Abstract
The cause of Crohn's disease (CD) remains poorly understood. Counterintuitively, these patients possess an impaired acute inflammatory response, which could result in delayed clearance of bacteria penetrating the lining of the bowel and predispose to granuloma formation and chronicity. We tested this hypothesis in human subjects by monitoring responses to killed Escherichia coli injected subcutaneously into the forearm. Accumulation of 111In-labeled neutrophils at these sites and clearance of 32P-labeled bacteria from them were markedly impaired in CD. Locally increased blood flow and bacterial clearance were dependent on the numbers of bacteria injected. Secretion of proinflammatory cytokines by CD macrophages was grossly impaired in response to E. coli or specific Toll-like receptor agonists. Despite normal levels and stability of cytokine messenger RNA, intracellular levels of tumor necrosis factor (TNF) were abnormally low in CD macrophages. Coupled with reduced secretion, these findings indicate accelerated intracellular breakdown. Differential transcription profiles identified disease-specific genes, notably including those encoding proteins involved in vesicle trafficking. Intracellular destruction of TNF was decreased by inhibitors of lysosomal function. Together, our findings suggest that in CD macrophages, an abnormal proportion of cytokines are routed to lysosomes and degraded rather than being released through the normal secretory pathway.
Crohn's disease (CD) is a chronic inflammatory disorder, primarily affecting the gastrointestinal tract. It arises through an aberrant interaction between the bowel contents and the immune system, although the mechanistic basis of disease pathogenesis remains poorly understood. To date, numerous hypotheses have been proposed including atypical infection, the presence of abnormal particulate material within the bowel, and autoimmunity (Marks and Segal, 2008). Most current theories center on disordered T lymphocyte activation (Xavier and Podolsky, 2007). Although T lymphocytes are important in the induction and maintenance of the chronic inflammatory phase, their involvement in the induction of CD lesions remains unproven.
Recent genome-wide association studies (GWAS) have generated considerable interest after the identification of >30 polymorphisms that increase susceptibility to CD (Wellcome Trust Case Control Consortium, 2007; Barrett et al., 2008). Several of these refocus attention on the innate immune system. The CARD15 (caspase-recruitment domain 15) gene remains the best studied and encodes a pathogen-recognition protein primarily expressed within phagocytes (Hugot et al., 2001; Ogura et al., 2001). Its product, NOD2, responds to bacteria-derived peptidoglycan by stimulating proinflammatory cytokine release, a function attenuated by the described polymorphisms. Other significantly replicated associations include the ATG16L1 and IRGM genes coding for proteins involved in autophagy.
Although these associations are highly significant when comparing very large population groups and are valuable in providing an indication of cellular processes and pathways involved in CD pathogenesis, the effect sizes at any individual locus are weak (the majority incurring odds ratios <1.6), and polymorphisms are common in healthy control (HC) subjects. For example, the incidence of one or two CD-related mutations in the gene for NOD2 in the general population is ∼15 and 0.5%, respectively, whereas in CD the figures are 28 and 8% (Cuthbert et al., 2002; Hugot et al., 2007). However, only 1 in 1,000 of the population develop CD, which means that in a general population of 100,000 individuals, roughly 15,000 will have one relevant mutation and 500 will have two such mutations. This population will have ∼100 cases of CD, in whom there will only be 28 single and 8 double mutations on the NOD2 gene. The numbers are even more dramatic when examining mutations at the ATG16L1 (autophagy) locus with 27,000 of the general population having the GG CD-related genotype as compared with 37 patients (Hampe et al., 2007). GWASs have been informative but have failed to explain the cause of the diseases under investigation (Donnelly, 2008). All 32 CD-associated genes combined have been calculated to contribute <20% of the heritable risk (Barrett et al., 2008), leading to the concept of the “missing heritability” (Maher, 2008).
It was realized >30 yr ago that the acute inflammatory response in respect to neutrophil recruitment was defective in CD (Segal and Loewi, 1976). More recently, we have documented defective neutrophil accumulation and cytokine levels at sites of intestinal and skin trauma (Marks et al., 2006). These defects in acute inflammation were independent of the CARD15 genotype. Further investigation demonstrated reduced blood flow in the skin upon bacterial challenge that could be partially corrected by the administration of sildenafil. We concluded that a general phenotypic abnormality can be revealed in CD when the innate immune system is stressed in vivo, namely that acute inflammation is generally and severely impaired. We propose that the consequence of a weak initial reaction to the ingress of gut bacteria into the tissues would be their defective removal and that this persistence could drive secondary chronic inflammation. A unique set of experiments were performed to test this hypothesis directly by measuring the accumulation of neutrophils and clearance of bacteria from the tissues of CD patients. In addition, in vitro studies were performed using monocyte-derived macrophages to identify a defective mechanism, which could explain the diminished acute inflammatory response in CD patients. In the absence of a single causative gene and, hence, an accurate animal model of CD, these experiments had to be conducted on human subjects.
RESULTS
Neutrophil recruitment to sites of bacterial injection
We first determined whether the previously observed impaired recruitment of neutrophils to sites of local trauma in CD extended to the inflammation induced by bacteria in the tissues (Segal and Loewi, 1976; Marks et al., 2006). Autologous neutrophils were labeled with 111indium (Segal et al., 1981) and injected intravenously at the same time as Escherichia coli were inoculated into the volar aspect of each forearm in HC (n = 6) and CD (n = 5) subjects. The labeled neutrophils accumulated at the sites of bacterial injection (Fig. 1 a) in addition to the previously described distribution to the spleen, liver, and bone marrow (Fig. 1 b).
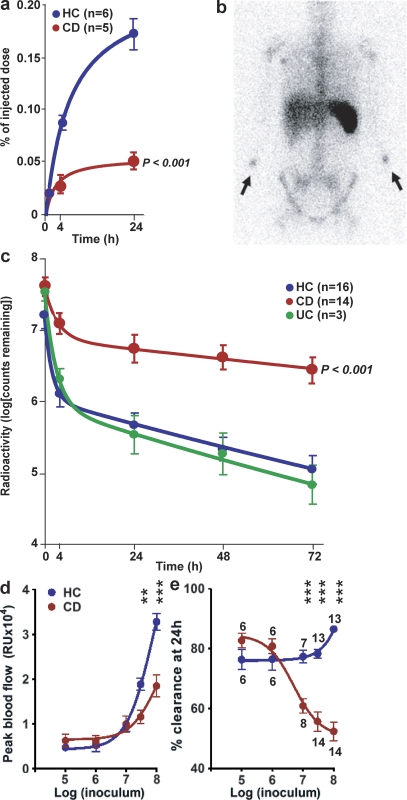
Figure 1.
Neutrophil accumulation and subsequent clearance of E. coli from the tissues is markedly delayed in a dose-dependent manner in CD. 111Indium-labeled autologous neutrophils were injected intravenously at the same time as killed E. coli were injected subcutaneously into each forearm. (a) Radioactivity measured over the injection sites showed a much smaller proportion of labeled cells accumulating in CD subjects. (b) γ-Camera image of a CD patient at 24 h after injection, demonstrating focal accumulations of radioactivity at bacterial injection sites (arrows) and confirming lack of bowel inflammation. (c) 32P-labeled killed E. coli were injected into the subcutaneous tissues of the forearm and radioactivity was measured at the skin surface. Clearance of radioactivity was much slower in CD than in HC or UC. Extrapolating these curves indicated that almost complete removal (99%) would take 10.2 and 7.1 d in HC and UC subjects, respectively, compared with 44.3 d in CD. (d and e) Effect of increasing bacterial dose from 105 to 108 on blood flow at injection site (d) and bacterial clearance (e). The numbers of subjects studied in the dose response experiment are depicted in e. All results are expressed as mean ± SEM (**, P < 0.01; ***, P < 0.001).
In HC subjects, significant cellular recruitment could be measured at 4 h (889 ± 53 cpm; n = 6), and this increased over a 24-h period (1,738 ± 98 cpm; n = 6), at which time ∼0.17 ± 0.014% of the injected dose of radioactivity was present at each injection site (Fig. 1 a). Assuming a whole body pool of roughly 1011 neutrophils (Cartwright et al., 1964), this represents ∼108 cells to deal with the 3 × 107 bacteria injected. The results were very different in CD subjects (P < 0.0001; Fig. 1 a), with less than one-third of the normal numbers of neutrophils accumulating at 4 h (290 ± 52 cpm; n = 5) and 24 h (555 ± 78 cpm; n = 5).
Clearance of 32P-labeled E. coli
A functional consequence of reduced neutrophil influx was assessed by measuring the clearance of 32P-labeled E. coli from the forearm in HC (n = 16), CD (n = 14), and ulcerative colitis (UC; n = 3) subjects. Immediately after inoculation, surface counts at each injection site were 2,688 ± 160 cpm (mean ± SEM). In all groups, clearance followed biphasic nonexponential kinetics, with a rapid initial phase followed by a slower secondary phase (Fig. 1 c). An overall test for equality demonstrated that clearance curves for HC and CD subjects were significantly different (P = 9 × 10−6), with a marked delay in CD compared with HC. UC subjects demonstrated clearance profiles similar to those of HC. Extrapolating clearance curves to a point where 99% of inoculated material would have been cleared gave total clearance times of 10.2 d (95% confidence interval: 8.3–13 d) in HC, 7.1 d (5.4–10.4 d) in UC, and 44.3 d (21.8–∞ d) in CD. Patients were subdivided into those with ileal or colonic involvement, as CD is a syndrome with both clinical and genetic evidence to suggest that precise pathogenic mechanisms operating in each region of the bowel may differ. Clearance appeared to be faster in the ileal (n = 5) than in the colonic (n = 9) CD subjects, but the differences were not statistically significant (Fig. S1 a). In addition, the presence of CD-associated mutations in CARD15 had no effect on bacterial clearance (Fig. S1 b).
The numbers of bacteria injected had an important effect on local blood flow and bacterial clearance. In HC subjects, blood flow increased about sevenfold between doses of 105 and 108 organisms (Fig. 1 d). Blood flow levels in CD patients were similar to those in HC between dosages of 105 and 107 bacteria. Doses >3 × 107 resulted in significantly reduced blood flow in CD subjects, with levels approximately half those recorded for HC. Bacterial clearance was dramatically different between the two groups (Fig. 1 e). In HC subjects, clearance rates were relatively stable between 105 and 3 × 107 organisms and actually increased at the highest dose, whereas in CD, although it was normal at 105 and 106, there was a dramatic and highly significant (P < 0.0001) drop in clearance at and above 107 bacteria.
Macrophage cytokine secretion
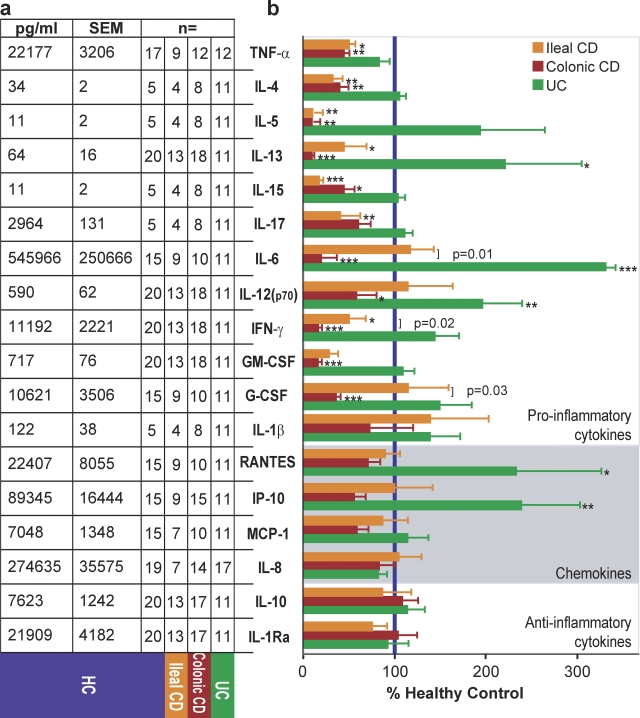
Dramatic differences were observed in the secretion of a panel of inflammation-related cytokines and chemokines by peripheral blood monocyte-derived macrophages after 24 h of stimulation with heat-killed E. coli (HkEc; Fig. 2). We observed distinct profiles in patients with ileal and colonic CD. The proinflammatory cytokines TNF, IL-4, IL-5, IL-13, IL-15, and IFN-γ were reduced in all CD patients, whereas IL-6, IL-12(p70), G-CSF, and GM-CSF were only low in colonic CD and IL-17 was only low in ileal CD. Direct comparisons between macrophages from ileal and colonic CD subjects identified IL-6, IFN-γ, and G-CSF as significantly different between the two phenotypes. All three were released at lower levels by colonic compared with ileal CD macrophages. In contrast, macrophages from patients with UC secreted proinflammatory cytokines at levels similar to, or greater than, HC subjects. All patients released normal levels of the antiinflammatory cytokines IL-1Ra and IL-10 and the chemokines RANTES, IP-10, MCP-1, and IL-8.
Figure 2.
Proinflammatory cytokine secretion by macrophages from CD patients is deficient in response to E. coli. Supernatants from macrophages stimulated for 24 h with HkEc were tested for the levels of cytokines and chemokines. (a) Macrophages from HC subjects released varying amounts of cytokines and chemokines after HkEc stimulation. (b) Cytokine and chemokine release expressed as a percentage of that secreted by HC cells (blue bar) from ileal and colonic CD patients. The numbers of subjects in each group are shown on left; each patient was used once. All results are expressed as mean ± SEM (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
In addition, we examined the secretion of IL-5, IL-6, IL-10, IL-17, GM-CSF, MCP-1, and IP-10 at 24 h by macrophages after stimulation with the TLR2 ligand Pam3CSK4. Secretion of IL-5 and GM-CSF were both reduced irrespective of CD phenotype, whereas secretion of IL-6 and IL-17 were low from patient macrophages with colonic or ileal CD, respectively (Fig. S2 a). Previously, we have extensively studied CD patients with NOD2 mutations using muramyl dipeptide and reported transcription profile and cytokine secretion abnormalities (Marks et al., 2006). We have now examined the influence of NOD2 status on TNF secretion by macrophages in response to TLR2 or TLR4 agonists. The presence of one or two CD-associated mutations in the NOD2 gene had no effect on TNF secretion (Fig. S2 b).
Differences in monocyte/macrophage populations and maturation were investigated in the three groups (Fig. S3). The initial M1 (CD14+/CD16−) and M2 (CD14+/CD16+) monocyte population were similar in the all three groups, as were messenger RNA (mRNA) levels of the macrophage maturation markers F4/80, L-selectin, and ICAM-1 after in vitro differentiation.
Macrophage mRNA transcription profiles
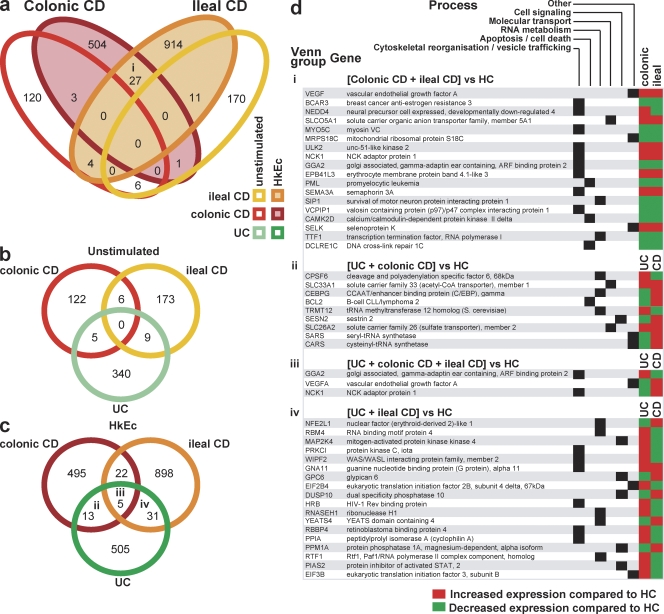
To account for impaired cytokine secretion, we examined global transcription profiles of macrophages. RNA was examined from macrophages from HC (n = 26), ileal (n = 10), colonic CD (n = 16), and UC (n = 20) subjects, before and after exposure to HkEc. The proinflammatory cytokines GM-CSF, IL-6, IFN-γ, and TNF, which had demonstrated diminished secretion, were tested by quantitative PCR and found to have mRNA levels equivalent to those of HC after HkEc stimulation (Table S1). However, disease-specific changes were observed and are presented in Fig. 3. A four-way Venn diagram shows the numbers of probe sets differing in expression level (P < 0.01 and fold change >1.2) before and after stimulation between either group of CD patients and HC subjects (Fig. 3 a). Significant changes from HC levels were observed in the expression of hundreds of genes in the different groups, which were internally consistent. Surprisingly little overlap was observed between profiles of HkEc-stimulated macrophages in ileal and colonic CD.
Figure 3.
Abnormal transcriptional profiles of CD macrophages on Affymetrix gene arrays. (a) Venn diagram shows the number of probe sets differing in expression (P < 0.01) between ileal and colonic CD versus HC macrophages in the unstimulated state and after exposure to HkEc. (b and c) As in a, but comparing differentially expressed probe sets between ileal and colonic CD and UC versus HC in unstimulated (b) and HkEc-stimulated (c) cells. (d) i-iv designate overlaps of differentially expressed genes of known function after HkEc stimulation between disease groups, as labeled in Venn diagrams.
The identities of the differentially regulated genes common to ileal and colonic CD, and the cellular processes in which they are involved, are shown in Fig. 3 (d and i). The 27 probe sets represent 18 genes of known function. Of these, nine have previously been reported as participating in vesicle trafficking and cytoskeletal organization, suggesting that the problem may lie in the posttranscriptional processing, storage, or secretion of cytokines. The data were also analyzed by determining the most significantly induced or repressed genes in individual groups, with false discovery rate (FDR) correction applied. These gave similar results to those shown in Fig. 3 (Fig. S4; Tables S2 and S3), demonstrating an overrepresentation of genes involved in vesicle trafficking and cytoskeletal organization.
To control for nonspecific effects resulting from chronic bowel inflammation, we conducted similar analyses on macrophages from UC patients. In UC, a total of 354 and 554 probe sets differed from HC, before and after HkEc stimulation, respectively. There was very little overlap between differentially expressed genes in CD and UC (Fig. 3, b and c). Of those genes common to both, a relatively large proportion was also involved in vesicle trafficking and cytoskeletal organization, as well as RNA metabolism and cellular signaling (Fig. 3 d, ii-iv). However, although most of the differentially expressed genes common to both CD groups were similarly up- or down-regulated in comparison with HC (Fig. 3 d, i), the majority of those common to UC and CD changed reciprocally (Fig. 3 d, ii-iv), highlighting the fact that UC and CD are distinct diseases, possibly at opposite ends of the inflammatory response (Marks and Segal, 2008).
Transcription, translation, and secretion of cytokines by macrophages
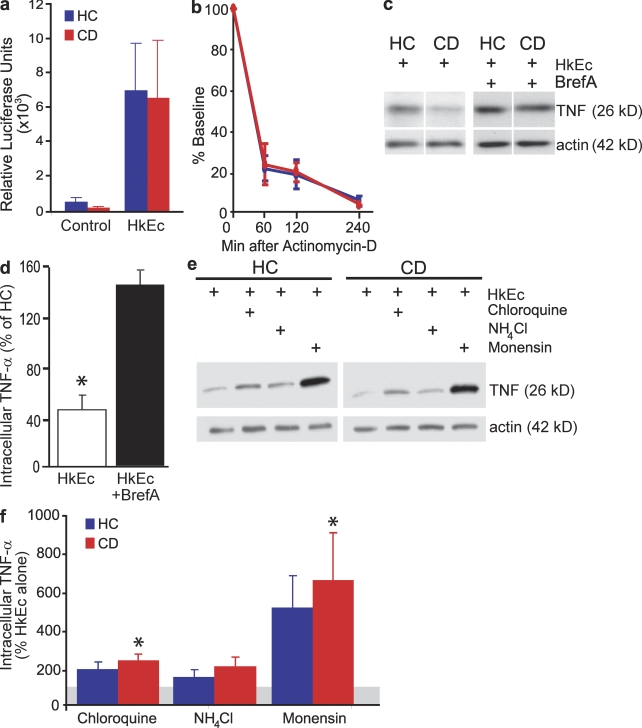
Our observations of diminished proinflammatory cytokine secretion, equivalent cytokine steady-state mRNA levels, and a high proportion of abnormally expressed genes associated with vesicle trafficking and cytoskeletal organization in CD led us to investigate further aspects of RNA metabolism and intracellular protein handling. Cytokine transcriptional activation was investigated using macrophages transfected with a TNF promoter–luciferase reporter. TNF gene transcription occurred in CD (n = 7) and HC (n = 8) macrophages after HkEc stimulation at equivalent levels (Fig. 4 a). TNF mRNA stability was determined using actinomycin-d (an inhibitor of RNA polymerase) and was identical between HC and CD macrophages after HkEc stimulation (Fig. 4 b).
Figure 4.
Intracellular levels of TNF are lower in CD, despite normal transcription and mRNA stability, but restored to normal in the presence of Bref-A and lysosome inhibitors. (a) Macrophages transfected with an adenoviral vector containing a TNF promoter and luciferase reporter demonstrated equivalent TNF transcription levels in HC (n = 8) and CD (n = 7) subjects. (b) TNF mRNA stability is comparable in HC and CD macrophages after stimulation with HkEc. (c) Intracellular levels of TNF after HkEc stimulation with or without Bref-A were determined by Western blotting. (d) CD contained significantly lower levels of intracellular TNF than HC macrophages after HkEc stimulation but returned to normal levels with the inclusion of Bref-A (n = 6 in all groups). (e) Intracellular levels of TNF after HkEc stimulation with or without lysosomal inhibitors were determined by Western blotting. (f) Inhibitors of lysosomal proteolysis increase intracellular TNF levels in HC and CD macrophages (n = 4 in all groups; gray line denotes level of HkEc alone). Significance levels are compared with HkEc alone. All patients in these studies had colonic CD and subjects were used once per assay. Results are shown as mean ± SEM (*, P < 0.05).
Cell lysates were prepared from macrophages from HC (n = 6) and colonic CD (n = 6) subjects, and intracellular TNF was quantitated by Western blotting (Fig. 4, c and d). The level of intracellular TNF in HkEc-stimulated macrophages was lower in CD (P < 0.05) than HC (Fig. 4, c and d). The inclusion of brefeldin A (Bref-A), a potent inhibitor of ER-to-Golgi protein transport during macrophage HkEc stimulation resulted in approximately equivalent levels of intracellular TNF in both CD and HC (Fig. 4, c and d). The results obtained with Bref-A demonstrated that TNF translation occurs normally in CD macrophages.
We investigated the role of the lysosome in intracellular cytokine trafficking with the inclusion of monensin, NH4Cl, or chloroquine during macrophage HkEc stimulation. NH4Cl and chloroquine partition into acidic compartments and elevate the pH, whereas monensin is a proton ionophore with similar but more potent effects. In HkEc-stimulated macrophages from colonic CD patients (n = 4), all three inhibitors of lysosomal function increased intracellular levels of TNF by between twofold and sevenfold (Fig. 4, e and f). Incubating macrophages with the proteosome inhibitor MG132 was without effect (unpublished data).
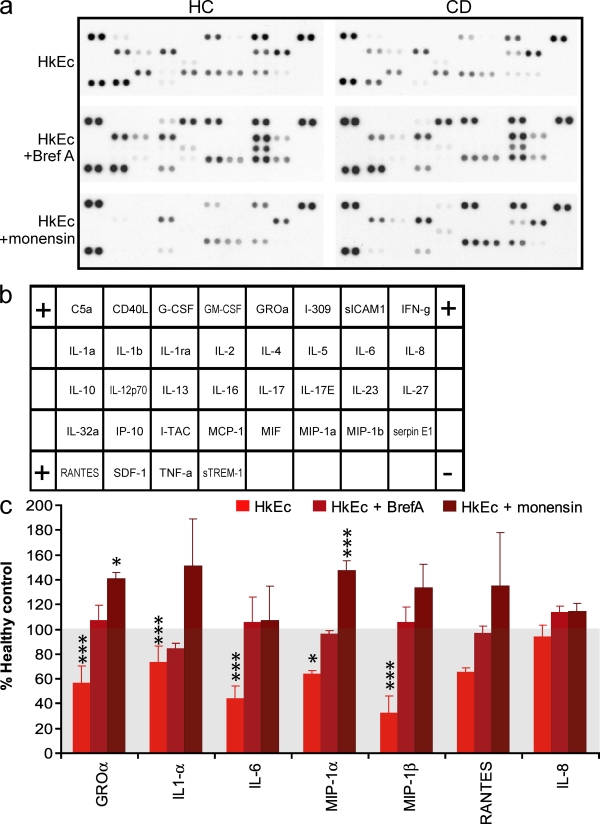
In addition to TNF, the intracellular levels of GRO-α, IL-1-α, IL-6, MIP-1α, and MIP-1β were also depressed in CD macrophages after HkEc stimulation (Fig. 5). The inclusion of Bref-A resulted in the normalization of intracellular levels of all the cytokines and chemokines tested (Fig. 5, a and c). In comparison, monensin either normalized the intracellular levels (IL-1α, IL-6, and MIP-1β) or resulted in a significant elevation compared with HC (n = 4; GRO-α and MIP-1α; Fig. 5, a and c).
Figure 5.
Intracellular levels of chemokines and cytokines after HkEc stimulation in the presence of vesicle trafficking and lysosomal inhibitors. (a) Intracellular cytokine array profiles obtained from macrophages stimulated with HkEc in the presence of absence of either Bref-A or monensin. (b) Cytokine array map. (c) Various chemokines and cytokines demonstrate reduced intracellular levels in CD macrophages after HkEc stimulation and are either normalized or elevated after Bref-A or monensin treatment (n = 4 in all groups). All patients studied had colonic CD and subjects were used once per assay. Results shown are mean ± SEM (*, P < 0.05; ***, P < 0.001).
DISCUSSION
In contrast to the relatively weak genetic effects identified in the GWAS (Wellcome Trust Case Control Consortium, 2007), a common major phenotypic abnormality becomes apparent in CD when the innate immune system is stressed in vivo. This is the result of a severely impaired acute inflammatory response in these individuals (Marks et al., 2006). We have suggested that the intensity of the inflammatory response in the whole population follows a normal Gaussian distribution. Individuals at the extreme lower tail of this spectrum, as a consequence of the additive effect of many gene variants, are at risk of CD. NOD2, and potentially other systems, provide a compensatory boost to acute inflammation, which is protective in this setting (Marks and Segal, 2008).
These studies provide the first demonstration in CD that there is a profound defect in the recruitment of neutrophils after the introduction of E. coli into the tissues and that the subsequent clearance of these organisms is grossly delayed in this condition. This bacterium was used in these studies because it is the major aerobic component of the bowel flora and present in those regions of the gut affected by CD (Finegold, 1969). E. coli have been implicated in the pathogenesis of CD (Rhodes, 2007), have been found within macrophages in Crohn's tissue (Liu et al., 1995), and have also been cultured from draining mesenteric lymph nodes (Ambrose et al., 1984b). Granulomas removed from CD tissue have been shown by PCR to contain E. coli DNA (Ryan et al., 2004), and an ileal CD E. coli isolate (LF82) was capable of inducing granuloma formation in vitro (Meconi et al., 2007). In addition, we had previously shown that there was an important abnormality of acute inflammation in response to E. coli in CD (Marks et al., 2006).
The observed abnormalities in CD did not occur as a consequence of the inflammatory process, drug therapy, or bowel ulceration. All the patients were in remission and most were not on therapy, with a minority receiving 5-aminosalicylic acid–containing drugs. Bacterial clearance was normal in the control group of subjects with UC, who were also in remission and on 5-aminosalicylic acid or no treatment.
The rate at which neutrophils accumulate at sites of bacterial ingress into the tissues is crucial to the final outcome. It has previously been demonstrated that in a three-dimensional tissue matrix, bacterial phagocytosis is directly related to neutrophil numbers (Li et al., 2004). In the absence of adequate neutrophil recruitment, monocytes and macrophages might function in a containing role, such that material remaining after inoculation is surrounded and only slowly degraded (Issekutz et al., 1981). This is supported by the recovery of bacterial products from Crohn's granulomata (Ryan et al., 2004).
The consequence of the delayed neutrophil accumulation is that foreign fecal material is not cleared adequately from the tissues, leading to granulomatous inflammation. Congenital monogenic innate immunodeficiencies, in which neutrophil dysfunction predominates, frequently manifest bowel inflammation that is indistinguishable from CD (Rahman et al., 2008). These disorders are also associated with major defects in the clearance of foreign material caused by failure of neutrophils to accumulate in sufficient numbers (congenital neutropenias and leukocyte adhesion deficiency), poor digestion within excessively acidic phagolysosomes (chronic granulomatous disease and glycogen storage disease-1b), and impaired vesicle trafficking and fusion of the granules and phagosomes (Chediak-Higashi and Hermansky-Pudlak syndromes). The genes underlying these disorders have not been detected by GWAS of CD because of their extreme rarity.
In the absence of a monogenic lesion in the immune system, CD patients might also be unduly susceptible to bacterial infection, although the necessary large-scale studies have not been performed (Kyle, 1980; Ambrose et al., 1984a; Kaplan et al., 2007). It becomes more difficult to assess the incidence of disease-specific infection once a diagnosis of CD has been made because of confounding factors including surgery, malnutrition, immunosuppressant therapy, and bowel-related complications such as abscesses and fistulae. Another consideration is bacterial load. Most acute infections originate from a small number of inoculating organisms, which even the dampened immunity in CD appears to be able to control. Our findings of the relationship between bacterial dose and clearance are important in this respect because they demonstrate that CD patients can deal efficiently with small numbers of organisms in the tissues but that the clearance mechanisms are overwhelmed by a large bolus of bacteria. CD occurs almost exclusively in the terminal ileum and colon, which respectively contain ∼108 and 1011 bacteria/ml, higher than the 105 bacteria/ml or less in other regions of the small bowel (Farthing, 2004). Reflux through the ileocaecal valve carries caecal contents into the terminal ileum, elevating the microbial levels in this region (Malbert, 2005). After surgery, recurrence is seen in ∼80% of cases, and where small bowel has been resected, recurrence generally occurs in the small bowel just proximal to the anastomosis. This indicates that recurrence is highly dependent on the proximity to the large bowel contents rather than a specific abnormality associated with the terminal ileum (Cameron et al., 1992). Therefore, the large bowel and adjacent ileum provide a unique environment in the body that contains a massive bacterial load, which can gain instant access to the tissues if the mucosal barrier is breeched.
The detrimental effect of fecal content in the tissue might not primarily result from the infectious nature of the organisms, as many are not particularly virulent, but by the quantity of organic material that must be removed before resolution can occur. Difficulty in clearing exogenous organic material could also account for the false-positive results in CD to Kveim tests, which were conducted to diagnose sarcoidosis by injecting a preparation of the spleen of a patient into the skin, which was subsequently examined for a granulomatous response (Mitchell, 1971).
Clearly, the retention of fecal material in the tissues of the wall of the bowel will have major local and systemic inflammatory, immunological, and constitutional consequences. Many of the immunological studies conducted on Crohn's patients, such as those implicating adaptive immunity in local tissue damage (Xavier and Podolsky, 2007), might be measuring secondary immunological responses to this foreign material.
We then turned our attention to identifying the mechanism underlying impaired neutrophil accumulation. CD is caused by the complex interplay of many genes coupled with bacterial exposure, which makes it difficult, if not impossible, to identify specific molecular lesions common to these patients, and GWASs clearly indicate that there are no dominant causative genes. However, the characteristic pathological features of the condition are suggestive of a common cellular pathology produced by the various additive molecular aetiologies. The neutrophils themselves appear functionally normal (Moráin et al., 1981), and their diminished migration to acute inflammatory sites in the skin or bowel has been attributed to defective local cytokine production (Marks et al., 2006). These mediators are primarily secreted by resident tissue macrophages (Medzhitov, 2008), which, in the bowel, are derived and continually replenished from peripheral blood monocytes (Smythies et al., 2006). We therefore measured the secretion of a panel of inflammatory cytokines and chemokines by macrophages in response to stimulation with E. coli or TLR ligands and observed the production of dramatically low levels of proinflammatory cytokines in cells from CD patients. In our previous study, we found local levels of IL-8 and IL-1β to be low after trauma to the skin and bowel (Marks et al., 2006). In the current study, we have found that macrophages secrete normal amounts of IL-8 after stimulation with bacteria. This discrepancy between tissue levels and isolated macrophages in response to E. coli could lie in the type of cells producing IL-8 in skin windows and bowel biopsies, and/or in a different agonist. Neutrophils themselves produce copious amounts of IL-8 and IL-1β, so the lower levels in skin windows and bowel biopsies in CD are consistent with the reduced recruitment at these sites (Fujishima et al., 1993; Greten et al., 2007). In the study by Marks et al. (2006), the release of IL-8 by macrophages from CD patients was diminished after stimulation with wound fluid, TNF, and C5a but unaltered after stimulation with the TLR4 ligand lipopolysaccaride. IL-8 release is normal after bacterial stimulation (whole E. coli as well as TLR2 stimulation), so the lower levels in the in vivo study could be the result of activation of macrophages by nonmicrobial agonists.
To explain the cause of impaired cytokine secretion by macrophages from CD patients, we examined the profiles of RNA expression against a chip containing fragments of genes representing the whole human transcriptome. The first important observation was that macrophages from patients with colonic or ileal CD and UC exhibited distinct expression profiles when compared with HC, identifying these as three separate disease clusters. Second, levels of mRNA encoding proinflammatory cytokines were normal in unstimulated and HkEc stimulated macrophages, providing evidence that the defective secretion was not at the level of gene transcription.
A possible mechanism responsible for diminished proinflammatory cytokine release was provided by examining the abnormally expressed genes common to both types of CD after HkEc stimulation. Approximately half of the genes had some association with cellular secretory systems, suggesting that impaired macrophage secretion is responsible for the diminished proinflammatory cytokine release. The failure of a secretory system would provide a generic mechanism for the impaired release of several functionally related cytokines. Hermansky-Pudlak syndrome and Chediak-Higashi disease, both of which are associated with noninfectious granulomatous enterocolitis, are the result of primary abnormalities in the cytoskeletal transport of vesicles (Rahman et al., 2008). These findings are particularly compelling given the recent descriptions of variants in autophagy genes which are associated with CD susceptibility (Hampe et al., 2007; Parkes et al., 2007; Rioux et al., 2007). Autophagy involves vesicle trafficking in the formation of double-membrane vesicles that deliver cytoplasmic material to the lysosomes (Levine and Deretic, 2007). Additionally, autophagy is required to kill and remove intracellular organisms in macrophages, so its dysfunction is consistent with the theme we have developed: that CD results from a failure to digest and remove microbial and other foreign material.
Interestingly, there was no other direct relationship between the collections of genes identified in this study in macrophages and the polymorphisms highlighted by the previous GWAS (Wellcome Trust Case Control Consortium, 2007; Barrett et al., 2008), with the exception of HLA-G and SPTLC2, which were up-regulated and down-regulated, respectively, in colonic CD (Table S3). This is not entirely unexpected because association studies interrogate the whole genome in a nonselective manner, whereas we have focused on RNA expression in a single cell type, which plays a pivotal role in inflammation, in resting cells and those after activation with a naturally occurring agonist.
Several microarray expression studies have previously been performed in CD and UC. However, their experimental approach differed greatly from ours, as did the results they obtained. This is not surprising in view of the very diverse cell populations and variable states of stimulation in their samples. Those on blood cells were performed on freshly isolated unstimulated mixed cell populations consisting of lymphocytes and monocytes with granulocyte contamination (Mannick et al., 2004; Burczynski et al., 2006). The majority used bowel biopsies, in which a wide spectrum of cells is present in noninflamed tissue. These are further supplemented by innate and adaptive immune cells when inflammation is present (Lawrance et al., 2001; Costello et al., 2005; Hughes, 2005).
To understand the failure of macrophages to secrete normal quantities of proinflammatory cytokines in CD, we used TNF as a model, because this molecule has been extensively studied and experimental tools are readily available. We found that gene transcription and mRNA stability are normal in CD macrophages. Bref-A prevents vesicular movement between the ER and the Golgi, resulting in accumulation of proteins within the former. Stimulation in the presence of Bref-A revealed normal cytokine synthesis by CD macrophages. Normal synthesis coupled with deficient secretion and low intracellular levels of the cytokines indicate accelerated degradation. The accumulation of normal amounts of TNF and other proinflammatory cytokines after the addition of lysosomal inhibitors to CD cells indicates that this aberrant breakdown occurs in the lysosomal compartment.
The observation that macrophage secretion of TNF in CD is abnormally low might at first seem paradoxical, given the therapeutic efficacy of TNF blockade in many of these patients (van Deventer, 1999). However, we are comparing two temporally distinct events, acute inflammation over a time course of hours with chronic inflammation over weeks to months. The acute inflammatory response, dependent on the acute release of proinflammatory cytokines, is important for the clearance of bacteria and fecal material from the tissues, which takes place over a few hours. The lack of TNF after the administration of therapeutic antagonists for the treatment of other chronic inflammatory diseases has been reported to precipitate the development of CD (Charach et al., 2008; Yazisiz et al., 2008).
Failure to clear foreign material from the tissues induces a chronic granulomatous reaction around the retained organic material over a much longer time frame. The accumulation of macrophages and T lymphocytes in these granulomata leads to local tissue damage and constitutional symptoms through the sustained secretion of cytokines (Bazzoni and Beutler, 1996). Even if the net production of cytokines by each cell were lower than normal, the overall number of cells is so great that damaging concentrations are produced. In this setting, drugs directed against TNF can prove beneficial by direct cytokine blockade, by inducing leukocyte apoptosis, and by stimulating the effect of Treg cells, which suppress inflammation (Wong et al., 2008). A clear example of the dichotomous effect of TNF is provided by the dextran sodium sulfate (DSS) bowel inflammation model in TNF knock-out mice. Contrary to initial expectations, mice deficient in this cytokine are more susceptible to the induction of bowel inflammation by DSS, whereas TNF inhibition is effective in ameliorating established DSS colitis in wild-type animals (Naito et al., 2003).
The relationship of impaired cytokine secretion to disordered packaging and vesicle transport, rather than defects in their production or stability, provides a novel insight into the mechanisms underlying the pathogenesis of CD. Similar mechanisms could lead to abnormalities in other relevant cell types, such as the Paneth cell (Cadwell et al., 2008). A basic abnormality in macrophage biology could also explain extraintestinal manifestations of CD, such as arthritis, and lesions in the eyes, skin, lungs, and other tissues (Ephgrave, 2007). There may be many other diseases that present with exuberant granulomatous inflammation as a result of an underlying failure of acute inflammation and innate immunity, leading to defective clearance of the initiating agent.
MATERIALS AND METHODS
Patients.
These studies were approved by the Joint University College London (UCL)/UCL Hospitals (UCLH) Committee for the Ethics of Human Research (project numbers 02/0324 and 04/Q0502/29) and the Administration of Radioactive Substances Advisory Committee (RPC597-790). Patients from the Gastroenterology outpatient clinic at UCLH who met inclusion criteria were recruited to participate in the study. All patients had definitive diagnoses of CD or UC, which were confirmed using standard diagnostic criteria, with quiescent disease (Harvey-Bradshaw or Mayo score <3; Harvey and Bradshaw, 1980; Schroeder et al., 1987). Patients on either no medication or a stable maintenance dose of 5-aminosalicylates (2.5 g/d) for the previous 3 mo were included. None of the patients had received corticosteroid, immunosuppressant, anti-TNF, or metronidazole therapy within 3 mo of enrollment. HC subjects approximately matched for age, sex, and smoking history were recruited from the Department of Medicine, UCL. Written informed consent was obtained from all volunteers. No subject was studied more than once in each of the different experiments. Details of patients and HC subjects included in in vivo experiments are provided (Table S4). CD patients were genotyped for the three common polymorphisms in CARD15 as previously described (Table S4; Marks et al., 2006). In some studies, we compared the results obtained from CD patients with ileal or colonic involvement, as there is evidence that different pathogenic mechanisms might operate in these regions of the bowel (Cuthbert et al., 2002). We excluded the ∼30% of CD patients with mixed ileocolonic disease to simplify phenotyping and analysis.
Neutrophil accumulation studies.
Details of controls and patients are provided in Table S4. No patient had received corticosteroid, immunosuppressant, anti-TNF, or nonsteroidal drug therapy within 3 mo of enrolling in the study, although patients on maintenance 5-aminosalicylates with no change in their dose in the previous 3 mo were included. Patients with a history of skin disease were not enrolled for any injection studies.
A fully antibiotic-sensitive clinical isolate of E. coli (NCTC 10418) was grown overnight at 37°C in DME (Sigma-ALdrich) and 1.25 g/liter sterile yeast extract (Oxoid). Bacteria were then killed by exposure to a UV source (ChemiDoc, trans-UV mode; Bio-Rad Laboratories) for 60 min, and washed multiple times in sterile saline. Bacterial concentrations were determined by optical density (OD600 = 0.365 equates to 108 E. coli/ml [Yourassowsky et al., 1989]). Sterility of the final sample was confirmed by multiple cultures.
Peripheral venous blood was collected from subjects into syringes containing 5 U/ml heparin. Separate blood samples were taken for full blood count and routine serum biochemistry. Neutrophils were isolated by centrifugation through Lymphoprep (Axis Shield), and then erythrocytes were removed by sedimentation with 10% dextran followed by hypotonic lysis. The neutrophils were then washed once in sterile injection-grade normal saline and incubated with 6 MBq 111In-oxine (GE Healthcare) for 20 min at room temperature (Segal et al., 1981). A mean ± SEM of 3.40 ± 0.35 × 107 cells labeled with 5.15 ± 0.17 MBq (n = 11) were resuspended in 5 ml of normal saline. These were injected intravenously through a cannula in a vein in the antecubital fossa, which was subsequently flushed with a further 10 ml of normal saline.
Immediately after neutrophil injection, 3 × 107 UV-killed E. coli suspended in 100 μl of normal saline were injected subcutaneously into the volar aspect of each forearm. Radioactivity was counted using a β counter in scaler mode (RadEye B20; Thermo Fisher Scientific) over these injection sites at baseline and at 1, 4, and 24 h after injection. These sites were shielded from the remainder of the body during counting to avoid detection of label accumulating in the liver or spleen. To control for background radioactivity, this was counted over a control site in the left calf and these counts were subtracted from counts over the injection sites to obtain specific accumulations of radioactivity produced by the bacterial injections. The absolute radioactivity at the injection sites was determined by calibration of the meter against serial dilutions of a sample of free 111In-oxine with known activity. The 24-h counts over the injection sites in the six HC subjects were 1,738 ± 98 cpm. We did not study control subjects with UC because we felt that the additional information we would obtain did not justify the theoretical risk of administering radioactivity systemically to these subjects.
Bacterial clearance studies.
Details of controls and patients included in each set of experiments are provided in Table S4. E. coli were grown, as before, overnight at 37°C in 9 ml of phosphate-free DME (Invitrogen) supplemented with 1 ml of phosphate-containing DME (Sigma-Aldrich), 1.25 g/liter of sterile yeast extract, and 18.5 MBq 32P-orthophosphate PBS (GE Healthcare). This isotope was chosen because it is a β emitter with a short half-life (∼14.7 d) but of sufficient energy to emit through skin and readily and stably incorporate into bacteria (Jordan, 1970). Labeled bacteria were killed by UV exposure for 60 min and washed multiple times in sterile saline to remove unbound radiolabel. A specific activity ± SEM of 6.39 ± 1.52 MBq per 108 bacteria was achieved. Unlabeled bacteria from the same reference sample were also grown and killed by UV exposure for 60 min, in the absence of radiolabel and in phosphate-containing DME supplemented with sterile yeast extract.
A preparation of E. coli containing 1 kBq of radioactivity was mixed with unlabeled bacteria to produce a dose range of 105, 106, 107, 3 × 107, and 108 bacteria in 100 μl of sterile saline for inoculation (different doses were used depending on the experiment) and injected into the volar aspect of both forearms. Surface radioactivity was counted over a 3-min period using a β counter in scaler mode (RadEye B203). Initial surface counts per minute at each site were 2,688 ± 160 (mean ± SEM). Counts were subsequently repeated at 4, 24, 48, and 72 h. Correlations between the readings in each arm were r2 = 0.67 (P < 0.005).
Radiation dose in bacterial clearance studies.
Using values published for 32P in the MIRDOSE 3.1 spherical tumor model (Stabin, 1996), with a presumed tissue dispersion volume of 0.1–3.9 ml, a dose of 1 kBq was calculated to deliver an absorbed dose of 0.06–1.00 Gy if no clearance occurred. If a biological half-life of 3 h was presumed, the absorbed dose was estimated at 0.5–10.0 mGy.
The threshold dose of 32P resulting in erythema after extravasation has been determined as 10 Gy (Castronovo et al., 1988). A dose of 1 kBq was therefore selected as producing sufficient emitted radioactivity to be recordable at the skin surface but falling well within acceptable established safety limits and ensuring that inflammation generated was solely the result of the presence of bacteria rather than isotope.
Macrophage isolation and culture and stimulation.
Peripheral venous blood was collected from subjects into syringes containing 5 U/ml heparin. Mononuclear cells were isolated by differential centrifugation (900 g for 30 min at 20°C) over Lymphoprep and washed twice with sterile PBS (Invitrogen) at 300 g (5 min at 20°C). Cells were resuspended in 10 ml RPMI-1640 medium (Invitrogen) supplemented with 100 U/ml of penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), and 20 mM Hepes, pH 7.4 (Sigma-Aldrich) and plated at a density of ∼5 × 106 cells/ml in 8 cm2 Nunclon Surface tissue culture dishes (Nunc) at 37°C in 5% CO2. After 2 h, nonadherent cells were discarded and 10 ml of fresh RPMI supplemented with 10% FBS (Sigma-Aldrich) was added to each tissue culture dish. Cells were then cultured for 5 d at 37°C in 5% CO2, with the addition of a further 10 ml of fresh 10% FBS/RPMI after 24 h.
Adherent cells were scraped on day 5 and replated in 96-well culture plates (Nunc) at equal densities (105/well) in X-Vivo-15 medium (Cambrex). These primary monocyte-derived macrophages were incubated overnight at 37°C in 5% CO2 to adhere and were then stimulated for up to 24 h, depending on the experiment, with 2.5 × 105 HkEc, prepared as previously described (Marks et al., 2006), 2 μg/ml Pam3CSK4 (Enzo Biochem, Inc.), or 200 ng/ml LPS (Enzo Biochem, Inc.).
Cytokine secretion assays.
Macrophage supernatants were collected after 24-h stimulation of primary monocyte-derived macrophages with HkEc as described in the preceding section. The expression profile of a panel of cytokines in macrophage supernatants was measured using the Beadlyte Bio-Plex human cytokine assay (Bio-Rad Laboratories) according to the manufacturer's instructions. Our assay was customized to detect and quantify IL-1Ra, IL-4, IL-5, IL-6, IL-10, IL-12, IL-13, IL-15, IL-17, G-CSF, GM-CSF, IFN-γ, IP-10, and MCP-1. IL-8 secretion was measured using an ELISA kit (R&D Systems). The assay was conducted according to the manufacturers' instructions with recombinant human standards.
TNF release was measured using a cytotoxicity bioassay (obtained from B. Beutler, The Scripps Institute, La Jolla, CA) as previously described (Aggarwal et al., 1985). Mouse L929 fibroblast cells were grown in DME (Invitrogen), supplemented with 10% FBS (Sigma-ALdrich), 100 U/ml of penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen) at 37°C in 5% CO2. A confluent monolayer of mouse L929 fibroblasts was trypsinized and resuspended to 4 × 105 cells/ml in DME. L929 cells were seeded into 96-well flat bottomed tissue culture plates (4 × 104 cells/well) and incubated overnight at 37°C in 5% CO2. After overnight culture, the medium was discarded, replaced by 50 μl DME containing 0.04 mg/ml cyclohexamide, and incubated for 20 min at 37°C in 5% CO2. 50 μl of cell-free supernatant (diluted 1:50 in DME), collected from primary macrophages as already described, were added to individual wells. Serially diluted recombinant human TNF (100–0 pg/ml; R&D Systems) was used to determine the standard curve for the assay.
Cytokine release in culture supernatants was normalized for the numbers of viable cells in each well, ascertained with the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide, tetrazolium salt) assay (Boehringer Ingelheim). 20 μl of 2.5 ng/ml MTT was added to each well and incubated for 4 h at 37°C in 5% CO2. Supernatants were carefully discarded and 100 μl/well of lysis solution (90% isopropanol, 0.5% SDS, 0.04 N HCl, and 10% H2O) was added to each well for 1 h at room temperature. The absorbance was read at 570 nm using a microplate reader (Anthos Labtec Instruments).
RNA purification.
Total RNA was prepared from monocyte-derived macrophages, either unstimulated or stimulated for 4 h with HkEc, using the RNeasy Mini kit with RNase-free DNase treatment (QIAGEN). Optical density readings were determined for OD260/OD280 and OD260/OD230 using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific) to assess protein and solvent contamination, respectively. Before GeneChip studies, RNA quality was further analyzed by assessing ribosomal RNA bands 28S/18S ratios using a Bioanalyzer (Agilent Technologies) high resolution electrophoresis system.
Whole genome GeneChip expression analysis.
RNA samples were anonymized and randomized by disease group, treatment, and gender into three balanced batches. Linear amplification of RNA samples was performed using the standard Ovation RNA Amplification System 2 (NuGEN) as one batch on two 96-well plates. Samples were then fragmented, biotin labeled, and hybridized in three separate batches to the human U133 plus 2 GeneChip (Affymetrix) for 16 h to generate the transcriptomic data. The microarrays were washed and scanned according to Affymetrix protocols. Quality was assessed using report files generated in GCOS (GeneChip Operating System; Affymetrix) and checked against in-house criteria (GlaxoSmithKline) for probe and hybridization quality by analysis of the data for background, percentage of present calls, background standard deviation, raw Q (noise), scaling factor, GAPDH 3′ to 5′ ratio, and β-actin 3′ to 5′ ratio (Heber and Sick, 2006). The data were assessed for homogeneity of quality control metrics by principal component analysis (PCA) of gene expression using Array Studio (OmicSoft Corporation). To ensure that all samples corresponded to the annotated gender, expression was analyzed for gender-specific genes (XIST and USP9Y). Data from a total of 66 microarrays were therefore analyzed: HC, 16 unstimulated and 12 HkEc; ileal CD, 9 unstimulated and 6 HkEC; and colonic CD, 13 unstimulated and 10 HkEc. Global gene expression of the normalized probe intensity data were initially analyzed using Array Studio for the visual assessment of key trends by PCA. Further analysis of the intensities used a General Linear Model (GLM) performed in Array Studio. The GLM was used to build comparisons of HCs versus each disease group by treatment. A second experiment was then performed in which comparisons were made between macrophages from HC (7 unstimulated and 5 HkEc) and UC (19 unstimulated and 20 HkEc). Genes were considered to be differentially expressed between groups where P < 0.01, fold change was ≥1.2, and expression level was greater than the median cut of 5.0 fluorescence units. Where appropriate, the p-value was corrected for multiple comparisons (FDR correction, Benjamini and Hochberg [1995] method). For the overlaps of the Venn diagrams where a significant result was obtained independently in two separate experiments, the FDR correction was not applied; for assessment of the most significantly regulated genes, both raw p-value and FDR-corrected p-value are presented. Microarray data in MIAME format can be found at http://www.ebi.ac.uk/microarray-as/ae/ under accession nos. E-TABM-733 and E-TABM-734.
TaqMan protocol.
Expression levels of selected genes found to be differentially regulated by the transcriptomic array data were validated by real-time RT-PCR TaqMan analysis using the ABI Prism 7900 Sequence Detector System (Applied Biosystems). 200 ng of total RNA were reverse transcribed to complementary DNA (cDNA) using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the standard protocol. The equivalent of 10 ng RNA per well was arrayed into 384-well plates using a Biomek FX robot (Beckman Coulter), and quantitative RT-PCR was performed using a 7900HT Sequence Detector System (Applied Biosystems) in a 10-μl reaction volume.
The following TaqMan primers and probes used: GMCSF (forward, 5′-AGCCTCACCAAGCTCAAGGG-3′; reverse, 5′-GGGTTGGAGGGCAGTGCT-3′; probe, 5′-CCCTTGACCATGATGGCCAGCC-3′), IL6 (forward, 5′-TGACCCAACCACAAATGCCA-3′; reverse, 5′-CATGTCCTGCAGCCACTGG-3′; probe, 5′-CTGTGCCTGCAGCTTCGTCAGCA-3′), IFN-γ (forward, 5′-CCAACGCAAAGCAATACATGAAC-3′; reverse, 5′-ACCTCGAAACAGCATCTGACTCC-3′; probe, 5′-TCATCCAAGTGATGGCTGAACTGTCGC-3′), and TNF (forward, 5′-TCCTCTCTGCCATCAAGAGCC-3′; reverse, 5′-GTCGGTCACCCTTCTCCAGC-3′; probe, 5′-TGGAAGACCCCTCCCAGATAGATGGGC-3′). The final optimized concentrations for the forward and reverse primers plus the probe were 900 nmol/l, 900 nmol/liter and 100 nmol/liter, respectively. A standard curve was generated using human genomic DNA (Promega). PCR parameters were the following: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. Data were acquired and processed with Sequence Detector 2.3 software (Applied Biosystems). A linear regression line calculated from the standard curves of serially diluted genomic DNA allowed relative transcript levels in RNA-derived cDNA samples to be calculated from the fluorescent signal in each run.
The data generated by the real-time PCR TaqMan were analyzed in Array Studio. The genes selected for RT-PCR were normalized using a covariate to account for any change in expression caused by RNA loading of the samples. This covariate was represented by the scores from the first principal component obtained from a PCA of the two housekeepers, β-actin and cyclophilin-α, and β2-microglobulin (identified from the microarray data analyses as having invariant expression across all samples). A general linear model was built to look at the comparisons of HCs versus each disease group by treatment, and unstimulated versus stimulated samples by disease group. An FDR correction was applied using the Benjamini and Hochberg (1995) methods.
TNF reporter studies.
Recombinant replication-deficient adenoviral constructs encoding pAdTrack-TNF 5′ promoter-luciferase-3′ UTR (AdTNF 5′/3′) and pAdTrack-TRF 5′ promoter-luciferase (AdTNF 5′ only) were generated as previously described (Horwood et al., 2003). Macrophages were plated in 96-well plates (Nunc) at 105 cells/well and allowed to express adenoviral transgenes for 24 h before stimulation with HkEc for 4 h. Luciferase reporter gene assays were performed as previously described (Palmer et al., 2008). Before lysis, viral infection rates were measured by GFP fluorescence (excitation, 485 nm; emission, 520 nm) using a FLUOstar Omega machine (BMG LABTECH) and software according the manufacturer's instructions, and luciferase activity was normalized to these levels.
mRNA stability.
Macrophages were stimulated with HkEc for 4 h followed by the addition of 2 μg/ml actinomycin D. Total RNA was purified as described in RNA purification at 0, 1, 2, and 4 h. TNF mRNA levels were determined by RT quantitative PCR and normalized to GAPDH and ribosomal 18S.
Cytokine protein arrays.
5 × 105 macrophages were incubated with HkEc alone or in combination with 2.5 μM Bref-A (Merck) or 2.5 μM monensin (Sigma-Aldrich) for 4 h. Cells were lysed in TBS containing 0.1% NP-40 and protease inhibitors, sonicated (3× 3-s bursts), and then interrogated with commercially available antibody arrays (Human Cytokine Array Panel A; R&D Systems), according to the manufacturer's instructions. Processed arrays were exposed to x-ray film, and the signal was determined and normalized to the internal positive control spots by densitometry using ImageJ software (National Institutes of Health).
Lysosomal inhibition and cytokine production.
2 × 105 macrophages from patients with CD and HC subjects were prepared as previously described and stimulated for 4 h with 5 × 105 HkEc plus either cell media, or media with 2.5 μM monensin, 10 mM NH4Cl, or 100 μM chloroquine. Macrophages were lysed and run on 10% tricine PAGE gel followed by Western blotting. Membranes were incubated with goat anti-TNF (R&D Systems) 1:1,500 overnight at 4°C. After washing, the membranes were incubated with anti–goat-HRP (Vector Laboratories) at 1:2,000 for 1 h at room temperature, washed again, and then visualized with ECL plus detection reagent (GE Healthcare). All gels were subjected to ImageJ software analysis and normalized to actin.
Online supplemental material.
Fig. S1 shows bacterial clearance in CD and HC subjects subdivided according to disease phenotype and CARD15 genotype. Fig. S2 shows macrophage cytokine release after 24-h stimulation with TLR2 and TLR4 ligands. Fig. S3 depicts macrophage phenotype analysis. Fig. S4 shows functional groupings of the most significantly up-regulated and down-regulated genes in macrophages stimulated with HkEc. Table S1 shows cytokine mRNA levels after HkEc stimulation. Table S2 depicts differentially expressed genes in ileal CD macrophages stimulated with HkEc. Table S3 depicts differentially expressed genes in colonic CD macrophages stimulated with HkEc. Table S4 provides demographics of subjects used in the injection studies. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091233/DC1.
Acknowledgments
We thank Sara McCartney and Louise Langmead for allowing us to study patients under their care, Paul Cutler, Kevin Lee, Andy Calver, and Annegret Pelchen-Matthews for helpful discussions, Mike Wren for microbiological assistance, Wendy Waddington and Peter Ell for assistance with radiation studies, Jane Tempero for technical assistance, and all volunteers for participating in these studies.
This work was supported by the Wellcome Trust and The Broad Medical Research Program.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: Bref-A, brefeldin A; CD, Crohn's disease; cDNA, complementary DNA; DSS, dextran sodium sulfate; FDR, false discovery rate; GWAS, genome-wide association studies; HC, healthy control; HkEc, heat-killed E. coli; mRNA, messenger RNA; UC, ulcerative colitis.
References
- Aggarwal B.B., Kohr W.J., Hass P.E., Moffat B., Spencer S.A., Henzel W.J., Bringman T.S., Nedwin G.E., Goeddel D.V., Harkins R.N. 1985. Human tumor necrosis factor. Production, purification, and characterization.J. Biol. Chem. 260:2345–2354 [PubMed] [Google Scholar]
- Ambrose N.S., Alexander-Williams J., Keighley M.R. 1984a. Audit of sepsis in operations for inflammatory bowel disease.Dis. Colon Rectum. 27:602–604 [DOI] [PubMed] [Google Scholar]
- Ambrose N.S., Johnson M., Burdon D.W., Keighley M.R. 1984b. Incidence of pathogenic bacteria from mesenteric lymph nodes and ileal serosa during Crohn's disease surgery.Br. J. Surg. 71:623–625 [DOI] [PubMed] [Google Scholar]
- Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. ; NIDDK IBD Genetics Consortium; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease.Nat. Genet. 40:955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni F., Beutler B. 1996. The tumor necrosis factor ligand and receptor families.N. Engl. J. Med. 334:1717–1725 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate - a practical and powerful approach to multiple testing.Journal of the Royal Statistical Society, Series B. 57:289–300 [Google Scholar]
- Burczynski M.E., Peterson R.L., Twine N.C., Zuberek K.A., Brodeur B.J., Casciotti L., Maganti V., Reddy P.S., Strahs A., Immermann F., et al. 2006. Molecular classification of Crohn's disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells.J. Mol. Diagn. 8:51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K., Liu J.Y., Brown S.L., Miyoshi H., Loh J., Lennerz J.K., Kishi C., Kc W., Carrero J.A., Hunt S., et al. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells.Nature. 456:259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J.L., Hamilton S.R., Coleman J., Sitzmann J.V., Bayless T.M. 1992. Patterns of ileal recurrence in Crohn's disease. A prospective randomized study.Ann. Surg. 215:546–551, discussion :551–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright G.E., Athens J.W., Wintrobe M.M. 1964. The kinetics of granulopoiesis in normal man.Blood. 24:780–803 [PubMed] [Google Scholar]
- Castronovo F.P., Jr., McKusick K.A., Strauss H.W. 1988. The infiltrated radiopharmaceutical injection: dosimetric considerations.Eur. J. Nucl. Med. 14:93–97 [DOI] [PubMed] [Google Scholar]
- Charach G., Grosskopf I., Weintraub M. 2008. Development of Crohn's disease in a patient with multiple sclerosis treated with copaxone.Digestion. 77:198–200 [DOI] [PubMed] [Google Scholar]
- Costello C.M., Mah N., Häsler R., Rosenstiel P., Waetzig G.H., Hahn A., Lu T., Gurbuz Y., Nikolaus S., Albrecht M., et al. 2005. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays.PLoS Med. 2:e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert A.P., Fisher S.A., Mirza M.M., King K., Hampe J., Croucher P.J., Mascheretti S., Sanderson J., Forbes A., Mansfield J., et al. 2002. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease.Gastroenterology. 122:867–874 [DOI] [PubMed] [Google Scholar]
- Donnelly P. 2008. Progress and challenges in genome-wide association studies in humans.Nature. 456:728–731 [DOI] [PubMed] [Google Scholar]
- Ephgrave K. 2007. Extra-intestinal manifestations of Crohn's disease.Surg. Clin. North Am. 87:673–680 [DOI] [PubMed] [Google Scholar]
- Farthing M.J. 2004. Bugs and the gut: an unstable marriage.Best Pract. Res. Clin. Gastroenterol. 18:233–239 [DOI] [PubMed] [Google Scholar]
- Finegold S.M. 1969. Intestinal bacteria. The role they play in normal physiology, pathologic physiology, and infection.Calif. Med. 110:455–459 [PMC free article] [PubMed] [Google Scholar]
- Fujishima S., Hoffman A.R., Vu T., Kim K.J., Zheng H., Daniel D., Kim Y., Wallace E.F., Larrick J.W., Raffin T.A. 1993. Regulation of neutrophil interleukin 8 gene expression and protein secretion by LPS, TNF-alpha, and IL-1 beta.J. Cell. Physiol. 154:478–485 [DOI] [PubMed] [Google Scholar]
- Greten F.R., Arkan M.C., Bollrath J., Hsu L.C., Goode J., Miething C., Göktuna S.I., Neuenhahn M., Fierer J., Paxian S., et al. 2007. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta.Cell. 130:918–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F.M., Briggs J., et al. 2007. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1.Nat. Genet. 39:207–211 [DOI] [PubMed] [Google Scholar]
- Harvey R.F., Bradshaw J.M. 1980. A simple index of Crohn's-disease activity.Lancet. 315:514. [DOI] [PubMed] [Google Scholar]
- Heber S., Sick B. 2006. Quality assessment of Affymetrix GeneChip data.OMICS. 10:358–368 [DOI] [PubMed] [Google Scholar]
- Horwood N.J., Mahon T., McDaid J.P., Campbell J., Mano H., Brennan F.M., Webster D., Foxwell B.M. 2003. Bruton's tyrosine kinase is required for lipopolysaccharide-induced tumor necrosis factor α production.J. Exp. Med. 197:1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L. 2005. Consistent across-tissue signatures of differential gene expression in Crohn's disease.Immunogenetics. 57:709–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cézard J.P., Belaiche J., Almer S., Tysk C., O'Morain C.A., Gassull M., et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease.Nature. 411:599–603 [DOI] [PubMed] [Google Scholar]
- Hugot J.P., Zaccaria I., Cavanaugh J., Yang H., Vermeire S., Lappalainen M., Schreiber S., Annese V., Jewell D.P., Fowler E.V., et al. ; for the IBD International Genetics Consortium 2007. Prevalence of CARD15/NOD2 mutations in Caucasian healthy people.Am. J. Gastroenterol. 102:1259–1267 [DOI] [PubMed] [Google Scholar]
- Issekutz T.B., Issekutz A.C., Movat H.Z. 1981. The in vivo quantitation and kinetics of monocyte migration into acute inflammatory tissue.Am. J. Pathol. 103:47–55 [PMC free article] [PubMed] [Google Scholar]
- Jordan B. 1970. Use of 33P as an indicator for 32P pulse labeling of nucleic acids in bacterial cultures.J. Bacteriol. 101:657–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan G.G., Pedersen B.V., Andersson R.E., Sands B.E., Korzenik J., Frisch M. 2007. The risk of developing Crohn's disease after an appendectomy: a population-based cohort study in Sweden and Denmark.Gut. 56:1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle J. 1980. Urinary complications of Crohn's disease.World J. Surg. 4:153–160 [DOI] [PubMed] [Google Scholar]
- Lawrance I.C., Fiocchi C., Chakravarti S. 2001. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes.Hum. Mol. Genet. 10:445–456 [DOI] [PubMed] [Google Scholar]
- Levine B., Deretic V. 2007. Unveiling the roles of autophagy in innate and adaptive immunity.Nat. Rev. Immunol. 7:767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Karlin A., Loike J.D., Silverstein S.C. 2004. Determination of the critical concentration of neutrophils required to block bacterial growth in tissues.J. Exp. Med. 200:613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., van Kruiningen H.J., West A.B., Cartun R.W., Cortot A., Colombel J.F. 1995. Immunocytochemical evidence of Listeria, Escherichia coli, and Streptococcus antigens in Crohn's disease.Gastroenterology. 108:1396–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. 2008. Personal genomes: The case of the missing heritability.Nature. 456:18–21 [DOI] [PubMed] [Google Scholar]
- Malbert C.H. 2005. The ileocolonic sphincter.Neurogastroenterol. Motil. 17:41–49 [DOI] [PubMed] [Google Scholar]
- Mannick E.E., Bonomolo J.C., Horswell R., Lentz J.J., Serrano M.S., Zapata-Velandia A., Gastanaduy M., Himel J.L., Rose S.L., Udall J.N., Jr, et al. 2004. Gene expression in mononuclear cells from patients with inflammatory bowel disease.Clin. Immunol. 112:247–257 [DOI] [PubMed] [Google Scholar]
- Marks D.J., Segal A.W. 2008. Innate immunity in inflammatory bowel disease: a disease hypothesis.J. Pathol. 214:260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks D.J., Harbord M.W., MacAllister R., Rahman F.Z., Young J., Al-Lazikani B., Lees W., Novelli M., Bloom S., Segal A.W. 2006. Defective acute inflammation in Crohn's disease: a clinical investigation.Lancet. 367:668–678 [DOI] [PubMed] [Google Scholar]
- Meconi S., Vercellone A., Levillain F., Payré B., Al Saati T., Capilla F., Desreumaux P., Darfeuille-Michaud A., Altare F. 2007. Adherent-invasive Escherichia coli isolated from Crohn's disease patients induce granulomas in vitro.Cell. Microbiol. 9:1252–1261 [DOI] [PubMed] [Google Scholar]
- Medzhitov R. 2008. Origin and physiological roles of inflammation.Nature. 454:428–435 [DOI] [PubMed] [Google Scholar]
- Mitchell D.N. 1971. The Kveim test in Crohn's disease.Proc. R. Soc. Med. 64:164–166 [PMC free article] [PubMed] [Google Scholar]
- Moráin C.O., Segal A.A., Walker D., Levi A.J. 1981. Abnormalities of neutrophil function do not cause the migration defect in Crohn's disease.Gut. 22:817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Takagi T., Handa O., Ishikawa T., Nakagawa S., Yamaguchi T., Yoshida N., Minami M., Kita M., Imanishi J., Yoshikawa T. 2003. Enhanced intestinal inflammation induced by dextran sulfate sodium in tumor necrosis factor-alpha deficient mice.J. Gastroenterol. Hepatol. 18:560–569 [DOI] [PubMed] [Google Scholar]
- Ogura Y., Bonen D.K., Inohara N., Nicolae D.L., Chen F.F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R.H., et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease.Nature. 411:603–606 [DOI] [PubMed] [Google Scholar]
- Palmer C.D., Mutch B.E., Workman S., McDaid J.P., Horwood N.J., Foxwell B.M. 2008. Bmx tyrosine kinase regulates TLR4-induced IL-6 production in human macrophages independently of p38 MAPK and NFkappB activity.Blood. 111:1781–1788 [DOI] [PubMed] [Google Scholar]
- Parkes M., Barrett J.C., Prescott N.J., Tremelling M., Anderson C.A., Fisher S.A., Roberts R.G., Nimmo E.R., Cummings F.R., Soars D., et al. ; Wellcome Trust Case Control Consortium 2007. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility.Nat. Genet. 39:830–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman F.Z., Marks D.J., Hayee B.H., Smith A.M., Bloom S.L., Segal A.W. 2008. Phagocyte dysfunction and inflammatory bowel disease.Inflamm. Bowel Dis. 14:1443–1452 [DOI] [PubMed] [Google Scholar]
- Rhodes J.M. 2007. The role of Escherichia coli in inflammatory bowel disease.Gut. 56:610–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. 2007. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis.Nat. Genet. 39:596–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P., Kelly R.G., Lee G., Collins J.K., O'Sullivan G.C., O'Connell J., Shanahan F. 2004. Bacterial DNA within granulomas of patients with Crohn's disease—detection by laser capture microdissection and PCR.Am. J. Gastroenterol. 99:1539–1543 [DOI] [PubMed] [Google Scholar]
- Schroeder K.W., Tremaine W.J., Ilstrup D.M. 1987. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study.N. Engl. J. Med. 317:1625–1629 [DOI] [PubMed] [Google Scholar]
- Segal A.W., Loewi G. 1976. Neutrophil dysfunction in Crohn's disease.Lancet. 308:219–221 [DOI] [PubMed] [Google Scholar]
- Segal A.W., Ensell J., Munro J.M., Sarner M. 1981. Indium-111 tagged leucocytes in the diagnosis of inflammatory bowel disease.Lancet. 318:230–232 [DOI] [PubMed] [Google Scholar]
- Smythies L.E., Maheshwari A., Clements R., Eckhoff D., Novak L., Vu H.L., Mosteller-Barnum L.M., Sellers M., Smith P.D. 2006. Mucosal IL-8 and TGF-beta recruit blood monocytes: evidence for cross-talk between the lamina propria stroma and myeloid cells.J. Leukoc. Biol. 80:492–499 [DOI] [PubMed] [Google Scholar]
- Stabin M.G. 1996. MIRDOSE: personal computer software for internal dose assessment in nuclear medicine.J. Nucl. Med. 37:538–546 [PubMed] [Google Scholar]
- van Deventer S.J. 1999. Review article: targeting TNF alpha as a key cytokine in the inflammatory processes of Crohn's disease—the mechanisms of action of infliximab.Aliment. Pharmacol. Ther. 13:3–8 [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls.Nature. 447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M., Ziring D., Korin Y., Desai S., Kim S., Lin J., Gjertson D., Braun J., Reed E., Singh R.R. 2008. TNFalpha blockade in human diseases: mechanisms and future directions.Clin. Immunol. 126:121–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier R.J., Podolsky D.K. 2007. Unravelling the pathogenesis of inflammatory bowel disease.Nature. 448:427–434 [DOI] [PubMed] [Google Scholar]
- Yazisiz V., Avci A.B., Erbasan F., Yildirim B., Terzioğlu E. 2008. Development of Crohn's disease following anti-tumour necrosis factor therapy (etanercept).Colorectal Dis. 10:953–954 [DOI] [PubMed] [Google Scholar]
- Yourassowsky E., Van der Linden M.P., Crokaert F. 1989. Correlation between growth curves and killing curves of Escherichia coli in the presence of fleroxacin and ampicillin.Chemotherapy. 35:423–430 [DOI] [PubMed] [Google Scholar]