Abstract
Protein splicing is a precise auto-catalytic process in which an intein excises itself from a precursor with the concomitant ligation of the flanking sequences. Protein splicing occurs through acid-base catalysis in which the ionization states of active site residues are crucial to the reaction mechanism. In inteins, several conserved histidines have been shown to play important roles in protein splicing, including the most conserved “B-block” histidine. In this study, we have combined NMR pKa determination with quantum mechanics/molecular mechanics (QM/MM) modeling to study engineered inteins from Mycobacterium tuberculosis (Mtu) RecA intein. We demonstrate a dramatic pKa shift for the invariant B-block histidine, the most conserved residue among inteins. The B-block histidine has a pKa of 7.3 ± 0.6 in a precursor and a pKa of < 3.5 in a spliced intein. The pKa values and QM/MM data suggest that the B-block histidine has a dual role in the acid-base catalysis of protein splicing. This histidine likely acts as a general base to initiate splicing with an acyl shift and then as a general acid to cause the breakdown of the scissile bond. The proposed pKa shift mechanism accounts for the biochemical data supporting the essential role for the B-block histidine and for the absolute sequence conservation of this residue.
Keywords: Intein, protein splicing, NMR, pKa, catalytic mechanism, QM/MM
Introduction
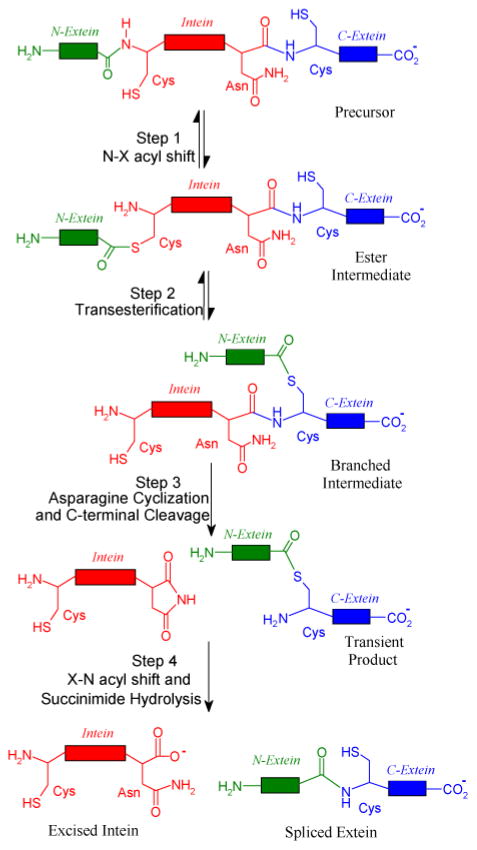
Protein splicing is an auto-catalytic process in which an in-frame protein fusion, called an intein, is excised from the precursor protein with the concomitant ligation of the two flanking polypeptides, the N- and C-exteins (Fig. 1).1 Described by Tom Muir as “Nature’s gift to the protein chemist”,2 inteins are widely used in protein engineering, protein labeling, protein purification and control of protein functions.3–6
Figure 1.
Four steps of protein splicing. During the four-step reaction, the intein (shown in red) is excised from the N-extein (shown in green) and C-extein (shown in blue), while the N-extein and C-exteins are ligated to form the mature protein.
The four steps of the protein splicing pathway have been well documented (Fig. 1).1,7 In the first step, N-X acyl shift (X=S or O), the side chain nucleophile of the first residue of intein (side chain S atom of a cysteine or O atom of a serine) attacks the carbonyl of the last residue of the N-extein,8,9 resulting in a linear ester intermediate. In the second step, transesterification, the nucleophile at the downstream splice junction attacks the linear ester. The N-extein is transferred to the side chain of the attacking nucleophile, forming a branched ester intermediate.10–13 In the third step, asparagine cyclization and C-terminal cleavage, the last residue of the intein, an asparagine, cyclizes. This is coupled to the cleavage of the branched ester and the release of the excised intein with an aminosuccinimide residue and the exteins joined by an ester bond.9,10,14,15 In the fourth step, X-N acyl shift and succinimide hydrolysis, the aminosuccinimide hydrolyzes and the ester linking the two exteins rearranges to form a peptide bond.14,16 For most of these steps, the mechanistic details are still lacking at the atomic level. For example, it is not known which residue activates the side chain nucleophile to initiate the N-X shift in the first step of splicing.
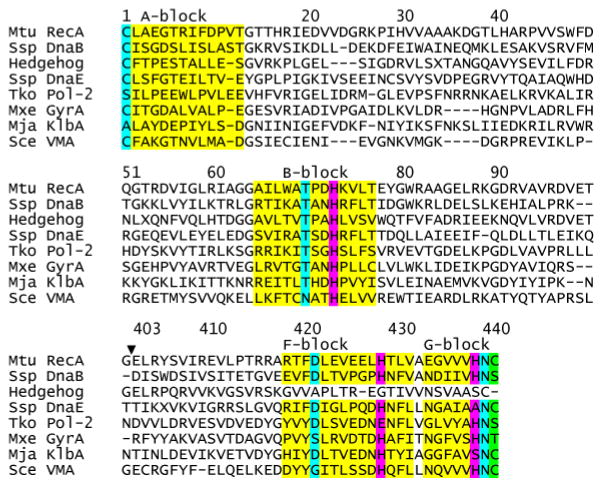
The intein sequence contains conserved sequence blocks (Fig. 2). Block A residues almost always start with a cysteine or a serine (Fig. 2), providing a nucleophile for the N-X acyl shift (Fig. 1). Block B contains the TXXH motif, where both T and H play important roles for the first step of protein splicing.17–19 The F block often contains an aspartate20 and a histidine that modulates C-terminal cleavage.21,22 The G block has a penultimate histidine and a C-terminal asparagine critical for C-terminal cleavage.9,10,14,15 The first residue of the C-extein is a C, S or T (Fig. 2), serving as the nucleophile for transesterification, the second step of the protein splicing reaction (Fig. 1).
Figure 2.
B-block histidine is the most conserved residue in the intein sequences. Structure-based multiple sequence alignment of ΔΔIhh-SM and other inteins was achieved the using DALI server. ΔΔIhh-SM is an engineered and minimized Mtu RecA intein (see text). The locations of the conserved blocks A, B, F, and G are indicated above the sequences and the corresponding residues are colored in yellow. The conserved histidines are colored in magenta while other key residues are colored in cyan. H73, H429, H439 are the conserved B-block, F-block and penultimate histidines, respectively. The first C-extein residue is colored in green. The gap (▼;) in the residue numbering for ΔΔIhh-SM results from the deletion of the endonuclease domain of Mtu RecA intein.
The B-block histidine is the most conserved residue in all intein sequences (Fig. 2).23,24 Mutagenesis studies have shown that the B-block histidine plays an essential role in splicing by catalyzing the N-X acyl shift.18,25,26 Crystal structures of inteins have shown that that the δ1 nitrogen of the B-block histidine is close to the amide nitrogen of the first residue of the intein, suggesting that the B-block histidine may promote N-X acyl shift by protonating the leaving group.7,17,19,27–30 The early crystal structure of the GyrA intein showed the scissile bond at the upstream splice junction to be in an unusual cis conformation, which may facilitate the N-X acyl shift by ground state destabilization.17 Consistent with this hypothesis, a clever NMR study revealed an unusually low 1JNC’ (12 Hz) of the N-terminal scissile bond 31 which reverted to a normal J-coupling with an alanine mutation of the B-block histidine. These results indicate that the B-block histidine may destabilize the ground state in a precursor. However, later crystal structures demonstrated a variety of conformations for the scissile bond, ranging from trans 19,22,29,32 to distorted trans conformation.28 Thus, the precise catalytic role of the invariant B-block histidine remains to be defined.
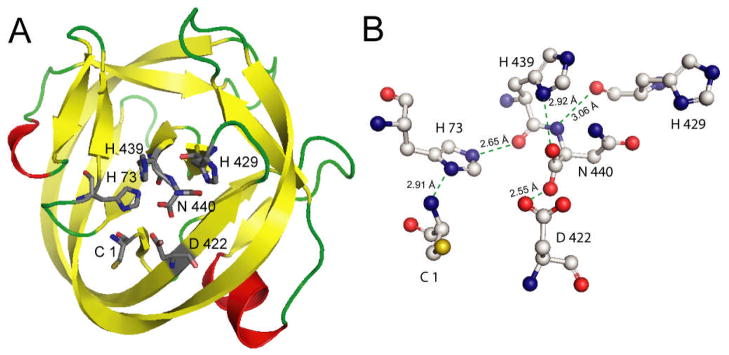
The ionization states of active site residues are crucial to the mechanism of acid-base catalysis in enzymes. Many catalytic residues have an elevated or depressed pKa.33 Solution NMR is the ideal tool for site-specific pKa measurement in enzymes. Although several NMR studies of inteins have been published,31,34–35 the pKa values of conserved histidines have not been determined in any intein. In this study, NMR pKa measurement and quantum mechanics/molecular mechanics (QM/MM) modeling have been carried out for engineered and minimized (139 aa residues) Mtu RecA inteins, ΔΔIhh-SM (splicing mutant with a V67L mutation) and ΔΔIhh-CM (cleavage mutant with V67L/D422G mutations).20 These inteins have been minimized by the deletion of the dispensable endonuclease domain20 and the replacement of a long, disordered loop with a short loop from the homologous hedgehog (hh) protein.36 The V67L mutation globally promotes the splicing reaction, while the additional D422G mutation enhances C-terminal cleavage.20 X-ray crystallography studies of these inteins demonstrate a typical HINT (hedgehog intein) horseshoe fold and close proximity of active site residues (Fig. 3).30
Figure 3.
Crystal structure of a minimized Mtu RecA intein, ΔΔIhh-SM (pdb code 2IN0). A. The overall horseshoe fold of HINT domains. Conserved residues, C1, H73, D422, H429, H439 and N440, are shown in stick representation. B. Hydrogen bond network between conserved residues.
In this study, we show that in intein-mediated protein splicing, the most conserved residue, the B-block histidine, experiences a large pKa shift and has a strikingly low pKa in the spliced intein. The pKa values of the B-block histidine and QM/MM modeling indicate that this histidine likely acts first as a general base with a high pKa then as a general acid with a low pKa in the first step of protein splicing.
Materials and Methods
In vivo splicing and cleavage assay
E. coli JM109 cells transformed with pMIC which encodes a fusion protein of maltose binding protein (MBP, M), ΔΔIhh-SM (I) and a small C-extein (C-terminal domain of I-TevI) were grown in 6 ml cultures to an OD600 of 0.5 −0.6 at 37 °C in Luria Bertani (LB) medium. Isopropyl β-D-thiogalactoside (IPTG) was added to a final concentration of 1 mM and the growth was continued for 3 h during which the expressed fusion proteins underwent splicing, C-terminal and/or N-terminal cleavage (Fig. S1). Cells were collected by centrifugation and lysed. The supernatant from the lysate was run on SDS-PAGE to assess the efficiency of protein splicing, C- and/or N-terminal cleavage based on the amount of the spliced and cleaved products. These products have been confirmed by Western blot with respective antibodies.30
Protein overexpression, purification and NMR sample preparation
NMR samples of ΔΔ Ihh-SM, ΔΔIhh-CM and ΔΔIhh-CM with a V67L mutation were prepared as described previously.30,37 The NI intein precursor is comprised of the intein itself (ΔΔIhh-SM, 139 residues, D24G, C1A), flanked by 17 N-extein residues (MAHHHHHHVGTGSNADP). The C1A mutation was introduced to prevent N-terminal cleavage. The NI precursor was expressed in the E. coli strain ER2566. Cells were grown at 37°C and induced with 1 mM IPTG at an OD600 of 0.3–0.4 at 20 °C, then grown for another 20–24 hours at 20 °C. Cell pellets were lysed by sonication in 50 mM Tris buffer, pH 8.0, with 0.5 M NaCl and 20 mM imidazole, and purified by affinity chromatography on Ni-NTA agarose (GE Healthcare) followed by gel filtration chromatography on a HiLoad 16/60 Superdex 200 preparation grade column (GE Healthcare). Isotope labeling was accomplished by growing cultures in M9 minimal medium containing either 1 g/L 15NH4Cl for a uniformly 15N-labeled sample, or 1 g/L 15NH4Cl and 1 g/L 13C6-D-glucose (Cambridge Isotope Laboratories) for a uniformly 15N,13C-labeled sample. Both procedures yielded ~20 mg of pure protein from 1 L of culture. The pure protein was concentrated and exchanged by ultrafiltration with Amicon Ultra Centrigugal Filter Devices (Millipore Cooperation) (10-kDa molecular weight cutoff) into 50 mM sodium phosphate buffer, pH 7.0, with 100 mM NaCl and 1 mM NaN3. The final protein concentration for NI was 1 mM with a volume of 500 μl.
NMR resonance assignment
All NMR experiments were carried out at 25 °C on either a Bruker 800 MHz (1H) or 600 MHz (1H) spectrometer, each equipped with a triple-resonance cryogenic probe. The complete assignment of ΔΔIhh-CM has been published37 and the assignment of ΔΔIhh-SM have been carried out in a similar manner. The assignments of histidines are based on the (Hβ)Cβ(CγCδ)Hδ spectrum (See supporting material Fig. S2-4), which connects backbone Cβ to Hδ in the imidazole ring.38
pH titration
For pH titration, we started with a pH 7 sample, which was split into two, one for pH titration in the range of 7 to 10.4 and the other for pH titration from 7 to 3.5. The pH values were measured with a Accumet XL25 pH-meter (Fischer Scientific, Fair Lawn, NJ, USA) equipped with an NMR electrode, calibrated with pH 4.0, 7.0, and 10.0 standard solutions (Fischer Scientific, Fair Lawn, NJ, USA). The pHs of the samples were adjusted with 0.1 or 0.2 M HCl or NaOH. 1H-15N HMQC spectra optimized for observing long range H-15N correlation39 were acquired with 2048×288 complex data points, spectral widths of 100 ppm in 15N and 16 ppm in 1H and 128 scans. The spectrum center was set as 200.0 ppm for 15N and 4.76 ppm for 1H. The observed 15N chemical shifts of the histidine imidazole ring, provided that the full titration curves were available, were plotted against pH and fit with nonlinear least-square regression analysis according to the Henderson-Hasselblach equations using the software Origin® (Microcal Origain, Northampton, MA, USA). The Henderson-Hasselbalch equation using a Hill parameter (nH) to account for nonideality is given by:
| (1) |
where δobs, δHA, and δA – are the chemical shifts for the observed, protonated, and deprotonated species, respectively. The pKa values for ΔΔIhh-CM and NI were determined in 100% D2O samples while pKa values for ΔΔIhh-SM were determined in 10% D2O samples. The pKa values were corrected for D2O according to Krezel et al.40
pKa estimation with limited number of titration points
The average chemical shift of histidine ring nitrogens is a good indication of its ionization state.41–43 When obtaining the full titration curve was not practical, the pKa of histidines in NI was estimated by using the following equation when the neutral histidine is in fast tautomer exchange:
| (2) |
where Ãobs = ½(δδ(obs)+ δε(obs)), δδ(obs) and δε(obs) being the 15N chemical shifts of histidine Nδ1 and Nε2 atoms measured by HMQC NMR spectrum at individual pH values; ÃH = ½(δδ + δεH) = ½(δε + δδH), δδ, δε, δδH, and δεH are the 15N chemical shifts of histidine (with either δ or ε position protonated) when the imidazole ring is neutral; ÃH+ = ½(δδH+ + δεH+), δδH+ and δεH+ is the 15N chemical shifts of histidine with both δ and ε position protonated in a positively charged imidazole ring. The derivation of the equation and details of pKa estimation are provided in the supporting information.
QM/MM modeling of reaction mechanism
The Mtu recA intein splicing mutant (SM) crystal structure (PDB code 2IMZ)30 was computationally appended to include both N- and C-exteins. The N-extein sequence consisted of Acetyl-VVKNK and the C-extein sequence consisted of CSPPF-N-methyl, both based on the native extein sequence. AMBER force field parameters44 were implemented with GROMACS code.45 This system was solvated with water molecules and equilibrated with classical molecular dynamics simulations with exteins present. MD simulations were carried out for 4 ns (0.5 ns equilibration, 3.5 ns production run) with temperature T = 298 K, pressure = 1 bar.
To test the N-S acyl shift and H73 acid/base behavior, multi-scale methods were implemented. Specifically, hybrid quantum mechanical and molecular mechanical (QM/MM) calculations46,47 were performed with Gaussian code,48 using the B3LYP functional49 with 6-31G(d,p) basis sets50 for the quantum region and the AMBER force field parameters for the classical region. This multi-scale method allows for bond breakage/formation within the QM active site while the remaining protein and solvent creates the structural backbone based on the folded protein and the electrostatic environment based on classical point charges. The protein system consisted of 2351 atoms and there were 1321 water molecules present, for a system total of 6314 atoms. The quantum mechanical region is comprised of the following residues: The N-terminal active site is based on the N-extein residue K(−1) and the intein residues C1-L2. Also included are T70, H73 and D422. From the G block and C-extein (C-terminal active site): V438-H439-N440-C(+1)-S(+2). At least three vicinal water molecules are included in the active site QM region. For non-mechanistic residues, those that affect the QM system via polarization, the entire amino acid may not be chosen for inclusion in the QM system: an example of such a residue is T70, where only the side chain is included. Similar density functional theory (DFT) methods have been tested in our own work51 and that of others52 and shown to provide accurate energetic and structural results.
Results and Discussion
B-block histidine H73 is essential for protein splicing in ΔΔIhh-SM
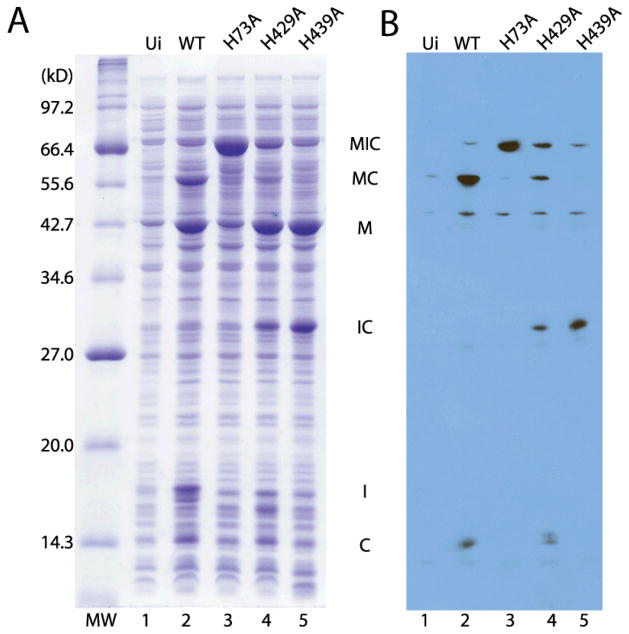
To show the functional significance of the B-block histidine H73 in minimized and engineered intein ΔΔIhh-SM, we tested the effect of an H73A mutation on splicing using an in vivo splicing assay. E. coli cells were induced to express an intein precursor MIC, composed of maltose binding protein (MBP) as N-extein, ΔΔIhh-SM intein and a short C-extein. The precursor underwent splicing, N- and C-terminal cleavage (Fig. S1) within the cells and the extent of splicing was assessed with SDS-PAGE and Western blot using an antibody against the C-extein (Fig. 4). With the WT ΔΔIhh-SM as the intein (H73), ligated exteins (MC) and large amount of I (intein) was generated by protein splicing and there was little pre-splicing precursor MIC remaining. Products of N-terminal cleavage (M and IC) and C-terminal cleavage (C) were also observed. Thus the engineered and minimized intein ΔΔIhh-SM can efficiently catalyze protein splicing and is a good model system for exploring the structural mechanism of protein splicing at atomic resolution.
Figure 4.
Inhibition of protein splicing by mutations of conserved histidines (B-block histidine H73A, F-block histidine H429A and penultimate histidine H439A). A. SDS-PAGE of in vivo splicing assay.B. Western blot with an anti-C-extein antibody. MW=Molecular weight marker; Ui=Uninduced; MIC=MBP-intein-C-extein fusion (71.5 kD); MC = MBP-C-extein fusion (56.1 kD); M=MBP (43.0 kD); IC-intein-C-extein fusion (28.6 kD) I =intein (15.4 kD).
Next we examined the effect of alanine mutation of the conserved histidines. In contrast to the wild type, in the H73A mutant, the splice products were completely absent and there was accumulation of large amount of MIC precursor (Fig. 4), indicating an essential role for B-block histidine in protein splicing. H73A also caused dramatic reduction in N-terminal cleavage product (M) (Fig. 4 lane 2 and 3), consistent with the notion that the B-block histidine plays a critical role in N-X acyl shift.18,25,26
The H429A mutation greatly reduced the amount of splice product (Fig., 4B lane 2 and 4), indicating an important but nonessential role for the F-block histidine in splicing. Large amount of N-terminal cleavage product M was still produced with H429A, suggesting F-block histidine does not contribute critically to N-terminal cleavage. The H429A mutation caused a reduction in the amount of C-terminal cleavage product C compared with WT, suggesting F-block histidine likely plays a role in C-terminal cleavage (Fig. 4B).
No splice product MC was generated with the H439A mutation, demonstrating the essential role of the penultimate histidine in protein splicing (Fig. 4A & B, lane 5). However, very little precursor protein MIC was observed with the H439A mutation. While large amount of N-terminal cleavage product M was produced, no C-terminal cleavage product C was generated. Significant amount of IC was accumulated without undergoing further C-terminal cleavage. These observations indicated that H439A blocked C-terminal cleavage.
Thus, not only is the B-block histidine the most conserved among three histidines, but also its mutation (H73A) causes the most dramatic inhibition of protein splicing and cleavage reactions.
B-block histidine has a strikingly low pKa in spliced intein product
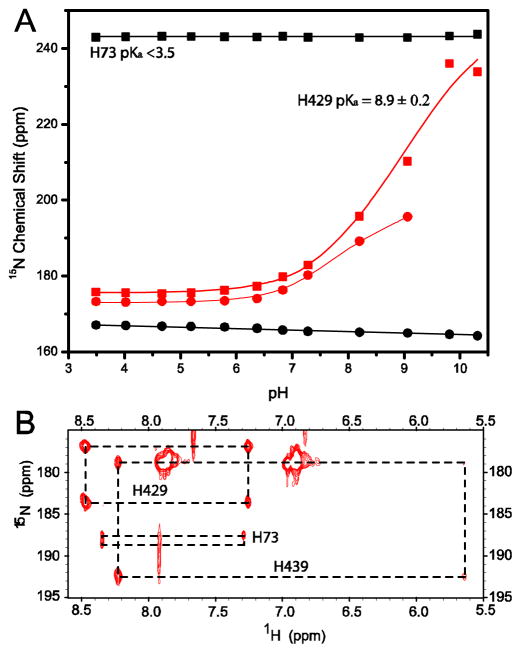
To explore the mechanistic role of the B-block histidine in protein splicing, we examined the ionization state of H73 in the spliced intein product ΔΔIhh-SM. Aromatic nitrogen resonances of H73 did not titrate in the experimental pH range of 3.5 to 10 (Fig. 5A), indicating that the pKa of H73 is significantly less than 3.5 and that H73 is a relatively strong acid in the intein product. It is known that acidic conditions favor the N-S (N-O) acyl shift in peptides.53 H73 may therefore facilitate the N-X acyl shift in the first step of protein splicing as a general acid to protonate the leaving nitrogen group. This catalytic role for the B-block histidine has been well-recognized7 and is supported by the short distance between the H73 ring and C1 amide nitrogen in the crystal structures of the Mtu RecA (Fig. 3) and other inteins.17,19,27–29 The depressed pKa for the B-block histidine is consistent with the essential role that this residue plays in the breakdown of the scissile bond at the N-terminal splicing junction, by acting as a general acid (Fig. 6).
Figure 5.
pKa determination of conserved histidines in spliced intein (A) and in a precursor with an N-extein, NI (B). (A) Plot of the ring 15N chemical shift versus pH for conserved histidines, B-block H73, F-block H429 in ΔΔIhh-SM. The squares and circles represent the chemical shifts of Nδ1 and Nε2, respectively. The pKa of H439 in the spliced intein is shown in Table I, along with pKas of other histidines in different constructs. (B) HMQC spectrum for intein precursor NI at pH 7.1. The average chemical shift of of H73 Nδ1 and Nε2 is 188 ppm, midway between the values expected for neutral (204.3 ppm) and charged histidines (176 ppm), suggesting that H73 has a pKa near 7 in the precursor protein.
Figure 6.
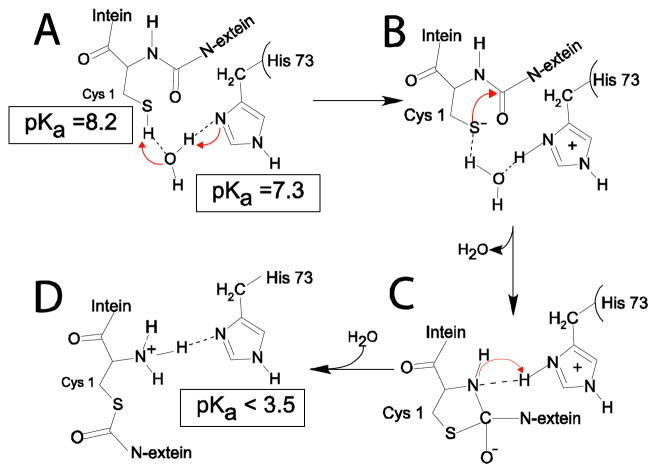
A pKa shift mechanism for B-block histidine H73 in the N-S acyl shift, the first step of protein splicing for the Mtu RecA intein. H73 acts as a general base to deprotonate the C1 thiol, initiating the NS acyl shift. Then H73 serves as a general acid to protonate the leaving nitrogen, breaking the scissile bond and completing the N-S acyl shift. The arrows indicate the routes of electron transfer and broken lines indicate hydrogen bonds. Intermediates A-D are described in the text.
Two factors could contribute to the low pKa of H73 in ΔΔIhh-SM: First, since H73 is 97% buried from solvent (calculated by the program molmol54), the energetic cost of a charged histidine ring could be high due to the low dielectric constant in the interior of a protein. However, we have evidence that the low pKa of H73 is not due to the inaccessibility of solvent. Although H73 appears buried in the static crystal structure, the hydrogen deuterium exchange was complete within minutes for the H73 backbone amide and the ε2 protons. Therefore the H73 ring has adequate access to solvent pH conditions through protein dynamics. Second, since H73 is proximal to the positively charged N-terminus (2.9 A between H73 Nδ1 and C1 amide N in the crystal structure; Fig. 3), a positively charged histidine ring will cause an unfavorable electrostatic interaction. This second factor lead us to hypothesize that in the absence of a nearby charged N-terminus in precursors with an N-extein, H73 is more likely to become protonated and should have a higher pKa. Thus, in a precursor, the higher pKa may enable the B-block histidine to act as a base, to deprotonate the C1 thiol group to initiate the first step of splicing (Fig. 6). We call this the pKa shift hypothesis in which the B-block histidine acts as a general base first to initiate N-X acyl shift and then serves as a general acid to complete the N-X acyl shift (Fig. 6).
B-block histidine has a pKa of 7.3 in pre-splicing precursor
To test our pKa shift hypothesis, we estimated the pKa of H73 in an intein precursor NI, composed of a short N-extein and ΔΔIhh-SM sequence with a C1A mutation (Fig. 5B) to prevent N-terminal cleavage. It is possible that the C1A mutation might modify H73 pKa in NI. However, as noted above, C1 influenced the pKa of H73 mainly through its amide group. Therefore mutating C1 side chain was unlikely to dramatically change H73 pKa in the precursor. In NI, full pH titration was not possible because NI precipitated at pH > 8 and aggregated at pH < 4 which resulted in severe line-broadening. When the histidine tautomer exchange is fast, the average chemical shift of Nδ1 and Nε2 reflects the effect of the pKa equilibrium.41–43 We therefore estimated the pKa of H73 in the precursor using the following equation:
where à is the average Nδ1 and Nε2 chemical shift measured from HQMC at a pH value close to pKa; ÃH and ÃH+ are the average Nδ1 and Nε2 chemical shift of the neutral imidazole ring and of the charged imidazolium ring, respectively. ÃH was calculated from the data in Fig. 5A to be 204.3 ppm, while ÃH+ was taken from the literature as 176 ppm.55 This method for pKa estimation has been validated using the pKa titration data in ΔΔIhh-SM where full titration curves are available (see supporting information). Using this method, we obtained a pKa value of 7.3 ± 0.6 for H73 in NI precursor. This result demonstrates a dramatic pKa shift, of at least 4 units, for the B-block histidine during protein splicing.
pKa shift mechanism for B-block histidine in protein splicing
The pKa value of the C1 thiol group has been determined by Paulus et al. to be 8.2,56 within 1 pH unit of the pKa values of H73 (7.3) in an intein precursor. Thus a proton transfer from the thiol group to the imidazole ring can occur at a reasonable rate.57 Given the proximity of their sidechains to the C1 thiol group (within 6 A) in the crystal structure of ΔΔIhh-SM, D422 and H73 are the most likely candidates to act as a general base to activate the C1 thiol group. However, the D422 sidechain is most likely to have a pKa near 4 in the intein precursor and would be much less efficient as a general base compared with H73 because of the large pKa difference between D422 and C1. Indeed, various mutations of D422 can still mediate N-terminal cleavage,58 suggesting D422 is not essential for the N-S acyl shift. In addition, D422 is not as highly conserved as the B-block histidine (Fig. 2). Thus, in the intein precursor, H73 is most likely the general base to deprotonate the C1 thiol to initiate the first step of splicing, the NS acyl shift (Fig. 6). The distance between H73 and C1 is 5.7 A in the crystal structure, too great for direct proton transfer. However, the proton transfer can occur through an intermediate water molecule. Indeed, a water molecule forming hydrogen bonds with both the C1 thiol group and the H73 ring nitrogen has been observed in our simulation with a precursor based on the crystal structure of ΔI-SM (Fig. 6A). However, such a water molecule has not been observed in any crystal structures of intein precursors. This could be due to mutations such as C1S or H73N in the precursor sequence used for crystallization or the intrinsic low sensitivity for observing water molecules by crystallography. Alternatively, a conformational change may occur in the intein precursor to bring the H73 δ1 nitrogen close to the C1 thiol group for a direct activation.
After the proton transfer between the C1 thiol group and H73, the positively charged H73 ring stabilizes the negatively charged C1 thiolate and oxythiazolidine intermediate (Fig. 6B). H73 will then protonate the nascent N-terminal amide group, causing the breakdown of the scissile peptide bond (Fig. 6C). An additional proton from water generates a positive charge at the intein N-terminus and lowers the pKa of H73 to below 3.5, stabilizing the endproduct of N-S acyl shift (Fig. 6D). In this pKa shift mechanism, H73 acts both as the catalytic base and acid for the N-S acyl shift, supported by the large pKa shift observed here. Similar pKa shifts associated with a dual role of a catalytic residue in acid/base catalysis have been observed in β-glucosidase.59
QM/MM modeling supports the pKa shift mechanism of the B-block histidine
Combined QM/MM modeling was applied to further evaluate the pKa shift mechanism in protein splicing. QM/MM is the method of choice for modeling chemical reactions catalyzed by enzymes.60 QM is required to describe the electron and proton transfers at the active site. However, the properties of the protein scaffold and solvent have a strong effect on enzyme reaction mechanism. For example, the pKa of a well-solvated histidine on the protein surface will be different from the pKa of a histidine buried in the hydrophobic core. Therefore atoms outside of the active site are modeled with classical MM. Atomic level modeling of the auto-catalytic splicing reaction of inteins involves understanding the role of protons as a function of tertiary structure. The protein backbone is semi-rigid, and with multiscale quantum and classical modeling (QM/MM), the structural and electrostatic environment can be a perturbation on the quantum mechanical active site. Currently full-protein QM simulations are computationally unfeasible, and with QM/MM multiscale methods we can model the position of protons and their catalytic role, which is intimately related to the pKa measurements presented in this work.
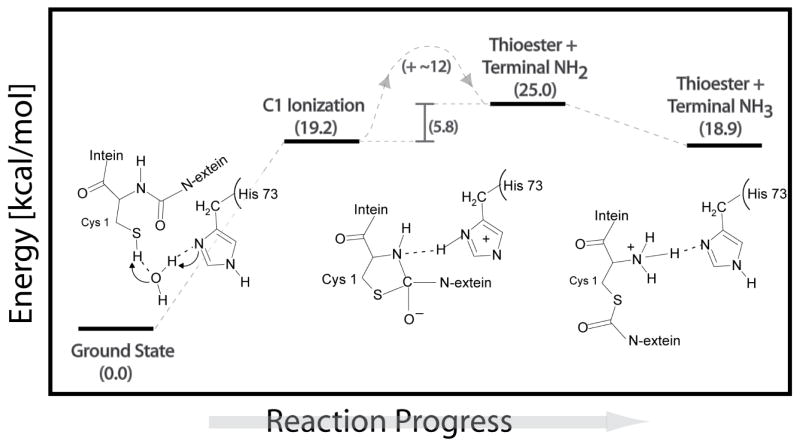
Multiscale QM/MM was used to create an energy profile for C1 ionization, N-S acyl shift, and for protonation of the nascent N-terminal amide (Fig. 7). The QM/MM modeling of the intein’s N-terminal rearrangement reaction corroborates the dual catalytic role of H73. H73 was first used as a catalytic base, removing a proton from the C1 thiol group via water, and then as an acid, by protonating the leaving nitrogen. The ionization energy for C1 by H73 via water was ~19.2 kcal/mol, comparable to the energy barrier of C-terminal cleavage in inteins.51,61 Once the proton has migrated from C1 to H73, the C1 thiolate group is activated to attack the peptide carbonyl carbon, forming a transitory ring structure, oxythiazolidine, with an energy 12 kcal/mol higher than the ionized C1 state. The oxythiazolidine intermediate is highly activated for N-protonation and C-N bond breakage. The newly formed C-S bond caused a decrease in resonance at the peptide bond and an increased bond length between C and amide N, resulting in a more nucleophilic scissile bond amide N. The recently protonated H73 was in position to directly and spontaneously donate a proton to the scissile peptide bond nitrogen, without the need for a transitory water molecule. The energy of the thioester with an –NH2 terminal group was 25.0 kcal/mol, and it was determined to be a structurally stable intermediate. A second protonation then occurred at the N-terminus, via solvent, resulting in a positively charged –NH3+ group that was electrostatically coupled to the neutral H73 (with energy 18.9 kcal/mol). Indeed, the average pKa for a solvent exposed N-terminus ranges from 8.8 to 10.8.62 The positive N-terminus considered in QM/MM simulations caused H73 to be energetically unable to accept a second proton, consistent with the extreme reduction of the H73 pKa in the intein product observed experimentally. The QM/MM simulations of the C1 ionization, the N-S acyl shift, and the N-terminal thioester formation are fully consistent with the observed pKa values of H73 before and after N-S acyl shift.
Figure 7.
Reaction profile for N-S acyl shift derived from QM/MM simulations.
Mechanistic implications of pKa values of other conserved histidines
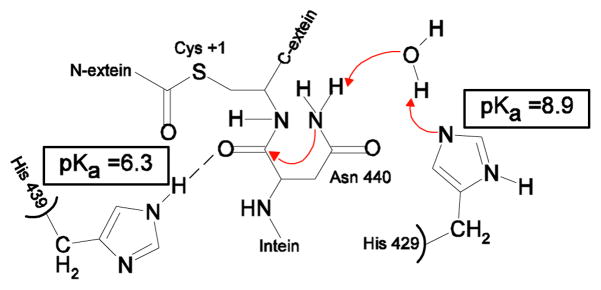
The conserved F-block histidine, H429 in Mtu recA, showed a higher than usual pKa in both the spliced product and precursor NI (Table I), which suggests that H429 may serve as a proton acceptor in the splicing reaction. Indeed, the F-block histidine was shown to play an important role in protein splicing, especially in C-terminal cleavage (Fig. 4).12 The crystal structure of ΔΔIhh-SM 30 showed that the imidazole ring of H429 can form a hydrogen bond with a water molecule, Wat15, which in turn has a hydrogen bond with the NH2 sidechain of N440. The F-block histidine may deprotonate the Nδ atom of N440 through this hydrogen bond network to activate the Nδ of N440, which then carries out the nucleophilic attack on the carbonyl of the scissile bond, resulting in asparagine cyclization and the breakdown of the scissile bond, as suggested by Sun et al. in Ssp DnaE intein.63 The pKa of H439 was determined to be ~6, indicating that the H439 ring is neutral at pH 7. H439 may act as a hydrogen bond donor to the scissile bond carbonyl, as shown by the crystal structure of ΔΔIhh-SM (Fig. 8).30
Table I.
pKa values of histidines in various intein constructs
| Histidine pKa | ΔΔIhh-SM | ΔΔIhh-CM | NI |
|---|---|---|---|
| B-block H73 | <3.5 | <4.1 | 7.3±0.6 |
| F-block H429 | 8.9±0.2 | 8.4±0.5 | 7.9±0.6 |
| G-block H439 | 6.2±0.2 | 6.2±0.2 | 7.3±0.6 |
| His 17 | 4.5±0.3 | 4.7±0.2 | 4.4±0.6 |
| His 30 | 7.1±0.2 | 6.6 ±0.4 | 6.6±0.6 |
| His 41 | 6.7±0.2 | 6.3±0.2 | 6.6±0.6 |
Figure 8.
Mechanistic role of the F-block histidine and penultimate histidine.
Histidine pKa values are similar in ΔΔIhh-SM and the cleavage mutant ΔΔIhh-CM (Table I); therefore the difference in splicing and cleavage activities between these two mutants is not due to the differences in the ionization state of histidines in the spliced product. As far as other histidines are concerned, H17 has a low pKa of ~4.5, likely because it is 89% buried from the solvent (calculated by molmol54 using the crystal structure of ΔΔIhh-SM). H30 and H41 have normal pKa values, which are the same in the precursor and the product. Thus, the nonconserved H17, H30 and H41 do not have distinctive roles in splicing, while the B-, F- and G-block histidines have important functions in intein catalysis with their characteristic pKa values.
Conclusions
In summary, we demonstrate a dramatic pKa shift for the B-block histidine, the most conserved intein residue, during protein splicing. The combined NMR and QM/MM data suggest that the B-block histidine has a dual role in acid-base catalysis of protein splicing. The B-block histidine likely acts as a general base to initiate N-X acyl shift then as a general acid to cause the breakdown of the scissile bond. The proposed pKa shift mechanism accounts for the absolute sequence conservation of the B-block histidine, and is in accord with biochemical data supporting the critical role of the B-block histidine in the N-X acyl shift and splicing. It is important to note that although this mechanism is plausible, other mechanisms may be at work in N-X acyl shift because The proposed mechanism will likely have important implications for the development of intein-based biotechnologies for protein purification, protein engineering and biosensing. Finally, it is important to note that proposed mechanism is likely one plausible mechanism for protein splicing rather than the only way in which splicing reaction operates.
Supplementary Material
Acknowledgments
We thank financial support from National Institute of Health (R01GM81408 to CW; R01GM44844 to MB), the National Science Foundation (CTS03-04055-NIRT to GB), and the Computational Center for Nanotechnology Innovations (CCNI) at RPI. P.T.S. acknowledges funding from the Interconnect Focus Center. Y.L. acknowledges the Cultivation Fund of the Key Scientific and Technical Innovation Project, Ministry of Education of China (No. 707036). B.P. acknowledges funding from NIH Biomolecular Science and Engineering training grant (GM067545). We would like to thank Shekhar Garde and Patrick Van Roey for helpful discussions.
Footnotes
Supporting Information Available. One table describing all the mutations used in this paper (Table S1), validation of the pKa determination using limited titration points (Table S2), complete Ref. 48, and 4 Figures (Protein splicing and its side reactions, assignment of histidine resonances in ΔΔIhh-SM, ΔΔIhh-CM, and NI) are provided, total 10 pages. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Paulus H. Chemical Society Reviews. 1998;27:375–386. [Google Scholar]
- 2.Belfort M. In: Homing Endonucleases and Inteins. Belfort M, Derbyshire V, Stoddard BL, Wood DW, editors. Vol. 16. Springer; Berlin Heidelberg: 2005. pp. 1–9. [Google Scholar]
- 3.Blaschke UK, Silberstein J, Muir TW. Methods Enzymol. 2000;328:478–96. doi: 10.1016/s0076-6879(00)28414-0. [DOI] [PubMed] [Google Scholar]
- 4.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. J Am Chem Soc. 2003;125:10561–9. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 5.Tyszkiewicz AB, Muir TW. Nat Methods. 2008 doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 6.Wood DW, Derbyshire V, Wu W, Chartrain M, Belfort M, Belfort G. Biotechnol Prog. 2000;16:1055–63. doi: 10.1021/bp0000858. [DOI] [PubMed] [Google Scholar]
- 7.Saleh L, Perler FB. The Chemical Record. 2006;6:183–193. doi: 10.1002/tcr.20082. [DOI] [PubMed] [Google Scholar]
- 8.Shao Y, Xu MQ, Paulus H. Biochemistry. 1996;35:3810–5. doi: 10.1021/bi952592h. [DOI] [PubMed] [Google Scholar]
- 9.Chong S, Shao Y, Paulus H, Benner J, Perler FB, Xu MQ. J Biol Chem. 1996;271:22159–68. doi: 10.1074/jbc.271.36.22159. [DOI] [PubMed] [Google Scholar]
- 10.Xu MQ, Comb DG, Paulus H, Noren CJ, Shao Y, Perler FB. Embo J. 1994;13:5517–22. doi: 10.1002/j.1460-2075.1994.tb06888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu MQ, Southworth MW, Mersha FB, Hornstra LJ, Perler FB. Cell. 1993;75:1371–7. doi: 10.1016/0092-8674(93)90623-x. [DOI] [PubMed] [Google Scholar]
- 12.Chong S, Xu MQ. J Biol Chem. 1997;272:15587–90. doi: 10.1074/jbc.272.25.15587. [DOI] [PubMed] [Google Scholar]
- 13.Nichols NM, Benner JS, Martin DD, Evans TC., Jr Biochemistry. 2003;42:5301–11. doi: 10.1021/bi020679e. [DOI] [PubMed] [Google Scholar]
- 14.Shao Y, Xu MQ, Paulus H. Biochemistry. 1995;34:10844–50. doi: 10.1021/bi00034a017. [DOI] [PubMed] [Google Scholar]
- 15.Xu MQ, Perler FB. Embo J. 1996;15:5146–53. [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Y, Paulus H. J Pept Res. 1997;50:193–8. doi: 10.1111/j.1399-3011.1997.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 17.Klabunde T, Sharma S, Telenti A, Jacobs WR, Jr, Sacchettini JC. Nat Struct Biol. 1998;5:31–6. doi: 10.1038/nsb0198-31. [DOI] [PubMed] [Google Scholar]
- 18.Mizutani R, Anraku Y, Satow Y. Journal of Synchrotron Radiation. 2004;11:109–112. doi: 10.1107/s0909049503023495. [DOI] [PubMed] [Google Scholar]
- 19.Mizutani R, Nogami S, Kawasaki M, Ohya Y, Anraku Y, Satow Y. J Mol Biol. 2002;316:919–29. doi: 10.1006/jmbi.2001.5357. [DOI] [PubMed] [Google Scholar]
- 20.Wood DW, Wu W, Belfort G, Derbyshire V, Belfort M. Nat Biotechnol. 1999;17:889–92. doi: 10.1038/12879. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh I, Sun L, Xu MQ. J Biol Chem. 2001;276:24051–8. doi: 10.1074/jbc.M011049200. [DOI] [PubMed] [Google Scholar]
- 22.Ding Y, Xu MQ, Ghosh I, Chen X, Ferrandon S, Lesage G, Rao Z. J Biol Chem. 2003;278:39133–42. doi: 10.1074/jbc.M306197200. [DOI] [PubMed] [Google Scholar]
- 23.Perler FB. Nucleic Acids Research. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietrokovski S. Protein Science. 1994;3:2340–50. doi: 10.1002/pro.5560031218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawasaki M, Nogami S, Satow Y, Ohya Y, Anraku Y. J Biol Chem. 1997;272:15668–15674. doi: 10.1074/jbc.272.25.15668. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh I, Sun L, Xu MQ. J Biol Chem. 2001;276:24051–24058. doi: 10.1074/jbc.M011049200. [DOI] [PubMed] [Google Scholar]
- 27.Duan X, Gimble FS, Quiocho FA. Cell. 1997;89:555–64. doi: 10.1016/s0092-8674(00)80237-8. [DOI] [PubMed] [Google Scholar]
- 28.Poland BW, Xu MQ, Quiocho FA. J Biol Chem. 2000;275:16408–13. doi: 10.1074/jbc.275.22.16408. [DOI] [PubMed] [Google Scholar]
- 29.Sun P, Ye S, Ferrandon S, Evans TC, Xu MQ, Rao Z. J Mol Biol. 2005;353:1093–105. doi: 10.1016/j.jmb.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Van Roey P, Pereira B, Li Z, Hiraga K, Belfort M, Derbyshire V. Journal of Molecular Biology. 2007;367:162–173. doi: 10.1016/j.jmb.2006.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanelli A, Shekhtman A, Cowburn D, Muir TW. Proc Natl Acad Sci U S A. 2004;101:6397–402. doi: 10.1073/pnas.0306616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anraku Y, Mizutani R, Satow Y. IUBMB Life. 2005;57:563–74. doi: 10.1080/15216540500215499. [DOI] [PubMed] [Google Scholar]
- 33.Fersht A. Structure and Mechanism in Protein Science: A guide to enzyme catalysis and protein folding. WH Freeman and Company; New York: 1999. [Google Scholar]
- 34.Johnson MA, Southworth MW, Herrmann T, Brace L, Perler FB, Wuthrich K. Protein Sci. 2007;16:1316–28. doi: 10.1110/ps.072816707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oeemig JS, Aranko AS, Djupsjobacka J, Heinamaki K, Iwai H. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.03.058. [DOI] [PubMed] [Google Scholar]
- 36.Hiraga K, Derbyshire V, Dansereau JT, Van Roey P, Belfort M. Journal of Molecular Biology. 2005;354:916–926. doi: 10.1016/j.jmb.2005.09.088. [DOI] [PubMed] [Google Scholar]
- 37.Du Z, Liu Y, McCallum SA, Dansereau JT, Derbyshire V, Belfort M, Belfort G, Van Roey P, Wang C. Biomolecular NMR Assignments. 2008;2:111–113. doi: 10.1007/s12104-008-9098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamazaki T, Forman-Kay JD, Kay LE. J Am Chem Soc. 1993;115:11054–11055. [Google Scholar]
- 39.Bax A, Summers MF. Journal Of The American Chemical Society. 1986;108:2093–2094. doi: 10.1021/ja00263a024. [DOI] [PubMed] [Google Scholar]
- 40.Krezel A, Bal W. J Inorg Biochem. 2004;98:161–6. doi: 10.1016/j.jinorgbio.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Bachovchin WW. Magnetic Resonance In Chemistry. 2001;39:S199–S213. [Google Scholar]
- 42.Bachovchin WW, Roberts JD. J Am Chem Soc. 1978;100:8041–8047. [Google Scholar]
- 43.Farr-Jones S, Wong WYL, Gutheil WG, Bachovchin WW. J Am Chem Soc. 1993;115:6813–6819. [Google Scholar]
- 44.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. Journal of the American Chemical Society. 1995;117:5179–97. [Google Scholar]
- 45.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. Journal of Computational Chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 46.Vreven T, Morokuma K, Farkas O, Schlegel HB, Frisch MJ. Journal of Computational Chemistry. 2003;24:760–769. doi: 10.1002/jcc.10156. [DOI] [PubMed] [Google Scholar]
- 47.Maseras F, Morokuma K. Journal of Computational Chemistry. 1995;16:1170–9. [Google Scholar]
- 48.Frisch MJ, et al. Gaussian 03, Revision C.02. Wallingford, CT: 2004. [Google Scholar]
- 49.Becke AD. Journal of Chemical Physics. 1993;98:5648–52. [Google Scholar]
- 50.Hehre WJ, Ditchfield R, Pople JA. Journal of Chemical Physics. 1972;56:2257–61. [Google Scholar]
- 51.Shemella P. PhD Disssertation. Renssealer Polytechnic Institute; 2008. [Google Scholar]
- 52.Elstner M, Frauenheim T, Suhai S. Theochem. 2003;632:29–41. [Google Scholar]
- 53.Iwai K, Ando T, Hirs CHW. Methods in Enzymology. Vol. 11. Academic Press; 1967. p. 263. [Google Scholar]
- 54.Koradi R, Billeter M, Wuthrich K. J Mol Graph. 1996;14:51–5. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 55.William WB. Magnetic Resonance in Chemistry. 2001;39:S199–S213. [Google Scholar]
- 56.Shingledecker K, Jiang S-q, Paulus H. Archives of Biochemistry and Biophysics. 2000;375:138–144. doi: 10.1006/abbi.1999.1645. [DOI] [PubMed] [Google Scholar]
- 57.Silverman DN, Tu C, Chen X, Tanhauser SM, Kresge AJ, Laipis PJ. Biochemistry. 1993;32:10757–62. doi: 10.1021/bi00091a029. [DOI] [PubMed] [Google Scholar]
- 58.Pereira B. PhD Disssertation. Renssealer Polytechnic Institute; 2008. [Google Scholar]
- 59.McIntosh LP, Hand G, Johnson PE, Joshi MD, Korner M, Plesniak LA, Ziser L, Wakarchuk WW, Withers SG. Biochemistry. 1996;35:9958–66. doi: 10.1021/bi9613234. [DOI] [PubMed] [Google Scholar]
- 60.Senn HM, Thiel W. Angew Chem Int Ed Engl. 2009;48:1198–229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 61.Shemella P, Pereira B, Zhang Y, Van Roey P, Belfort G, Garde S, Nayak SK. Biophys J. 2007;92:847–53. doi: 10.1529/biophysj.106.092049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voet D, Voet JG. Biochemistry. 3. 2004. [Google Scholar]
- 63.Sun P, Ye S, Ferrandon S, Evans TC, Xu MQ, Rao Z. Journal of Molecular Biology. 2005;353:1093–1105. doi: 10.1016/j.jmb.2005.09.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.