Abstract
Specific inhibitors of mitogen-activated protein kinase/extra-cellular signal-regulated kinase (ERK) kinase (MEK) have been developed that efficiently inhibit the oncogenic RAF-MEK-ERK pathway. We used a systems-based approach to identify breast cancer subtypes particularly susceptible to MEK inhibitors and to understand molecular mechanisms conferring resistance to such compounds. Basal-type breast cancer cells were found to be particularly susceptible to growth inhibition by small-molecule MEK inhibitors. Activation of the phosphatidylinositol 3-kinase (PI3K) pathway in response to MEK inhibition through a negative MEK-epidermal growth factor receptor-PI3K feedback loop was found to limit efficacy. Interruption of this feedback mechanism by targeting MEK and PI3K produced synergistic effects, including induction of apoptosis and, in some cell lines, cell cycle arrest and protection from apoptosis induced by proapoptotic agents. These findings enhance our understanding of the interconnectivity of oncogenic signal transduction circuits and have implications for the design of future clinical trials of MEK inhibitors in breast cancer by guiding patient selection and suggesting rational combination therapies.
Introduction
The RAS-RAF-mitogen-activated protein kinase (MAPK)/extra-cellular signal-regulated kinase (ERK) kinase (MEK)-ERK and phosphoinositide 3-kinase (PI3K)-PTEN-AKT signaling pathways play central roles in the signal transduction networks promoting tumor initiation and tumor progression. This is highlighted by the high frequency of mutations of RAS, RAF, PI3KCA, AKT, and PTEN as well as amplification of AKT present in a broad spectrum of human malignancies (1–5). Because signal transduction networks integrate multiple upstream inputs, targeting pathways downstream of the receptors could conceivably result in greater therapeutic efficacy and broader applicability. For example, abolishing signal transduction through MEK offers the potential advantage of inhibiting both proliferation-promoting and antiapoptotic signals originating from either activated cell surface receptors or mutant RAS and B-RAF. In breast cancer, activation of the RAF-MEK-ERK signaling cascade is associated with increased metastasis risk (6). Furthermore, cross-talk between the RAF-MEK-ERK pathway and the estrogen receptor (ERα) might mediate tamoxifen resistance (7). Although these findings validate the RAF-MEK-ERK pathway as a therapeutic target in breast cancer, clinical studies of MEK inhibitors have shown only limited antitumor activity (8, 9). The mechanisms underlying the poor clinical response to inhibition of MEK remain unclear and no markers of susceptibility or resistance to MEK inhibitors suitable for guiding patient selection have been identified.
The availability of advanced genomic and proteomic technologies has facilitated capturing the complex molecular responses to defined stimuli at a systems level. For example, reverse-phase protein array (RPPA) represents an emerging proteomic technology allowing for comprehensive assessment of signal transduction pathways at the protein and phosphoprotein level (10, 11). Here, we pursued a systems-based experimental strategy to create an in-depth understanding of the responses of breast cancer cells to MEK inhibition. We found basal-type breast cancers to be particularly sensitive to such agents. Furthermore, a potent negative feedback loop between the RAF-MEK-ERK and PI3K pathways was discovered, which counteracts effects of MEK inhibition on cell cycle and apoptosis induction. In agreement with this finding, combined treatment with MEK and PI3K inhibitors resulted in marked tumor cell growth inhibition.
Materials and Methods
Reagents
Chemical synthesis and characterization of PI3K inhibitors PI103 and PIK90 are described by Knight and colleagues (12). The following other reagents were used: rapamycin (Calbiochem), U0126 (Promega), and epidermal growth factor (EGF; Millipore). CI1040 (PD 184352) was kindly provided by Dr. Jenny Bain (University of Dundee, Dundee, United Kingdom). All drugs were diluted in DMSO except for PIK90, which was diluted in 1:1 (v/v) DMSO:H2O. The list of antibodies used in this study is provided in Supplementary Table S6.
Cell culture
Breast cancer cell lines used in this study were described in detail previously (13). The serum used throughout the course of this study was FetalClone III (HyClone), which is not supplemented with EGF.
Cell growth inhibition assay and data analysis
Cell viability was determined using the CellTiter-Glo assay (Promega). Cells were treated with MEK inhibitors CI1040 or U0126 dissolved in DMSO. Nine dilutions of each drug were made in 1:5 serial dilution. The cell growth was determined using CellTiter-Glo assay at days 0 and 3 of drug exposure. Growth inhibition was calculated as described by the National Cancer Institute/NIH Developmental Therapeutics Program7 (14). The 50% growth-inhibiting concentration (GI50), total growth-inhibiting concentration (TGI), and 50% lethal concentration (LC50) were calculated using a standardized algorithms. Drug combination studies were designed according to Chou and Talalay (15). CalcuSyn software (Biosoft) was used to calculate the combination index (CI). CI < 1 indicates synergy, a CI about 1 shows an additive effect, and CI > 1 indicates antagonism. Combined drugs were used at fixed molar ratios.
Identification of predictors of response to MEK inhibition based on mRNA expression
SplineMarker (a variant of the linear splines method allowing for modeling of nonlinear relationships between molecular markers and response; ref. 16) was used to identify the mRNA predictors of response to the MEK inhibitors CI1040 and UO126 using existing mRNA expression profiles from 51 breast cancer cell lines (13). A false discovery rate of 10% was used to identify transcripts associated with responsiveness to MEK inhibition. Baseline expression data for the MAPK-related gene predictors were hierarchically clustered with XCluster8 using a noncentered metric and Pearson correlation. We visualized the data with Java TreeView.9 Model variables are learned by performing least squares fit between the molecular profile and sensitivity profile for each molecular variable. In SplinMarker algorithm, the model is determined via leave-one-out cross-validation. The goodness of fit is assessed by evaluating a P value corresponding to the F statistic for the fit (16). P values are corrected for multiple hypotheses testing using the false discovery rate method (17). Ingenuity pathway enrichment analysis was performed using the Ingenuity Knowledge Base Database10 separately for the sensitivity and resistance mRNA predictors. P values were computed using Fisher’s exact test, with Affymetrix HT_Hgu133A as a reference set. We used a threshold of P < 0.01 to identify significant pathways.
Identification of protein predictors of response to MEK inhibitors
Protein expression profiles were generated in 35 breast cancer cell lines using conventional Western blot analysis for the detection of 64 proteins. In addition, expression of 34 proteins was assessed by RPPA. Proteins predicting response to MEK inhibition were identified using the SplineMarker algorithm.
Proteomic analysis of MEK inhibition by RPPA
MDAMB231 cells were transferred to low serum conditions 24 h before treatment. The cells were pretreated with U0126 (10 µmol/L) or DMSO (control) for 30 min, after which EGF was added at a final dose of 10 ng/mL. Protein lysates were collected at 1, 4, and 24 h after EGF stimulation, denatured in SDS sample buffer, and spotted onto nitrocellulose-coated FAST slides (Whatman) using a GeneTAC G3 arrayer (Genomic Solutions). Proteins were detected using a set of validated antibodies (Supplementary Materials and Methods) and quantified as described before (18–20).
Preparation of protein lysates and Western blots
The cells were treated with drugs either in low serum or in full serum conditions as indicated in the figures. For the low serum conditions, cells were washed in the medium containing 0.1% fetal bovine serum (FBS) and incubated in this medium for 24 h. Drugs or DMSO (control) was added directly to this medium, and 30 min later, EGF was added at a final dose of 10 ng/mL. Cells were harvested at time intervals after EGF stimulation. For the full serum conditions, cells were grown in their normal growth medium and, at 48 h after plating, treated with the drugs. Protein lysates were prepared from cells at 70% to 90% confluency. The cells were washed in ice-cold PBS and then extracted in the lysis buffer containing 1% Triton X-100, 50 mmol/L HEPES (pH 7.5), 150 mmol/L NaCl, 25 mmol/L β-glycerophosphate, 25 mmol/L NaF, 5 mmol/L EGTA, 1 mmol/L EDTA, protease cocktail inhibitor set III, and phosphatase cocktail inhibitor set II (Calbiochem). The lysates were clarified by centrifugation for 13,000 rpm for 10 min on ice and frozen at −80°C. Protein concentrations were determined using the bicinchoninic acid protein assay kit (Pierce Biotechnology). Protein extract (20 µg per lane) was electrophoresed, transferred to polyvinylidene difluoride membranes (Millipore), and probed with specific antisera using standard techniques. Bound antibodies on immunoblots were detected by enhanced chemiluminescence (Amersham).
Cell cycle and apoptosis analyses
Cells were treated with drugs 24 h after plating and harvested for apoptosis or cell cycle assays at 72 h after treatment. Apoptosis was measured in live cells by Annexin V-FITC and propidium iodide (PI) labeling using Apoptosis Detection Kit I (BD Pharmingen) and quantified by flow cytometry (FACSCalibur, Becton Dickinson) with CellQuest software. For cell cycle analysis, cells were fixed in 70% ethanol, treated with PI (30 µg/mL) and RNase A (100 µg/mL), and subjected to fluorescence-activated cell sorting (FACS) analysis. Data analysis was performed using ModFit LT cell cycle analysis software (Verity Software House). All treatments were done in triplicates and at least 40,000 cells were acquired from every sample for the FACS analysis.
Results
Basal-type breast cancer cell lines are particularly susceptible to MEK inhibition
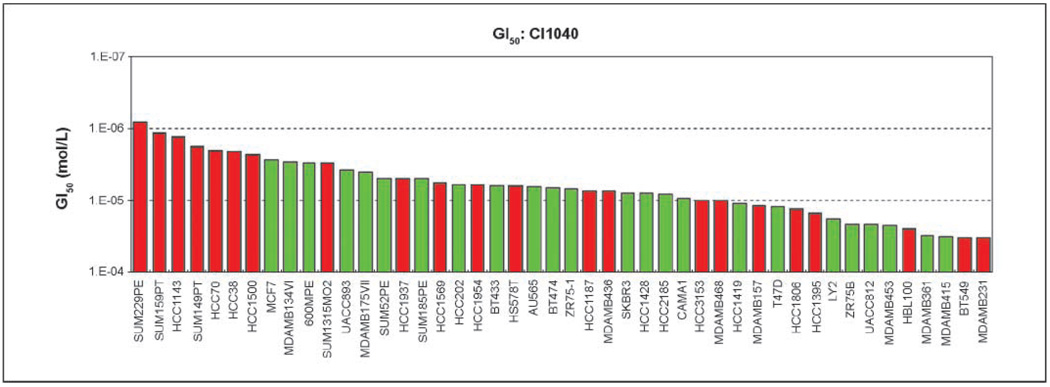
We ascertained the effect of pharmacologic inhibition of MEK in a large panel of breast cancer cell lines that was described before (13). Cells were treated with the specific MEK inhibitors CI1040 or U0126 and cell viability was determined. A wide spectrum of growth inhibition was observed. For U0126, the GI50 ranged from 0.8 µmol/L (HCC1187 cells) to 60 µmol/L (MDAMB231 cells), whereas the GI50 for CI1040-treated cells ranged from 0.8 µmol/L (SUM229PE) to 33 µmol/L (MDAMB231 cells). Compared with luminal-type cell lines, basal-type breast cancer cells showed more frequent growth inhibition in response to MEK inhibitor treatment, particularly following treatment with CI1040 (Fig. 1; Supplementary Materials and Methods). For the majority of cell lines, however, the LC50 was not reached. Only nine cell lines treated with U0126 and eight cell lines treated with CI1040 were killed at concentrations of equal or less than 50 and 25 µmol/L, respectively (data not shown).
Figure 1.
Comparison of sensitivity of breast cancer cell lines to MEK inhibitor CI1040. Forty-six basal or luminal breast cancer cell lines were treated with increasing doses of CI1040 and cell viability was assessed at 0 h (time of drug addition) and at 72 h after treatment. GI50 was calculated from the dose-response curves as described in Materials and Methods. Data are presented as GI50 (mol/L) of cell lines in the order of most sensitive to most resistant (from left to right). Red and green columns, basal and luminal cellular phenotype, respectively. All data for GI50, TGI, and LC50 for CI1040 and U0126 response of cell lines are summarized in Supplementary Table S1.
MAPK pathway genes predict responsiveness to MEK inhibitors in human breast cancer cell lines and tumors
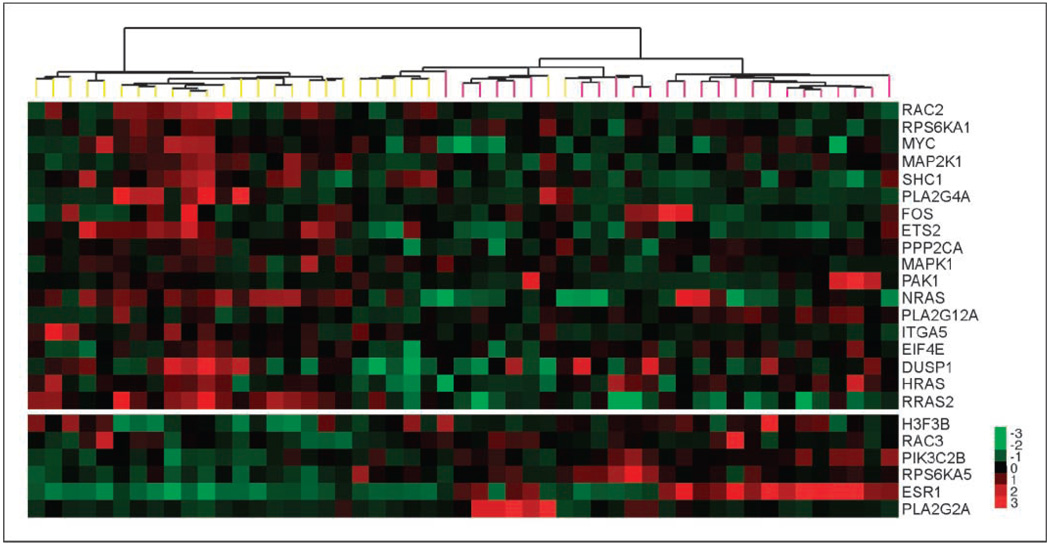
To identify genes and pathways predicting sensitivity to MEK inhibitors, we performed a correlation analysis of mRNA expression data from the cell line panel with the above-mentioned growth inhibition data. The previously reported mRNA microarray data of the cell line set were used (13). Using the SplineMarker algorithm, we identified 1,548 (883) gene transcripts associated with sensitivity to CI1040 (UO126), and 790 (671) transcripts were associated with resistance. Among these, 292 and 127 predictors, associated with sensitivity and resistance, respectively, were common to both drugs. Pathway enrichment analysis revealed that the predictive mRNA markers are significantly enriched in four pathways for U0126 and eight for CI1040 (Supplementary Table S2). These included ERK/MAPK signaling, purine metabolism, p53 signaling, and metabolism of xenobiotics by cytochrome P450. Genes involved in ERK/MAPK signaling were predictive of sensitivity, whereas PI3K pathway components were associated with resistance to MEK inhibitors (Fig. 2). Hierarchical cluster analysis revealed that genes predictive of susceptibility to MEK inhibitors were relatively overexpressed in basal-type cell lines, whereas resistance markers were highly expressed in luminal cells (Fig. 2). A multivariate model of response to CI1040 consisting of 26 mRNA predictors from the Ingenuity MAPK pathway gene set was confirmed as involved in this pathway by at least one additional database (KEGG, BioCarta, GeneGo, and GSEA). We trained the model on a randomly selected set of 31 breast cancer cell lines and tested the accuracy of the model on the remaining 10 cell lines. The best model consisted of only one gene (RAC2). The sensitivity predicted by the model was strongly correlated with the measured sensitivity on the test set: r = 0.69 (P = 0.027). We applied this in vitro molecular predictor to forecast sensitivity to CI1040 of previously reported 118 tumors (21). The model predicted that the basal tumors are more sensitive to CI1040 than the luminal tumors. The difference in predicted sensitivities between these two groups was statistically significant (P = 6.7e–03).
Figure 2.
Breast cancer cell lines can be hierarchically clustered according to their expression of the MAPK-related gene predictors. Each row represents the relative transcript abundance (in log2 space) for one gene; each column represents data from one cell line. Expression of genes in the top panel predicts sensitivity to MEK inhibitors, whereas expression of those in the bottom panel predicts resistance. Within each panel, genes are ordered by q value. In the top panel, the most significant predictors are at the top; in the bottom panel, the most significant predictors are at the bottom. In the tree, yellow end nodes denote basal cell lines and pink end nodes denote luminal cell lines.
Protein predictors of susceptibility to MEK inhibition
Because the MAPK pathway is regulated predominantly through protein modification, particularly phosphorylation, we ascertained the differences in protein expression levels as predictors of susceptibility to MEK inhibition in the panel of 53 breast cancer cell lines by performing Western blot analysis for the detection of 64 proteins. In addition, expression and phosphorylation of 34 proteins was assessed by RPPA technology. Fifteen phosphoproteins common to both approaches were included in the analysis. The correlation between protein expression levels in the breast cancer cell panel and GI50 of CI1040 as well as U0126 was calculated using SplineMarker (Supplementary Table S3). Ten proteins were significantly correlated with susceptibility to MEK inhibition for both drugs (Supplementary Table S4). Cytokeratin-5/6, a well-established marker for the basal breast cancer subtype, showed the strongest correlation with GI50 of the MEK inhibitors. Expression of cytokeratin-18 and ERα, both luminal subtype markers, was correlated with resistance to MEK inhibitors. These findings clearly support the notion that basal-type breast cancer cell lines are particularly susceptible to MEK inhibitors.
Temporal proteomic analysis reveals a novel negative feedback loop between MEK and PI3K mediated by the EGF receptor
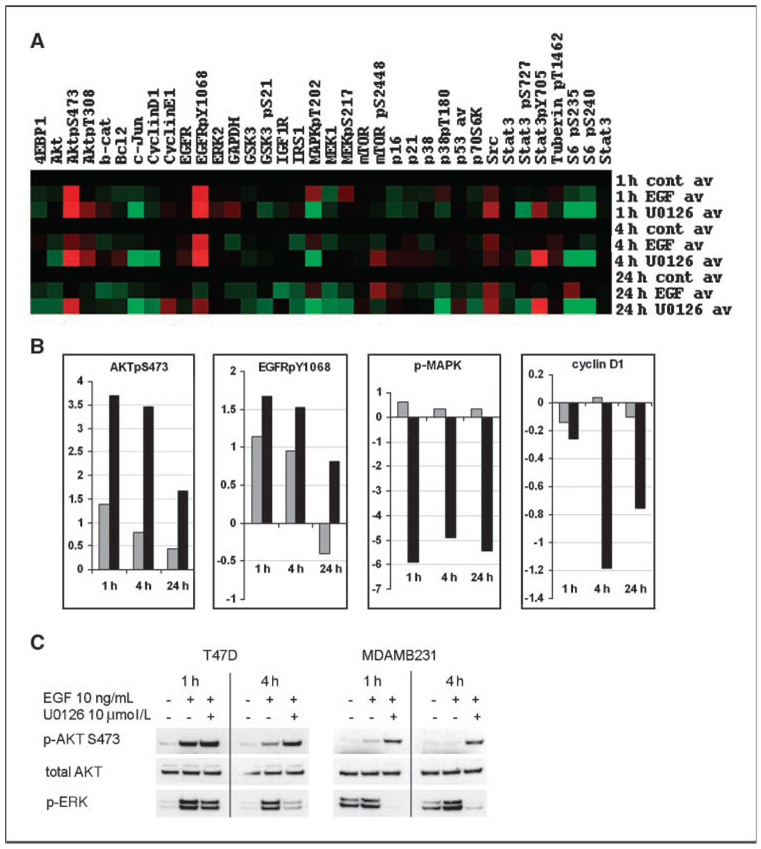
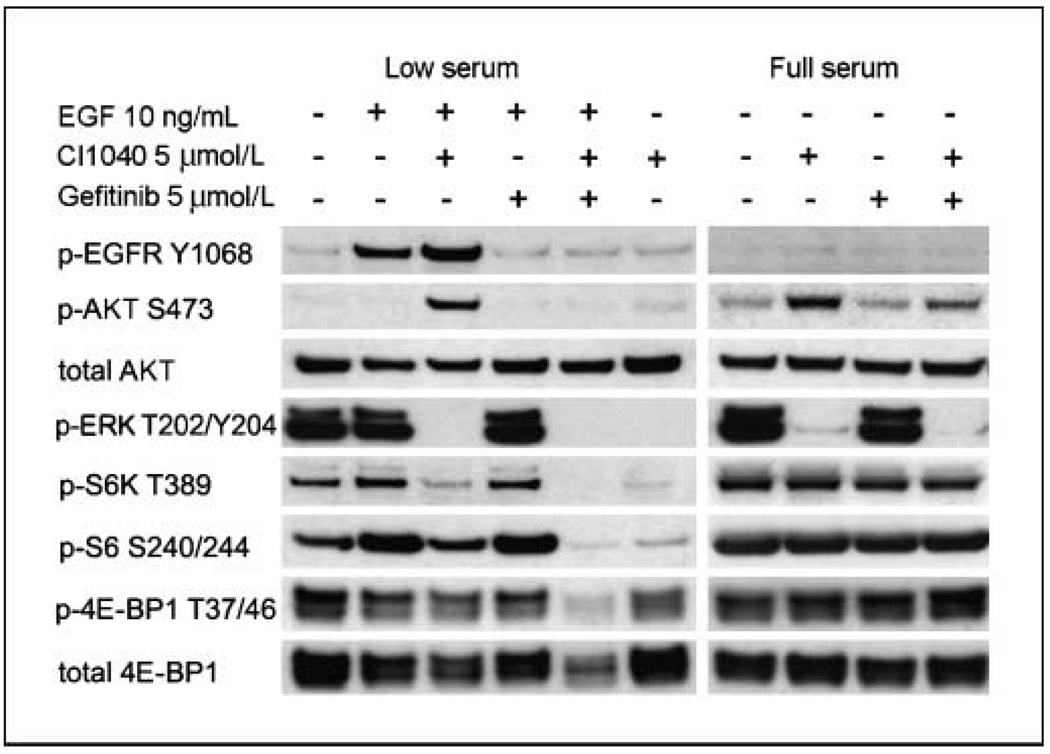
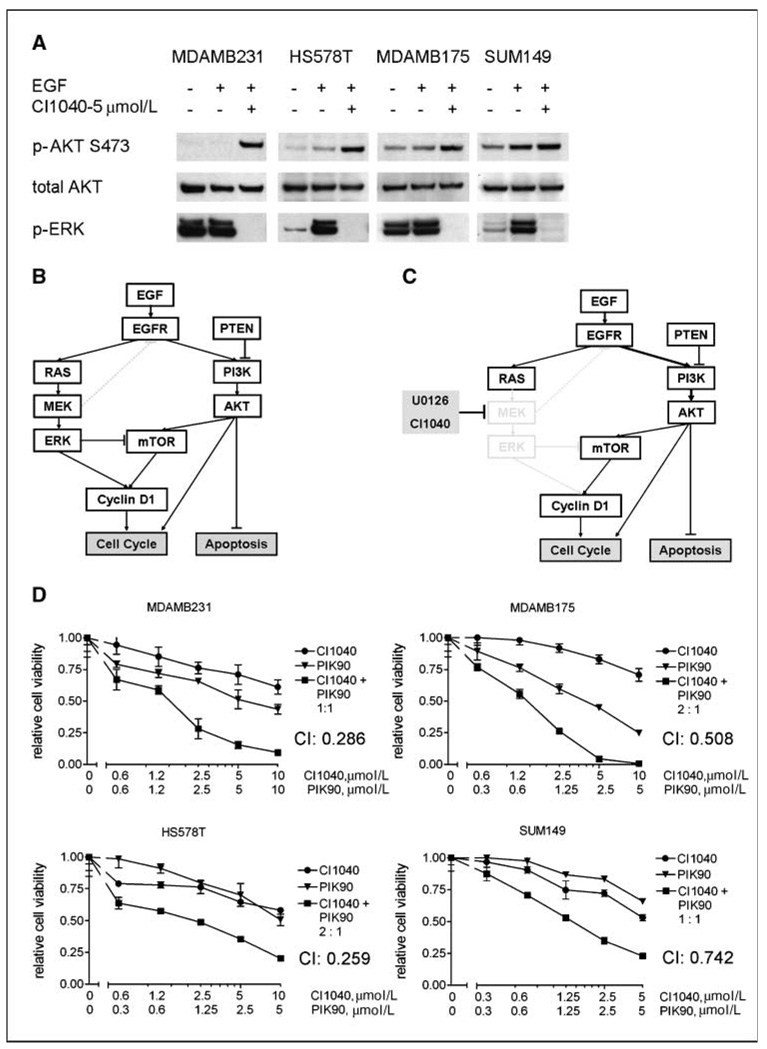
To further understand the response of signal transduction pathways to MEK inhibition, we analyzed temporal changes in protein expression and phosphorylation by RPPA technology. As expected, a strong reduction in phosphorylated ERK (p-ERK) levels in response to MEK inhibitor treatment was observed, along with down-regulation of effectors of ERK, such as cyclin D1 (Fig. 3A and B). Additionally, EGF treatment induced phosphorylation of the EGF receptor (EGFR) and downstream effectors, such as AKT. Unexpectedly, MEK inhibition led to markedly enhanced phosphorylation of EGFR and activation of the PI3K pathway, as determined by phosphorylated AKT (p-AKT) at S473. We confirmed key findings by conventional Western blotting in MDAMB231 and T47D cells using two different MEK inhibitors, CI1040 and U0126 (Fig. 3C; data not shown), showing that activation of AKT in response to MEK inhibition is not limited to a single cell line or a result of off-target effects of one drug. Overactivation of EGFR and AKT was most striking in the presence of EGF in the medium, as determined by Western blot analysis (Fig. 4). To test whether the increased phosphorylation of AKT was dependent on overactivation of EGFR, we treated MDAMB231cells with the specific EGFR inhibitor gefitinib in the presence or absence of CI1040. We found that in low serum conditions in the presence of EGF, MEK inhibitor–induced activation of AKT was fully abolished by the treatment with gefitinib. In 10% serum, the effect of gefitinib was still observed, although to a lesser degree. To rule out that MEK-dependent activation of AKT being a cell line–specific phenomenon, we assessed p-AKT in eight cell lines treated with CI1040 under low serum conditions and found increased p-AKT levels in five cell lines: MDAMB231, T47D, HS578T, MDAMB175, and Sum149 (Fig. 5A; data not shown). Thus, a negative regulatory feedback loop exists between MEK and AKT that depends on activation of the EGFR and thus represents a signal-amplifying mechanism (Fig. 5B and C).
Figure 3.
Proteomic analysis of MEK inhibition in breast cancer cells. A, heat map of protein and phosphoprotein expression profiles. MDAMB231 cells were treated with MEK inhibitor U0126 (10 µmol/L) as described in Materials and Methods, and 30 min later, they were stimulated with EGF (10 ng/mL). The protein lysates were collected at 1, 4, and 24 h after EGF addition and analyzed by RPPA. Values are expressed as log2 fold difference from control (untreated) samples at each time point. B, relative expression changes of AKTpS473, EGFRpY1068, phosphorylated MAPK (p-MAPK), and Cyclin D1 as detected by RPPA. Gray columns, EGF treatment; black columns, EGF + U0126 treatment. Control = 0. Values for phosphoproteins were normalized by corresponding total protein levels. C, conventional Western blot analysis of p-AKT and p-ERK (p-MAPK) expression in response to U0126 treatment in T47D and MDAMB231 cell lines. Cells were treated with MEK inhibitor in the same conditions as for RPPA experiment and the protein lysates were collected at 1 and 4 h after EGF stimulation.
Figure 4.
Western blot analysis of the biochemical response of MDAMB231 cells treated with MEK inhibitor CI1040, EGFR inhibitor gefitinib, or their combination in low (0.1% FBS) and full (10% FBS) serum conditions. Cells were harvested at 4 h after drug treatment. In low serum conditions and in the presence of EGF, MEK inhibition results in strong phosphorylated EGFR (p-EGFR) and p-AKT up-regulation, which is abrogated by EGFR inhibitor.
Figure 5.
Synergistic response of breast cancer cell lines to a combination of MEK and PI3K inhibitors correlates with their up-regulated p-AKT status induced by MEK inhibition. A, Western blot analysis of p-AKT and p-ERK expression in response to CI1040 treatment in four breast cancer cell lines: MDAMB231, MDAMB175, HS578T, and SUM149. The cells were treated with MEK inhibitor in low serum conditions, and in 30 min, they were stimulated with EGF and the protein lysates were collected at 4 h after EGF. B and C, schematic summary of the RAS-RAF-MEK-ERK and PI3K pathway interconnectivity in the absence (B) and presence (C) of MEK inhibitors. D, synergistic effect of combination of MEK and PI3K inhibitors on cell viability of cell lines displaying MEK inhibitor–induced AKT phosphorylation. Dose/effect curves for single inhibitors CI1040 and PIK90 and their combinations at fixed molar ratio are presented. Cell viability was measured at 72 h after treatment with the drugs using ATP-based cell viability assay (Promega). Relative cell viability of drug-treated cells was calculated as a fraction of control. Points, mean of triplicates; bars, SD. CIs at 50% dose response are calculated using CalcuSyn software.
Synergistic cellular effects of inhibition of MEK and PI3K in breast cancer cells with feedback activation of AKT
Because suppression of apoptosis is among the known biological effects mediated by the PI3K pathway, we hypothesized that the feedback activation of the pathway in response to MEK inhibition contributes to resistance of breast cancer cells to MEK inhibitors. We therefore hypothesized that inhibitors of PI3K should synergize with MEK inhibitors in cell lines showing feedback activation of AKT. We tested this using inhibitors of PI3K specific for the p110α isoform, PIK90 and PI103 (12). This PI3K isoform is frequently mutated in cancer and a crucial signal transducer in cancer cells (22–24). Synergistic growth inhibition following PI3K and MEK inhibition occurred in 4 cell lines out of 11 tested (Fig. 5D). The calculated CI values ranged from 0.259 (HS578T) to 0.742 (SUM149 cells), which are considered strong and moderate synergism, respectively. These cell lines had shown strong activation of AKT following MEK inhibition. However, only MDAMB175 and SUM149 cell lines showed an increase in apoptosis rates when treated with a combination of MEK and PI3K inhibitors (Supplementary Table S5; data not shown). In contrast, in other cell lines, including MDAMB231 cells, the combination of CI1040 or UO126 with PI3K inhibitors induced a complete G1 arrest (Fig. 6A). Interestingly, cell lines responding with apoptosis to the dual-inhibitor treatment harbor wild-type p53, whereas those showing synergistic effects on cell cycle distribution contain mutant p53.
Figure 6.
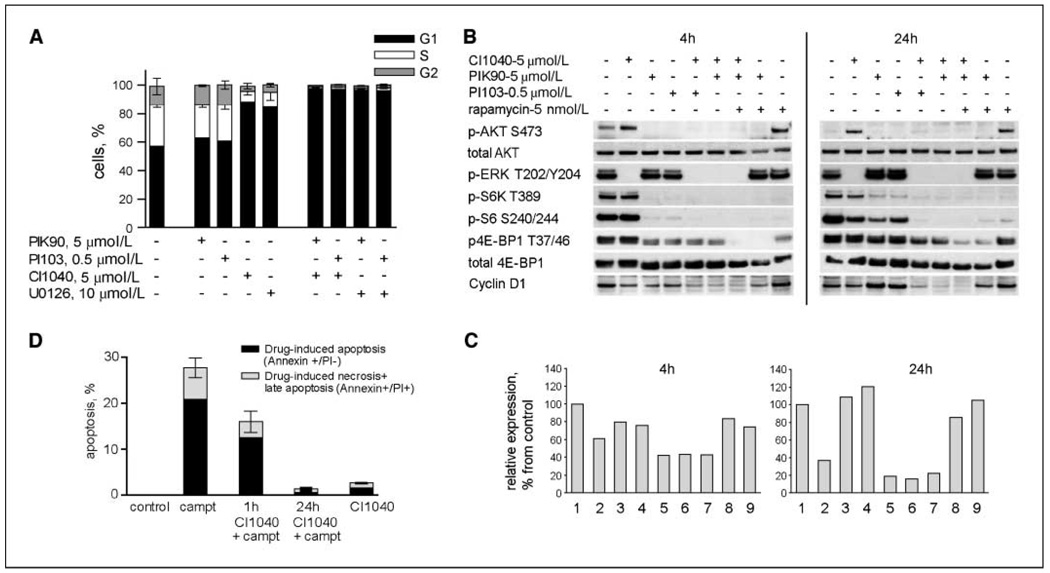
Biological effects of MEK inhibitors in combination with PI3K inhibitors and camptothecin. A, FACS-based analysis was used to determine percentage of MDAMB231 cells in G1, S, and G2 phases of the cell cycle at 72 h after treatment with the single drugs and with their combinations at indicated doses. Whereas PI3K inhibitors PIK90 and PI103 do not affect cell cycle distribution, their addition to MEK inhibitors CI1040 or U0126 results in complete G1 arrest. Columns, mean of duplicates; bars, SD. For each duplicate, 40,000 cells were acquired. The result was confirmed in two independent experiments. B, Representative Western blot analysis of MDAMB231 cells treated with the indicated drugs and their combinations for 4 or 24 h. The cells were treated with drugs in full serum conditions and harvested at 4 and 24 h after drug addition for protein and RNA isolation. C, cyclin D1 gene expression changes induced by the same drug treatments as in panel B: 1, control; 2, CI1040 (5 µmol/L); 3, PIK90 (5 µmol/L); 4, PI103 (0.5 µmol/L); 5, CI1040 + PI103; 6, CI1040 + PIK90; 7, CI1040 + PIK90 + rapamycin; 8, PIK90 + rapamycin; 9, rapamycin (5 nmol/L). Gene expression was analyzed by Taqman assay; the results are averaged from duplicates. Relative cyclin D1 expression is presented as the percentage of that of control (untreated) cells. D, G1 arrest induced by MEK inhibition rescues the cells from apoptosis induced by cytotoxic agent. MDAMB231 cells were treated with camptothecin (2.5 µmol/L) for 48 h, which resulted in 20% of apoptotic cells. Pretreatment of cells with CI1040 for 1 h reduced the amount of apoptotic cells, whereas pretreatment of cells for 24 h completely eliminated apoptosis induced by camptothecin. Columns, mean of duplicates; bars, SD. For each duplicate, 40,000 cells were acquired. The results were confirmed in two independent experiments.
Cooperative G1 growth arrest results from synergistic inhibition of cyclin D1
We sought to further understand the cell cycle effects induced by inhibition of MEK and PI3K and analyzed the response of proteins downstream of these molecules. This analysis included mammalian target of rapamycin (mTOR) kinase, an important regulator of protein translation. It is required for activation of S6 kinase (S6K) and 4E-binding protein 1 (4E-BP1), which regulates cap-dependent mRNA translation. Despite the inhibition of MEK by CI1040 in MDAMB231 cells, phosphorylation of both phosphorylated S6K and phosphorylated 4E-BP1 did not change (Fig. 6B), whereas inhibition of PI3K with PIK90 led to a significant reduction in S6K phosphorylation and a moderate inhibition of 4E-BP1 phosphorylation. The combination of CI1040 and PIK90, in contrast, further inhibited activation of S6K and caused a marked decrease in 4E-BP1 phosphorylation (Fig. 6B). Expression of cyclin D1, a target of translational regulation by the mTOR pathway (25), was slightly reduced after MEK inhibition, whereas treatment with PIK90 led to increased levels at the 24-hour time point. In contrast, the combination of both inhibitors resulted in a strong down-regulation of cyclin D1 protein, which was paralleled by reduction in the corresponding mRNA levels (Fig. 6C). Thus, regulation of cyclin D1 gene expression drives the expression levels of cyclin D1 protein, whereas translational regulation plays a minor role in this context. Indeed, knockdown of cyclin D1 expression by small interfering RNAs resulted in G1 arrest (data not shown), showing the effect of cyclin D1 levels on G1-S cell cycle progression in these cells. Based on these data, it seems that cyclin D1 represents a major intersection point between the RAF-MEK-ERK and PI3K pathways, mediating cell cycle arrest in response to pharmacologic inhibition of these pathways. Interestingly, dual inhibition of PI3K and mTOR (with PIK90 and rapamycin or the dual PI3K/mTOR inhibitor PI103), which acts downstream of both the MAPK and PI3K pathways, was not sufficient to induce a significant reduction of cyclin D1 protein or mRNA levels (Fig. 6B and C). Thus, reduction of MEK-dependent and mTOR-independent regulatory factors is required for the observed repression of cyclin D1.
MEK inhibition counteracts apoptotic cell death induced by camptothecin
Because it has been described by others that cells arresting in the G1 phase of the cell cycle might be protected from apoptosis induction, we assessed whether MEK inhibition would alter sensitivity to apoptosis induced by chemotherapy in MDAMB231 cells. As shown in Fig. 6D, we treated MDAMB231 cells for 1 or 24 hours with CI1040 or DMSO (control) before adding the chemotherapeutic agent camptothecin, which strongly induces apoptosis in these cells. In agreement with our prediction, apoptosis induction was inhibited after 1 hour of CI1040 pretreatment. After 24 hours of pretreatment with CI1040, the proapoptotic activity of camptothecin was completely abolished. Thus, MEK inhibition not only did not induce apoptosis but also acted as a cytoprotective when breast cancer cells were treated with proapoptotic agents.
Discussion
In this study, we examine the molecular features of breast cancer cells that determine sensitivity to pharmacologic inhibition of the MEK-ERK signal transduction pathway. We used a large set of breast cancer cell lines as a model system. Previously, we showed that these cell lines retain molecular abnormalities characteristic for human breast cancers (13) and we used this system to define molecular predictors for susceptibility to lapatinib and for EGFR and Her2/Neu. These were validated in a clinical study of lapatinib in combination with paclitaxel.11 Similar approaches were previously used to identify molecular signatures of susceptibility to conventional chemotherapeutic agents (26). Thus, the validity of cell line–based approaches for the preclinical development and optimization of treatment strategies using targeted therapeutics has been well established.
A major finding of this study is the particular sensitivity of basal-type cell lines to inhibitors of MEK. This has implications for our understanding of the role of the MAPK pathway in basal-type breast cancer as well as for the further development of MEK inhibitors clinically. The predictive nature of basal-type features was strongly supported by our analysis of correlation of protein expression patterns and sensitivity to MEK inhibitors. The most predictive protein marker was cytokeratin-5/6, a well-established marker of basal-type breast cancer (27–29). Additional subtype-defining markers were found to be correlated with response to MEK inhibitors (EGFR with sensitivity, ERα, and cytokeratin-18 with resistance). High expression levels of EGFR and lack of expression of ERα and cytokeratin-18 are characteristic for basal-type breast cancers (30). Our analysis of gene expression predictors of response revealed a strong enrichment of genes encoding for MAPK pathway components. Furthermore, we found increased expression of these gene predictors in basal-type cell lines, showing that basal-type cells use this pathway to a greater extent than luminal cells and depend on its activity for proliferation. Interestingly, gene expression of the Rho-like GTPase RAC2 was highly predictive of responsiveness to MEK inhibition in breast cancer cell lines and in human tumors. Activation of the MAPK pathway has been shown as a central feature of basal-type breast cancers (31). Overexpression of EGFR, amplification and mutation of the KRAS oncogene, and overexpression of αB-crystallin have been shown to lead to activation of this pathway in basal-type breast cancer. αB-crystallin is a MEK-activating heat shock protein that is overexpressed in basal-type breast cancers and associated with poor outcome (32). Luminal-type cells might use the MEK-ERK pathway to a lesser extent and seem to be more dependent on the PI3K pathway, shown by the preferential occurrence of PI3K mutations in this subtype (33) and our finding that AKT protein levels predict resistance to MEK inhibitors.
The role of the PI3K pathway in conferring resistance to MEK inhibitors is highlighted by our discovery of a negative feedback loop activating AKT in response to MEK inhibition in an EGFR-dependent fashion, thus amplifying EGF signals (Fig. 5B and C). Cross-talk between the MAPK and PI3K pathways has been described before. A negative feedback loop has been shown from ERK to the Grb2-associated binder 1 protein, a scaffold protein that propagates EGFR signals to the PI3K and the RAS/MAPK pathways (34, 35). Because the PI3K pathway is a known promoter of the cell cycle and negative regulator of apoptosis, we reasoned that feedback activation of AKT could counteract therapeutically relevant effects of MEK inhibition. Indeed, dual inhibition of MEK and PI3K led to synergistic down-regulation of cyclin D1 mRNA and protein levels, as well as growth inhibition in cell lines showing strong feedback activation of AKT following MEK inhibition. The role of cyclin D1 in mediating efficacy of MEK inhibitors is further highlighted by our finding that cyclin D1 protein levels were correlated with resistance to MEK inhibition, whereas levels of glycogen synthase kinase 3 (a negative regulator of cyclin D1) were correlated with sensitivity to MEK inhibitors. Only a subset of our cell lines responded with cell killing to the dual inhibition, whereas the remainder of cell lines showed synergistic inhibition of the cell cycle. Similar observations were shown by others previously (36). It is conceivable that the reduced levels of E2F1 as a consequence of loss of cyclin D1 expression could result in protection from apoptosis because known E2F1-dependent proapoptotic genes exist that are inhibited by PI3K (37). The E2F1-driven apoptotic mechanism is p53 dependent. Interestingly, wild-type p53 is present in cell lines undergoing apoptosis following MEK/PI3K inhibition, whereas p53-mutant cell lines showed synergistic cell cycle arrest. Moreover, in MDAMB231 cells, protection from apoptosis resulted from MEK inhibition in cells treated with camptothecin. These findings suggest that MEK inhibition might have antiapoptotic effects and thus counteract potential therapeutic effects of other agents administered in combination.
These results are relevant for the design of future clinical trials. The MEK inhibitors CI1040 and AZD6244 have shown only modest antitumor activity in early clinical trials (8, 9). In one study that included breast cancer patients, tumor biopsies showed reduction of p-ERK levels and reduction of the proliferation marker KI-67 in tumor biopsies (9). This finding corresponds well with our observation of predominant cell cycle effects of MEK inhibitors in breast cancer. Our findings suggest focusing further development efforts of MEK inhibitors in breast cancer on patients with basal-type cancers, which are representing ∼ 15% of all sporadic and >60% of BRCA1-related breast cancers (38, 39). Currently, treatment options for these patients are limited and MEK inhibitors might represent a promising therapy. Furthermore, inhibiting the MEK-EGFR-PI3K feedback loop is likely to result in therapeutic synergism, particularly in cases with wild-type p53.
Acknowledgments
Grant support: Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231 and NIH, National Cancer Institute grants P50 CA 58207, U54 CA 112970 (J.W. Gray), and P30 CA82103 (F. McCormick).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
In preparation.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Tenorio G, Alkhori L, Olsson B, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007;13:3577–3584. doi: 10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- 3.Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 4.Rochlitz CF, Scott GK, Dodson JM, et al. Incidence of activating ras oncogene mutations associated with primary and metastatic human breast cancer. Cancer Res. 1989;49:357–360. [PubMed] [Google Scholar]
- 5.Brugge J, Hung MC, Mills GB. A new mutational AKTivation in the PI3K pathway. Cancer Cell. 2007;12:104–107. doi: 10.1016/j.ccr.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Adeyinka A, Nui Y, Cherlet T, Snell L, Watson PH, Murphy LC. Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clin Cancer Res. 2002;8:1747–1753. [PubMed] [Google Scholar]
- 7.Thomas RS, Sarwar N, Phoenix F, Coombes RC, Ali S. Phosphorylation at serines 104 and 106 by Erk1/2 MAPK is important for estrogen receptor-α activity. J Mol Endocrinol. 2008;40:173–184. doi: 10.1677/JME-07-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinehart J, Adjei AA, Lorusso PM, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 9.Adjei AA, Cohen RB, Franklin W, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishizuka S, Charboneau L, Young L, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci U S A. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics Oxford England. 2007;23:1986–1994. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 12.Knight ZA, Gonzalez B, Feldman ME, et al. A pharmacological map of the PI3-K family defines a role for p110α in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monks A, Scudiero D, Skehan P, et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst. 1991;83:757–766. doi: 10.1093/jnci/83.11.757. [DOI] [PubMed] [Google Scholar]
- 15.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 16.Das D, Nahle Z, Zhang MQ. Adaptively inferring human transcriptional subnetworks. Mol Sys Biol. 2006;2 doi: 10.1038/msb4100067. 2006.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 19.Sheehan KM, Calvert VS, Kay EW, et al. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4:346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Murph MM, Smith DL, Hennessy B, et al. Individualized molecular medicine: linking functional proteomics to select therapeutics targeting the PI3K pathway for specific patients. Adv Exp Med Biol. 2008;622:183–195. doi: 10.1007/978-0-387-68969-2_15. [DOI] [PubMed] [Google Scholar]
- 21.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 23.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 24.Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 25.Rosenwald IB, Kaspar R, Rousseau D, et al. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem. 1995;270:21176–21180. doi: 10.1074/jbc.270.36.21176. [DOI] [PubMed] [Google Scholar]
- 26.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 27.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 28.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bocker W, Moll R, Poremba C, et al. Common adult stem cells in the human breast give rise to glandular and myoepithelial cell lineages: a new cell biological concept. Lab Invest. 2002;82:737–746. doi: 10.1097/01.lab.0000017371.72714.c5. [DOI] [PubMed] [Google Scholar]
- 30.Rakha EA, Reis-Filho JS, Ellis IO. Impact of basal-like breast carcinoma determination for a more specific therapy. Pathobiology. 2008;75:95–103. doi: 10.1159/000123847. [DOI] [PubMed] [Google Scholar]
- 31.Hoadley KA, Weigman VJ, Fan C, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moyano JV, Evans JR, Chen F, et al. αB-crystallin is a novel oncoprotein that predicts poor clinical outcome in breast cancer. J Clin Invest. 2006;116:261–270. doi: 10.1172/JCI25888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollestelle A, Elstrodt F, Nagel JH, Kallemeijn WW, Schutte M. Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol Cancer Res. 2007;5:195–201. doi: 10.1158/1541-7786.MCR-06-0263. [DOI] [PubMed] [Google Scholar]
- 34.Yu CF, Liu ZX, Cantley LG. ERK negatively regulates the epidermal growth factor-mediated interaction of Gab1 and the phosphatidylinositol 3-kinase. J Biol Chem. 2002;277:19382–19388. doi: 10.1074/jbc.M200732200. [DOI] [PubMed] [Google Scholar]
- 35.Kiyatkin A, Aksamitiene E, Markevich NI, Borisov NM, Hoek JB, Kholodenko BN. Scaffolding protein Grb2-associated binder 1 sustains epidermal growth factor-induced mitogenic and survival signaling by multiple positive feedback loops. J Biol Chem. 2006;281:19925–19938. doi: 10.1074/jbc.M600482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torbett NE, Luna A, Knight ZA, et al. A chemical screen in diverse breast cancer cell lines reveals genetic enhancers and suppressors of sensitivity to PI3K isotype-selective inhibition. Biochem J. 2008;415:97–110. doi: 10.1042/BJ20080639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abd El-Rehim DM, Ball G, Pinder SE, et al. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer. 2005;116:340–350. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- 39.Lakhani SR, Reis-Filho JS, Fulford L, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]