along Bergmann glial fibers in the developing CNS (Ma et al., 2004). By immunocytochemistry, Myosin IIB localizes to the leading process of migrating CGNs in vitro and in vivo (Figure 4B and C, see Figure S3 for a comparison of Myosin IIB and Myosin Va localization). We also used fluorescent myosin light chain (MLC) as a reporter for the localization of Myosin II motors (Barros et al., 2003; Mittal et al., 1987). MLC-Venus accumulates in the leading process in migrating neurons in vitro and in situ. The zone of intense MLC-Venus fluorescence expands and grows smaller in a cyclical manner during migration (Figure 4D, E and F, Movies 3 and 4). As the soma moves forward, the area of MLC reporter fluorescence decreases, consistent with a contraction within the MLC-labeled domain (Figure 4G for area measurement of Figure 4F). These data indicate that the spatial distribution of Myosin IIB protein is similar to that of f-actin described above. Myosin II motors are polarized and enriched in the leading process during glial-guided neuronal migration. Moreover, the oscillations in MLC reporter fluorescence suggest cyclical contraction in the leading process as the neuron migrates.
Figure 4. The proximal leading process contains an acto-myosin contractile domain.
(A) Myosin II activity cycle. (B) Immunostaining of Myosin Heavy Chain IIB (MHCIIB) in CGNs. (C) Myosin IIB also localizes to the leading process of neurons in organotypic cerebellar slices. Red: Myosin IIB Heavy Chain, Green: EGFP to highlight cell boundaries. (D) MLC-Venus (MLC9-Venus) reveals a Myosin II contractile structure in the proximal leading process (Bracket in Frame 1) of a CGN migrating in culture. The inset highlights the contraction of the MLC-labeled domain. (E) MLC-Venus reveals a Myosin II based contractile structure in the proximal leading process (bracket) of a CGN migrating in a cerebellar slice. (F) The MLC labeled domain the leading process decreases in area before somal movement, strongly suggesting leading process contraction. (G) Immunostaining of Myosin heavy chain IIB (MHCIIB), phosphorylated MLC (pMLC) and γ-tubulin (γTub) reveals that active Myosin II motors surround the centrosome. (H) Venus-MLCK co-localizes with f-actin forward of the soma just before somal translocation, indicating Myosin II motors are active in the leading process. Bracket highlights region of high overlap. lp leading process.
To determine whether active Myosin II motors are present in the leading process of neurons migrating along glial fibers, we examined the localization of phosphorylated MLC and the spatiotemporal dynamics of MLC kinase (MLCK). The phosphorylation of MLC on Ser19, which controls Myosin II activation and the assembly of contractile filaments (Moussavi et al., 1993), is positively regulated by MLCK or Rho-kinase (Rock) and negatively regulated by MLC phosphatase (MLCP) (Lo et al., 2004; Vicente-Manzanares et al., 2007). Staining with anti-phospho Ser19 specific antibody, used to detect phosphorylated MLC (pMLC), revealed labeling in the leading process (Figure 4G). In addition, the presence of Venus-MLCK in the leading process at all stages of the migration cycle suggests that activation of Myosin II occurs in this region during migration (Figure 4H, Movie 5). In neurons co-expressing Venus-MLCK and the moesin C-terminus-CFP actin reporter, Venus-MLCK transiently co-localized with actin in the proximal leading process just prior to somal translocation (Figure 4H, Movie 6). Taken together, the profile of active Myosin II suggests that the leading process may be a significant site of acto-myosin contractility within migrating neurons.
Rapid f-actin turnover and Myosin II motor activity are critical for leading process actin flow
The leading process is the major site for actin turnover and f-actin flow within migrating neurons (Figure 1, 2, 3 and 5B). In order to address whether actin turnover is required for actin flow in this region of the neuron, we performed FRAP and pulse-chase analysis on neurons treated with 1 μM Jasplakinolide. Jasplakinolide is a drug that hyper-stabilizes actin filaments and thus prevents dynamic turnover (Cramer, 1999). In the FRAP experiments using EGFP-UTRCH-ABD the proximal leading process displayed the highest actin turnover in control migrating neurons (Figure 5A). Jasplakinolide treatment led to a near three-fold increase in the recovery time of EGFP-UTRCH-ABD (n=15). In pulse-chase experiments using PA-EGFP-UTRCH-ABD, the PA-EGFP-UTRCH-ABD signal moves from the proximal leading process toward the distal portion of the leading process in control cells (Figure 5B). In contrast, there is little f-actin translocation away from the photo-activated region in neurons treated with 1 μM Jasplakinolide (Figure 5C). The average speed of forward actin translocation in Jasplakinolide treated cells was 0.012±0.002 μm/sec (n=7). Taken together, these findings confirm the presence of high actin dynamics in the leading process and show that actin turnover is required for actin flow away from the proximal region of the leading process in the direction of migration. These data further support the idea that the proximal leading process is a region of rapid actin remodeling.
Myosin IIB is present and active in the leading process of migrating neurons (Figure 4). We next examined how Myosin II motor function affects actin turnover and flow in the proximal leading process. Myosin II motor function was inhibited using the small molecule inhibitor Blebbistatin (Straight et al., 2003). The proximal leading process was the focus of these studies since this region displayed the highest actin turnover in control migrating neurons. In a set of FRAP experiments, EGFP-UTRCH-ABD was photobleached in regions of interest in the proximal leading process of neurons treated with Blebbistatin at a concentration that halts migration and centrosomal motility (50 μM). Blebbistatin treatment led to a near three-fold increase in the recovery time of EGFP-UTRCH-ABD (Figure 5A, n=15). In pulse-chase experiments using PA-EGFP-UTRCH-ABD, photo-activated EGFP-UTRCH-ABD signal translocates from the proximal region toward the distal portion of the leading process in control cells (Figure 5B). In contrast, pulse-chase time-lapse imaging of neurons treated with 50 μM Blebbistatin revealed little f-actin translocation away from the photo-activated region (Figure 5D). The average speed of forward actin translocation in Blebbistatin treated cells was 0.011±0.001 μm/sec (n=4). Taken together, these results show that Myosin II motor activity is required for high actin turnover and anterograde actin flow in the proximal portion of the leading process during migration.
Centrosome motility and actin cytoskeletal dynamics are correlated during the initial phase of the migration cycle
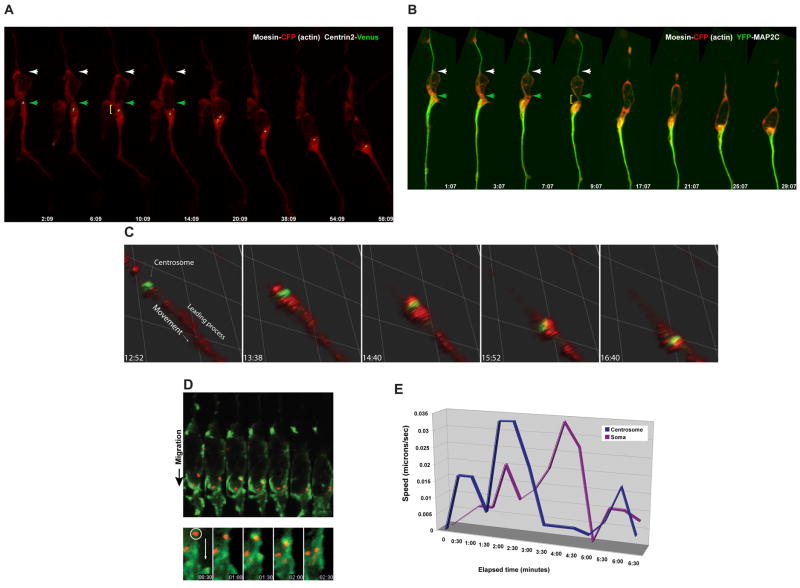
The centrosome polarizes and localizes to the proximal portion of the leading process just forward of the nucleus during glial-guided neuronal migration (Solecki et al., 2004). Given the concentration of f-actin, high actin turnover, and anterograde actin flow in this region of migrating neurons, we examined the relationship between centrosome motility and actin dynamics in the neuronal leading process. At all stages of the migration cycle, the centrosome, visualized using Centrin2-Venus, was embedded within the actin, visualized by moesin C-terminus-CFP, in the proximal leading process (Figure 6A, Movie 7). Forward movement of the centrosome occurs when the leading process widens (Figure 6A, compare 2:09 to 6:09). Interestingly, a constriction appears in the proximal leading process (brackets in 6:09 and 10:10) prior to somal translocation. Closer examination of f-actin and centrosome localization using Centrin2-Venus and the f-actin reporter RFP-UTRCH-ABD showed that f-actin accumulates in the vicinity of the centrosome when this organelle translocates forward during migration (Figure 6C). Indeed, line-scanning quantitation shows a transient peak of f-actin signal near the centrosomal peak when the centrosome moves forward (Figure S4). These results confirm that the centrosome is located in the vicinity of f-actin in the proximal leading process, and that forward movement of the centrosome during the motility cycle appears to be correlated with an accumulation of f-actin and contraction of the actin cytoskeleton.
Figure 6. Actin and Myosin Light Chain Kinase accumulate near the centrosome during the centrosomal movement phase of the migration cycle.
(A) The centrosome (labeled by Centrin2-Venus, Green) is embedded within the f-actin contractile domain (Moesin-CFP, Red) in the leading process of live neurons. Note the centrosome enters the leading process (time points 6:09 and 10:09) when the proximal leading process increases in volume. (B) Microtubules (labeled by Map2C-YFP, Green) are embedded within leading process f-actin (Moesin-CFP, Red) of a live neurons. Note microtubules translocate into the leading process (time points 7:07 and 9:07) when the proximal leading process increases in volume. (C) The centrosome (labeled by Centrin2-Venus, Green) is embedded within the f-actin (labeled by RFP-UTRCH-ABD, Red) contractile domain in the leading process of live neurons. Note the centrosome enters the leading process when it dilates. (D) Kymograph of a migrating neuron expressing Venus-MLCK and Centrin2-mCherry. Bottom: Close-up of image sequences focusing on region around the centrosome. MLCK (green) accumulates near the centrosome (red) at 1:30, before movement of the centrosome. (E) Velocity plotted against time of centrosome and somal movement for the time-lapse sequence in (D). A spike in speed occurs at 2:00, thirty seconds after MLCK first accumulates at the centrosome. A deceleration occurs at 2:30 seconds when MLCK no longer surrounds the centrosome.
Interactions between the actin and microtubule cytoskeletons control mechanical processes in cells, ranging from dynamic changes in cell shape to retrograde microtubule flow during cell and growth cone motility (Cai et al., 2006; Gomes et al., 2005; Gupton et al., 2002; Gupton and Waterman-Storer, 2006; Rosenblatt et al., 2004; Schaefer et al., 2002). Since centrosomes enter the leading process prior to somal translocation, and they are microtubule organizing centers, we examined the localization and dynamics of the microtubule and actin networks by co-expressing YFP-tagged Map2C, a microtubule and f-actin cross linker (Roger et al., 2004), and the actin reporter moesin C-terminus-CFP. Map2C labeled microtubules are most abundant in the leading process (Figure 6B, Movie 10). As was seen for centrosomes, bulk forward movement of Map2C labeled microtubules occurs when the proximal leading process widens (Figure 6B, time point 9:07). These results demonstrate for the first time that microtubules translocate in a directed manner during the first stages of the migration cycle and are consistent with our observations that the centrosome enters the leading process prior to somal translocation. Thus the microtubule network undergoes forward movement during the period of neuronal migration that occurs before somal translocation.
Dynamic f-actin turnover and Myosin II motor activity are required for centrosome motility and coordinated translocation of the centrosome and soma in migrating neurons
The spatiotemporal correlation between f-actin accumulation and centrosome motility during an apparent contraction of the proximal leading process, led us to examine the relationship between centrosomes and Myosin II. Migrating neurons were immunostained with antibodies against Myosin IIB, pMLC as an indicator of Myosin II activity, and the centrosomal marker γ-tubulin. The centrosome is present in the region that contains both Myosin IIB and pMLC (Figure 4G). We also examined the localization of Venus-MLCK, an activator of Myosin II, in relation to that of Centrin2-Venus. In some cases, Venus-MLCK accumulates near the centrosome as this organelle translocates toward the leading process (Figure 6D and E, Movies 8 and 9). These experiments suggest that the centrosome is located in close proximity acto-myosin when it moves during migration.
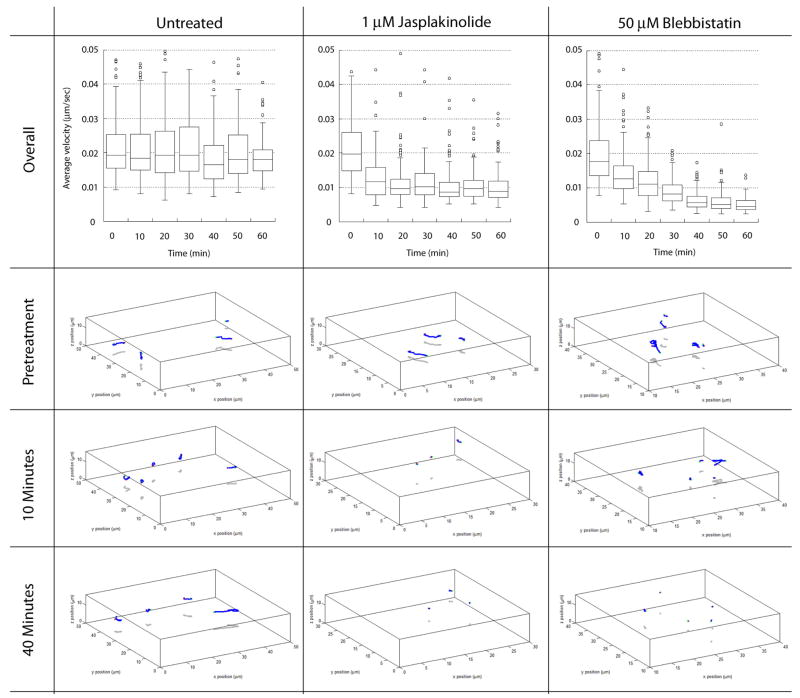
In stationary neurons, centrosomes undergo random positioning events that can be used to assay the forces necessary for basal centrosome motility. To determine whether actin dynamics and Myosin II activity are required for centrosome motility, we followed centrosome movement in stationary neurons before and after the addition of Jasplakinolide, which hyper-stabilizes actin filaments, or Blebbistatin, which inhibits Myosin II motor activity. A custom algorithm written by the Image Science & Machine Vision Group in the Measurement Science & Systems Engineering Division at Oak Ridge National Laboratory (ORNL) was used to automatically plot and track the 3D position of all centrosomes imaged over time, essentially producing the first large scale 4D analysis of centrosome motility. Consistent with our model, both Jasplakinolide and Blebbistatin potently reduced centrosomal motility (Figure 7, see Movie 11 for one Blebbistatin Example). The average velocity of control or pretreatment centrosomes was 0.022 μm/second. Jasplakinolide quickly inhibited centrosomal motility, reducing velocity to 0.011 μm/second within 20 minutes. Blebbistatin was even more potent, reducing centrosome velocity to 0.006 μm/second by 50 minutes. Taken together, these results show that centrosome motility is dependent upon actin dynamics and Myosin II motor activity.
Figure 7. Dynamic actin and Myosin II motor activity is required for centrosomal motility.
Centrosomes were imaged over the course of one hour in control, Jasplakinolide-treated or Blebbistatin-treated neurons. Total displacement was measured before or at various time points after the addition of cytoskeletal drugs using a specialized centrosome-tracking algorithm developed by the ORNL Image Science & Machine Vision Group. Average centrosome displacement is unaffected in control neurons. 4-dimensional volume tracks of representative centrosome in control experiments at time point 0, 10 and 40. The initial centrosome position is marked by a green circle, the final position marked by a red circle and 4D path marked by blue line. Average centrosome displacement rapidly declines after the addition of 1 μM Jasplakinolide and shorter path lengths in the 4D tracking reflect the reduction in velocity. Average centrosome displacement rapidly declines after the addition of 50 μM Blebbistatin and shorter path lengths in the 4D tracking reflect the reduction in velocity.
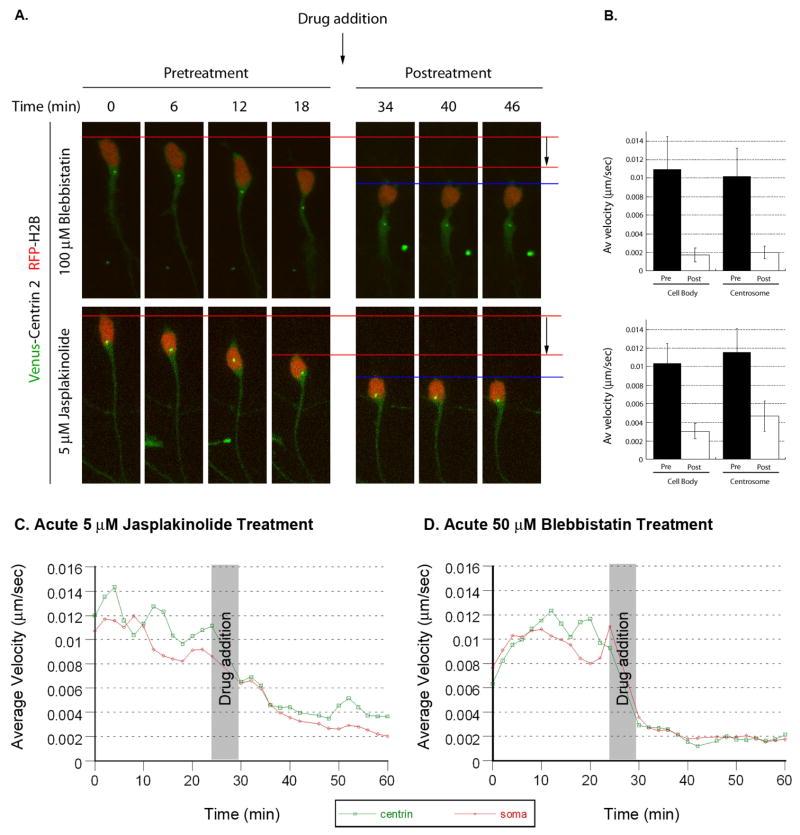
Having established that acto-myosin contractility is needed for basal centrosomal motility, we examined whether actin dynamics and Myosin II activity are required for coordinated forward movement of the centrosome and soma. Centrosomal movement was visualized using Centrin2-Venus and somal translocation was visualized using Histone-2B mCherry to highlight the nucleus. CGNs migrating along glial fibers were imaged before and after the addition of 5 μM Jasplakinolide or 100 μM Blebbistatin. Addition of either Jasplakinolide or Blebbistatin rapidly halts both centrosomal motility and somal translocation (Figure 8, See Movie 12 for one example). The average pre-treatment velocities of both centrosome and soma were 0.01 μm/second. Jasplakinolide reduces average centrosome velocity to 0.0045 μm/second and average somal velocity to 0.003 μm/second. Blebbistatin reduces average centrosome velocity to 0.002 μm/second and average somal velocity to 0.0019 μm/second. Kinetic analysis of these data reveal that centrosomes and neuronal cell bodies halt movement nearly simultaneously when Jasplakinolide or Blebbistatin are added to the bath. It should be noted that centrosomes that had entered the proximal portion of the leading process also stopped moving, showing that motile centrosomes were not just “passengers” in neuronal cell bodies (Figure 9A for one example). Conversely, treatment with Calyculin A, a bulk activator of acto-myosin contractility, stimulated centrosomal and somal translocation of stationary neurons (Figure S5, Movie 13). Taken together, these results show that actin dynamics and Myosin II activity are required for centrosome and somal translocation during glial-guided neuronal migration.
Figure 8. Dynamic actin and Myosin II motor activity is required for coordinated movement of centrosome and cell body.
CGNs were transfected with expression vectors encoding Centrin2-Venus (centrosomal label, green) and H2B-mCherry (nuclear label, red). Time-lapse imaging was used to track centrosomal or somal velocity in migrating neurons. After cells migrated, 100 μM Blebbistatin or 5 μM Jasplakinolide was added to the culture and imaged for a further 28 minutes (time lapse images are displayed in (A)). Addition of either drug potently inhibits forward movement of both centrosome and cell body. (B) Graphs of velocity of the cell body and centrosome before and after drug treatment (Blebbistatin n=20, Jasplakinolide n=22). Velocity kinetics of the centrosome (red) or soma (green) in Jasplakinolide treated (C) or Blebbistatin (D) treated cells.
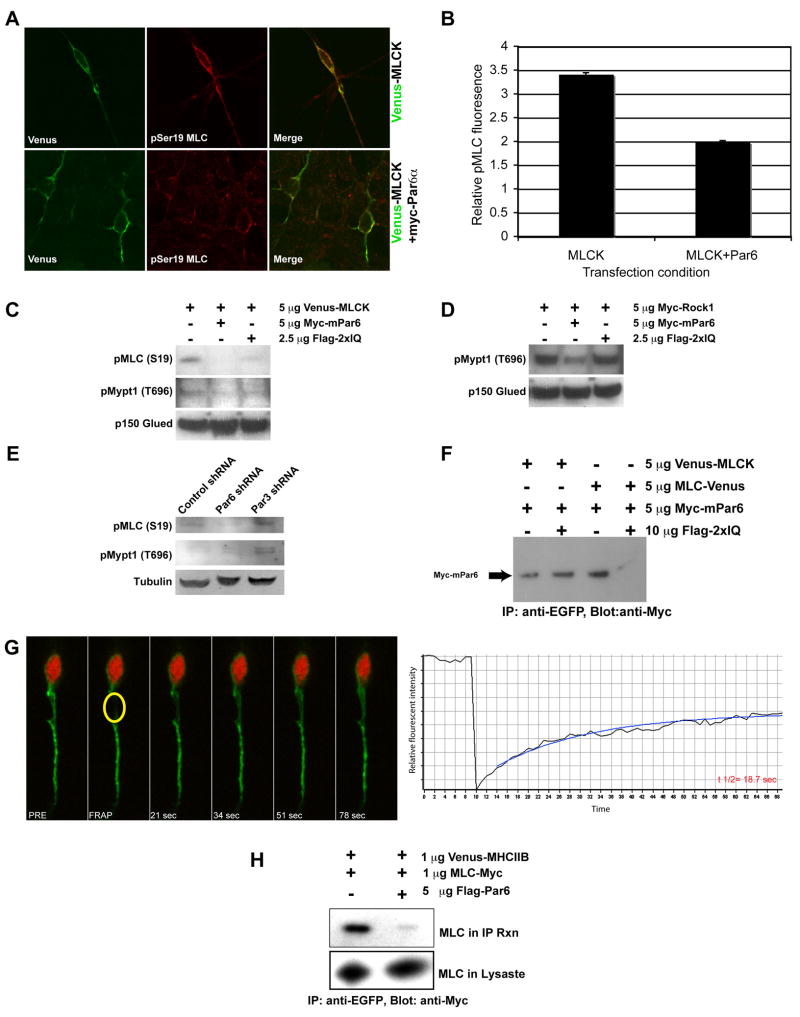
Figure 9. Par6α regulates Myosin II motor activity and leading process f-actin dynamics.
(A) Ectopic expression of Par6α inhibits MLC phosphorylation in CGNs. Co-expression of Myc-Par6α with Venus-MLCK reduced MLC phosphorylation detected by immunostaining with an anti-phospho Ser19 antibody. (B) Quantitation of average MLC Ser19 fluorescence intensity in CGNs reveals a decrease in MLC phosphorylation upon expression of Myc-Par6α (n=25 for Venus-MLCK or Venus-MLCK and Myc-Par6α expressing cells). (C) Ectopic expression of Par6α inhibits MLC and Mypt1 phosphorylation. HEK293T cells were transfected with a Venus-MLCK construct to boost MLC phosphorylation. Ectopic expression of Myc-Par6α reduced MLC and Mypt1 phosphorylation detected by immunoblotting with anti-MLC phospho-Ser19 or anti-Mypt1 Thr696 antibodies. Ectopic expression of the IQ domain only blocks MLC phosphorylation. (D) Ectopic expression of Par6α inhibits Rock1-mediated phosphorylation of Mypt1. HEK293T cells were transfected with a Myc-Rock1 construct. Ectopic expression of Myc-Par6α reduced Mypt1 phosphorylation detected by immunoblotting with anti-Mypt1 Thr696 antibody. Ectopic expression of the IQ domain does not block Mypt1 Thr696 phosphorylation. (E) Silencing of Par6α with a Par6α shRNA reduces MLC Ser19 phosphorylation while mildly increasing affecting Mypt1 Thr696 phosphorylation. (F) Ectopic expression of the IQ domain blocks Par6α binding to MLC but does not effect a Par6/MLCK interaction. (G) Par6α IQ domain over-expression slows leading process f-actin dynamics. CGNs were nucleofected with the EGFP-UTRCH-ABD (green) and the Par6α IQ domain expression vectors. Time-lapse image frames and the corresponding FRAP recovery curve highlight the slow recovery time of f-actin in a CGN over-expressing the IQ domain. The average t1/2 for 14 separate replicates was 21±1.3 seconds. (H) Ectopic expression of Par6α inhibits MLC interaction with Myosin Heavy Chain. Venus-MHCIIB and MLC-Myc were transfected into HEK293T cells in the presence or absence of ectopic Par6α expression. Venus-MHCIIB was immunoprecipitated with anti-EGFP antibody and the amount of bound MLC detected by anti-Myc staining.
Par6α regulates Myosin II motor activity
Previous work from our laboratory identified Par6α as a key regulator of centrosome motility and somal translocation during glial-guided neuronal migration (Solecki et al., 2004). Given that the PAR polarity complex also controls actin cytoskeletal dynamics in a number of systems (Mertens et al., 2006), we wondered whether Par6α regulates actin dynamics required for neuronal migration. Importantly, Par6α is present in the leading process of migrating CGNs (Solecki et al., 2004), and inhibition of Myosin II motor activity phenocopies perturbation of the PAR complex with regard to centrosome and somal motility. We therefore examined whether Par6α regulates Myosin II motor activity. Ectopic expression of Par6α decreases pMLC levels in HEK293 cells and CGNs (Figure 9A, B, C). These results show that Par6α regulates MLC phosphorylation, which is required for Myosin II activity and acto-myosin contractility.
A global reduction in MLC phosphorylation could be due to either a decrease in MLC-directed kinase activity or an increase in Myosin Light Chain Phosphatase (MLCP) activity. MLCK and Rock1 are both kinases capable of directly phosphorylating MLC (Kamm and Stull, 2001; Kimura et al., 1996), while MLCP dephosphorylates MLC on Ser19 (Matsumura, 2005). Rock1 also phosphorylates Thr696 of the Mypt1 myosin binding subunit of MLCP, inhibiting MLCP dephosphorylation of MLC Ser19 (Kimura et al., 1996). We found that ectopic expression of Par6α inhibits both basal and Rock1 mediated phosphorylation of MLCP Thr696 (Figure 9C, D), indicating that Par6α decreases myosin activity, at least in part, through increased MLCP activity. These results suggest that Par6α signaling regulates Myosin II motor activity in the proximal leading process through a pathway that involves the RhoA GTPase effector Rock1.
As expected, silencing of Par6α, and its binding partner Par3, led to a slight increase in MLCP Thr696 phosphorylation. However, silencing of Par6α reduces basal MLC phosphorylation (Figure 9E), suggesting that Par6α may regulate MLC phosphorylation by additional mechanisms. The Par6α amino acid sequence contains a previously unidentified IQ motif between the PB1 and CRIB domains of Par6α (Figure S6). Calmodulin or EF hand domain proteins, such as MLC, bind to IQ motifs and are important regulators of acto-myosin contractility (Bahler and Rhoads, 2002). Immunoprecipitation studies reveal that full-length Par6α binds to MLC and MLCK by coimmunoprecipitation, and that overexpression of the Par6α IQ motif interferes with the Par6α and MLC interaction (Figure 9F). Interestingly, ectopic expression of the IQ domain decreases MLC Ser19 phosphorylation without affecting MLCP Thr696, an activity similar to Par6α silencing (Figure 9C and D). Moreover, ectopic expression of the Par6α IQ domain in CGNs decreases leading process actin dynamics to a similar magnitude as Blebbistatin, as measured by FRAP of EGFP-UTRCH-ABD (Figure 9G; average t1/2 in IQ expressing CGNs is 21.2±1.3 seconds n=14). These experiments show that Par6α binds to myosin-regulatory molecules, and that Par6α binding to MLC appears to be important for leading process actin dynamics in CGNs. Over-expression of Par6α may titrate away or delocalize myosin regulatory molecules, thus interfering with Myosin II activation and acto-myosin contractility. Consistent with this hypothesis, ectopic expression of full-length Par6α inhibits MLC binding to Myosin Heavy Chains (Figure 9H). These studies suggest a model where Par6α binds MLC through the IQ domain and recruits myosin regulatory molecules to sites of acto-myosin contractility (Figure S6).
Discussion
Our prior studies showed that the Par6α polarity complex localizes to the centrosome of migrating CNS neurons and orchestrates the saltatory movement of neurons along glial fibers. Using time-lapse imaging of live CGNs migrating along glial fibers, we examined the precise spatiotemporal coordination of actin dynamics during this specialized mode of migration. The proximal portion of the leading process is a region of high actin turnover, and during forward movement there is a cycle of f-actin accumulation in this region. F-actin also flows from the proximal to the distal regions of the leading process in the direction of migration. Myosin II motors are present along the length of the leading process and oscillations in Myosin II fluorescence signal during the motility cycle suggest the occurrence of acto-myosin contractile events in this region. Pharmacological stabilization of f-actin or inhibition of Myosin II motors blocks f-actin dynamics and disrupts forward migration, suggesting that rapid f-actin turnover, which is powered by Myosin II motor activity, functions in the forward movement of the centrosome and provides the force to translocate the cell soma during glial guided migration. Importantly, the Par6α polarity protein, which regulates centrosome and somal motility during migration, also regulates Myosin II motor activity. Together, these results provide insights into cytoskeletal regulation and organization that affect the saltatory movement cycle of migrating neurons. We propose that acto-myosin contractility in the leading process acts to pull cytoskeletal components and the soma forward during migration.
Actin Dynamics Near the Cell Body of Migrating Cells
One of the principal differences between the generalized mode for motility of fibroblasts and epithelial cells and the locomotion of neurons along thin glial fibers is the absence of a “leading edge” domain in which integrin-based adhesion provides the tractile force for forward movement (Ridley et al., 2003). While “leading edge” actin dynamics associated with protrusive events have been studied extensively (Ponti et al., 2004; Schaefer et al., 2002; Schaefer et al., 2008), relatively little is known about the dynamic organization of the actin cytoskeleton in the leading process and soma in migrating neurons. This is hardly surprising given the leading process is only 1–2 μm across, about half the width of the thin veil of protrusive actin in fibroblast lamellipodia (Ponti et al., 2004). Our studies show the proximal leading process is a significant site for actin cytoskeletal remodeling during neuronal migration along glial fibers. 4D volumetric analysis and pulse-chase photoactivation experiments show an apparent transfer of actin between the soma and proximal leading process before somal translocation, as well as an anterograde flow of f-actin toward distal regions of the leading process.
Recently, fluorescent speckle microscopy (FSM) has been used to define f-actin flow in migrating epithelial cells (Ponti et al., 2004; Salmon et al., 2002). These studies divide cellular actin into different zones based upon the vector of f-actin flow. Similar to what we observe in migrating neurons, f-actin flows in the direction of migration in the region of epithelial cells just forward of the nucleus. Interestingly, these studies also noted anterograde co-movement of f-actin and microtubules suggesting that forward actin flow may regulate the positioning of the microtubule cytoskeleton (Gupton et al., 2002; Salmon et al., 2002). Our experiments confirm anterograde actin flow near the cell body in migrating cells and suggest that actin dynamics in this region coordinates centrosomal and somal motility.
Location of Myosin II motors in neurons migrating along glial fibers
Myosin II motors are required for the migration of neurons both in vitro and in vivo (Ma et al., 2004; Schaar and McConnell, 2005). The current time-lapse imaging studies, using multiple vital probes for both f-actin and Myosin II motor components, suggest that an acto-myosin contractile region is present in the leading process of neurons migrating along glial fibers. Immunolabeling with antibodies recognizing Myosin IIB heavy chain and Ser19 phosphorylated MLC reveal the presence of Myosin II motors in the leading process of cultured granule neurons or cells migrating across the molecular layer in sections of early postnatal cerebellum. These staining profiles are nearly identical to the localizations of our f-actin, MLC and MLCK live imaging probes. Interestingly, time-lapse imaging revealed oscillations in the area of leading process MLC-Venus fluorescent signal both in culture and cerebellar slices, suggesting a cycle of contraction/decontraction occurs within this region of the migrating neuron. Pharmacological studies using Blebbistatin, a cell permeable inhibitor specific for Myosin II, show that Myosin II motor activity is essential for proximal leading process actin dynamics, suggesting that not only are Myosin II motors present near a region of actin cytoskeletal dynamics but also that motor activity drives actin dynamics in the proximal leading process.
While Myosin II motors have long been implicated in contractile events in the posterior region of crawling cells, like fibroblasts and epithelial cells, there is growing evidence for Myosin II motor function anterior to the nucleus, similar to that we observed for neurons migrating on radial glia. For example, Meshel et al. report that Myosin II is localized to the rear of fibroblasts migrating on 2 dimensional substrates, while it is localized to the front of the same cells migrating on collagen fibers, a geometry highly similar to neurons migrating along glial fibers (Meshel et al., 2005). Recently, Doyle et al. extended these results by showing that fibroblasts migrating along a thin “1 dimensional” micro-patterned surface migrate with a mode, cadence, lack of a broad “leading edge” lamellipodia and acto-myosin organization similar to that of neurons migrating along glial fibers (Doyle et al., 2009). In conclusion, our model strongly suggests that the majority of acto-myosin is located forward of the nucleus and may pull the centrosome and soma forward during migration. We should note that bath application of Blebbistatin inhibits Myosin II motor activity in all regions of the cell, thus, we cannot completely discount that a minor fraction Myosin II motor activity residing within the trailing process contributes toward locomotion along the glial fiber.
The acto-myosin cytoskeleton and centrosome motility
As the centrosome is an organizer of the microtubule cytoskeleton, microtubule based mechanisms are usually invoked to control its position. While cytoplasmic dynein and microtubule plus end anchoring at the cell cortex are evolutionarily conserved mechanisms regulating the position of microtubule arrays (Dujardin et al., 2003; Dujardin and Vallee, 2002), there accumulating evidence that the actin cytoskeleton is an active participant in centrosome motility. Our live imaging analysis of migrating CGNs details that centrosomes move forward with timing similar to the actin rearrangements occuring in the proximal leading process. Functional studies using Blebbistatin, Jasplakinolide and Calyculin A further suggest that the acto-myosin cytoskeleton contributes towards centrosome positioning. This has not been reported previously for migrating interphase cells. Our conclusion is supported by studies on the movement of daughter centrioles towards the mid-body during mitosis (Piel et al., 2001) and the observation that cortical Myosin II motors are essential for mitotic spindle rotation (Rosenblatt et al., 2004).
The importance of Myosin II in centrosome positioning suggested in this study does not diminish of the critical role of cytoplasmic dynein in positioning microtubule arrays or centrosomes in migrating neurons (Dujardin et al., 2003; Dujardin and Vallee, 2002; Tsai et al., 2007). In fact, Myosin II and dynein may cooperate to position the centrosome during migration. This view is supported by studies that document the interaction of cytoplasmic dynein with the actin-rich cell cortex. The actin related protein 1 (Arp1), an essential component of dynactin (a cargo adaptor for dynein), can directly bind to spectrins and other known cortical actin interacting factors (Garces et al., 1999; Holleran et al., 2001; Holleran et al., 1996). During slow anterograde axonal transport, cytoplasmic dynein requires an intact actin cytoskeleton to transport microtubule fragments towards the periphery of neuronal cells, suggesting that in some cases dynein and acto-myosin may cooperate to organize the neuronal cytoskeleton (Myers et al., 2006). As dynein accumulation has been reported in the leading process of migrating neurons (Tsai et al., 2007), it may well provide a link between an anterograde f-actin flow and the apparent forward flux of microtubules that we observe just prior to forward movement of the somal and nucleus in migrating neurons. It will be important to determine if the proximal portion of the leading process is a site of high turnover or movement of dynein or its cargo adaptor dynactin and whether rapid actin remodeling or Myosin II motor activity contributes to dynein/dynactin dynamics in the proximal leading process.
Polarity signaling and acto-myosin contractility
Our previous studies identified the conserved PAR polarity-signaling complex as a key regulator of centrosome motility and somal translocation in migrating neurons. Our new findings suggest that in addition to regulating centrosome positioning and function (Solecki et al., 2004), the Par6 polarity complex also regulates acto-myosin contractility in migrating neurons. How does the PAR complex coordinately regulate the actin and microtubule cytoskeletons to control the polarization of migrating neurons? As key regulators of the actin and microtubule cytoskeletons, the Rho GTPases provide an ideal link between the PAR complex and the cytoskeleton. While the PAR complex acting in cooperation with CDC42 and Rac1 is likely to regulate actin polymerization and protrusion in the leading process (Nishimura et al., 2005; Tsai et al., 2005; Zhang and Macara, 2006), interplay between the PAR complex and RhoA/Rock signaling must be crucial to fine tune acto-myosin contractility needed to power centrosome and somal positioning, the most labor intensive portions of the motility cycle. We show that ectopic expression of Par6α inhibits Rock1 phosphorylation of Thr696 of Mypt1, which leads to enhanced MLCP dephosphorylation of MLC and ultimately a reduction in the acto-myosin contractility that drives neuronal migration. Our finding that Par6α negatively regulates the RhoA signaling pathway is consistent with a recent study by Zhang and Macara that identified an interaction between Par6/aPKC and p190RhoGAP that inhibits RhoA signaling to promote dendritic spine biogenesis (Zhang and Macara, 2008).
We also provide mechanistic insight into the cellular and molecular targets of the PAR complex in the control of neuronal polarity and morphogenesis. We show that Par6α interacts with the Myosin II regulators MLC and MLCK. Disruption of Par6-MLC binding via over-expression of the IQ-like domain of Par6α inhibits MLC phosphorylation and increases the turnover time of leading process f-actin in granule neurons. Taken together our findings suggest that Par6α regulates Myosin II activity by modulating MLC phosphorylation through two distinct pathways: Rock1 inhibition of MLCP and through a direct interaction with MLC itself. Perhaps Par6α binding to MLC and MLCK actively regulates their recruitment to sites of acto-myosin contractility.
Elucidating the signaling pathways that regulate cytoskeletal dynamics and the adhesion of neurons to glia is crucial to understanding the cellular mechanisms of directed neuronal migration during corticogenesis. In the future, it will be of great interest to determine if RhoA regulates centrosome positioning during neuronal migration, as active RhoA has been reported in the leading process of migrating CGNs (Guan et al., 2007), and whether Par6α regulates Rock1 directly in migrating neurons or interferes with the RhoA pathway through an upstream interaction with p190RhoGAP (Zhang and Macara, 2008). Moreover, it is critical to identify the extrinsic signals that regulate local assembly of acto-myosin within the leading process as these factors may act to guide granule neurons to their final destination within the developing cerebellar cortex.
Methods
Preparation of CGNs, nucleofection and imaging
CGNs were prepared as described (Hatten, 1985). Briefly, cerebella were dissected from the brains of P6 mice. After the pial layer was peeled away, the tissue was treated with trypsin and triturated into a single cell suspension, using fine bore Pasteur pipettes. The suspension was layered onto a discontinuous Percoll gradient and separated by centrifugation. The small cell fraction was then isolated. The resulting cultures routinely contain 95% CGNs and 5% glia. For imaging experiments, expression vectors encoding fluorescently labeled cytoskeletal proteins were introduced into granule neurons via Amaxa nucleofection, using the Amaxa Mouse Neuron Nucleofector kit per manufacturer’s instructions and program A30. A range of 1–5 μg of pCIG2 expression vector was used to express the fusion protein of interest. After cells recovered for 10 minutes from the nucleofection, they were plated in movie dishes (Mattek) coated with low concentrations of poly-D-lysine or poly-L-ornithine to facilitate the attachment of neurons to glial processes (according to methods established by (Edmondson and Hatten, 1987)). For Myosin II activation or inhibition experiments, cells were imaged for 10 minutes prior to the addition of drug, and then the indicated amount of Calyculin A or Blebbistatin was added to the bath, and the cultures were imaged for an additional 20 minutes.
CGN cultures were imaged with a Carl Zeiss Axiovert 200M equipped with a 63×, 1.4 NA, PlanApochromat objective. A PerkinElmer Wallac UltraView confocal head with 514-nm excitation filter and Orca ER cooled CCD camera (Hamamatsu) were used for high resolution imaging at near real-times speeds. Z-stacks were collected (2–3 μm z-stacks, 4–5 sections per stack) every 15 seconds during imaging. Images were processed and analyzed using MetaMorph (Universal Imaging Corp.) or Slidebook (Intelligent Imaging innovations). Cells were usually imaged for 45–60 minutes. Cells that failed to migrate during image acquisition were termed stationary neurons, while cells that moved were termed migrating neurons. Some cultures were imaged using a 6-line Ultraview confocal microscope or a Marianas Workstation (Intelligent Imaging Innovations), with image collection parameters identical to the Metamorph driven system.
For ectopic-expression experiments, purified CGNs were plated into Lab-Tek 16 well slides coated with poly-D-lysine and Matrigel. Transfection mixtures containing a Venus-MLCK, alone or in combination with Myc-Par6α, were introduced using Amaxa nucleofection. Cultures were incubated for 36–48 hrs to allow for neurite extension and neuron migration, and then fixed and processed for immunocytochemistry. Cultures were imaged with a Radiance 2000 confocal laser-scanning microscope.
FRAP and Photoactivation of migrating CGNs
For FRAP and Photoactivation Studies CGNs were respectively nucleofected with either an EGFP-UTRCH-ABD or a combination of PA-EGFP-UTRCH-ABD and RFP-UTRCH-ABD. 18 hours post-nucleofection, cultures were imaged with a Marianas spinning disk confocal microscope (Intelligent Imaging Innovations) equipped with a Micropoint Laser Illumination & Ablation System (Photonics Instruments) fitted to emit 440 nm light needed to photobleach EGFP or photoactivate PA-EGFP. For FRAP experiments regions of interest were bleached by 30 iterations of a 440 nm laser line (35% laser power) and an image acquired at one second intervals after bleaching. For Photoactivation experiments regions of interest were activated by 15 iterations of a 440 nm laser line (20% laser power) and an z-stack encompassing the entire neuron (of both PA-EGFP-UTRCH-ABD and RFP-UTRCH-ABD) image acquired at 20 seconds intervals. All image processing and measurements were carried out using Slidebook (Intelligent Imaging Innovations).
Line scanning and Volumetric Analysis of Migrating Neurons
For line scanning of migrating neuron time-lapse sequences, projection of each frame (time-point) was produced using Slidebook (v4). Using the ruler tool, a spline was drawn from the growth cone of the leading process to the tip of the trailing process. This spline was given a radius of 10 microns, which encompassed the entire width of the cell. The intensity along the length of the line was then measured in the red (free-mCherry) and green (EGFP-UTRCH-ABD) channels.
For Volumetric analysis of migrating neuron time-lapse sequences 3D images at each time-point were used to investigate the change in distribution of EGFP-UTRCH-ABD and the cytoplasm during the migration of the cell. The mask function in Slidebook was utilized to create a mask encompassing the entire cell. This mask was then subdivided 4 different regions using the Boolean mask function. The cell body and trailing process were isolated first. We then measured the leading process and divided this into two to get the proximal and distal region of the leading process. This was repeated for each time point. Once all masks were produced for each time points, we then extracted the data for several criteria e.g. the volume of each mask, the average intensity and the sum intensity in each of the four regions of the cell.
Imaging protocol for centrosome tracking
4D time-lapse images of 15 μm stacks were obtained over the course of a time-lapse sequence using Slidebook v4 (Intelligent Imaging Innovations inc.) Z-stacks were converted to maximum projections and a Laplasian 2D filter was applied to the image at each time-point to emphasize the contrast at the edge of objects such as the centrosome. Then using Slidebooks’s automated particle tracking protocol all objects that moved contiguously over 15 frames were tracked and the interval velocities, average velocities and total displacements of each object were attained. Each object was then manually identified as a centrosome that was present within the cell soma for at least 1 frame over the duration of observation before being included within the appropriate dataset. A detailed description of the Oak Ridge National Laboratory centrosome-tracking algorithm is described in text Supplemental Text.
Additional Methods
A description of the expression vectors and the protocols for Western blotting and immunocytochemistry can be found in the Supplemental Materials.
Supplementary Material
Acknowledgments
We are grateful to Drs. Linda Van Aelst and Bob Adelstein for critically reading the manuscript, Dr. Rick Horwitz for helpful discussions and Dr. Jakub Famulski for aiding with the shRNA experiments. We are indebted to Dr. Bill Bement for providing the panel of UTRCH-ABD vectors and Dr. Atsushi Miyawaki for providing the Venus reporter. We also thank Dr. Anne-Bresnick for the MLCK cDNA, Dr. Shuh Narumiya for the Rock1 cDNA, Dr. Gary Banker for the Map2C cDNA, Dr. Ken Jacobson for the Paxillin cDNA and Greg Law (Perkin Elmer) for the extended use of an Ultraview confocal microscope. We also thank Dr. Regan Baird and Intelligent Imaging Innovations Inc. for setting up our Marianas Workstation and developing Slidebook tools for Volumetric and Line Scanning analysis. Supported by NCI 2 P30CA021765-30 (DJS), a March of Dimes Basil O’Connor Starter Scholar Research Award (DJS) and NIH grants R01-NS15429-26 (MEH) and R01 NS051778-02 (MEH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bahler M, Rhoads A. Calmodulin signaling via the IQ motif. FEBS Lett. 2002;513:107–113. doi: 10.1016/s0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
- Barros CS, Phelps CB, Brand AH. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev Cell. 2003;5:829–840. doi: 10.1016/s1534-5807(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Bellenchi GC, Gurniak CB, Perlas E, Middei S, Ammassari-Teule M, Witke W. N-cofilin is associated with neuronal migration disorders and cell cycle control in the cerebral cortex. Genes Dev. 2007;21:2347–2357. doi: 10.1101/gad.434307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvindrah R, Graus-Porta D, Goebbels S, Nave KA, Muller U. Beta1 integrins in radial glia but not in migrating neurons are essential for the formation of cell layers in the cerebral cortex. J Neurosci. 2007;27:13854–13865. doi: 10.1523/JNEUROSCI.4494-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Biais N, Giannone G, Tanase M, Jiang G, Hofman JM, Wiggins CH, Silberzan P, Buguin A, Ladoux B, Sheetz MP. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys J. 2006;91:3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer LP. Role of actin-filament disassembly in lamellipodium protrusion in motile cells revealed using the drug jasplakinolide. Curr Biol. 1999;9:1095–1105. doi: 10.1016/s0960-9822(99)80478-3. [DOI] [PubMed] [Google Scholar]
- Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163:1205–1211. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB. Dynein at the cortex. Curr Opin Cell Biol. 2002;14:44–49. doi: 10.1016/s0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- Edmondson J, Liem R, Kuster J, Hatten M. Astrotactin: a novel neuronal cell surface antigen that mediates neuron-astroglial interactions in cerebellar microcultures. J Cell Biol. 1988;106:505–517. doi: 10.1083/jcb.106.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson JC, Hatten ME. Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J Neurosci. 1987;7:1928–1934. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O’Connell CB, Wang Y, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Feng Y, Olson EC, Stukenberg PT, Flanagan LA, Kirschner MW, Walsh CA. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–679. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- Fishell G, Hatten ME. Astrotactin provides a receptor system for CNS neuronal migration. Development. 1991;113:755–765. doi: 10.1242/dev.113.3.755. [DOI] [PubMed] [Google Scholar]
- Garces JA, Clark IB, Meyer DI, Vallee RB. Interaction of the p62 subunit of dynactin with Arp1 and the cortical actin cytoskeleton. Curr Biol. 1999;9:1497–1500. doi: 10.1016/s0960-9822(00)80122-0. [DOI] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Gregory WA, Edmondson JC, Hatten ME, Mason CA. Cytology and neuron-glial apposition of migrating cerebellar granule cells in vitro. J Neurosci. 1988;8:1728–1738. doi: 10.1523/JNEUROSCI.08-05-01728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan CB, Xu HT, Jin M, Yuan XB, Poo MM. Long-range Ca2+ signaling from growth cone to soma mediates reversal of neuronal migration induced by slit-2. Cell. 2007;129:385–395. doi: 10.1016/j.cell.2007.01.051. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Salmon WC, Waterman-Storer CM. Converging populations of f-actin promote breakage of associated microtubules to spatially regulate microtubule turnover in migrating cells. Curr Biol. 2002;12:1891–1899. doi: 10.1016/s0960-9822(02)01276-9. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Hatten ME. Neuronal regulation of astroglial morphology and proliferation in vitro. J Cell Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. New directions in neuronal migration. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- Holleran EA, Ligon LA, Tokito M, Stankewich MC, Morrow JS, Holzbaur EL. beta III spectrin binds to the Arp1 subunit of dynactin. J Biol Chem. 2001;276:36598–36605. doi: 10.1074/jbc.M104838200. [DOI] [PubMed] [Google Scholar]
- Holleran EA, Tokito MK, Karki S, Holzbaur EL. Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J Cell Biol. 1996;135:1815–1829. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- Kamm KE, Stull JT. Dedicated myosin light chain kinases with diverse cellular functions. J Biol Chem. 2001;276:4527–4530. doi: 10.1074/jbc.R000028200. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Adelstein RS. Chicken nonmuscle myosin heavy chains: differential expression of two mRNAs and evidence for two different polypeptides. J Cell Biol. 1991;112:915–924. doi: 10.1083/jcb.112.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholmanskikh SS, Koeller HB, Wynshaw-Boris A, Gomez T, Letourneau PC, Ross ME. Calcium-dependent interaction of Lis1 with IQGAP1 and Cdc42 promotes neuronal motility. Nat Neurosci. 2006;9:50–57. doi: 10.1038/nn1619. [DOI] [PubMed] [Google Scholar]
- Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Komuro H, Rakic P. Distinct modes of neuronal migration in different domains of developing cerebellar cortex. J Neurosci. 1998;18:1478–1490. doi: 10.1523/JNEUROSCI.18-04-01478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- Litman P, Amieva MR, Furthmayr H. Imaging of dynamic changes of the actin cytoskeleton in microextensions of live NIH3T3 cells with a GFP fusion of the F-actin binding domain of moesin. BMC Cell Biol. 2000;1:1. doi: 10.1186/1471-2121-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CM, Buxton DB, Chua GC, Dembo M, Adelstein RS, Wang YL. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol Biol Cell. 2004;15:982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Kawamoto S, Hara Y, Adelstein RS. A point mutation in the motor domain of nonmuscle myosin II-B impairs migration of distinct groups of neurons. Mol Biol Cell. 2004;15:2568–2579. doi: 10.1091/mbc.E03-11-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Mertens AE, Pegtel DM, Collard JG. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 2006;16:308–316. doi: 10.1016/j.tcb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Meshel AS, Wei Q, Adelstein RS, Sheetz MP. Basic mechanism of three-dimensional collagen fibre transport by fibroblasts. Nat Cell Biol. 2005;7:157–164. doi: 10.1038/ncb1216. [DOI] [PubMed] [Google Scholar]
- Mittal B, Sanger JM, Sanger JW. Visualization of myosin in living cells. J Cell Biol. 1987;105:1753–1760. doi: 10.1083/jcb.105.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussavi RS, Kelley CA, Adelstein RS. Phosphorylation of vertebrate nonmuscle and smooth muscle myosin heavy chains and light chains. Mol Cell Biochem. 1993:127–128. 219–227. doi: 10.1007/BF01076773. [DOI] [PubMed] [Google Scholar]
- Myers KA, He Y, Hasaka TP, Baas PW. Microtubule transport in the axon: Re-thinking a potential role for the actin cytoskeleton. Neuroscientist. 2006;12:107–118. doi: 10.1177/1073858405283428. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, Hoshino M, Kaibuchi K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol. 2005;7:270–277. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- O’Rourke NA, Dailey ME, Smith SJ, McConnell SK. Diverse migratory pathways in the developing cerebral cortex. Science. 1992;258:299–302. doi: 10.1126/science.1411527. [DOI] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus Rhesus. J Comp Neurol. 1971;141:283–312. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rivas RJ, Hatten ME. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J Neurosci. 1995;15:981–989. doi: 10.1523/JNEUROSCI.15-02-00981.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin MW, Itoh K, Adelstein RS, Bridgman PC. Localization of myosin II A and B isoforms in cultured neurons. J Cell Sci. 1995;108(Pt 12):3661–3670. doi: 10.1242/jcs.108.12.3661. [DOI] [PubMed] [Google Scholar]
- Roger B, Al-Bassam J, Dehmelt L, Milligan RA, Halpain S. MAP2c, but not tau, binds and bundles F-actin via its microtubule binding domain. Curr Biol. 2004;14:363–371. doi: 10.1016/j.cub.2004.01.058. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J, Cramer LP, Baum B, McGee KM. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- Salmon WC, Adams MC, Waterman-Storer CM. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J Cell Biol. 2002;158:31–37. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AW, Kabir N, Forscher P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J Cell Biol. 2002;158:139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AW, Schoonderwoert VT, Ji L, Mederios N, Danuser G, Forscher P. Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev Cell. 2008;15:146–162. doi: 10.1016/j.devcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007 doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeshima H, Hirano T, Kengaku M. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc Natl Acad Sci U S A. 2007;104:16182–16187. doi: 10.1073/pnas.0708047104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Choi CK, Horwitz AR. Integrins in cell migration--the actin connection. J Cell Sci. 2009;122:199–206. doi: 10.1242/jcs.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176:573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–226. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.