Abstract
Allele-specific gene silencing by RNA interference (RNAi) is therapeutically useful for specifically suppressing the expression of alleles associated with disease. To realize such allele-specific RNAi (ASPRNAi), the design and assessment of small interfering RNA (siRNA) duplexes conferring ASP-RNAi is vital, but is also difficult. Here, we show ASP-RNAi against the Swedish- and London-type amyloid precursor protein (APP) variants related to familial Alzheimer′s disease using two reporter alleles encoding the Photinus and Renilla luciferase genes and carrying mutant and wild-type allelic sequences in their 3′-untranslated regions. We examined the effects of siRNA duplexes against the mutant alleles in allele-specific gene silencing and off-target silencing against the wild-type allele under heterozygous conditions, which were generated by cotransfecting the reporter alleles and siRNA duplexes into cultured human cells. Consistently, the siRNA duplexes determined to confer ASP-RNAi also inhibited the expression of the bona fide mutant APP and the production of either amyloid β 40- or 42-peptide in Cos-7 cells expressing both the full-length Swedish- and wild-type APP alleles. The present data suggest that the system with reporter alleles may permit the preclinical assessment of siRNA duplexes conferring ASP-RNAi, and thus contribute to the design and selection of the most suitable of such siRNA duplexes.
Keywords: RNAi, allele-specific gene silencing, amyloid precursor protein, Swedish mutation, London mutation, reporter allele
INTRODUCTION
RNA interference (RNAi) is a powerful tool for suppressing the expression of a gene of interest (Dykxhoorn et al, 2003; Meister and Tuschl, 2004; Mello and Conte, 2004). In mammals, RNAi can be induced by direct introduction of synthetic small interfering RNA (siRNA) duplexes into cells or generation of siRNA duplexes using short-hairpin RNA expression vectors and its application is expanding to various fields of science; therapeutic use of RNAi in medical science and pharmacogenesis is particularly promising (Caplen, 2004; Dykxhoorn et al, 2003; Hannon and Rossi, 2004; Karagiannis and El-Osta, 2005; Wood et al, 2003). Allele-specific gene silencing by RNAi (allele-specific RNAi: ASP-RNAi) is an advanced application of RNAi techniques, by which the expression of an allele of interest can be inhibited (Victor et al, 2002). Accordingly, ASP-RNAi is thought to be therapeutically useful, i.e., it can specifically suppress the expression of alleles causing disease without inhibiting the expression of corresponding wild-type alleles. To realize and control such ASP-RNAi, the following issues must be addressed: selection of competent siRNA duplexes that strongly induce ASPRNAi; and qualitative and quantitative evaluation of allele-specific gene silencing.
In this article, we describe an easy assay system for assessment of ASP-RNAi with mutant and wild-type reporter alleles encoding the Photinus and Renilla luciferase genes. Using the amyloid precursor protein (APP) variants (the Swedish- and London-type variants) related to familial Alzheimer′s disease (Goate et al, 1991; Mullan et al, 1992) as model mutant alleles, we determined the effects of siRNA duplexes against the mutant APP on allele-specific silencing as well as off-target silencing against the wild-type allele. The siRNA duplexes having the potential to specifically suppress the expression of the mutant reporter allele consistently inhibited the expression of the bona fide mutant APP as well as amyloid β 40- and 42-peptides in Cos-7 cells expressing both the full-length Swedish- and wild-type APP alleles. These observations suggest that the present system could permit the selection of siRNA duplexes having the potential to confer ASP-RNAi.
MATERIALS AND METHODS
Preparation of oligonucleotides
DNA and RNA oligonucleotides were obtained from INVITROGEN and TAKARA, respectively. For preparation of duplexes, sense- and antisense-stranded oligonucleotides (20 μM each) were mixed and annealed as described previously (Hohjoh, 2002). The sequences of synthesized oligonucleotides are shown in Tables 1 and 2. Non-silencing siRNA duplex (siControl; Qiagen) was used as a negative control.
Table 1.
Synthetic DNA oligonucleotides
| Name | Sequence (5′---------------3′) |
|---|---|
| ssAPPwt(Sw) | CTAGCATGCAGGAGATCTCTGAAGTGAAGATGGATGCAGAATTCCGACA |
| asAPPwt(Sw) | GGCCTGTCGGAATTCTGCATCCATCTTCACTTCAGAGATCTCCTGCATG |
| ssAPP(K670N-M671L) | CTAGCATGCAGGAGATCTCTGAAGTGAATCTGGATGCAGAATTCCGACA |
| asAPP(K670N-M671L) | GGCCTGTCGGAATTCTGCATCCAGATTCACTTCAGAGATCTCCTGCATG |
| ssAPPwt(Lo) | CTAGCATGCTGTCATAGCGACAGTGATCGTCATCACCTTGGTGATGCTGA |
| asAPPwt(Lo) | GGCCTCAGCATCACCAAGGTGATGACGATCACTGTCGCTATGACAGCATG |
| ssAPP(V717I ) | CTAGCATGCTGTCATAGCGACAGTGATCATCATCACCTTGGTGATGCTGA |
| asAPP(V717I ) | GGCCTCAGCATCACCAAGGTGATGATGATCACTGTCGCTATGACAGCATG |
| ssAPP(V717F ) | CTAGCATGCTGTCATAGCGACAGTGATCTTCATCACCTTGGTGATGCTGA |
| asAPP(V717F ) | GGCCTCAGCATCACCAAGGTGATGAAGATCACTGTCGCTATGACAGCATG |
| ssAPP(V717G) | CTAGCATGCTGTCATAGCGACAGTGATCGGCATCACCTTGGTGATGCTGA |
| asAPP(V717G ) | GGCCTCAGCATCACCAAGGTGATGCCGATCACTGTCGCTATGACAGCATG |
Table 2.
Synthetic siRNAs used in this study. Sense- and antisense-stranded siRNA elements are indicated by ‘-ss’ and ‘-as’, respectively.
| siRNAs against the Swedish APP mutant | |
|---|---|
| Name | Sequence (5′---------------3′) |
| si(T7/C8)-ss | AGUGAAUCUGGAUGCAGAAUUU |
| si(T7/C8)-as | AUUCUGCAUCCAGAUUCACUUU |
| si(T8/C9)-ss | AAGUGAAUCUGGAUGCAGAAUU |
| si(T8/C9)-as | UUCUGCAUCCAGAUUCACUUUU |
| si(T9/C10)-ss | GAAGUGAAUCUGGAUGCAGAUU |
| si(T9/C10)-as | UCUGCAUCCAGAUUCACUUCUU |
| si(T10/C11)-ss | UGAAGUGAAUCUGGAUGCAGUU |
| si(T10/C11)-as | CUGCAUCCAGAUUCACUUCAUU |
| si(T11/C12)-ss | CUGAAGUGAAUCUGGAUGCAUU |
| si(T11/C12)-as | UGCAUCCAGAUUCACUUCAGUU |
| si(T12/C13)-ss | UCUGAAGUGAAUCUGGAUGCUU |
| si(T12/C13)-as | GCAUCCAGAUUCACUUCAGAUU |
| siRNAs against the London APP mutants | |
|---|---|
| Name | Sequence (5′---------------3′) |
| si(A8)-ss | AGUGAUCAUCAUCACCUUGUU |
| si(A8)-as | CAAGGUGAUGAUGAUCACUUU |
| si(A9)-ss | CAGUGAUCAUCAUCACCUUUU |
| si(A9)-as | AAGGUGAUGAUGAUCACUGUU |
| si(A10)-ss | ACAGUGAUCAUCAUCACCUUU |
| si(A10)-as | AGGUGAUGAUGAUCACUGUUU |
| si(A11)-ss | GACAGUGAUCAUCAUCACCUU |
| si(A11)-as | GGUGAUGAUGAUCACUGUCUU |
| si(A12)-ss | CGACAGUGAUCAUCAUCACUU |
| si(A12)-as | GUGAUGAUGAUCACUGUCGUU |
| si(T8)-ss | AGUGAUCUUCAUCACCUUGUU |
| si(T8)-as | CAAGGUGAUGAAGAUCACUUU |
| si(T9)-ss | CAGUGAUCUUCAUCACCUUUU |
| si(T9)-as | AAGGUGAUGAAGAUCACUGUU |
| si(T10)-ss | ACAGUGAUCUUCAUCACCUUU |
| si(T10)-as | AGGUGAUGAAGAUCACUGUUU |
| si(T11)-ss | GACAGUGAUCUUCAUCACCUU |
| si(T11)-as | GGUGAUGAAGAUCACUGUCUU |
| si(T12)-ss | CGACAGUGAUCUUCAUCACUU |
| si(T12)-as | GUGAUGAAGAUCACUGUCGUU |
| si(G8)-ss | GUGAUCGGCAUCACCUUGGUU |
| si(G8)-as | CCAAGGUGAUGCCGAUCACUU |
| si(G9)-ss | AGUGAUCGGCAUCACCUUGUU |
| si(G9)-as | CAAGGUGAUGCCGAUCACUUU |
| si(G10)-ss | CAGUGAUCGGCAUCACCUUUU |
| si(G10)-as | AAGGUGAUGCCGAUCACUGUU |
| si(G11)-ss | ACAGUGAUCGGCAUCACCUUU |
| si(G11)-as | AGGUGAUGCCGAUCACUGUUU |
| si(G12)-ss | GACAGUGAUCGGCAUCACCUU |
| si(G12)-as | GGUGAUGCCGAUCACUGUCUU |
Cell culture
HeLa, T98G and Cos-7 cells were grown at 37°C in Dulbecco′s modified Eagle′s medium (Wako) supplemented with 10% fetal bovine serum (Sigma), 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma) in 5% CO2-humidified chamber. T98G cells (Registry No. IFO50295) were obtained from the Health Science Research Resources Bank.
Construction of reporter and expression plasmids
In order to construct plasmids carrying reporter alleles, the phRL-TK (Promega) and pGL3-TK (Ohnishi et al., 2005) plasmids encoding the Renilla and Photinus luciferase genes, respectively, both of which were driven by the same herpes simplex virus thymidine kinase (TK) promoter, were digested with Xba I and Not I, and were subjected to ligation with synthetic oligonucleotide duplexes corresponding to the Swedish-, London- and wild-type APP alleles (sequences of the oligonucleotides used are indicated in Table 1). The resultant plasmids carry allelic APP sequences in the 3′-untranslated regions (UTRs) of the luciferase genes (Figure 1A). Expression plasmids, pAPP695WT and pAPP695SWE encoding full-length cDNAs of the wild- and Swedish-type APP alleles, respectively, were kindly provided by Dr Tanahashi (Tanahashi and Tabira, 2001).
Figure 1.
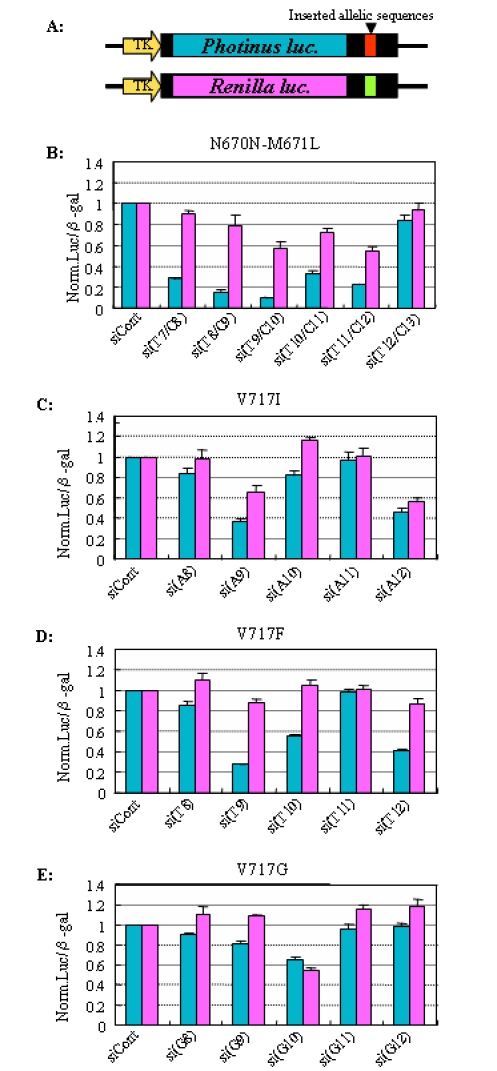
Assessment of ASP-RNAi with reporter alleles. (A) Schematic drawing of reporter alleles. Reporter alleles were constructed based on the Photinus and Renilla luciferase reporter genes driven by the same TK promoter, and allelic sequences of wild-type and mutant (synthetic oligonucleotides) were inserted into the 3′-UTRs of the reporter genes, i.e., the reporter alleles encode luciferase reporter genes carrying artificially inserted allele sequences of interest. Assessment of siRNA duplexes on the induction of ASP-RNAi against the Swedish APP mutant (B) and against the London APP mutants (C-E) was carried out. Synthetic siRNA duplexes against the mutants indicated were cotransfected with the mutant and wild-type reporter alleles and the β-galactosidase gene (control) into HeLa cells. The Photinus and Renilla luciferase genes carry the mutant and wild-type allelic sequences, respectively. Twenty-four hours after transfection, dual-luciferase and β-galactosidase assays were carried out. The levels of either Photinus (blue boxes) or Renilla (pink boxes) luciferase activity was normalized against the levels of β-galactosidase activity, and the ratios of mutant and wild-type luciferase activities in the presence of siRNA duplexes were normalized against the control ratio obtained in the presence of the siControl duplex (siCont). Data are averages of at least three independent determinations. Error bars represent standard deviations.
Transfection and reporter assay
The day before transfection, cells were trypsinized, diluted with fresh medium without antibiotics, and seeded into 24-well culture plates (approximately 0.5 × 105 cells/well). Cotransfection of synthetic siRNA duplexes with reporter plasmids was carried out using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer′s instructions, and to each well, 0.24 μg (40 nM) of siRNA duplexes, 0.2 μg of pGL3-TK-backbone plasmid, 0.05 μg of phRL-TK-backbone plasmid and 0.1 μg of pSV- -Galactosidase control vector (Promega) were applied. Twenty-four hours after transfection, cell lysate was prepared and expression levels of luciferase and β-Galactosidase were examined by the Dual-Luciferase reporter assay system (Promega) and Beta-Glo assay system (Promega), respectively, according to the manufacturer′s instructions. In the case of transfection of siRNA duplexes and expression plasmids (pAPP695WT and pAPP695SWE) into Cos-7 cells, 0.4 μg of each plasmid and 0.24 μg of siRNA duplexes were applied. Forty-eight hours after transfection, culture media was collected and cell lysate was prepared.
Western blotting and ELISA
Culture media and cell lysate prepared from transfected Cos-7 cells were examined by western blotting as described previously (Lesne et al., 2003). Equal amounts of proteins were separated by SDS-PAGE and electrophoretically blotted onto PVDF membranes (Millipore). Membranes were blocked for 1 h in blocking solution (5 % (v/w) fat-free milk and 0.05 % (v/v) Tween-20 in PBS) and were incubated with anti-APP antibody 22C11 (Chemicon) or anti- -tubulin antibody DM1A (Sigma) followed by washing in PBS and further incubation with horseradish peroxidaseconjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories). Antigen-antibody complexes were visualized using ECL chemiluminescent reagent (Amersham). Levels of A 40 and A 42 production in culture media were examined by human/rat β amyloid 40 and 42 ELISA kits (Wako) according to the manufacturer′s instructions.
RT-PCR
Total RNA extraction, including treatment with DNase I (Ambion) twice followed by reverse transcription, were carried out as described previously (Sago et al., 2004). The resultant cDNAs were examined by real-time (RT)-PCR using the ABI PRISM 7300 sequence detection system (APPlied Biosystems) with a SYBER green PCR master mix (Applied Biosystems) according to the manufacturer′s instructions. PCR primers used were as follows:
- For detection of the Renilla luciferase transcript:
- renilla-F; 5′-GTTCTTTTCCAACGCTATTG-3′
- renilla-R; 5′-GAAGCTCTTGATGTACTTAC-3′
- For detection of the Photinus luciferase transcript:
- photinus-F; 5′-TTTGATATGTGGATTTCGAG-3′
- photinus-R; 5′-ATCGTATTTGTCAATCAGAG-3′
RESULTS
Assessment of siRNAs in heterozygous model system
In this study, the Swedish- and London-type mutants of the APP gene, which are involved in familial Alzheimer′s disease, were used as model mutant alleles. The Swedish-and London-type APP mutants carry double and single nucleotide substitutions, respectively, which are followed by amino acid substitutions (K670N-M671L in the Swedish APP; V717I, V717F or V717G in the London APP) (Goate et al, 1991; Mullan et al, 1992). The resultant amino acid sequences in the Swedish and London-type APPs are preferably digested by β- and β-secretase, respectively, resulting in accumulation of A 40 and A 42 peptides, which are the key factors of Alzheimer′s disease (Cai et al, 1993; Citron et al, 1992; Mattson, 2004; Suzuki et al, 1994).
Mutant and wild-type reporter alleles were constructed as described in Materials and Methods. The resultant reporter alleles (Figure 1A), synthetic siRNA duplex against the mutant allele and the β-galactosidase gene (control) were cotransfected into human cells. Note that the transfected cells are artificially heterozygous with the mutant and wild-type APP reporter alleles; thus, the effects of test siRNA duplexes on suppression of both the mutant and wild-type alleles can be simultaneously examined.
ASP-RNAi against the Swedish-type APP allele
When the Renilla and Photinus luciferase genes were regarded as the Swedish and wild-type reporter alleles, respectively, the effects of the si(T7/C8) - si(T12/C13) duplexes against the Swedish mutant on allele-specific gene silencing were examined in HeLa cells. The results are shown in Figure 1B. The siRNA duplexes, except for the si(T12/C13) duplex, appeared to induce inhibition of mutant (Photinus) allele expression, while little or moderate inhibition of wild-type (Renilla) allele expression was seen, suggesting that the siRNA duplexes were able to discriminate the mutant reporter allele from the wild-type reporter allele. The si(T12/C13) duplex appeared to yield little or no RNAi activity. Considering the influence of the siRNA duplexes on the expression of the wild-type allele, the si(T8/C9) duplex appears to be the most suitable siRNA duplex conferring ASP-RNAi against the mutant allele. As for the si(T9/C10) and si(T11/12) duplexes inducing moderate levels of inhibition of wild-type allele expression, further analyses were carried out (Figure 4). Similar results were also obtained when the luciferase genes were exchanged between the mutant and wild-type reporter alleles, i.e., the Photinus and Renilla luciferase genes carried the wild-type and Swedish allele sequences, respectively (data not shown). In addition, when T98G cells, a human glioblastoma cell line, and Cos-7 cells were used instead of HeLa cells, results similar to those obtained in HeLa cells were observed (data not shown).
Figure 4.
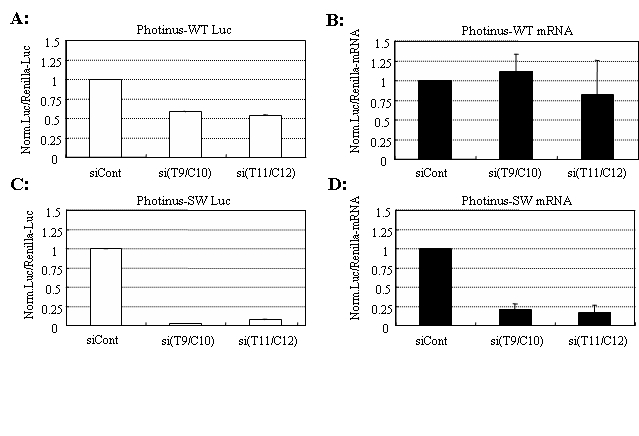
Possible translation inhibition and off-target silencing of wild-type reporter allele by siAPP duplexes. The si(T9/C10) or si(T11/C12) duplexes against the Swedish mutant allele together with either wild or mutant reporter allele plasmid carrying Photinus luciferase and the phRL-TK plasmid encoding Renilla luciferase (control) were introduced into HeLa cells. Twenty-four hours after transfection, dual-luciferase assay and isolation of total RNA were carried out. Off-target (to wild-type reporter allele) (A) and on-target (RNAi; to mutant reporter allele) (C) gene silencing were assessed based on luciferase activities. Ratios of normalized target (Photinus) luciferase activity to control (Renilla) luciferase activity are indicated: the ratios of luciferase activity determined in the presence of the si(T9/C10) or si(T11/C12) duplexes were normalized against the ratios obtained in the presence of the siControl duplex (siCont). Isolated RNAs in (B) and (D) corresponding to (A) and (C), respectively, were subjected to reverse transcription to synthesize first-stranded cDNAs. The resultant cDNAs were examined by real-time PCR with specific primers for Photinus and Renilla luciferase. RNA expression levels for Photinus luciferase are normalized against those of Renilla luciferase, and the ratios of Photinus luciferase RNA expression levels in the presence of the si(T9/C10) or si(T11/C12) duplexes are normalized against the ratios obtained in the presence of the siControl duplex. Data are averages of at least three independent determinations. Error bars represent standard deviations.
ASP-RNAi against London-type APP alleles
Because the London-type mutant possesses three types of single nucleotide change involved in amino acid substitution at position 717 (V717I, V717F and V717G), three mutant reporter alleles and corresponding wild-type reporter allele were constructed, and the effects of synthetic siRNA duplexes against the London-type mutants on suppression of the expression of either the target mutant allele or wild-type allele were examined under the present system. As shown in Figure 1C-E, various levels of gene silencing were observed and some of the siRNA duplexes, si(T9) and si(T12) (Figure 1D), appeared to discriminate the mutant alleles from the wild-type allele to some degree, resulting in ASP-RNAi; however, the other siRNA duplexes examined yielded less significant ASP-RNAi. Compared with the results for ASPRNAi against the Swedish allele (Figure 1B), the induction and activation of ASP-RNAi against the London alleles appeared to be inferior to those against the Swedish mutant.
Western blot analyses of wild-type and Swedish APP in ASP-RNAi
We further investigated ASP-RNAi of siAPP duplexes against the Swedish mutant with full-length cDNAs of the Swedish and wild-type APP alleles, which were transiently expressed in Cos-7 cells. The pAPP695SWE and/or pAPP695WT expression plasmids encoding full-length cDNAs of the Swedish and wild-type APP alleles, respectively, and siRNA duplexes targeting the Swedish mutant were cotransfected into Cos-7 cells, and expression of wild-type APP (APPWT) and Swedish APP (APPSWE) was examined by Western blotting. As shown in Figure 2, under homo(or hemi)zygous-like conditions, in which either APPWT or APPSWE was expressed, the signal intensity of sAPPSWE (secreted APP) and cAPPSWE (celluar APP) was apparently decreased in the presence of the si(T8/C9), si(T9/C10) and si(T11/C12) duplexes. In contrast, signals for either sAPPWT or cAPPWT were detected in the presence of any of the siRNA duplexes examined, which is consistent with the data for the reporter alleles described above. When APPSWE and APPWT were both expressed in the cells (heterozygous-like conditions), signals for APP were seen in the presence of any of the siRNA duplexes. Based on the results under homozygous-like conditions, the signals for APP in the presence of the si(T8/C9), si(T9/C10) and si(T11/C12) duplexes were most likely derived from APPWT.
Figure 2.
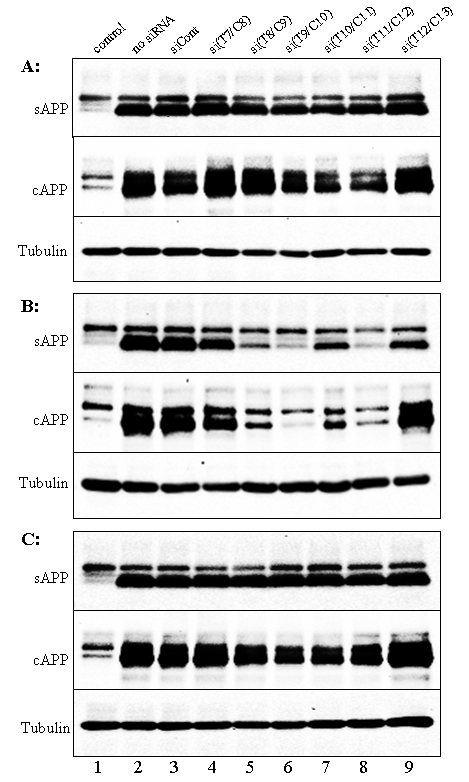
Expression of APPWT and APPSWE polypeptides under ASP-RNAi. Either the pAPP695WT (A) and pAPP695SWE (B) expression plasmids or the plasmids (C) together with the indicated siRNA duplexes against the Swedish mutant were introduced into Cos-7 cells, and expressed APP polypeptides in culture media (secreted APP: sAPP) and in cells (cellar APP: cAPP) were examined by Western blotting. Lane 1 (control) shows no transfected Cos-7 cells, in which endogenous APP is detectable. Lanes 2-9 are cells transfected with expression plasmid(s), and lanes 3-9 are cotransfected cells with the indicated siRNA duplexes. Expression of α-tubulin (control) is also shown.
The utility of ASP-RNAi using the siRNA duplexes assessed here in medical treatment can be demonstrated by confirming a significant decrease in A peptides, which are a key factor in the development of Alzheimer′s disease under heterozygous conditions expressing both APPSWE and APPWT. We thus determined the production levels of A 40 and A 42 peptides by means of ELISA. As shown in Figure 3, significant decreases in the production of either A 40 or A 42 peptide by RNAi (Figure 3A-C) and ASP-RNAi (Figure 3D-F) with the evaluated siRNA duplexes, particularly si(T8/C9), si(T9/C10) and si(T11/C12), was confirmed under homozygous and heterozygous conditions, respectively. Therefore, these results suggest the potential utility of such siRNA duplexes as therapeutic agents.
Figure 3.
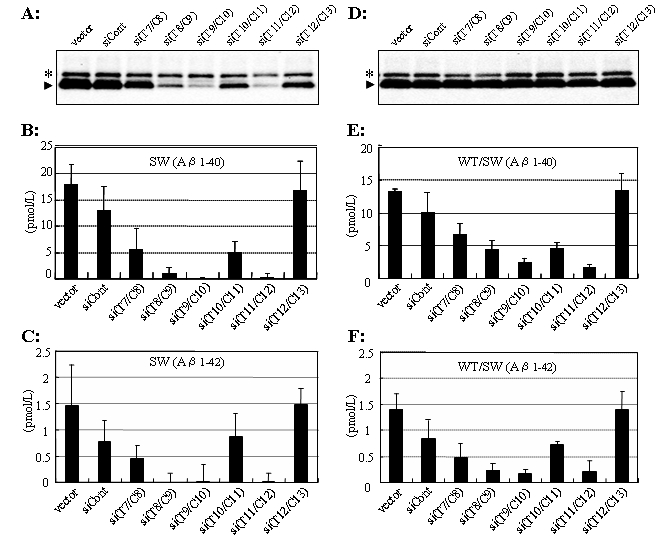
Production of A 40 and A 42 peptides under ASP-RNAi. The pAPP695SWE (A-C) plasmid and both the pAPP695SWE and pAPP695WT (D-F) plasmids together with the indicated siRNA duplexes against the Swedish mutant were cotransfected into Cos-7 cells, and expressed sAPP polypeptide and A 40 and A 42 peptides in culture media were examined by western blotting (A, D) and ELISA (B, C, E, F), respectively. “Vector” indicates cells transfected with only plasmid(s). Endogenous and exogenous (expressed) sAPPs are indicated by asterisks and arrow heads, respectively. ELISA data are averages of three independent determinations. Error bars represent standard deviations.
DISCUSSION
While ASP-RNAi is believed to be a useful technique, to realize and control ASP-RNAi, it is vital to design and select competent siRNA duplexes conferring ASP-RNAi; however, this is rather difficult without a procedure for assessing such siRNA duplexes. The system we present here could allow assessment, if designed siRNA duplexes have the potential for specifically inhibiting the expression of target alleles without suppressing the expression of other alleles. From a series of experiments with the Swedish- and London-type APP variants as model mutant alleles, we were able to determine potential siRNA duplexes for inducing ASP-RNAi. With regard to siRNA duplexes targeting the Swedish mutant, we further demonstrated that the si(T8/C9), si(T9/C10) and si(T11/C12) siRNA duplexes were able to significantly decrease the production of either A 40 or A 42 peptide in Cos-7 cells expressing both the full-length Swedish- and wild-type APP alleles. Accordingly, such competent siRNA duplexes conferring ASP-RNAi against mutant alleles likely hold utility as therapeutic agents.
In contrast to the Swedish mutant, there were difficulties in suppressing the London-type mutants carrying single nucleotide substitutions from the wild-type allele by ASPRNAi. The difference between ASP-RNAi activities against the Swedish- and London-type mutants may have been caused by the number of base substitutions: the former and latter mutants carry double and single base substitutions, respectively. Another important point to note in the results for the London-type mutant is that different substitutions showed different ASP-RNAi activities, suggesting that the type of base change between the mutant and wild-type alleles could influence ASP-RNAi. With regard to the V717I (Figure 1C) and V717G (Figure 1E) mutants, a possible wobble base pair between siRNA and the wild-type mRNA (Du et al, 2005) and high GC content of siRNA used(Ui-Tei et al, 2004), respectively, might have negatively influenced the induction of ASP-RNAi; these possibilities require further examination in the future.
To further progress ASP-RNAi, it is necessary to design competent siRNA duplexes conferring strong allele-specific gene silencing. Chemical modifications (Chiu and Rana, 2003; Hall et al, 2004) and structural devices in siRNAs are considered to be applicable for improving ASP-RNAi, and assessment of such siRNAs is feasible using the system we presented here. Altogether, it is suggested that the present assay system may contribute to the design and selection of the most suitable of siRNA duplexes conferring ASP-RNAi.
Finally, we add data indicating the possible inhibition of wild-type allele translation by the present siRNA duplexes. Because si(T9/C10) and si(T11/C12) exhibited moderate levels of inhibition of the expression of wild-type reporter allele (Figure 1B), we further investigated RNA levels of the wild-type allele by RT (real-time)-PCR. As shown in Figure 4, the levels of RNA expression of the wild-type allele in the presence of si(T9/C10) were similar to those in the presence of siControl, suggesting the possible inhibition of translation of the wild-type allele by the si(T9/C10) duplex. This may be due to a microRNA-like effect (Poy et al, 2004; Tang, 2005), and further study into this possibility remains necessary. With regard to the si(T11/C12) duplex, because a decrease trend in the levels of wild-type allele transcript was seen, it is possible that off-target gene silencing (Jackson et al, 2003) of the wild-type allele may occur in the presence of the duplex. Consequently, it is conceivable that the present system could further contribute to studies into off-target gene silencing and the function of microRNAs.
CONCLUSIONS
The present assay system with wild-type- and mutant-reporter alleles could permit assessment of siRNA duplexes having the potential for specifically inhibiting the expression of the mutant allele without inhibiting the expression of the wild-type allele, and thus contribute to the design and selection of siRNA duplexes suitable for allele-specific gene silencing.
ACKNOWLEDGEMENTS
This work was supported in part by research grants from the Ministry of Health, Labor, and Welfare in Japan and by Promega KK. We would like to thank Dr. H. Tanahashi for providing the pAPP695SWE and pAPP695WT plasmids and Drs. Y. Wang and K. Wada for providing Cos-7 cells. We also thank Y. Tamura, T. Sakai, K. Omi and Dr. A. Hasegawa for their helpful cooperation.
LIST OF ABBREVIATIONS
- ASP-RNAi
Allele-specific RNA interference
- APP
Amyloid precursor protein
- TK
Thymidine kinase
- UTR
Untranslated region
- sAPP
Secreted APP
- cAPP
Celluar APP
- Aβ
myloid β
STATEMENT OF COMPETING INTERESTS
Corresponding author has a pending patent on the method of this paper.
REFERENCES
- Cai XD, Golde TE, Younkin SG. Release of excess amyloid beta protein from a mutant amyloid beta protein precursor. Science. 1993;259:514–516. doi: 10.1126/science.8424174. [DOI] [PubMed] [Google Scholar]
- Caplen NJ. Gene therapy progress and prospects. Downregulating gene expression: the impact of RNA interference. Gene Ther. 2004;11:1241–1248. doi: 10.1038/sj.gt.3302324. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron M, Oltersdorf T, Haass C, et al. Mutation of the beta-amyloid precursor protein in familial Alzheimer′s disease increases beta-protein production. Nature. 1992;360:672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer′s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Hall AH, Wan J, Shaughnessy EE, Ramsay Shaw B, Alexander KA. RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucleic Acids Res. 2004;32:5991–6000. doi: 10.1093/nar/gkh936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Hohjoh H. RNA interference (RNAi) induction with various types of synthetic oligonucleotide duplexes in cultured human cells. FEBS Lett. 2002;521:195–199. doi: 10.1016/s0014-5793(02)02860-0. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Karagiannis TC, El-Osta A. RNA interference and potential therapeutic applications of short interfering RNAs. Cancer Gene Ther. 2005;12:787–795. doi: 10.1038/sj.cgt.7700857. [DOI] [PubMed] [Google Scholar]
- Lesne S, Docagne F, Gabriel C, et al. Transforming growth factor-beta 1 potentiates amyloid-beta generation in astrocytes and in transgenic mice. J Biol Chem. 2003;278:18408–18418. doi: 10.1074/jbc.M300819200. [DOI] [PubMed] [Google Scholar]
- Mattson M. P. Pathways towards and away from Alzheimer′s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- Mullan M, Crawford F, Axelman K, et al. A pathogenic mutation for probable Alzheimer′s disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet. 1992;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Tokunaga K, Hohjoh H. Influence of assembly of siRNA elements into RNA-induced silencing complex by fork-siRNA duplex carrying nucleotide mismatches at the 3′- or 5′-end of the sense-stranded siRNA element. Biochem Biophys Res Commun. 2005;329:516–521. doi: 10.1016/j.bbrc.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Sago N, Omi K, Tamura Y, et al. RNAi induction and activation in mammalian muscle cells where Dicer and eIF2C translation initiation factors are barely expressed. Biochem Biophys Res Commun. 2004;319:50–57. doi: 10.1016/j.bbrc.2004.04.151. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Cheung TT, Cai XD, et al. An increased percentage of long amyloid beta protein secreted by familial amyloid beta protein precursor (beta APP717) mutants. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- Tanahashi H, Tabira T. Three novel alternatively spliced isoforms of the human beta-site amyloid precursor protein cleaving enzyme (BACE) and their effect on amyloid beta-peptide production. Neurosci Lett. 2001;307:9–12. doi: 10.1016/s0304-3940(01)01912-7. [DOI] [PubMed] [Google Scholar]
- Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Takahashi F, et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor M, Bei Y, Gay F, et al. HAT activity is essential for CBP-1-dependent transcription and differentiation in Caenorhabditis elegans. EMBO Rep. 2002;3:50–55. doi: 10.1093/embo-reports/kvf006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MJ, Trulzsch B, Abdelgany A, Beeson D. Therapeutic gene silencing in the nervous system. Hum Mol Genet. 2003;12:R279–284. doi: 10.1093/hmg/ddg275. Spec No 2. [DOI] [PubMed] [Google Scholar]


